Abstract
Three bacteria namely Bacillus luciferensis K2, Bacillus amyloliquefaciens K12 and Bacillus subtilis BioCWB possessing plant growth promotion and biocontrol potential against phytopathogens and rice leaf folder were identified from organic soils of Sikkim, India. The results revealed significant higher production of phytohormones IAA (97.1 μg mL−1) and GA3 (10.6 μg mL−1) was found in K2, whereas BioCWB had higher phosphate solubilization (570.0 μg mL−1) efficacy and also possessed nitrogen fixation ability (5.34 log copy number mL−1 culture). All these bacteria had higher antagonistic activities against phytopathogens viz. Rhizoctonia solani, Fusarium proliferatum, Athelia rolfsii and Colletotrichum gloeosporioides and also had higher larvicidal activity against rice leaf folder Cnaphalocrocis medinalis (Guenne) under in vitro conditions. Molecular insights into the antagonistic mechanisms of Bacillus strains deciphered the presence of several antimicrobial peptides (ericin, subtilin, surfactin, iturin, bacilysin, subtilosin, fengycin and bacillomycin), volatiles (dimethyl disulphide, methyl-Furan, acetic acid, Z-1,3-pentadiene and 3-hexyn-2-ol) and soluble metabolites (9-octadecenamide, E-15-heptadecenal, E-3-eicosene and 5-octadecene). Furthermore, liquid microbial inoculum prepared using the bacterial strains (K2, K12 and BioCWB) were evaluated under glass house (rice) and field condition (capsicum), which significantly enhanced plant growth in rice and yield in capsicum compared to control. The present study revealed the combination of Bacillus spp. (K2, K12 and BioCWB) can be used as bio-inoculants for improving agricultural production in Sikkim. Moreover, for the first time, we demonstrated plant growth promoting (PGP) traits, antifungal and insecticidal properties of B. luciferensis.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1938-7) contains supplementary material, which is available to authorized users.
Keywords: PGP traits, Biocontrol, Bacillus spp., Sikkim organic soil
Introduction
Sikkim an agrarian state of India situated at 27°N–28°N latitude and 88°E to 89°E longitude with terrestrial elevations ranging from 300 to 7000 feet from mean sea level in the Eastern Himalayan range, is among the highest and richest biodiversity spots of the world (Maharana et al. 2000). Organic farming is predominantly practiced across this region (Yadav et al. 2013). Generally, nutrient depletion in soil due to leaching, volatilization and fixation are some of the problems related to decreased plant growth and development (Pathak et al. 2010). Several previous reports have shown soil bacteria to play an important role in plant growth and development by producing phytohormones such as indole-3-acetic acid (IAA) and gibberellic acid (GA3) (Egamberdieva et al. 2017), nutrient solubilization (Kumar et al. 2012) and nitrogen fixation (Fisher 1999). In addition to plant growth promoting (PGP) potential, several endospore forming Bacillus species such as B. subtilis, B. amyloliquefaciens, B. thuringiensis and B. megaterium have been reported for their effectiveness in suppressing activity of several plant pathogens and insect pests (Prabhukarthikeyan et al. 2014; Elanchezhiyan et al. 2018). Among the different modes of antagonism, antimicrobial peptides (AMPs) such as surfactin, iturin, bacillomycin and fengycin produced by Bacillus spp. have demonstrated to play important role in suppressing several pathogens in plant (Koumoutsi et al. 2004; Elanchezhiyan et al. 2018). Apart from the AMPs, bacteria secreted metabolites such as volatile organic compounds (VOCs) and soluble metabolites play key roles in plant growth regulation, development of stress resistance and suppression of diseases (Tyc et al. 2017). Specifically, VOCs are known to confer swift action in plants rather than soluble metabolites due to their low molecular weights (< 300 Da) and high vapor pressure (10 Pa at 20 °C) that readily evaporates and diffuses through cell surfaces (Schulz and Dickschat, 2007). A detailed description of different types of VOCs secreted by bacteria has been well documented by Thorn and Greenman (2012).
India is one of the major agrarian Asian country comprising 29 states, of which Sikkim has become the first organic state with the adoption of 100% organic farming (Kumar et al. 2018a, b). It has been reported that climate variability and erratic precipitation are the major drivers of pest incidence (Macfadyen et al. 2018). The prevalence of plant diseases caused by rice blast fungus (Sridhar and Singh, 2001), sheath blight, stem rot fungus (Kumar et al. 2017a) and soil-borne fungal pathogens have brought severe constraints in the productivity of organic rice and horticultural crops in this area. The insect pest particularly rice leaf folder Cnaphalocrocis medinalis (Guenne) has turned into major pest causing leaf defoliation which has been shown to cause 61.9% yield loss in rice under epidemic situations. Apart from cereals, Sikkim is one of the major repositories of the “Capsicum annum complex” which comprises three cultivated species namely C. annuum, C. frutescens, and C. chinense of which C. annuum is widely cultivated. One of the major limitations in the cultivation of C. annuum has been the poor development of morphological characters largely due to nutrient deficiency (Jha and Saha 2017).
Currently, there are separate management practices available for improving plant growth and disease management, especially in Sikkim’s organic farming. However, the development of an effective microbial strategy comprising insecticidal, fungicidal properties with plant growth promotion abilities could be a viable option for plant health management practices. Additionally, as supported by previous studies it is vivid that indigenous microbes are better at promoting plant growth and managing pest and diseases than alien microbes (Kumar and Gopal 2015). Keeping this in mind, attempts were made in the present study to identify some of the novel multi-potential PGP bacteria from soils of Sikkim and assess their antagonistic potential against disease-causing phytopathogens.
Materials and methods
Sample collection and isolation of plant growth promoting bacteria
The soil sampling was conducted at 23 different agricultural fields (cropping system of rice followed by vegetable crops) across Sikkim, India. Ten soil samples were collected from each site at depth of 15–20 cm, completely homogenized and transported to laboratory in cool pack for further analysis. The collected soil samples were passed through 2 mm sieve to remove coarse rock and plant material, thoroughly mixed for uniformity and stored at 4 °C. Bacteria from collected soil samples were isolated by serial dilution technique using different agar media viz. Nutrient, Pikovskaya’s, Jensen’s and King’s B agar medium and preserved in respective media at 4 °C. The fungal pathogens used in this study viz. Rhizoctonia solani (MH60071), Fusarium proliferatum (MF033170) and Athelia rolfsii (MH636611), which causes sheath blight, sheath rot and seedling blight, respectively in rice were collected from Crop Protection Division, NRRI, Cuttack and Colletotrichum gloeosporioides a pathogen causing anthracnose in Capsicum plants was collected from microbial repository at NRRI, Cuttack and preserved in potato dextrose agar medium at 4 °C. The third instar rice leaf folder Cnaphalocrocis medinalis (Guenne) was collected from NRRI research farm, Cuttack.
Estimation of IAA and GA3 production
Production of IAA by bacteria was quantified by spectrophotometric method (Gordon and Weber 1951), wherein the bacterial cultures were grown in tryptophan (100 mg L−1) amended nutrient broth, incubated at 30 ± 2 °C for 5 days in the dark. Following which IAA was estimated by determining the colour change using Salkowski reagent. GA3 production by bacteria was assessed as per the spectrophotometric method described by Borrow et al. (1955).
