Abstract
Genetic variants conferring risk for schizophrenia (SCZ) have been extensively studied, but the role of posttranscriptional mechanisms in SCZ is not well studied. Here we performed the first genome-wide microRNA (miRNA) expression profiling in serum-derived exosome from 49 first-episode, drug-free SCZ patients and 46 controls and identified miRNAs and co-regulated modules that were perturbed in SCZ. Putative targets of these SCZ-affected miRNAs were enriched strongly for genes that have been implicated in protein glycosylation and were also related to neurotransmitter receptor and dendrite (spine) development. We validated several differentially expressed blood exosomal miRNAs in 100 SCZ patients as compared with 100 controls by quantitative reverse transcription-polymerase chain reaction. The potential regulatory relationships between several SCZ-affected miRNAs and their putative target genes were also validated. These include hsa-miR-206, which is the most upregulated miRNA in the blood exosomes of SCZ patients and that previously reported to regulate brain-derived neurotrophic factor expression, which we showed reduced mRNA and protein levels in the blood of SCZ patients. In addition, we found 11 miRNAs in blood exosomes from the miRNA sequence data that can be used to classify samples from SCZ patients and control subjects with close to 90% accuracy in the training samples, and approximately 75% accuracy in the testing samples. Our findings support a role for exosomal miRNA dysregulation in SCZ pathophysiology and provide a rich data set and framework for future analyses of miRNAs in the disease, and our data also suggest that blood exosomal miRNAs are promising biomarkers for SCZ.
Keywords: exosome, schizophrenia, miRNA, brain-derived neurotrophic factor, biomarker
Introduction
Schizophrenia (SCZ) is a chronic, severe mental disorder that affects approximately 1% of general population globally and is associated with substantial morbidity and mortality.1 The disease is characterized by positive symptoms (delusions and hallucinations), negative symptoms (apathy, anhedonia, and social withdrawal) and cognitive impairments.2,3 Although genetic and environmental factors are considered to be implicated in the onset and development of SCZ,4 the etiology of the disease is still far from being understood. A relatively recent study combined all genome-wide association studies for SCZ and reported 108 independent genomic loci that exceed genome-wide significance. However, those associations were accompanied by a small effective size, and the effect size has not been found to be causal.5
In addition to genetic factors, increasing evidence suggest that epigenetics maybe involved in the pathogenesis of SCZ.6,7 Of the known epigenetic markers, microRNAs (miRNAs) have generated great interest in the past decade because of their widespread and global mechanisms of action.8,9 miRNAs have been shown to play critical roles in neurodevelopment and maintenance of central nervous system, such as neural differentiation, synaptic plasticity, and cognitive functions.10–12 Moreover, a number of studies have found differential expression profile of circulating miRNAs in neuropsychiatric diseases as compared with healthy control (HC) subjects; these include Alzheimer’s disease,13 major depression,14 bipolar disorder,15 and SCZ.16 These results provide valuable information for us to better understand the nature of the devastating disorders, and also suggest the potential roles of miRNAs as biomarkers for various neuropsychiatric diseases.
miRNAs are found to be present in blood, cerebrospinal fluid, saliva, and urine, and they can be released into circulation as free form or packaged inside exosomes.17 Exosomes are small vesicles that typically have a size of 40–100 nm in diameter and are secreted by most types of cells (including neurons and astrocytes).17 Of particularly interesting findings are that exosomes release from cells of origin into circulation and to reach neighboring and distant cells, where the contents of exosomes including miRNAs are delivered to the target cells, and eventually modulate the phenotype of recipient cells.18 Furthermore, it has been demonstrated that exosomal miRNAs are involved in various biological and pathological processes, such as inflammatory response,19 cardiomyocyte hypertrophy,20 senescence, and aging.21 Circulating exosomal miRNAs have also suggested to be potential biomarkers for cancer diagnosis and prognosis.22 However, studies on exosomal miRNAs in neuropsychiatric diseases are very limited, therefore preventing us to further understand these disorders.
In this study, we performed genome-wide profiling of blood exosomal miRNA expression in SCZ patients and controls. We recruited 2 sets of participants, with a training set for predictor discovery and a testing set for class prediction. We found differential expression profiling of blood exosomal miRNAs in SCZ and identified a cluster of miRNAs that have the potential to be used for the diagnosis of SCZ. Using bioinformatics and gene network analyses, we were able to identify several coexpression modules of exosomal miRNA that were significantly associated with the disease, and link these perturbations with transcriptional changes in SCZ.
Materials and Methods
Subjects and Samples
All the SCZ patients were recruited from the Third People’s Hospital of Foshan, Guangdong, China. Age- and sex-matched healthy participants were recruited as control subjects through advertisements. The Structured Clinical Interview for DSM-IV and International Classification of Diseases 10 were used for the diagnosis of SCZ by experienced psychiatrists, and the psychiatrists assessed the psychopathological status of the SCZ patients using the Positive and Negative Syndrome Scale (PANSS) questionnaire scores (total score, positive symptom score, and negative symptom score). SCZ patients with medical illness were excluded from this study. The demographic and clinical characteristics from the patients and controls are presented in the (abcdefghlink)supplementary table S1(abcdefghxref).
