Abstract
Background:
Birds are considered as a reservoir for pathogenic and non-pathogenic fungi. Pigeon droppings have the potential for spreading these fungi to the environment. Cryptococcus species are important fungi associated with pigeon droppings. In this regard, there are many types of yeast associated with guano that is important for human and animal health.
Aims:
The main objective of this study is the identification of non-Cryptococcus yeasts isolated from pigeon dropping in Shiraz, Southern Iran.
Methods:
A total of 100 unknown yeasts, which were previously screened and identified as non-Cryptococcus from pigeon guano through the conventional methods, were used in this study. Identification of the isolates was performed based on conventional methods and DNA sequence analysis of internal transcribed spacer (ITS) rDNA gene region. The sequence results were deposited in NCBI database using the Basic Local Alignment Search Tool (BLAST).
Results:
A total of 16 species belonging to 7 genera were identified as Candida spp. 51% (8 species), Rhodotorula sp. 24%, Trichosporon spp. 21% (3 species), Rhodosporidium 2%, Saccharomyces 1%, Rhizoctonia 1%, and Meyerozyma 1%. The predominant isolates were Rhodotorula rubra (24%), Candida famata (20%), and Trichosporon asahii (13%). The other species were Rhodosporidium kratochvilovae 2 (2%), Saccharomyces cerevisiae 1 (1%), Rhizoctonia solani 1 (1%), and Meyerozyma caribbica 1 (1%).
Conclusion:
Pigeon excreta examined in this study were associated with several kinds of opportunistic yeasts which could cause diseases in prone human and animals.
Key Words: Candida, DNA sequence, Non-Cryptococcus, Pigeon
Introduction
Avian dropping is known as the reservoir of different pathogenic and opportunist organisms (Chee and Lee, 2005 ▶). Pigeons are reported as a major carrier of Cryptococcus species including C. neoformans, which is the most deadly opportunist yeast known (Costa et al., 2010 ▶; Soltani et al., 2013 ▶), and is considered as reservoir of zoonotic yeasts and other pathogenic fungi (Medina et al., 2017 ▶).
In an urban area, pigeons could have a threating role in public health, as they live close to people in public parks, on the roof of hospital buildings, rooms near the air handler units, and even in sacred places where they are fed for religious reasons (Haag-Wackernagel and Moch, 2004 ▶).
On average, a well-fed pigeon could deposit 25 pounds of guano a year (Haag-Wackernagel and Moch, 2004 ▶). Naturally, by the wind and during raining or after some disasters like flooding, their droppings disseminate and come into close contact with people, especially the susceptible ones.
Although Cryptococcus has received much attention in medicine, it is not the only pathogen that affects human health. Recently, other opportunistic fungi such as Candida spp., Trichosporon spp., Rhodotorula spp., Geotrichum spp., Mucor spp., and Aspergillus spp. have been isolated from pigeon droppings (Khosravi, 1997 ▶; Lanzafame et al., 2001 ▶; Costa et al., 2010 ▶; Abulreesh et al., 2015 ▶; Lee et al., 2017 ▶). In recent years, opportunistic fungal infections by uncommon yeast species are increasing (Lanzafame et al., 2001 ▶; Munoz et al., 2005 ▶). Candidiasis is known as the most common fungal infection in human and birds of prey (Deem, 2003 ▶). Several cases of candidemia with uncommon Candida species such as C. fumata and C. lusitania have been reported, which are less known concerning their epidemiological features and ways of transition (Chen et al., 2009 ▶). Identification of these species helps to understand the importance of these fungi and risk of encountering and provides useful information for epidemiological and ecological studies.
Although the yeast composition of pigeon faeces has been reported by many studies from different regions of Iran and around the world, transmission of the pathogens depends on climatic condition, stability, in the environment, and environmental factors such as temperature and humidity (Tsiodras et al., 2008 ▶).
Although there are many studies conducted based on culture-dependent methods, they are not precise enough to identify the yeast species (Rastogi and Sani, 2011 ▶). In this regard, the molecular-based method is more specific and sensitive than conventional methods. Taxonomic identification of fungi based on DNA sequence analysis is one of the best techniques applied to identify non-coding rDNA spacer regions.