Phosphate solubilization, nitrogen fixation, and siderophore production
Phosphate solubilization potential of the bacterial isolates was estimated in NBRIP medium enriched with tri-calcium phosphate as an insoluble phosphate source. For the qualitative estimation of phosphate solubilization, 24 h old bacterial culture was spotted in NBRIP agar medium, plates were incubated at 30 ± 2 °C for 7 days and efficacy was assessed based on clear zone formation. Similarly, quantitative estimation of phosphate solubilization by bacterial isolates was assessed using NBRIP liquid medium (Panda et al. 2016) and soluble P content in the supernatant was estimated as per methodology adopted from Murphy and Riley (1962). The nitrogen-fixing ability of bacterial isolates was assessed based on their growth on N-free Jensen’s medium by repeated culturing (Kumar et al. 2017b). Further, siderophore production of bacteria isolates was qualitatively determined by the method suggested by Schwyn and Neilands (1987).
Ammonia, HCN production, protease and chitinase activity
Ammonia production by bacterial isolates was qualitatively determined using Nessler’s reagent which was added to culture supernatant of five-day old bacteria from peptone broth and development of brown to yellow colour indicated the production of ammonia (Cappuccino and Sherman 1992). HCN production by bacteria was detected using the filter paper method as described by Bakker and Schippers (1987). Protease activity was qualitatively determined by visualizing halo zone formation around bacteria colonies in 3% agar skim milk medium, incubated at 30 ± 2 °C for 4 days (Abo-Aba et al. 2006). Similarly, halo zone formation around bacteria colonies using colloidal chitin agar medium was used for determining chitinase activity (Hsu and Lockwood 1975).
Assessment of antifungal activities of bacterial isolates
The antifungal assay was performed by dual culture technique using potato dextrose agar medium (Lahlali and Hijri 2010). The following fungal pathogens viz., R. solani, F. proliferatum, A. rolfsii and C. gloeosporioides were used for the assay. Agar disc (5 mm) of phytopathogens from 5 days old culture was placed at one pole of the Petri’s dish and 24 h old bacteria culture was streaked on the opposite pole. Antifungal activity of bacterial strains was determined by comparison with control plates inoculated only with fungus. The fungal growth was monitored for 7 days at 30 ± 2 °C and three replications per isolate were maintained. Fungal growth (colony diameter) was measured and percentage inhibition calculated according to the formula described by Lahlali and Hijri (2010).
where Cd colony diameter (mm) of the control and Td colony diameter (mm) of the test plate
Antagonistic activity assay was also performed under broth conditions wherein initially the mycelia dry weight was calculated from which the percentage inhibition by bacteria was calculated according to the formula described by Lahlali and Hijri 2010.
where Cw mycelia weight (g) in control and Tw mycelia weight (g) in the treatment broth
Screening for larvicidal activities of bacterial isolates
Bacterial isolates were cultured in 100 mL nutrient broth, incubated at 30 ± 2 °C for 24 h at 150 rpm, and was further subjected to centrifugation at 10,000 rpm for 10 min to obtain cell pellets. The cell pellets were washed three times with sterile distilled water and the cell loads were adjusted to 101–106 bacterial cells per mL in sterile distilled water and used to calculate LC50 against C. medinalis (Guenne) third instar larvae using leaf piece bioassay (Panneerselvam et al. 2018).
DNA extraction and amplification of the antimicrobial peptides and 16S rRNA genes of bacterial isolates
Genomic DNA was extracted using the bacterial DNA isolation kit manufactured by Zymo™, USA. Approximately 5 mL of 24 h old bacterial culture grown on nutrient broth was centrifuged at 8000 rpm for 15 min and DNA extraction from cell pellet was carried as per protocol available in the kit. The quantity and purity of isolated DNA were determined by UV spectrophotometer (Nanodrop; Thermo Scientific). 27f (5′-AGAGTTTGATCCTGGCTCAG) and 1492r (5′-GGTTACCTTGTTACGACTT) primers were used to amplify 16S rRNA (Suzuki and Giovannoni 1996) and the purified PCR samples were sequenced through Sanger sequencing by Eurofins, Bengaluru, India. Contigs were generated from the resulting forward and reverse read sequences using CAP3 assembly program (Huang and Madan 1999) and the presence of chimera was checked using DECIPHER v2.0 (Wright 2016). Further, sequences were submitted to NCBI database after identifying the bacteria using nBLAST. The phylogenetic tree was constructed using the molecular evolution and genetic analysis (MEGA) software version 10 (Kumar et al. 2018b). Initially, MUSCLE multiple alignment program was used for the alignment of the sequences and Neighbor-Joining phylogenetic tree was constructed using the following parameters viz. 1000 bootstrap replications, maximum composite likelihood substitution model and uniform rate of nucleotide changes with partial deletion.
The genes of antimicrobial peptides viz. ericin, subtilin, surfactin, iturin, bacilysin, subtilosin, fengycin, bacillomycin, mycosubtilin and mersacidin were amplified using specific primers provided in supplementary Table 1. A 50 μL reaction mixture containing approximately 200 μM each dNTPs, 20 pmol each of both forward and reverse primers, 0.5 U of Taq DNA polymerase (NEB) and 50 ng of total DNA was used for PCR amplification in thermocycler. The PCR program followed was: initial denaturation at 96 °C (3 min), followed by 35 cycles of denaturation at 96 °C (1 min), annealing at 55 °C (1 min) and extension at 72 °C (3 min) with final 10 min extension at 72 °C (Vinodkumar et al. 2017). The nitrogen-fixation abilities of bacterial isolates were compared with Azotobacter tropicalis and Azotobacter chroococcum by quantifying nifH gene abundance using the qPCR technique as reported by Kumar et al. (2017b).
Estimation of bacterial volatiles through TD-GC/MS
The nutrient broth was used for growing bacterial isolates in 500 mL flasks having 70% headspace. Volatiles emitted by bacterial isolates were captured using Tenax TA coated stainless steel desorbing column (Perkin Elmer HO244966) that was inserted through rubber stoppers. Uninoculated nutrient broth served as the negative control. Volatiles captured were analyzed with GC–MS fitted with Thermal Desorber turbomatrix 150 (Perkin Elmer, USA) as per conditions suggested by (Mishra et al. 2017) and the detected compounds were identified using mass spectral database (NIST, 2014).
Estimation of bacterial metabolites through GC/MS
The bacterial cultures were grown in 250 mL nutrient broth and subjected to centrifugation at 10,000 rpm for 10 min, and the pH of the supernatant was adjusted to 2.0. After pH adjustment, supernatant was mixed with an equal quantity of ethyl acetate and kept for overnight agitation in a shaker. The solvent fraction was separated and dried in a vacuum flash evaporator, during which the non-volatile compounds present in the solvent phase was concentrated. The crude extract was dried overnight in sterile Petri plate and extracted using 1.0 ml methanol and filtered through 0.2 µm syringe filter for GC–MS analysis as per the method described by Anantha et al. (2016).