All the participants included in this study gave written informed consent. The study protocol has been approved by the ethics committee at The Third People’s Hospital of Foshan, Guangdong, China, and the experiments were conducted in accordance with the Declaration of Helsinki.
The blood exosome isolation and validation methods are presented in the (abcdefghlink)Supplementary Methods(abcdefghxref). The miRNA library construction and sequencing, western blots, and quantitative RT-PCR methods are also presented in the (abcdefghlink)supplementary methods(abcdefghxref).
Differential Expression Analysis
miRNAs with mean expression ≥ 10 transcipts per million (TPM) across all samples were selected to do differential expression analysis between control group and case group using DESeq2.23 Significantly differentially expressed miRNAs (DE miRNAs) were reported at classic Benjamini–Hochberg multiple test corrected P values (Q values, false discovery rate [FDR]) < .05.24,25
The other bioinformatic analyses including random forest classifier, weighted gene coexpression network analysis (WGCNA), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and gene ontology enrichment analysis are presented in the (abcdefghlink)supplementary methods(abcdefghxref>.]
BDNF ELISA Assay
Serum BDNF protein levels were measured by enzyme-linked immunosorbent assay (ELISA) kit purchased from Biosensis, and we followed the manufacturer’s instructions to detect BDNF concentrations. Significant difference was defined as P < .05 by 2-tailed t test.
Results
Differential Expression of Blood Exosomal miRNAs in SCZ
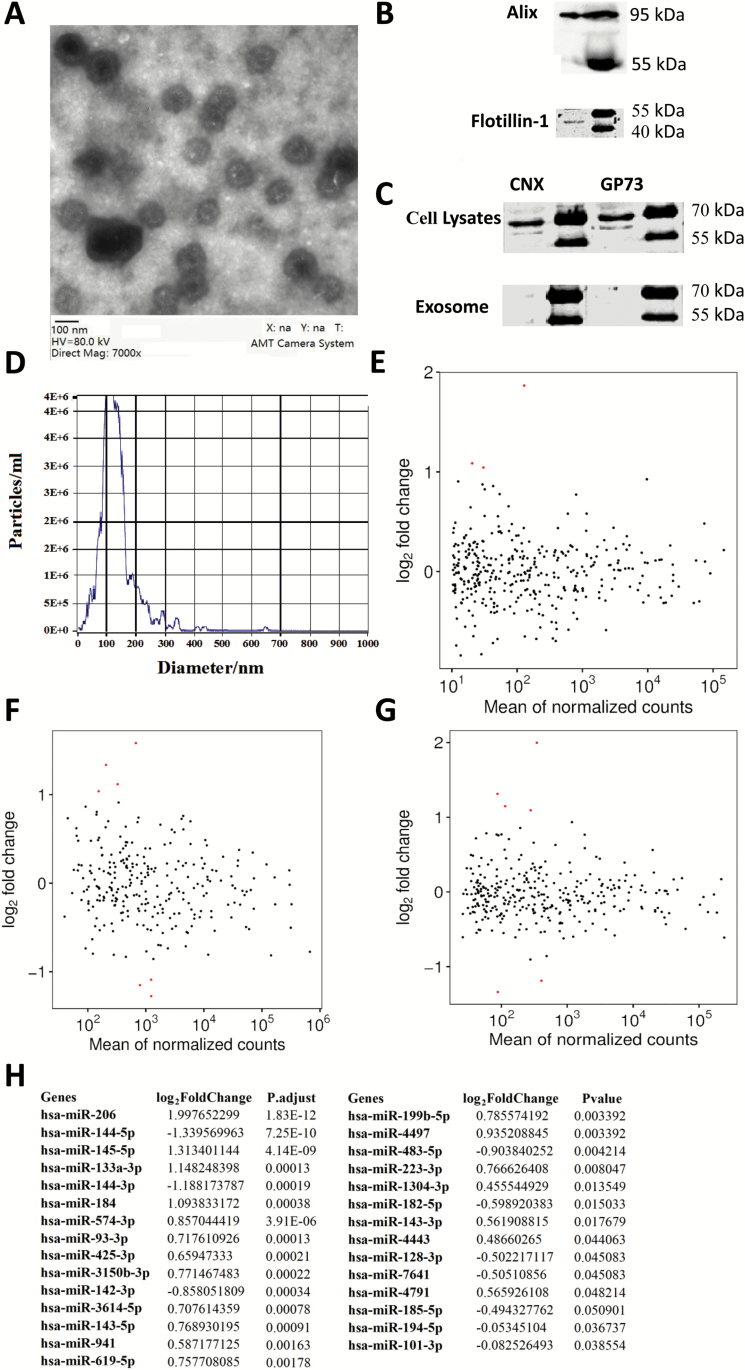
We collected blood exosomes (figure 1A–D) from 2 sets of participants for the analyses of miRNA profile in first-episode, drug-free SCZ patients as compared with controls. The training set of participants included 23 SCZ patients and 23 HC subjects, and the miRNA expression profiles of blood exosomes were analyzed using Illumina HiSeq 2500 high-throughput sequencing (miRNA-seq). The miRNA-seq identified thousands of miRNAs that were existed in the blood exosomes. To focus on the highly expressed miRNAs, we removed the miRNAs with sequences less than 10 reads (mean TPM < 10), which led to the selection of 353 miRNAs for differential analyses between cases and controls (figure 1E). We identified 18 miRNAs in the blood exosomes showing significant (FDR < 0.05) expression changes between SCZ patients and HC subjects: 12 were upregulated and 6 were downregulated. Of the 18 miRNAs, hsa-miR-206, hsa-miR-145-5p, and hsa-miR-133a-3p had 2-fold increase in SCZ patients when compared with controls (figure 1E and (abcdefghlink)supplementary table S3(abcdefghxref)).