The internal transcribed spacer (ITS) includes the ITS1 and ITS2 noncoding regions that are located between the 18S rRNA and 28S rRNA genes and are quite variable among related species. By amplification and sequencing of these regions, we can achieve a higher resolution compared to other methods (Bellemain et al., 2010 ▶). Using this method, Cafarchia could identify the yeasts species isolated from hens faeces (Cafarchia et al., 2018 ▶) but there is less data regarding identification of yeasts associated with bird droppings by molecular techniques.
In our previous study, we identified many yeast isolates as Cryptococcus species by molecular and basic conventional methods (Pakshir et al., 2018 ▶). In the present work, we also isolate many unknown yeast species associated with pigeon guano. The main aim of this study is to identify the pathogenic and non-pathogenic yeasts isolates associated with pigeon dropping in Shiraz, Southern Iran by ITS sequence analysis.
Materials and Methods
Isolates preparation
In this study, 100 unknown frozen yeast stock isolates from pigeon guano - previously identified as non-Cryptococcus (our previous study) - were analyzed (2014). The samples were subcultured on Sabouraud’s Dextrose Agar pH = 5.6 ± 0.2 (Merck, Germany), supplemented with chloramphenicol (50 mg/L), incubated at 25°C for 48 h, and inspected daily for growth.
Conventional identification methods
Primary identification of isolates was based on micromorphological analysis of yeast shape using the teased mount preparation results (round, ovoid or rectangular shape), growth on Corn Meal Agar Tween 80 (Merck, Germany) for detection of pseudohypha and chlamydoconidia, production of germ tube in serum and colony color on CHROMagar Candida media (Paris, France).
Molecular method
DNA preparation
Genomic DNA was extracted through the boiling method described by Makimura et al. (1994) ▶ with small modifications. Briefly, a small amount of the yeast colony was suspended in 100 µL of lysis buffer containing 100 mM Tris-HCl, 0.5% sodium dodecyle sulfate (SDS), and 30 mM ethylenediaminetetraacetic acid (EDTA) and boiled for 15 min at 100°C. A solution of potassium acetate (2.5 M) was then added to the lysate, held on ice for 1 h, centrifuged at 16125 × g for 5 min, and the supernatant was transferred to a new tube. The yeast DNA in the supernatant was washed twice with ethanol and air dried and re-suspended in 50 µL of distilled water (DW) prior to use for polymerase chain reaction (PCR). The purity and quantity of DNA were evaluated by absorbance ratio of A260/A280, and also nano-drop reading results.
Amplification and sequencing of the ITS regions
A set of universal primers (ITS1 5´-TCC GTA GGT GAA CCT GCG G-3´ and ITS4 5´-TCC TCC GCT TAT TGA TAT GC-3´) (Meta-Bion International, Martinsried, Germany) were employed for amplification of ITS region of an ITS1-5.8S-ITS2 segment of the ribosomal DNA gene. PCR amplification was carried out in a final volume of 50 μL consisting of 5 μL of 10 × PCR buffer, 1.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates (0.2 mM each), 1.2 U of Taq DNA polymerase (Roche Molecular Biochemicals, Mannheim, Germany), 0.5 μM of each primer, 2 µL of DNA template, and eventually with DW to a final volume (50 µL). An initial denaturation step at 95°C for 5 min was followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 50 s, and extended at 72°C for 1 min, with a final extension step at 72°C for 7 min and kept at 4°C for 5 min. Negative controls were also used in each set of reactions. The PCR product was electrophoresed on 1.2% agarose gel and stained with ethidium bromide. Then, the PCR products were purified and sequenced (Bioneer Company, South Korea). The sequence results were processed using the web-based blasting program and Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST), and the data were compared with those in the NCBI/Genebank database.
Results
Germ tube formation was observed in eight isolates of Candida albicans (8%). Candida albicans, C. krusei, and C. glabrata present green, white-pink, and purple colony colors respectively and the other yeast species had no color and remain white.
ITS region of extracted DNA was amplified in all samples (Fig. 1) and sequence results of PCR products demonstrate 16 species of fungi, belonging to 7 genera (Table 1), and compared with the other data (Table 2).