Antagonistic potential of Bacillus spp. against rice sheath blight caused by R. solani under glasshouse condition
Pot experiment studies were conducted in the glasshouse at ICAR-NRRI, India and evaluated in two rice cropping season’s trial-I (kharif) and trial-II (rabi). Surface-sterilized seeds of Oryza sativa var. Naveen (rice) were used in this experiment. Liquid-based bioformulation was prepared using three Bacillus spp. (K2, K12, BioCWB) using the modified method of Manikandan et al. (2010), wherein the selected bacterial isolates were grown separately in nutrient broth for 48 h and then equally mixed along with osmoprotectants (glycerol and polyvinylpyrrolidone). The surface-sterilized rice seeds were soaked for 24 h separately in sterile distilled water containing bacterial formulation (2.0 × 109 CFU ml−1) at the rate of 5 ml L−1. After soaking, the seeds were dried and sown in pots (45 × 60 cm) filled with sterilized soil (121 °C, 15 psi for 2 h for 2 consecutive days) collected from ICAR-NRRI, research farm. Twenty days rice seedlings were uprooted and their roots were dipped in the liquid formulation (5 ml L−1) for about 2 h, which were later transplanted in plastic pots with three hills per pot. After 35 days of transplanting, R. solani was inoculated by placing agar disc (5 mm) from 7 days old fungal culture on the sheath of rice plant which was secured by wrapping absorbent cotton and sealed using parafilm. After 24 h of pathogen inoculation, plants were sprayed with bacterial liquid formulation (5 ml L−1). The fungicide treated seeds was maintained as the negative control, wherein seed was treated with carbendazim (2 g kg−1) followed by plant foliar spray (0.1% w/v). Untreated plants served as the positive control. The percentage disease index (PDI) was calculated using the formula:
whereas the disease grade was given with the following descriptions: 0, no infection observed; 1, lesions limited to lower 20% of the plant height; 3, (20–30%); 5, (31–45%); 7, (46–65%); 9, more than 65% (IRRI 2013).
Evaluation of plant growth promoting potential of Bacillus spp. in rice plant
The plant growth promoting potential of all the three Bacillus spp. were evaluated in rice variety Naveen. Bacterial inoculations and rice transplantation in pots were carried as per previously described methodology. After 60 DAT (days after transplanting), the agronomic parameters particularly plant height, shoot length, root length, total biomass and tiller number were measured to understand the plant growth promoting potential of Bacillus spp.
Evaluation of plant growth promoting potential of Bacillus spp. in Capsicum plant
The field experiment was conducted for one cropping season at Ralap Farm (27.3°N 88.6°E), East Sikkim, India. The C. annuum var. Krishna was used in this experiment. The field experiment was laid out in complete randomized block design with the plot size of 3.0 × 1.0 m2 area. The following five treatments viz., T1, uninoculated control; T2, B. subtilis (BioCWB); T3, B. amyloliquefaciens (K12); T4, B. luciferensis (K2) and T5, combination of BioCWB + K12 + K2 were imposed and replicated five times. For the experiment, 25 days old seedlings were transplanted with the spacing of 30 × 40 cm in a raised bed. Bacterial inoculants (2.0 × 109 CFU ml−1) were applied after 1 week at the rate of 2.0 L acre−1. Before application, microbial cultures were suspended in water at the rate of 1.0 L per 100 L of water and drenched basally. The data on plant growth parameters i.e. plant height, fresh and dry weight of root and shoots, fruits number were observed at regular intervals. In this experiment, farm yard manure (FYM) 10 tonnes ha−1 and vermicompost 5 tonnes ha−1 were applied uniformly.
Statistical analysis
The data obtained were statistically analyzed using Web-Based Agricultural Statistical Software Package (WASP 2.0) developed by Central Coastal Agricultural Research Institute (ICAR), Ela Goa (www.ccari.res.in/waspnew.html). PCA analysis was performed using R software (Team 2000). Calculation of LC50 was based on probit analysis carried using Polo Plus, LeOra software version 2.0 (LeOra 2007). The data generated on the antagonistic potential of bacterial strains against rice sheath blight was analyzed using IRRISTAT v.92-1 program developed by International Rice Research Institute (IRRI), Philippines. The larval mortality was verified by using Abbott’s formula (Abbott 1925) and the difference between treatments was evaluated using the least significant difference (LSD) at p < 0.05.
Results and discussion
Plant growth promoting traits and biochemical characterization
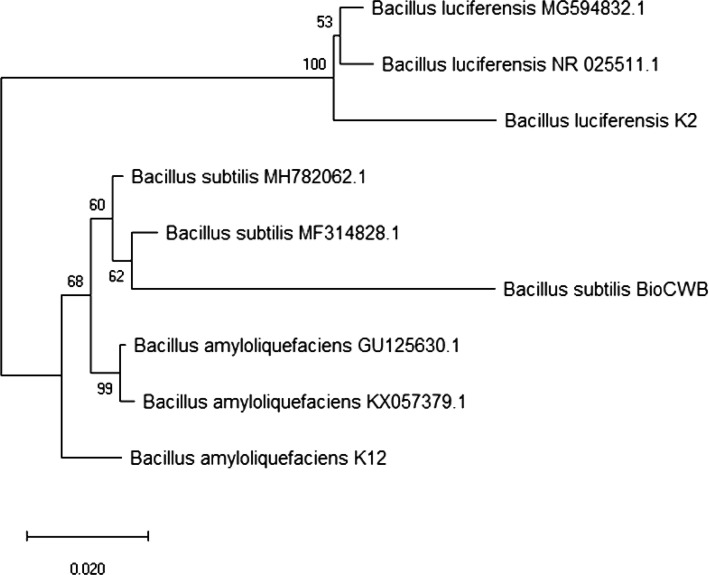
In this study, 63 bacteria were isolated from Sikkim’s long-term organic farming soil, of which three efficient isolates were selected based on their PGP traits, in particular, the production of phytohormones such as IAA and GA3, phosphate solubilization, nitrogen fixation and siderophore production (Supplementary Table 2). The selected three isolates were identified as B. luciferensis K2, B. amyloliquefaciens K12 and B. subtilis BioCWB with the following Genbank accession numbers MN087224, MG490140 and MN087225, respectively (Fig. 1). Additionally, these three bacterial strains have also been deposited at microbial culture repository at ICAR-National Bureau of Agriculturally Important Microorganisms (ICAR-NBAIM), Mau, UP, India having the following accession numbers viz. B. subtilis strain BioCWB (NAIMCC-B-02285), B. luciferensis strain K2 (NAIMCC-B-02286) and B. amyloliquefaciens strain K12 (NAIMCC-B-02288). The quantitative estimation of IAA and GA3 production (Table 1) indicated that B. luciferensis K2 produced significantly higher GA3 (10.6 µg mL−1) and IAA (97.1 µg mL−1) compared to other strains. Besides, phosphate solubilization studies showed that B. subtilis BioCWB could solubilize significantly higher phosphate (570 µg mL−1) than B. luciferensis K2 (417.3 µg mL−1) and B. amyloliquefaciens K12 (364.1 µg mL−1). However, out of three Bacillus spp., only B. subtilis BioCWB had nitrogen-fixing ability (Table 1). Further, all the three strains exhibited positive results for siderophore production, ammonia production, HCN production, protease and chitinolytic activities (Table 1). There are multiple previous research findings which have proved the PGP traits of Bacillus through the production of phytohormones such as gibberellin (Bottini et al. 2004) and IAA (Idris et al. 2007). In our present study, based on the growth of B. subtilis BioCWB after repeated streaking (15 times) in nitrogen-free Jensen’s medium, it was presumed to have nitrogen fixing capabilities, which was further confirmed by detecting the presence of nifH gene through nifH specific primers. Another comparative analysis based on nifH gene abundance in B. subtilis BioCWB, Azotobacter tropicalis and A. chroococcum showed gene abundance was 5.34 log copy number mL−1 in B. subtilis BioCWB as compared to A. tropicalis and A. chroococcum which had 5.97 and 6.92 log copy number mL−1 culture, respectively. Previous workers have used nifH gene to underpin the nitrogen-fixing potential of Bacillus species (Ding et al. 2005) and have proved the mechanism of nitrogen fixation in B. subtilis (Fisher 1999). Although we isolated N-fixing Bacillus species, the N-fixing ability was presumed to be lower than that of other diazotrophs, based on the nifH gene abundances.