Fig. 1.
Blood exosomal miRNA changes in patients with SCZ. (A) Serum exosome observation by electron microscopy and ×7000 magnification. (B) Western blots show the exosomal marker Alix and Flotillin-1 were present in the exosomes extracted from blood. (C) Western blots show that endoplasmic reticulum marker CNX and Golgi marker GP-73 were not present in the peripheral blood exosomes. (D) Serum exosome validation by nanoparticle tracking device—ZetaView. Note that the particles peak around at 100 nm. (E) MA-plot displaying miRNA differences between cases and controls for the training set of participants. (F) MA-plot for the testing set of participants. (G) MA-plot for pooled set of participants. (H) Significantly changed blood exosomal miRNA expression in the 49 cases as compared with 46 controls. CNX, Calnexin; GP-73, Golgi protein-73.
To validate the blood exosomal miRNA expression profile in SCZ, we profiled the miRNA expression in the blood exosomes from the testing set of participants, which included 26 SCZ patients and 23 HC subjects. A total of 265 miRNAs were subjected to differential analyses after removing miRNAs with mean TPM below 10 (figure 1F), and 29 miRNAs were found to show significant expression changes between cases and controls. In addition, 7 out of the 29 miRNAs had more than 2-fold change in SCZ patients compared with controls, and these include hsa-miR-206, hsa-miR-145-5p, and hsa-miR-133a-3p, indicating the reproducibility of the regulation of these miRNAs in SCZ patients (figure 1F and (abcdefghlink)supplementary table S4(abcdefghxref)).
We then pooled the data from the 2 sets of participants for analyses (49 cases and 46 controls). The analyses showed that 26 miRNAs were significantly associated with SCZ, and 6 miRNAs (hsa-miR-206, hsa-miR-145-5p, hsa-miR-133a-3p, hsa-miR-144-5p, hsa-miR-144-3p, and hsa-miR-184) had more than 2-fold change in the SCZ patients as compared with controls (figure 1G and H and (abcdefghlink)supplementary table S5(abcdefghxref)).
Exosomal miRNAs as Biomarkers for SCZ
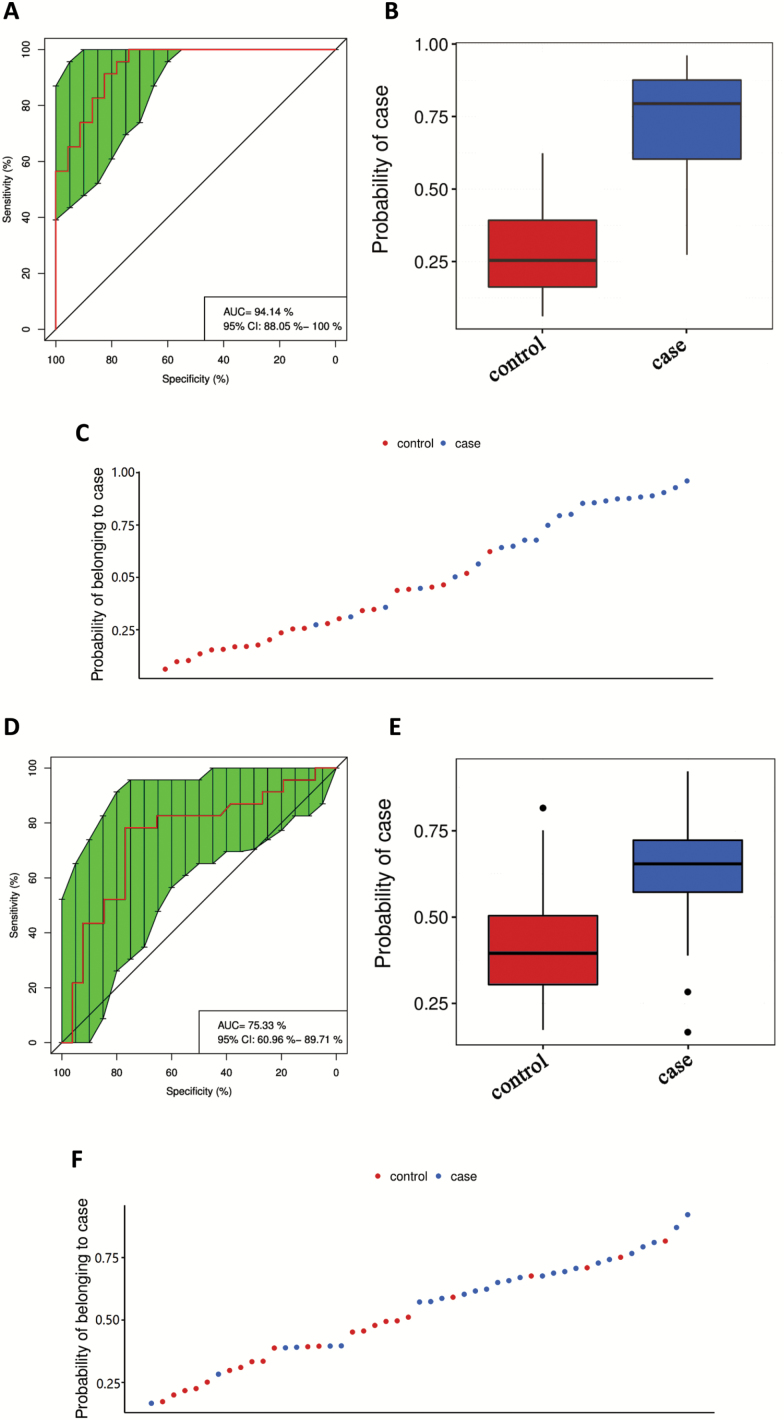
Given the robust associations of some exosomal miRNAs with SCZ, we therefore explored whether miRNAs could be served as biomarkers to differentiate between SCZ patients and HC subjects. A total of 353 miRNAs from the training set of participants were subjected to random forest classifier for potential miRNA biomarker analyses, and a set of 11 miRNAs ((abcdefghlink)supplementary methods(abcdefghxref)) were selected as the