Fig. 1.
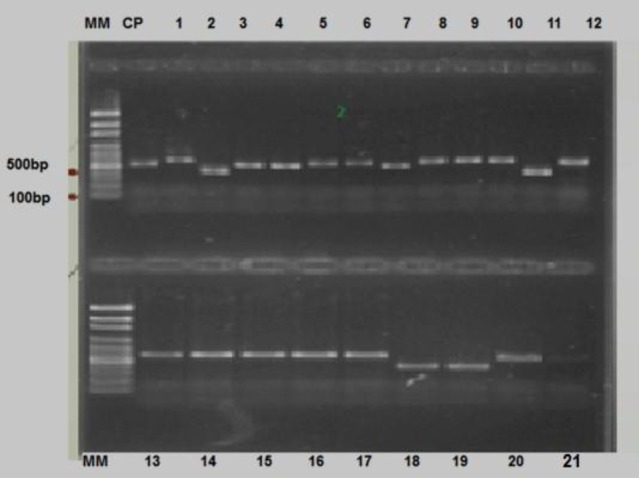
Results of PCR products electrophoresis. Line MM: 100 bp molecular marker, Line CP: Standard positive control, Lines 1, 5, 6: Rhodotorula rubra, Lines 2, 11, 18, 19: Candida cutenulate, Line 3: Saccharomyces cerevisiea, Lines 4, 7: Trichosporon asahii, Lines 8-10 and 12-17: Candida famata, Line 20: Cryptococcus albidus, and Line 21: Negative control (distilled water)
Table 1.
Result of yeast species identified from pigeon dropping in Shiraz
| Yeast species | Number | Percent | Yeast species | Number | Percentage |
|---|---|---|---|---|---|
| Candida famata | 20 | 20% | Rhodotorula rubra | 24 | 24% |
| Candida albicans | 8 | 8% | Trichosporon asahii | 13 | 13% |
| Candida glabrata | 8 | 8% | Trichosporon coremiiforme | 6 | 6% |
| Candida cutenulata | 7 | 7% | Rhodosporidium kratochvilovae | 2 | 2% |
| Candida krusei | 2 | 2% | Trichosporon moniliiforme | 1 | 1% |
| Candidia colliculosa | 2 | 2% | Saccharomyces cerevisiea | 1 | 1% |
| Candida lusitania | 1 | 1% | Rhizoctonia solani | 1 | 1% |
| Pichia anomala | 3 | 3% | Meyerozyma caribbica | 1 | 1% |
| Total | 100 |
Table 2.
Comparison of the most common yeast species isolated from pigeon dropping in Iran and the other countries
| Yeast species | Iran | Other countries |
|---|---|---|
| Cryptococcus | Yes | Yes |
| Candida | Yes | Yes |
| Trichosporon | Yes | Yes |
| Rhodotorula | Yes | Yes |
| Saccharomyces | Yes | Yes |
| Geotrichum | Yes | Yes |
| Pichia | Yes | Yes |
| Rhodosporidium | Yes | No |
| Rhizoctonia | Yes | No |
| Meyerozyma | Yes | No |
| Torulopsis | No | Yes |
| Malassezia | No | Yes |
| Wallemia | No | Yes |
| Debaromyces | No | Yes |
| Zygoseccharomyces | No | Yes |
Genus Candida has the highest prevalence and Rodotorula rubra 24%, Candida famata 20%, and Trichosporon asahii 13% were the most dominant species isolated from pigeon extra.
Discussion
Pigeon droppings, which are found in public areas such as streets, parks and gardens, old buildings and hospitals, and pigeon towers, are considered as a possible source of infections to human and animals. Nitrogen-rich environment contaminated with birds dropping provides a suitable condition for bacteria and fungi growth (Harrigan, 1998 ▶). The chemical properties and composition of pigeon faeces (pH, uric acid, and nitrogen) create an excellent substrate for fungal spores to propagate. In this regard, the abundance of pathogenic fungi has been linked to weather (humidity, temperature, and radiation), vegetation, and microbes associated with guano (Lee et al., 2017 ▶). Excreta of pigeons also were reported as an ideal environment for yeast replication, especially Candida and Trichosporon species (Medina et al., 2017 ▶).