Fig. 1.
A phylogenetic neighbor-joining tree showing the relationship of Bacillus strains. The bar represents five nucleotide substitutions per 1000 nucleotides of the 16S rRNA gene sequence
Table 1.
Biochemical estimations and plate assays of Bacillus strains
| Strain | IAA (µg mL−1) | GA (µg mL−1) | Phosphate solubilization (µg mL−1) | N-fixing activity | Siderophore production | HCN production | Ammonia production | Protease activity | Chitinase activity |
|---|---|---|---|---|---|---|---|---|---|
| K2 | 97.1 ± 1.4a | 10.6 ± 0.5a | 417.3 ± 4.3b | − | + | + | + | + | + |
| K12 | 49.9 ± 1.1b | 4.6 ± 0.8b | 364.1 ± 2.1c | − | + | + | + | + | + |
| BioCWB | 10.4 ± 0.7c | 3.9 ± 0.5c | 570.0 ± 2.6a | + | + | + | + | + | + |
| CD (0.05) | 12.3 | 0.56 | 23.4 | NA | NA | NA | NA | NA | NA |
Lowercase letters represent significant variations among the data at p < 0.05
+ positive, − negative
Antagonistic activity against plant pathogenic fungi
Both the dual plate and broth culture studies revealed that B. subtilis BioCWB had higher antagonistic activity against R. solani (79.6–84.5%), F. proliferatum (76.6–86.7%), A. rolfsii (86.7–87.8%) and C. gloeosporioides (78.0–83.2%) followed by B. luciferensis K2 and B. amyloliquefaciens K12 (Tables 2, 3). In addition, biocontrol potential of Bacillus spp. were evaluated against rice sheath blight under glasshouse studies and results showed (Table 4) that application of B. subtilis BioCWB significantly suppressed (69.5–72.6%) rice sheath blight incidence followed by B. luciferensis K2 (52.2–54.4%) and B. amyloliquefaciens K12 (26.2–28.7%). In chemical treatment studies, there was 82.6–83.55% disease suppression whereas the control treatment recorded 82.0–85.2% rice sheath blight incidence. Many studies have proved that Bacillus is known for biocontrol of plant diseases by different mechanisms, which include antibiosis, competition for nutrients and induction of defense responses in the host plant (Prabhukarthikeyan et al. 2014; Stein 2005). Further, compatibility assays conducted on nutrient agar medium deciphered B. luciferensis K2, B. amyloliquefaciens K12 and B. subtilis BioCWB to have no antagonistic effect on each other.
Table 2.
Antagonistic activity of Bacillus strains against R. solani, F. proliferatum, A.rolfsii and C. gloeosporioidesunder dual culture plate method
| Strain | R. solani | F. proliferatum | A. rolfsii | C. gloeosporioides | ||||
|---|---|---|---|---|---|---|---|---|
| Mycelial growth diameter (cm) | Percent inhibition over control | Mycelial growth diameter (cm) | Percent inhibition over control | Mycelial growth diameter (cm) | Percent inhibition over control | Mycelial growth diameter (cm) | Percent inhibition over control | |
| K2 | 2.0c | 75.0a | 1.3c | 83.8a | 1.1c | 86.3 | 2.5b | 68.7c |
| K12 | 6.2b | 22.9b | 2.2b | 72.5b | 1.4b | 82.9 | 2.2b | 72.2b |
| BioCWB | 1.6d | 79.6a | 1.1c | 86.7a | 1.1c | 86.7 | 1.7c | 78.0a |
| Control | 8.0a | – | 8.0a | – | 8.0a | – | 8.0a | – |
| CD (0.05) | 0.322 | 3.420 | 0.228 | 3.224 | 0.224 | NS | 0.221 | 2.36 |
Lowercase letters represent significant variations among the data at p < 0.05
Table 3.
Antagonistic activity of Bacillus strains against R. solani, F. proliferatum, A.rolfsii and C. gloeosporioides under broth assay
| Strain | R. solani | F. proliferatum | A. rolfsii | C. gloeosporioides | ||||
|---|---|---|---|---|---|---|---|---|
| Mycelial dry weight (g) | Percent inhibition over control | Mycelial dry weight (g) | Percent inhibition over control | Mycelial dry weight (g) | Percent inhibition over control | Mycelial dry weight (g) | Percent inhibition over control | |
| K2 | 0.20c | 80.8b | 0.32c | 75.9a | 0.42c | 78.0b | 0.33c | 82.1a |
| K12 | 0.79b | 25.3c | 0.46b | 65.9b | 0.68b | 65.1c | 0.39b | 78.9b |
| BioCWB | 0.15d | 84.5a | 0.27c | 76.6a | 0.25d | 87.8a | 0.31c | 83.2a |
| Control | 1.04a | – | 1.34a | – | 1.93a | – | 1.85a | – |
| CD (0.05) | 0.042 | 1.983 | 0.059 | 2.497 | 0.045 | 1.608 | 0.0521 | 2.731 |
Lowercase letters represent significant variations among the data at p < 0.05
Table 4.
Evaluation of antagonistic potential of Bacillus strains against R. solani in potted rice plants
| Treatments | Trial I | Trial II | ||
|---|---|---|---|---|
| PDIa | % Reduction over control | PDIa | % Reduction over control | |
| K2 | 40.6 (39.58)c | 52.2 | 37.35 (37.69)c | 54.5 |
| K12 | 62.8 (52.43)d | 26.2 | 58.50 (49.90)d | 28.7 |
| BioCWB | 23.3 (28.86)b | 72.6 | 25.00 (29.99)b | 69.5 |
| Chemical | 14.81 (22.63)a | 82.6 | 13.50 (21.56)a | 83.5 |
| Control | 85.2 (67.51)e | 82.10 (65.07)e | ||
Lowercase letters represent significant variations among the data at p < 0.05
aValues are the mean of five replications. Values in the parenthesis are arscine transformed values. Means followed by a common letter are not significantly different at 5% level by DMRT
Larvicidal activities of Bacillus spp. against rice leaf folder
The larvicidal potential of Bacillus species against third instar leaf folder larvae (Table 5) showed B. luciferensis treated leaves showed 50% (LC50) leaf folder larval mortality with the cell loads of 2.00 × 104 CFU ml−1, whereas B. subtilis and B. amyloliquefaciens treated leaves showed 50% larval mortality with 2.10 × 104 CFU ml−1 and 2.97 × 104 CFU ml−1 inoculum load, respectively. However, there was no significant variation in larvicidal activity among the Bacillus spp. Additionally, in the present study, Bacillus spp. were identified to produce siderophore, ammonia, HCN, protease, and antimicrobial metabolites, which indicates the combined action of enzymatic activity along with other antimicrobial components might be responsible for the strong larvicidal activity against rice leaf folder (Panneerselvam et al. 2018).