optimal set of miRNAs to diagnose SCZ. We used the 11 miRNAs to draw the receiver operating characteristic curve, and the area under curve (AUC) was 0.94 (95% CI, 0.88–1.0), which yielded a sensitivity of 0.783 and specificity of 0.957 to diagnose SCZ (figure 2A–C). We next applied the 11 miRNAs to the testing set of participants for class prediction; these miRNAs yielded a sensitivity of 0.769 and specificity of 0.783 to diagnose SCZ, and the AUC was 0.753 (95% CI, 0.61–0.90; figure 2D–F). These results suggest that blood exosomal miRNAs have the potential to be biomarkers for diagnosis of SCZ, although this is a relatively small sample size and would need further testing on the exosomal miRNAs to be considered as biomarker candidates for clinical use.
Fig. 2.
Blood exosomal miRNAs as biomarkers to differentiate between SCZ patients and controls. (A) ROC curves were utilized to evaluate the accuracy of a cluster of 11 miRNAs for the diagnosis of SCZ in a training set of participants. Boxplot (B) and scatter plot (C) of probability of participants belonging to cases by the 11 miRNAs in the training set. (D) ROC curves were utilized to evaluate the accuracy of a cluster of 11 miRNAs for the diagnosis of SCZ in a testing set of participants. Boxplot (E) and scatter plot (F) of probability of participants belonging to case in the testing set. AUC, area under curve; ROC, receiver operating characteristic.
Perturbation of Exosomal miRNA Coexpression Modules in SCZ
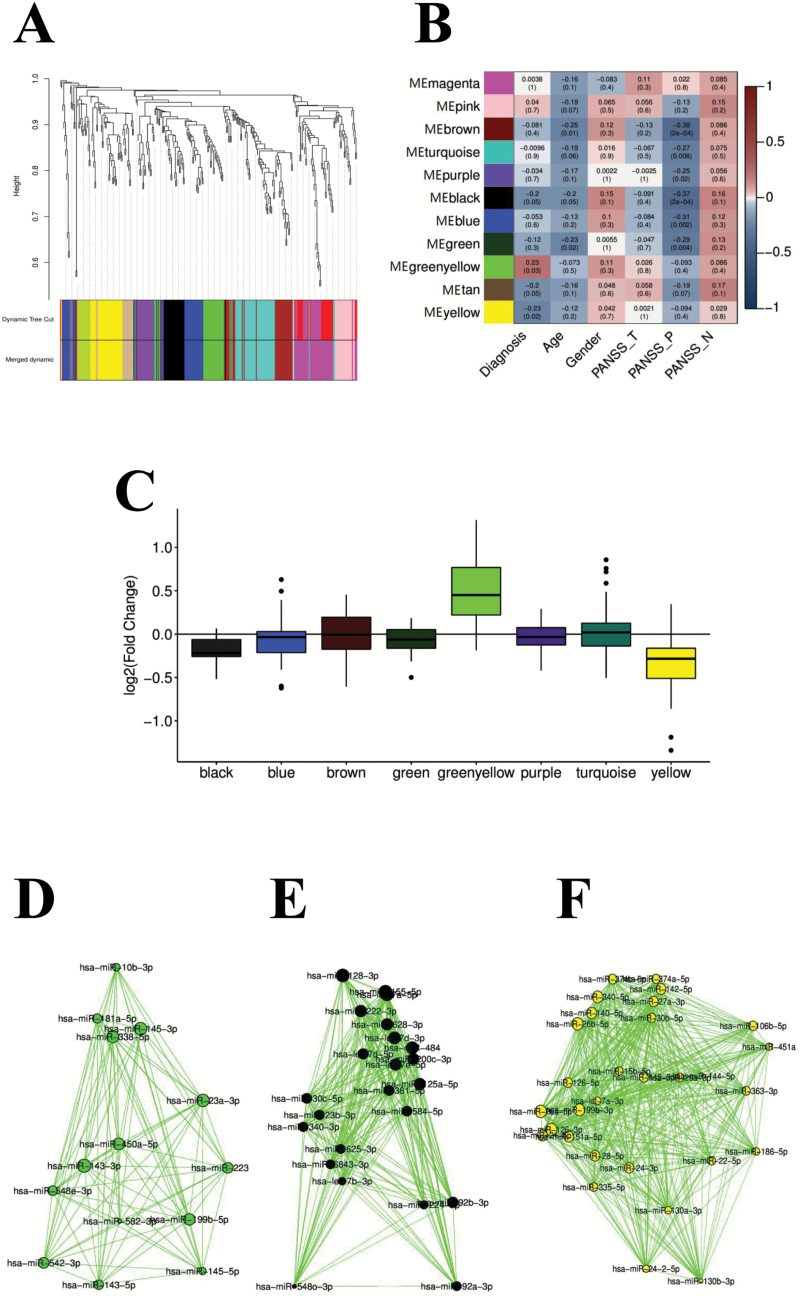
To better understand the relationship between blood exosomal miRNA changes and disease status at a systems level, 95 samples (49 cases and 46 controls) were subjected to WGCNA to assign individual miRNAs to coexpression modules, which resulted in the identification of 11 modules (figure 3A). We then calculated correlation between the first principal component of each module and disease status, and identified 3 modules that were significantly correlated with disease status (FDR < 0.05): one upregulated (greenyellow) and 2 downregulated (black and yellow; figure 3B–F).
Fig. 3.
miRNA coexpression modules dysregulated in blood exosomes of SCZ patients. (A) Dendrogram showing miRNA coexpression modules defined in 95 samples. Color bars indicate dynamic module assignments (Dynamic Tree Cut) and merged module assignments (Merged dynamic). (B) Pearson’s correlation coefficient (and P value in parentheses) between disease status, age, gender, disease severity, and module eigengene. (C) Log2 transformed fold change distribution of miRNAs in the module black, blue, brown, green greenyellow, purple, turquoise, and yellow. (D–F) Coexpression network plots for greenyellow, black, and yellow modules. Node size is proportional to node connectivity, and edge indicates coexpression of connected nodes with intramodular connectivity >0.5.