If there was any evidence for transmission of the pathogen to humans from the avian species, it could classify as direct/indirect transmission. Although there is little known about the actual direct/indirect transmission of these pathogens, the risk exists through ingestion of water contaminated from faeces or exposure to inanimate surfaces contaminated by bird secretions or droppings (Tsiodras et al., 2008 ▶). Pigeons are an important source in the spread and maintenance of C. neoformans. In addition, various species of pathogenic/non-pathogenic yeasts are associated with pigeons that are important in human and animal health (Nweze et al., 2015 ▶; Simi et al., 2018 ▶). To date, approximately 48 species of potentially pathogenic fungi from 28 genera have been isolated from pigeons globally (Abulreesh et al., 2015 ▶; Lee et al., 2017 ▶).
Among various diagnostic methods, molecular techniques such as DNA sequence analysis of ITS region are the most accurate techniques that have been used in many countries like Korea and China (Cafarchia et al., 2006 ▶; Lee et al., 2017 ▶). Using this method, we could identify 17 species belonging to 8 genera. In Iran, many studies (by non-molecular techniques) report that the highest prevalence of non-Cryptococcus yeasts associated with pigeon dropping belongs to genus Candida (Khosravi, 1997 ▶; Soltani et al., 2013 ▶). Similar to those reports, in our study, Candida spp. was the most prevalent genus isolate but among the species, C. famata was the predominant one; however, this data was not in agreement with the other data reported from Iran, Egypt, and Brazil (Khosravi, 1997 ▶; Costa et al., 2010 ▶; Soltani et al., 2013 ▶; Abulreesh et al., 2015 ▶).
In this study, the other uncommon Candida species associated with pigeons were C. catenulata, C. colliculosa, and C. lusitania. These species are responsible for candidiasis but diseases occurred by these organisms are less frequently encountered. Candida colliculosa is found in fruits and dairy products like grapes and milk and was responsible for fungal endocarditis in a Turkish gardener patient (Kaygusuz et al., 2003 ▶). Candida catenulata and C. famata also were isolated from droppings of captive birds in Italy (Mancianti et al., 2002 ▶). Elsewhere, C. catenulata was reported as the causative agent of fungemia in a patient with gastric cancer (Radosavljevic et al., 1999 ▶).
Pichia anumala (anamorphic stage of Candida pelliculosa) was another yeast identified in our study. Murphy et al. (1986) ▶ report nosocomial outbreaks of fungemia due to P. anomala in Liverpool. Another study has reported the same species responsible for an outbreak of nosocomial fungemia in the pediatric wards of a hospital in India (Chakrabarti et al., 2001 ▶). Candida species was also reported as main causative infections in different animals like dogs, buffalo, cattle, horse and goat (Jadhav and Pal, 2013 ▶).
Rhodotorula rubra was the second most frequent isolate from pigeon excreta in this study. This genus is mostly associated with bird droppings. Cafarchia et al. (2006) ▶ reported this fungus as the highest number of isolates from cloacae of migratory wild birds in Italy while Costa et al. (2010) ▶ reported it in Brazilian pigeons. Although this yeast is not as dangerous for humans, it could occasionally cause diseases like meningitis in hospitalized patients (Lanzafame et al., 2001 ▶). There are several reports of skin infections in chickens, lung infection in sheep, dermatitis in cat and respiratory tract in dog all of which are caused by Rodotorola species (Wirth and Goldani, 2012 ▶; Biegańska et al., 2018 ▶).
Trichosporon sp. was the third most frequent genus isolate including T. asahii (13%), T. coremiiforme (6%), and T. moniliforme (1%). This genus is also isolated from pigeon droppings in Egypt and Brazil (Costa et al., 2010 ▶; Abulreesh et al., 2015 ▶). Trichosporon species is considered as one of the most important etiological causes of nosocomial infections (Ghiasian et al., 2006 ▶; Miceli et al., 2011 ▶) and has been reported as the second- or third-most-common agent of yeast fungemia (Chagas-Neto et al., 2009 ▶).