Table 5.
Toxicity of Bacillus strains against third instars larva (n = 30) of rice leaf folder C. medinalis (Guenee)
| Bacteria | X2 (DF)a | Slope | LCb50 | LC50 confidence interval | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| K2 | 0.69 (4) | 2.60 ± 0.4 | 2.00 × 104 | 4.22 × 103 | 2.22 × 105 |
| K12 | 3.21 (4) | 3.47 ± 0.5 | 2.97 × 104 | 5.68 × 103 | 1.01 × 105 |
| BioCWB | 2.98 (4) | 4.40 ± 0.6 | 2.10 × 104 | 1.01 × 104 | 1.09 × 105 |
aDegree of freedom, bLC50 of one treatment is significant different if the both lower and upper fiducial limit does not include LC50 value of other treatment
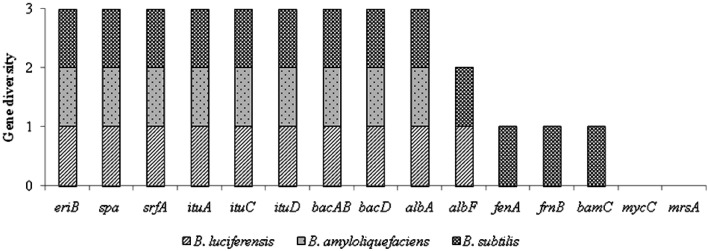
Antimicrobial peptide genes present in Bacillus spp
To understand the mechanisms of the antagonistic potential of Bacillus spp., specific genes responsible for antimicrobial peptides (AMPs) production were assessed through PCR. Among the ten antimicrobial peptides (AMP) assayed (Fig. 2), B. subtilis strain BiocWB was found to possess the maximum of eight AMPs namely ericin, subtilin, surfactin, iturin, bacilysin, subtilosin, fengycin and bacillomycin except mycosubtilin and mersacidin. The isolate B. amyloliqufaciens strain K12 and B. luciferensis strain K2 were found to possess six AMPs viz., ericin, subtilin, surfactin, iturin and bacilysin and subtilosin, respectively. In a previous study Vater et al. (2002) opined that antibiotics production with wide antimicrobial spectrum provided strong active biocontrol activities against plant fungal pathogens, for examples cyclic lipopeptides production by Bacillus spp. such as bacillomycin, fengycin, iturin, and surfactin, dipeptides (bacilysin) and lantibiotic (subtilin) played a major role in bio-control activity (Stein 2005; Bongers et al. 2005).
Fig. 2.
Results of PCR amplification of AMP genes from Bacillus strains
Volatiles and soluble metabolites released by Bacillus spp
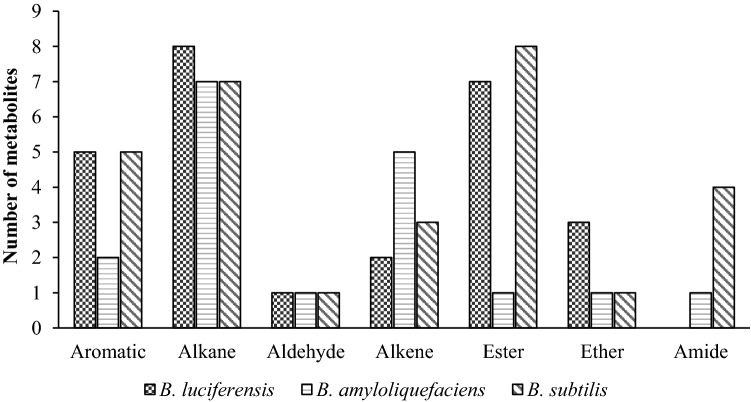
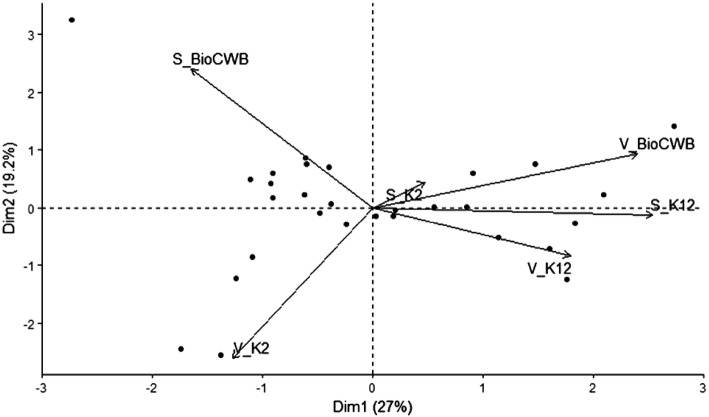
The release of volatiles by microorganisms such as bacteria and fungi has been studied by several researchers (Kai et al. 2009). In our study, many aliphatic, aromatic, sulfur-containing and nitro compounds were characterized using TD-GC–MS technique and the results showed that 29, 26 and 23 different volatile compounds were observed in B. subtilis BioCWB, B. luciferensis K2 and B. amyloliquefaciens K12, respectively (Fig. 3). The compounds having maximum area percentage in B. subtilis BioCWB were 1,3-Pentadiene, (Z)-(17.98%), 3-Hexyn-2-ol (15.44%) and disulfide dimethyl (10.25%). Similarly, abundant volatiles quantified in B. luciferensis K2 were 2-methyl-Furan (17.56%), acetic acid (14.94%) and disulfide dimethyl (8.18%), whereas B. amyloliquefaciens K12 had only two abundant volatiles namely disulfide dimethyl (15.79%) and 3-methyl-Furan (11.24%). Huang et al. (2012) reported that VOCs produced by Bacillus spp. play important roles in the plant, such as preventing the colonization of fungal phytopathogens, triggering induced systemic resistance (ISR) by activating NPR1 and SA/ethylene mediated pathways (Rudrappa et al. 2010) and participating in growth regulations of the plant (Ryu et al. 2003). Among the soluble metabolites, disulfide dimethyl (DMDS) has been previously reported to play important role in control of several fungal diseases by triggering induced systemic resistance (ISR) in plant (Huang et al. 2012) and also improves plant growth by modifying root architecture thereby making this VOC an alternative to agricultural chemicals (Tyagi et al. 2019). Additionally, DMDS has been used as an alternative to methyl bromide which has been used as a fumigant (Fritsch 2004) for controlling phytopathogenic fungi (Zou et al. 2007) and nematodes (Coosemans 2004). Although reports have claimed regarding the production of VOCs by the rhizobacteria B. subtilis and B. amyloliquefaciens and their involvement in plant growth stimulation and ISR (Farag et al. 2006), this is the first report of DMDS quantification in B. luciferensis. Similarly, the soluble metabolites secreted by bacteria were quantified by GC–MS which showed the presence of aromatic, alkane, aldehyde, alkene, ester, ether and amide compounds. In our analysis, higher amount of alkane and ester compounds were present in B. subtilis BioCWB and B. luciferensis K2 as compared to B. amyloliquefaciens K12 (Fig. 4). Also B. subtilis BiocWB had 30 metabolites with 9-octadecenamide (23.256%) being the most abundant followed by 5-octadecene (6.074%) and 3-eicosene (5.735%) (Supplementary Fig. 1). Based on previous studies it was found the compound 9-Octadecenamide was reported to have antimicrobial properties (Gumgumjee and Hajar 2015). The other strain, B. luciferensis K2 had 26 metabolites among which E-15-Heptadecenal (9.89%) was found to be maximum followed by E-3-Eicosene (9.31%) (Supplementary Fig. 2). Similarly, B. amyloliquefaciens strain K12 recorded least number of 18 metabolites with E-15-heptadecenal (9.20%) being the maximum followed by E-3-eicosene (8.41%) (Supplementary Fig. 3). The compound E-15-heptadecenal had higher relative abundance in comparisons to other compounds as found in B. luciferensis and B. amyloliquefaciens which have earlier been reported to also have antimicrobial properties (Yogeswari et al. 2012). From the PCA analysis, it was observed that the volatiles released by B. luciferensis K2 had more variations in comparisons with B. subtilis BioCWB and B. amyloliquefaciens K12 whereas soluble metabolites had more variations in B. subtilis BioCWB as compared to B. amyloliquefaciens K12 and B. luciferensis K2 (Fig. 5).