WGCNA allows direct assessment of the relationship of modules to potential clinical moderators, including sex, age, and disease severity. Greenyellow and yellow modules were not significantly associated with age, gender, PANSS total score, PANSS positive score, and PANSS negative score. However, the black module showed a significant association with PANSS positive score, but not with the other clinical variables. In addition, brown, turquoise, purple, blue, and green modules were all significantly associated with PANSS positive score (figure 3B).
Pathway Enrichment Analysis of miRNA-Targeted Genes
To better understand the SCZ etiology, we next explored the potential target genes of the DE miRNA and SCZ-associated modules. Interestingly, the top DE miRNA hsa-miR-206 has been reported to regulate the expression of BDNF,26 which is a well-known neurotrophin that plays a key role in the neurodevelopment and maintenance of nervous system. We then applied the well-recognized algorithm TagertScan to bioinformatically predict the mRNA targets of the SCZ-associated miRNA modules, and performed KEGG pathway analyses to find significantly enriched pathways for the greenyellow, black, and yellow miRNA modules. mRNA targets of the upregulated greenyellow miRNA module are only significantly enriched for mucin type O-Glycan biosynthesis pathway, and this pathway enrichment was also found in the downregulated black and yellow miRNA modules. Moreover, the targets of the SCZ-affected yellow miRNA module are enriched for extracellular matrix (ECM)–receptor interaction pathway and mammalian target of rapamycin (mTOR) signaling pathway ((abcdefghlink)supplementary table S6(abcdefghxref)).
We then used ClusterProfiler to perform Gene Ontology (GO) enrichment analysis for targets of SCZ-affected miRNA modules, which assessed the shared functions. Targets of the downregulated black miRNA module are mostly enriched for genes that are related to neurotransmitter transporter, whereas the top GO terms for the targets of the downregulated yellow miRNA module are related to dendritic spine development and dendrite development ((abcdefghlink)supplementary figure S1(abcdefghxref)).