Trichosporon asahi species that accounted for the highest percentage of systemic invasive infection in susceptible patients, as well as T. coremiiforme and T. moniliforme, cause superficial skin infections (Rodriguez-Tudela et al., 2005 ▶; Asada et al., 2006 ▶). This fungi could also cause diseases in dogs and cats (Karnik et al., 2009 ▶; Biegańska et al., 2018 ▶).
Another yeast identified in our study was Saccharomyces cerevisiae. This is a useful yeast that is used as a probiotic in the bakery, food, and brewing industries around the world. However, S. cerevisiae was reported as the causative agent of nosocomial infections that can cause a wide range of clinical syndromes such as peritonitis, cellulitis, and liver abscess. Munoz et al. (2005) ▶ reported three cases of fungemia in an intensive care unit of a hospital in Spain and present another 57 cases of S. cerevisiae fungemia in their literature review.
The yeast Meyerozyma caribbica was another isolate in our study that belongs to genus Colletotrichum. These fungi in this genus are phytopathogens and responsible for fruit rot such as mango. The yeast M. caribbica was used against this genus as a biological control but there is no evidence of pathogenicity in human being yet (Bautista-Rosales et al., 2013 ▶).
The yeast Rhizoctonia solani isolated from a skin lesion of a patient in India (Kaore et al., 2012 ▶) but human mycosis by this fungus is rare and less common. Finally, most of the yeasts associated with pigeon droppings in Iran were similar with those reported from the other countries as mentioned in result section.
In this study, we identified many species of yeasts associated with pigeon excreta in Shiraz city and more than 90% of the isolates had a history of diseases in the human and animals as opportunistic fungi. Pigeons are important reservoirs and carriers for zoonotic yeast in the environment.
Acknowledgements
This study was funded by Deputy of Research and Technology of Shiraz University of Medical Sciences, Shiraz, Iran (Grant No. 7220) and was extracted from an MSc thesis by Z. Zareshahrabadi.
Conflict of interest
The authors report no conflicts of interest.
References
- Abulreesh HH, Organji1 SR, Elbanna K, Haridy Osman GE, Almalki1 MHK, Abdel-Mallek AY. First report of environmental isolation of Cryptococcus neoformans and other fungi from pigeon droppings in Makkah, Saudi Arabia and in vitro susceptibility testing. Asian Pac. J. Trop. Dis. 2015;5:622–626. [Google Scholar]
- Asada N, Uryu H, Koseki M, Takeuchi M, Komatsu M, Matsue K. Successful treatment of breakthrough Trichosporon asahii fungemia with voriconazole in a patient with acute myeloid leukemia. Clin. Infec. Dis. 2006;43:e39–e41. doi: 10.1086/505970. [DOI] [PubMed] [Google Scholar]
- Bautista-Rosales PU, Calderon-Santoyo M, Servín-Villegas R, Ochoa-Álvarez NA, Ragazzo-Sánchez JA. Action mechanisms of the yeast Meyerozyma caribbica for the control of the phytopathogen Colletotrichum gloeosporioides in mangoes. Biol. Control. 2013;65:293–301. [Google Scholar]
- Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegańska MJ, Rzewuska M, Dąbrowska I, Malewska-Biel B, Ostrzeszewicz M, Dworecka-Kaszak B. Mixed infection of respiratory tract in a dog caused by Rhodotorula mucilaginosa and Trichosporon jirovecii: a case report. Mycopathologia. 2018;183:637–644. doi: 10.1007/s11046-017-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafarchia C, Camarda A, Romito D, Campolo M, Quaglia NC, Tullio D, Otranto D. Occurrence of yeasts in cloacae of migratory birds. Mycopathologia. 2006;161:229–234. doi: 10.1007/s11046-005-0194-z. [DOI] [PubMed] [Google Scholar]