Fig. 3.
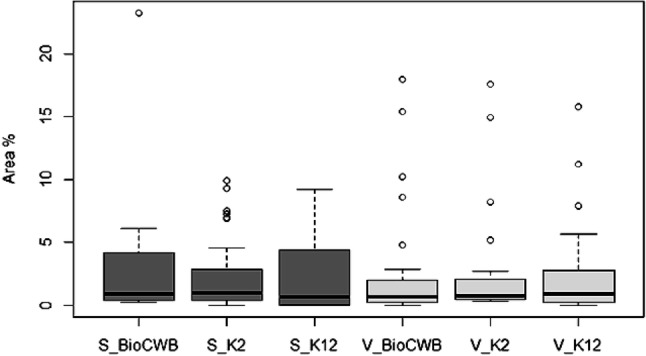
Comparisons of volatiles and soluble metabolites released by Bacillus spp. The figure represents total number of soluble metabolites (denoted “S”; dark-grey) and volatile metabolites (denoted “V”; light-grey) released by B. subtilis BioCWB, B. luciferensis K2 and B. amyloliquefaciens K12
Fig. 4.
Different types of soluble metabolites detected in individual Bacillus spp
Fig. 5.
PCA analysis of volatiles and soluble metabolites from Bacillus spp.The principal component analysis separates the distribution of volatile and soluble metabolites across two dimensions namely first principal component (Dim 1) and first principal component (Dim 2) representing 27% and 19.2% respectively. Soluble released by bacterial isolates, are denoted by prefix “S” whereas volatiles are denoted by prefix “V”
Evaluation of plant growth promoting potential of Bacillus spp. in rice and Capsicum plants
Based on agronomic growth parameters, there was a significant difference in all plant growth parameters except tiller number in rice plants treated with Bacillus spp. as compared to uninoculated control. All three Bacillus spp. (K2, K12 and BioCWB) significantly (p = 0.05) improved plant dry biomass in rice (6.2–7.3 g per plant) as compared to uninoculated control (Table 6). However, among the three bacterial strains, B. luciferensis K2 inoculated rice plants showed significantly higher plant biomass (7.3 g per plant) as compared to Bacillus spp. The combined PGP traits viz., phytohormone production and nutrient solubilization abilities observed in Bacillus spp. (K2, K12 and BioCWB) might have improved plant growth promotion in rice. The present results are corroborated with the earlier reports of Govindasamy et al. (2010) and Posada et al. (2018), who have described plant growth promoting potentials of Bacillus spp. Further, the evaluation of Bacillus spp. (K2, K12 and BioCWB) in Capsicum plants under field conditions in Sikkim (Table 7 and Fig. 6) clearly shows their plant growth promoting abilities and suitability other than cereals. In Capsicum plants, B. luciferensis K2 recorded significantly higher root dry weight, plant dry biomass and fruit number, followed by B. amyloliquefaciens K12 and B. subtilis BioCWB. Further, the combination of three strains was also carried in Capsicum plant and was found superior in enhancing plant height (48.3 cm), dry shoot (10.0 g), root weight (0.7 g), plant biomass (10.7 g) and fruit number (14.3) as compared to individual strains. In general, application of Bacillus spp. either individually or in combination significantly increased plant growth parameters as compared to uninoculated control. Many previous studies have proved that, species of Bacillus spp. like B. amyloliquefaciens, B. subtilis, B. thringiensis and B. megaterium (Jamal et al. 2018; Kour et al. 2018) possess PGP traits and enhance plant growth in several agricultural and horticultural crops.
Table 6.
Effect of Bacillus spp. inoculation on plant growth promotion in rice after 60 days of transplanting
| Treatment | Plant height (cm) | Root length (cm) | Fresh shoot wt. plant−1 (g) | Fresh root wt.plant−1 (g) | Dry shoot wt. plant−1 (g) | Dry root dry wt. plant−1 (g) | Total dry biomass plant−1 (g) | Tiller number plant−1 |
|---|---|---|---|---|---|---|---|---|
| K2 | 117.3 ± 3.12b | 19.3 ± 1.26a | 26.5 ± 0.42a | 2.5 ± 0.09b | 6.2 ± 0.06a | 1.1 ± 0.03a | 7.3 ± 1.15a | 7.8 ± 1.14 |
| K12 | 131.2 ± 1.46a | 16.9 ± 0.71b | 24.3 ± 0.18b | 2.3 ± 0.12c | 5.7 ± 0.29c | 1.03 ± 0.02c | 6.7 ± 0.47c | 8.2 ± 1.34 |
| BioCWB | 129.8 ± 1.21a | 19.1 ± 0.35a | 25.6 ± 0.19a | 2.8 ± 0.09a | 6.0 ± 0.13b | 1.09 ± 0.05b | 7.1 ± 0.93ab | 8.7 ± 0.89 |
| Control | 119.4 ± 2.05b | 16.7 ± 0.50b | 22.6 ± 0.28b | 2.2 ± 0.05c | 5.2 ± 0.10d | 1.01 ± 0.01c | 6.2 ± 0.37d | 7.0 ± 0.81 |
| CD (0.05) | 2.7 | 1.07 | 0.38 | 0.12 | 0.23 | 0.04 | 0.25 | NS |
Lowercase letters represent significant variations among the data at p < 0.05
Table 7.
Evaluation of Bacillus strains at field level in capsicum plant
| Treatments | Plant height (cm) | Fruits number | Dry shoot weight (g) | Dry root weight (g) | Total plant dry biomass (g) |
|---|---|---|---|---|---|
| T1—uninoculated control | 29.3 ± 2.6c | 6.0 ± 0.5c | 5.3 ± 0.2d | 0.4 ± 0.1c | 6.0 ± 0.5d |
| T2—BioCWB | 33.7 ± 3.5bc | 6.0 ± 0.5c | 7.0 ± 0.5c | 0.7 ± 0.1ab | 7.4 ± 0.6c |
| T3—K12 | 46.0 ± 3.1ab | 10.7 ± 1.5b | 6.9 ± 0.2c | 0.6 ± 0.2b | 7.5 ± 0.5c |
| T4—K2 | 41.3 ± 2.3abc | 8.7 ± 1.0bc | 8.2 ± 0.5b | 0.7 ± 0.1a | 8.9 ± 0.5b |
| T5—BioCWB + K12 + K2 | 48.3 ± 3.7a | 14.3 ± 0.5a | 10.0 ± 0.5a | 0.7 ± 0.1a | 10.7 ± 0.7a |
| CD (0.05) | 17.1 | 3.2 | 0.3 | 0.9 | 0.3 |
Lowercase letters represent significant variations among the data at p < 0.05
Fig. 6.