qRT-PCR Validation of miRNA Sequence Data
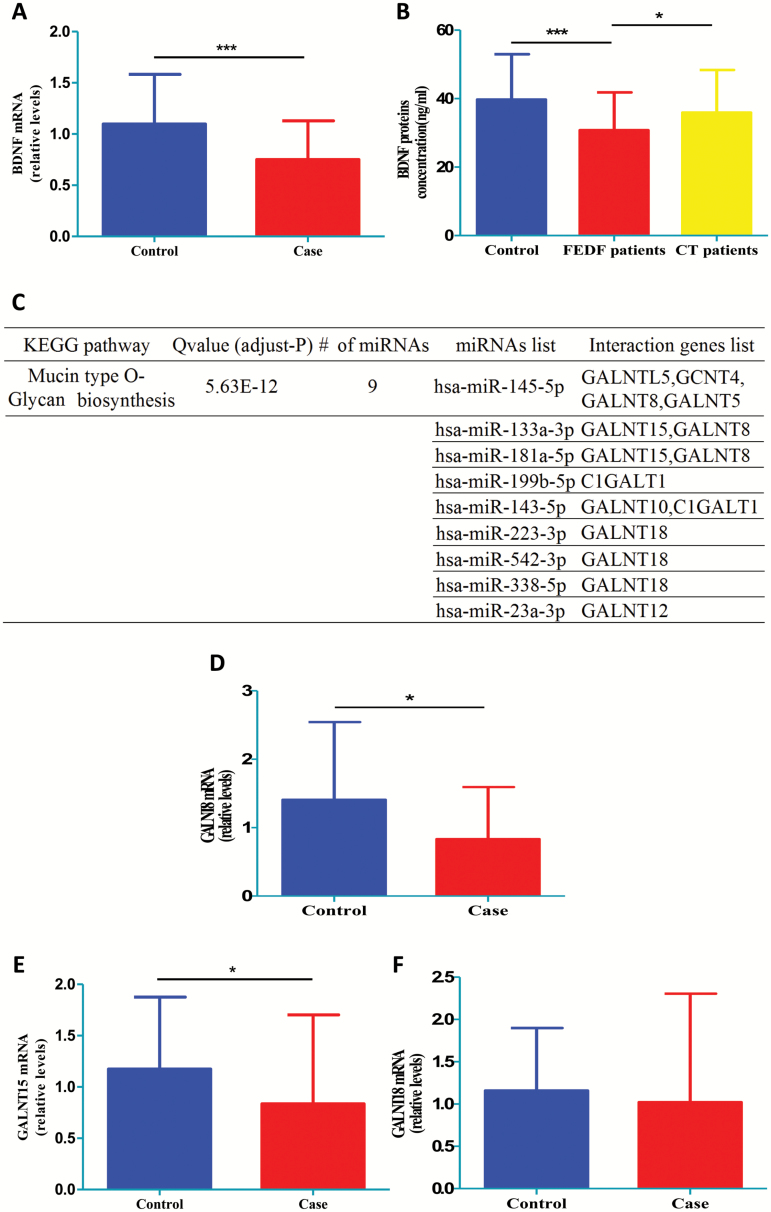
To further validate the sequence data, we used quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to analyze the differential expression miRNA levels with a larger sample size. We included 100 SCZ patients (57 first-episode, drug-free patients and 43 chronically treated patients) and 100 controls, and the results showed that hsa-miR-206 levels from blood exosomes were increased in the SCZ patients, which was consistent with the miRNA-seq data (figure 4A). However, the chronically medicated SCZ patients did not show significantly increased hsa-miR-206 levels as compared with controls (figure 4B). Consistently, the hsa-miR-144-3p levels were decreased in the SCZ patients, but restored to normal levels after long-term medication (figure 4C and D). Interestingly, the qRT-PCR results showed that the increased hsa-miR-619-5p levels found in the SCZ patients were not affected by medication (figure 4E and F).
Fig. 4.
Quantitative reverse transcription-polymerase chain reaction validation of differentially expressed miRNAs in the blood exosomes of SCZ patients. hsa-miR-206 (A), hsa-miR-144-3p (C), and hsa-miR-619-5p (E) expression in the blood exosomes of 100 cases and 100 controls. hsa-miR-206 (B), hsa-miR-144-3p (D), and hsa-miR-619-5p (F) in the blood exosomes of 57 FEDF SCZ patients, 43 CT SCZ patients, and 100 controls. FEDF, first-episode, drug-free; CT, chronically treated. **P < .01, ***P < .001.
Bioinformatic Predication Validation of the miRNA Targets
Next, to assess the validity of the bioinformatic predictions, we analyzed several potential targets of SCZ-associated miRNAs. We first analyzed the mRNA expression of BDNF, which is a potential target of the top upregulated miRNA (hsa-miR-206) in SCZ. Results from qRT-PCR showed decreased BDNF gene expression in the whole blood of first-episode, drug-free SCZ patients as compared with controls (figure 5A). Furthermore, ELISA assay revealed that serum BDNF protein levels were reduced in first-episode, drug-free SCZ patients, whereas antipsychotics increased the BDNF protein levels in the patients (figure 5B), suggesting a potential function of hsa-miR-206 to BDNF pathway in the SCZ pathology.
Fig. 5.
Validation of bioinformatic prediction of miRNA target genes. (A) Whole-blood BDNF mRNA levels between 45 FEDF SCZ patients and 45 controls. (B) Serum BDNF protein levels between 53 FEDF SCZ patients, 51 CT SCZ patients, and 47 controls. (C) KEGG pathway enrichment result for the SCZ-affected upregulated yellowgreen module. (D) GALNT8, (E) GLANT15, and (F) GLANT18 whole-blood mRNA levels between 45 FEDF SCZ patients and 45 controls. CT, chronically treated; FEDF, first-episode, drug-free. *P < .05, ***P < .001.