- Cafarchia C, Iatta R, Danesi P, Camarda A, Capelli G, Otranto D. Yeasts isolated from cloacal swabs, feces, and eggs of laying hens. Med. Mycol. 2018;57:340–345. doi: 10.1093/mmy/myy026. [DOI] [PubMed] [Google Scholar]
- Chagas-Neto TC, Chaves GM, Melo ASA, Colombo AL. Bloodstream infections due to Trichosporon spp: species distribution, Trichosporon asahii genotypes determined on the basis of ribosomal DNA intergenic spacer 1 sequencing, and antifungal susceptibility testing. J. Clin. Microbiol. 2009;47:1074–1081. doi: 10.1128/JCM.01614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Singh K, Narang A, Singhi S, Batra R, Rao KLN, Ray P, Gopalan S, Das S, Gupta V, Gupta AK, Bose SM, McNeil MM. Outbreak of Pichia anomala infection in the pediatric service of a tertiary-care center in Northern India. J. Clin. Microbiol. 2001;39:1702–1706. doi: 10.1128/JCM.39.5.1702-1706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee HY, Lee KB. Isolation of Cryptococcus neoformans var grubii (serotype A) from pigeon droppings in Seoul, Korea. J. Microbiol. 2005;43:469–472. [PubMed] [Google Scholar]
- Chen SCA, Marriott D, Playford EG, Nguyenb Q, Ellis D, Meyer W, Sorrella TC, Slavin M. Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin. Microbiol. Infec. 2009;15:662–669. doi: 10.1111/j.1469-0691.2009.02821.x. [DOI] [PubMed] [Google Scholar]
- Costa AK, Sidrim JJC, Cordeiro RA, Brilhante RSN, Monteiro AJ, Rocha MFG. Urban pigeons (Columba livia) as a potential source of pathogenic yeasts: a focus on antifungal susceptibility of Cryptococcus strains in Northeast Brazil. Mycopathologia. 2010;169:207–213. doi: 10.1007/s11046-009-9245-1. [DOI] [PubMed] [Google Scholar]
- Deem SL. Fungal diseases of birds of prey. Vet. Clin. North Am. Exot. Anim. Pract. 2003;6:363–376. doi: 10.1016/s1094-9194(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Ghiasian SA, Maghsood AH, Mirhendi SH. Disseminated, fatal Trichosporon asahii infection in a bone marrow transplant recipient. J. Microbiol. Immunol. Infect. 2006;39:426. [PubMed] [Google Scholar]
- Haag-Wackernagel D, Moch H. Health hazards posed by feral pigeons. J. Infect. 2004;48:307–313. doi: 10.1016/j.jinf.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Harrigan, WF. Laboratory methods in food microbiology. London, UK: Gulf Professional Publishing; 1998. [Google Scholar]
- Jadhav VJ, Pal M. Human and domestic animal infections caused by Candida albicans. J. Mycopathol. Res. 2013;51:243–249. [Google Scholar]
- Kaore NM, Atul AR, Khan MZ, Ramnani VK. A rare case of human mycosis by Rhizoctonia solani. Indian J. Med. Microbiol. 2012;30:361–363. doi: 10.4103/0255-0857.99508. [DOI] [PubMed] [Google Scholar]
- Karnik K, Reichle JK, Fischetti AJ, Goggin JM. Computed tomographic findings of fungal rhinitis and sinusitis in cats. Vet. Radiol. Ultrasound. 2009;50:65–68. doi: 10.1111/j.1740-8261.2008.01491.x. [DOI] [PubMed] [Google Scholar]
- Kaygusuz I, Mulazimoglu L, Cerikcioglu N, Toprak A, Oktay A, Korten V. An unusual native tricuspid valve endocarditis caused by Candida colliculosa. Clin. Microbiol. Infec. 2003;9:319–322. doi: 10.1046/j.1469-0691.2003.00511.x. [DOI] [PubMed] [Google Scholar]
- Khosravi AR. Isolation of Cryptococcus neoformans from pigeon (Columba livia) droppings in northern Iran. Mycopathologia. 1997;139:93–95. doi: 10.1023/a:1006863705759. [DOI] [PubMed] [Google Scholar]
- Lanzafame M, De Checchi G, Parinello A, Cattelan MTAM. Rhodotorula glutinis-related meningitis. J. Clin. Microbiol. 2001;39:410. doi: 10.1128/JCM.39.1.410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WD, Fong JJ, Eimes JA, Lim YW. Diversity and abundance of human-pathogenic fungi associated with pigeon faeces in urban environments. Mol. Ecol. 2017;26:4574–4585. doi: 10.1111/mec.14216. [DOI] [PubMed] [Google Scholar]