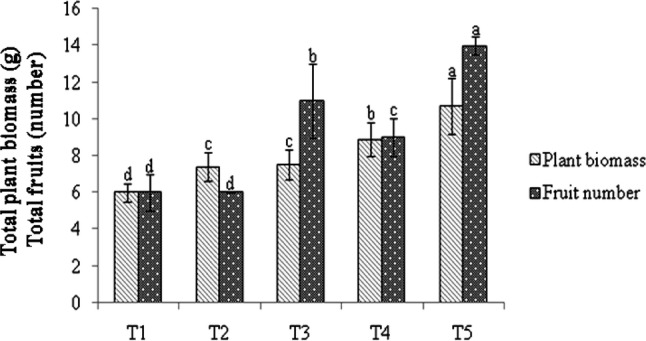
Evaluation of Bacillus spp. at field level in Capsicum plant. The following treatments were applied T1—uninoculated control, T2—B. subtilis BioCWB, T3—B. amyloliquefaciens K12, T4—B. luciferensis K2, T5—B. subtilis BioCWB + B. amyloliquefaciens K12 + B. luciferensisK2
Conclusion
The following native bacterial strains viz., B. subtilis BioCWB, B. luciferensis K2 and B. amyloliquiefaciens K12 could be used as bio-inoculants for enhancement of growth and yield of Capsicum under organic cultivation in Sikkim, India. Further, in the present study, the documented antifungal and insecticidal properties of bacterial strains demonstrated possibilities of application as biocontrol agents for organic cultivation of agricultural crops. For the first time, the novelties of multifunctional plant growth promotion and biocontrol potential associated with B. luciferensis K2 were documented from organic soils of Sikkim, India.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The first author thanks Director, ICAR-National Rice Research Institute, Odisha, India for providing all the support for carrying out this research work and special thanks to DBT, India for financial support.
Funding
We are grateful to the Department of Biotechnology, Government of India for promotion of Biotechnology in the North Eastern Region of India through Biotech Consortium India Limited and giving grants (BT/PR16291/NER/95/185/2015) for this research work.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abbott WS. A method for computing the effectiveness of an insecticide. J Econ Entomol. 1925;18:265–267. doi: 10.1093/jee/18.2.265a. [DOI] [Google Scholar]
- Abo-Aba SEM, Soliman EAM, Nivien AA. Enhanced production of extra cellular alkaline protease in Bacillus circulance through plasmid transfer. Res J Agric Biol Sci. 2006;16:526–530. [Google Scholar]
- Anantha PS, Deventhiran M, Saravanan P, Anand D, Rajarajan S. A comparative GC-MS analysis of bacterial secondary metabolites of Pseudomonas species. The Pharm Innov. 2016;5(part B 4):84. [Google Scholar]
- Bakker AW, Schippers B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas SPP-mediated plant growth-stimulation. Soil Biol Biochem. 1987;19(4):451–457. [Google Scholar]
- Bongers RS, Veening JW, Van Wieringen M, Kuipers OP, Kleerebezem M. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl Environ Microbiol. 2005;71(12):8818–8824. doi: 10.1128/AEM.71.12.8818-8824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow A, Brian PW, Chester VE, Curtis PJ, Hemming HG, Henehan C, Jeffreys EG, Lloyd PB, Nixon IS, Norris GL, Radley M. Gibberellic acids a metabolic product of the fungus Gibberella fujikuroi some observations on its production and isolation. J Sci Food Agric. 1955;6:340–348. [Google Scholar]
- Bottini R, Cassán F, Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol. 2004;65(5):497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- Cappuccino JG, Sherman N (1992) Serial dilution agar plating procedure to quantitate viable cells. Microbiology: a laboratory manual, 3rd edn. The Benjamin Cummings Publishing Co., Inc, Bedwood, pp 77–82
- Coosemans J (2004) Dimethyl disulphide (DMDS): a potential novel nematicide and soil disinfectant. In: VI International symposium on chemical and non-chemical soil and substrate disinfestation-SD2004 698, pp 57–64
- Ding Y, Wang J, Liu Y, Chen S. Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region. J Appl Microbiol. 2005;99(5):1271–1281. doi: 10.1111/j.1365-2672.2005.02738.x. [DOI] [PubMed] [Google Scholar]
- Egamberdieva D, Wirth SJ, Alqarawi AA, Abd-Allah EF, Hashem A. Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol. 2017;8:2104. doi: 10.3389/fmicb.2017.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanchezhiyan K, Keerthana U, Nagendran K, Prabhukarthikeyan SR, Prabakar K, Raguchander T, Karthikeyan G. Multifaceted benefits of Bacillus amyloliquefaciens strain FBZ24 in the management of wilt disease in tomato caused by Fusarium oxysporum f. sp. lycopersici. Physiol Mol Plant Pathol. 2018;103:92–101. [Google Scholar]
- Farag MA, Ryu CM, Sumner LW, Paré PW. GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry. 2006;67(20):2262–2268. doi: 10.1016/j.phytochem.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Fisher SH. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32(2):223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- Fritsch J (2004) Dimethyl disulfide as a new chemical potential alternative to methyl bromide in soil disinfestation in France. In: VI international symposium on chemical and non-chemical soil and substrate disinfestation-SD2004 698 (pp 71–76)
- Gordon SA, Weber RP. Colorimetric estimation of indole acetic acid. Plant Physiol. 1951;26(1):192. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindasamy Venkadasamy, Senthilkumar Murugesan, Magheshwaran Vellaichamy, Kumar Upendra, Bose Pranita, Sharma Vikas, Annapurna Kannepalli. Plant Growth and Health Promoting Bacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. Bacillus and Paenibacillus spp.: Potential PGPR for Sustainable Agriculture; pp. 333–364. [Google Scholar]
- Gumgumjee NH, Hajar AS. Antibacterial activities and GC-MS analysis of phytocomponents of Ehretia abyssinica R. br. ex. Fresen. Int J Applied Biol Pharm Technol. 2015;6(2):236–241. [Google Scholar]
- Hsu SC, Lockwood JL. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol. 1975;29(3):422–426. doi: 10.1128/am.29.3.422-426.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tsay JF, Chang SY, Yang HP, Wu WS, Chen CY. Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag Sci. 2012;68(9):1306–1310. doi: 10.1002/ps.3301. [DOI] [PubMed] [Google Scholar]
- Idris EE, Iglesias DJ, Talon M, Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact. 2007;20(6):619–626. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- IRRI, Standard Evaluation System (SES) of Rice (Revised) (2013) 5th Edition, Manila, Philippines
- Jamal Q, Lee YS, Jeon HD, Kim KY. Effect of plant growth-promoting bacteria Bacillus amyloliquefaciens y1 on soil properties, pepper seedling growth, rhizosphere bacterial flora and soil enzymes. Plant Protect Sci. 2018;54(3):1–9. [Google Scholar]
- Jha TB, Saha PS. Characterization of some Indian Himalayan Capsicums through floral morphology and EMA-based chromosome analysis. Protoplasma. 2017;254(2):921–933. doi: 10.1007/s00709-016-1001-z. [DOI] [PubMed] [Google Scholar]
- Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81(6):1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186(4):1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kour R, Ambardar S, Vakhlu J. Plant growth promoting bacteria associated with corm of Crocus sativus during three growth stages. Lett Appl Microbiol. 2018;67(5):458–464. doi: 10.1111/lam.13042. [DOI] [PubMed] [Google Scholar]