We next selected several targets of the upregulated greenyellow miRNA module, given this is the top DE miRNA module, and these selected genes are predicted to be regulated by multiple miRNAs (figure 5C). Of the 3 genes selected, the blood mRNA expression of GALNT8 (figure 5D) and GALNT15 (figure 5E) was significantly reduced in the first-episode, drug-free SCZ patients when compared with controls, whereas no significant association was found between blood GALNT18 mRNA and SCZ (figure 5F). These results therefore validated the credibility of the bioinformatic predictions.
Discussion
Our genome-wide, interactive analysis of data from blood exosome provided novel insights into the role of miRNAs for the onset and/or development of SCZ. A differential expression profile of blood exosome-derived miRNA was observed in the patients with SCZ. In the bioinformatic analyses on the targets of the SCZ-affected miRNAs and miRNA coexpression modules, we found enrichment for genes that have been associated with protein glycosylation, neurodevelopment, neurotransmission, and synaptic plasticity; these genes include BDNF, GALNT15, CDC42, and DISC1. Given that genetic mutations only partially explain the etiology of SCZ, we hypothesize that miRNA dysfunction provides an alternative pathway to disturb the functions of genes at the transcriptional levels, thereby contributing to the pathophysiology of the devastating disease. This hypothesis is supported by our experimental validation showing downregulation of mRNA targets by the SCZ-affected upregulated greenyellow miRNA module, and also by the downregulation of BDNF (which is a potential target for hsa-miR-206) at the transcriptional and translational levels in SCZ patients. Taken together, our results for the first time demonstrate the miRNA dysregulation in the blood exosome of patients with SCZ, therefore providing new insights into the molecular pathways that confer the vulnerability to the development of SCZ.
Previous studies have also examined miRNA levels in the blood of SCZ patients, but results were not consistent among studies.16,27–32 The inconsistency could be partially due to the heterogeneity nature of the disease. However, another possibility of the inconsistent data is the multiple origins of the miRNAs from blood, given that circulation miRNAs can be found in apoptotic bodies, microvesicles, exosomes, or present as free miRNAs,18 thereby contributing to the between-study heterogeneity. In addition, several studies recruited small sample sizes and/or assessed limited numbers of miRNAs.27,29,30,32 Here, our genome-wide analysis of miRNAs from blood exosomes with a relatively large sample size, and using the bioinformatic predictions and validations, provides the most comprehensive evaluations of miRNA dysregulation in SCZ. Notably, of all the DE miRNAs in the blood exosomes, hsa-miR-206 was the most upregulated one in the patients with SCZ. It has been reported that hsa-miR-206 interacted with BDNF mRNA directly, leading to the decreased expression of this gene, which then negatively affected cognitive functions of animals.26 In consistent with the previous finding on the hsa-miR-206-induced disruption of BDNF expression, our study showed that both BDNF mRNA and protein levels were downregulated in the blood of patients with SCZ. These are interesting findings because the neurotrophin hypothesis of SCZ has generated great interests over the last decade, and it postulated that the changes in the brains of SCZ patients are the result of disturbances of developing processes involving neurotrophic factors.3,33 This hypothesis is mainly supported by the preclinical and clinical studies showing close associations between BDNF and SCZ, especially the decreased blood BDNF levels found in the patients with SCZ.34–36 However, the mechanism underlying the BDNF dysregulation in patients with SCZ was poorly understood. Here, we showed significantly increased has-miR-206 levels and decreased BDNF levels in the patients with SCZ, and antipsychotics restored the dysregulations of has-miR-206 and BDNF in the patients. Our data therefore suggest that upregulation of exosomal has-miR-206 may contribute to the dysfunction of BDNF in SCZ. This finding further supports the neurotrophin hypothesis of SCZ, and miR-206 to BDNF signaling may provide a novel pathway for the intervention of SCZ development.
In addition to the identification of exosomal has-miR-206 as a potential upstream regulator of BDNF in SCZ, mucin-type O-Glycan biosynthesis pathway has been predicted to be strongly associated with SCZ by our bioinformatic analyses. Mucin-type O-glycosylation is a highly conserved type of protein modification present on membrane-bound and secreted proteins, which plays essential roles in development, organogenesis, and tissue homeostasis.37 Moreover, dysfunctions of O-glycosylation were found to be involved with various human diseases, but have not been linked to neuropsychiatric diseases. In this study, aberrations in enzymes responsible for O-glycosylation of proteins have been observed to be strongly associated with SCZ, therefore revealed a potential novel pathway that contributes to the etiology of the disease. This new association found in our study is not surprising, given the proper secretion of ECM proteins responsible for mediating between-cell communications has been proposed to require O-glycosylation,37 and previous studies showed components of ECM were substantially affected in the patients with SCZ, including chondroitin sulfate proteoglycans and Reelin (which is a glycoprotein).38,39 Interestingly, a recent exome array study of families with SCZ (included 118 cases and 223 controls) showed that genes harboring variants only in the SCZ patients were enriched (FDR = 0.05) for the ECM-receptor interaction pathway.40 Bioinformatic analyses of our data also predicated the targets of the SCZ-affected yellow miRNA module are enriched for ECM–receptor interaction pathway, adding to evidence that processes affecting the ECM components were linked to SCZ. Taken together, we hypothesize that exosomal miRNA dysfunctions lead to the O-glycosylation-associated abnormalities of ECM, resulting in the disrupted synaptic plasticity, connectivity, and neuronal migration, which then contribute to the pathophysiology of SCZ.