- Makimura K, Murayama SY, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- Mancianti F, Nardoni S, Ceccherelli R. Occurrence of yeasts in psittacines droppings from captive birds in Italy. Mycopathologia. 2002;153:121. doi: 10.1023/a:1014576304894. [DOI] [PubMed] [Google Scholar]
- Medina IR, Fuentes LR, Arteaga MB, Valcárcel FR, Arbelo FA, Castillo DP, Suárez SD, Quintana OF, Gutiérrez BV, Sergent FS, Acosta-Hernández B. Pigeons and their droppings as reservoirs of Candida and other zoonotic yeasts. Rev. Iberoam. Micol. 2017;34:211–214. doi: 10.1016/j.riam.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011;11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- Munoz P, Bouza E, Cuenca-Estrella M, Eiros JM, Jesús Pérez M, Sánchez-Somolinos M, Rincón C, Hortal J, Peláez T. Saccharomyces cerevisiae fungemia: an emerging infectious disease. Clin. Infect. Dis. 2005;40:1625–1634. doi: 10.1086/429916. [DOI] [PubMed] [Google Scholar]
- Murphy N, Damjanovic V, Hart CA, Buchanan CR, Whitaker R, Cooke RWI. Infection and colonisation of neonates by Hansenula anomala. Lancet. 1986;327:291–293. doi: 10.1016/s0140-6736(86)90827-5. [DOI] [PubMed] [Google Scholar]
- Nweze EI, Kechia FA, Dibua UE, Charles EZE, Uwakwe SONOJA. Isolation of Cryptococcus neoformans from environmental samples collected in Southeastern Nigeria. Rev. Inst. Med. Trop. S. Paulo. 2015;57:295–298. doi: 10.1590/S0036-46652015000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakshir K, Fakhim H, Vaezi A, Meise JF, Mahmoodi M, Zomorodian K, Javidnia J, Ansari S, Hagen F, Badali H. Molecular epidemiology of environmental Cryptococcus species isolates based on amplified fragment length polymorphism. J. Mycol. Med. 2018;28:599–605. doi: 10.1016/j.mycmed.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Radosavljevic M, Koenig H, Letscher-Bru V, Waller J, Maloisel F, Lioure B, Herbrecht R. Candida catenulata fungemia in a cancer patient. J. Clin. Microbiol. 1999;37:475–477. doi: 10.1128/jcm.37.2.475-477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi, G, Sani, RK. Molecular techniques to assess microbial community structure, function, and dynamics in the environment. In: Ahmad, I, Ahmad, F, Pichtel, J, editors. Microbes and microbial technology. 1st Edn. New York: Springer-Verlag; 2011. pp. 29–57. [Google Scholar]
- Rodriguez-Tudela JL, Diaz-Guerra TM, Mellado E, Cano V, Tapia C, Perkins A, Gomez-Lopez A, Rodero L, Cuenca-Estrella M. Susceptibility patterns and molecular identification of Trichosporon species. Antimicrob. Agents Chemother. 2005;49:4026–4034. doi: 10.1128/AAC.49.10.4026-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simi W, Leite-J DP, Paula CR, Hoffmann-Santos HD, Takahara DT, Hahna RC. Yeasts and filamentous fungi in psittacidae and birds of prey droppings in midwest region of Brazil: a potential hazard to human health. Braz. J. Biol. 2018;79:414–422. doi: 10.1590/1519-6984.181192. [DOI] [PubMed] [Google Scholar]
- Soltani M, Bayat M, Hashemi SJ, Zia MA, Pestechian N. Isolation of Cryptococcus neoformans and other opportunistic fungi from pigeon droppings. J. Res. Med. Sci. 2013;18:56. [PMC free article] [PubMed] [Google Scholar]
- Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, E. Falagasde M. Human infections associated with wild birds. J. Infect. 2008;56:83–98. doi: 10.1016/j.jinf.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth F, Goldani LZ. Epidemiology of Rhodotorula: an emerging pathogen. Interdiscip. Perspect. Infect. Dis. 2012;2012:465717. doi: 10.1155/2012/465717. [DOI] [PMC free article] [PubMed] [Google Scholar]