- Kumar BL, Gopal DS. Effective role of indigenous microorganisms for sustainable environment. 3 Biotech. 2015;5(6):867–876. doi: 10.1007/s13205-015-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Dubey RC, Maheshwari DK. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res. 2012;167(8):493–499. doi: 10.1016/j.micres.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Kumar S, Devi EL, Sharma SK, Ansari MA, Phurailatpam S, Ng TC, Singh TS, Prakash N, Kumar R, Kumawat N, Mandal D. Rice breeding strategies of North Eastern India for resilience to biotic and abiotic stresses: a review. ORYZA-An Int J Rice. 2017;54(1):1–12. [Google Scholar]
- Kumar U, Panneerselvam P, Govindasamy V, Vithalkumar L, Senthilkumar M, Banik A, Annapurna K. Long-term aromatic rice cultivation effect on frequency and diversity of diazotrophs in its rhizosphere. Ecol Eng. 2017;101:227–236. [Google Scholar]
- Kumar J, Pradhan M, Singhm N. Sustainable organic farming in Sikkim: an inclusive perspective, advances in smart grid and renewable energy. Singapore: Springer; 2018. pp. 367–378. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlali R, Hijri M. Screening, identification and evaluation of potential biocontrol fungal endophytes against Rhizoctonia solani AG3 on potato plants. FEMS Microbiol Lett. 2010;311(2):152–159. doi: 10.1111/j.1574-6968.2010.02084.x. [DOI] [PubMed] [Google Scholar]
- LeOra S (2007) Polo Plus, probit and logit analysis: user´s guide computer program version 2.0. LeOra Software, Berkeley, CA
- Macfadyen S, McDonald G, Hill MP. From species distributions to climate change adaptation: knowledge gaps in managing invertebrate pests in broad-acre grain crops. Agric Ecosyst Environ. 2018;253:208–219. [Google Scholar]
- Maharana I, Rai SC, Sharma E. Environmental economics of the Khangchendzonga National Park in the Sikkim Himalaya, India. GeoJournal. 2000;50(4):329–337. [Google Scholar]
- Manikandan R, Saravanakumar D, Rajendran L, Raguchander T, Samiyappan R. Standardization of liquid formulation of Pseudomonas fluorescens Pf1 for its efficacy against Fusarium wilt of tomato. Biol Control. 2010;54(2):83–89. [Google Scholar]
- Mishra Vineet Kumar, Passari Ajit Kumar, Chandra Preeti, Leo Vincent Vineeth, Kumar Brijesh, Uthandi Sivakumar, Thankappan Sugitha, Gupta Vijai Kumar, Singh Bhim Pratap. Determination and production of antimicrobial compounds by Aspergillus clavatonanicus strain MJ31, an endophytic fungus from Mirabilis jalapa L. using UPLC-ESI-MS/MS and TD-GC-MS analysis. PLOS ONE. 2017;12(10):e0186234. doi: 10.1371/journal.pone.0186234. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Murphy JAMES, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–36. [Google Scholar]
- Panda B, Rahman H, Panda J. Phosphate solubilizing bacteria from the acidic soils of Eastern Himalayan region and their antagonistic effect on fungal pathogens. Rhizosphere. 2016;2:62–71. [Google Scholar]
- Panneerselvam P, Kumar U, Sahu S, Mohapatra SD, Dangar TK, Parameswaran C, Jahan A, Senapati A, Govindharaj GPP. Larvicidal potential of Skermanella sp. against rice leaf folder (Cnaphalocrosis medinalis Guenee) and pink stem borer (Sesamia inferens Walker) J Invertebr Pathol. 2018;157:74–79. doi: 10.1016/j.jip.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Pathak H, Mohanty S, Jain N, Bhatia A. Nitrogen, phosphorus, and potassium budgets in Indian agriculture. Nutr Cycl Agroecosyst. 2010;86(3):287–299. [Google Scholar]
- Posada LF, Álvarez JC, Romero-Tabarez M, de-Bashan L, Villegas-Escobar V. Enhanced molecular visualization of root colonization and growth promotion by Bacillus subtilis EA-CB0575 in different growth systems. Microbiol Res. 2018;217:69–80. doi: 10.1016/j.micres.2018.08.017. [DOI] [PubMed] [Google Scholar]
- Prabhukarthikeyan R, Saravanakumar D, Raguchander T. Combination of endophytic Bacillus and Beauveria for the management of Fusarium wilt and fruit borer in tomato. Pest Manag Sci. 2014;70(11):1742–1750. doi: 10.1002/ps.3719. [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Biedrzycki ML, Kunjeti SG, Donofrio NM, Czymmek KJ, Paul WP, Bais HP. The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun Integrative Biol. 2010;3(2):130–138. doi: 10.4161/cib.3.2.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci. 2003;100(8):4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24(4):814–842. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Sridhar R, Singh UD. Genetic and pathogenic diversity of the rice blast pathogen, in major fungal diseases of rice. Dordrecht: Springer; 2001. pp. 1–7. [Google Scholar]
- Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbial. 2005;56:4845–4857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62(2):625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC (2000) R language definition. Vienna, Austria: R foundation for statistical computing
- Thorn RMS, Greenman J. Microbial volatile compounds in health and disease conditions. J Breath Res. 2012;6(2):024001. doi: 10.1088/1752-7155/6/2/024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Kim K, Cho M, Lee KJ. Volatile dimethyl disulfide affects root system architecture of Arabidopsis via modulation of canonical auxin signaling pathways. Environ Sustain. 2019;2019:1–6. [Google Scholar]
- Tyc O, Song C, Dickschat JS, Vos M, Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25(4):280–292. doi: 10.1016/j.tim.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Vater J, Kablitz B, Wilde C, Franke P, Mehta N, Cameotra SS. Matrix-assisted laser desorption ionization-time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl Environ Microbiol. 2002;68(12):6210–6219. doi: 10.1128/AEM.68.12.6210-6219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinodkumar S, Nakkeeran S, Renukadevi P, Malathi VG. Biocontrol potentials of antimicrobial peptide producing Bacillus species: multifaceted antagonists for the management of stem rot of carnation caused by Sclerotinia sclerotiorum. Front Microbiol. 2017;8:446. doi: 10.3389/fmicb.2017.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright Erik,S. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. The R Journal. 2016;8(1):352. [Google Scholar]
- Yadav SN, Chandra R, Khura TK, Chauhan NS. Energy input–output analysis and mechanization status for cultivation of rice and maize crops in Sikkim. Agric Eng Int CIGR J. 2013;15(3):108–116. [Google Scholar]
- Yogeswari S, Ramalakshmi S, Neelavathy R, Muthumary J. Identification and comparative studies of different volatile fractions from Monochaetia kansensis by GCMS. Glob J Pharmacol. 2012;6(2):65–71. [Google Scholar]
- Zou CS, Mo MH, Gu YQ, Zhou JP, Zhang KQ. Possible contributions of volatile-producing bacteria to soil fungistasis. Soil Biol Biochem. 2007;39(9):2371–2379. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.