Although it is known that exosomes cross blood-brain barrier easily,41 the population of brain-derived exosomes in the peripheral blood is unclear, and this is largely due to the difficulty of isolating brain-derived exosomes from blood. Here we hypothesize that there is a distinct population of brain-derived exosomes in the peripheral blood of SCZ patients. This is because our bioinformatic analyses on the targets of the SCZ-affected miRNAs and miRNA coexpression modules found enriched genes associated with neurodevelopment, neurotransmission, and synaptic plasticity, which suggest the changes in exosomal miRNAs in the peripheral blood are at least partially due to the brain pathophysiology at the onset of SCZ manifestations, and one way brain exosomes could get into peripheral blood is via resorption of cerebrospinal fluid (as exosomes are present in cerebrospinal fluid) into venous circulation. In addition, the changes in the peripheral blood exosomal miRNA content could reflect the pathophysiology of brain as well as peripheral system, given that the peripheral systems such as immune system and hematopoietic system also release exosomes into blood. Moreover, a study suggested that the exosome-mediated communication between the hematopoietic system and the brain under inflammation had physiological function.42 Nevertheless, the origin of exosomes in the blood and the potential differential roles of blood exosomes from different sources in SCZ patients require further investigations.
Collectively, our genome-wide, integrative analysis provides a framework for evaluating the functional involvement of miRNA from blood exosome in SCZ, and a rich set of SCZ-DE miRNAs for further study. By bioinformatic analysis of target genes of the miRNAs and correlating with our mRNA expression data, we provide several lines of evidence for a functional role of blood exosomal miRNA dysregulation in SCZ. Our analyses also identify a cluster of miRNAs from blood exosomes as potential biomarkers for SCZ, and future studies are necessary to translate the potential biomarkers from blood exosomes into benefit of SCZ patients.
Funding
National Natural Science Foundation of China (81703492); Beijing Natural Science Foundation (7182092); Minzu University Research Fund (2018CXTD03); Minzu University of China 111 project.
Supplementary Material
Acknowledgments
Cheng conceived the study; Dr Du and Dr Cheng designed the research; and Du, Yu, Hu, Li, Wei, and Zheng conducted the research. All the authors analyzed and interpreted the data. Cheng drafted the manuscript with critical revisions from all the authors. The authors have no biomedical financial interests or potential conflict of interest to declare.
References
- 1. Keller WR, Kum LM, Wehring HJ, Koola MM, Buchanan RW, Kelly DL. A review of anti-inflammatory agents for symptoms of schizophrenia. J Psychopharmacol. 2013;27(4):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cassoli JS, Guest PC, Santana AG, Martins-de-Souza D. Employing proteomics to unravel the molecular effects of antipsychotics and their role in schizophrenia. Proteomics Clin Appl. 2016;10(4):442–455. [DOI] [PubMed] [Google Scholar]
- 3. Qin XY, Wu HT, Cao C, Loh YP, Cheng Y. A meta-analysis of peripheral blood nerve growth factor levels in patients with schizophrenia. Mol Psychiatry. 2017;22(9):1306–1312. [DOI] [PubMed] [Google Scholar]
- 4. Schmidt-Kastner R, van Os J, Esquivel G, Steinbusch HW, Rutten BP. An environmental analysis of genes associated with schizophrenia: hypoxia and vascular factors as interacting elements in the neurodevelopmental model. Mol Psychiatry. 2012;17(12):1194–1205. [DOI] [PubMed] [Google Scholar]
- 5. Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. Jul 24 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cariaga-Martinez A, Saiz-Ruiz J, Alelú-Paz R. From linkage studies to epigenetics: what we know and what we need to know in the neurobiology of Schizophrenia. Front Neurosci. 2016;10:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hannon E, Dempster E, Viana J, et al. . An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 9. Mingardi J, Musazzi L, De Petro G, Barbon A. miRNA editing: new insights into the fast control of gene expression in health and disease. Mol Neurobiol. 2018;55:7717–7727. [DOI] [PubMed] [Google Scholar]
- 10. Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity—a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun E, Shi Y. MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp Neurol. 2015;268:46–53. [DOI] [PubMed] [Google Scholar]
- 12. Woldemichael BT, Mansuy IM. Micro-RNAs in cognition and cognitive disorders: potential for novel biomarkers and therapeutics. Biochem Pharmacol. 2016;104:1–7. [DOI] [PubMed] [Google Scholar]
- 13. Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp Neurol. 2012;235(2):491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gururajan A, Naughton ME, Scott KA, et al. . MicroRNAs as biomarkers for major depression: a role for let-7b and let-7c. Transl Psychiatry. 2016;6(8):e862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maffioletti E, Cattaneo A, Rosso G, et al. . Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord. 2016;200:250–258. [DOI] [PubMed] [Google Scholar]
- 16. Wei H, Yuan Y, Liu S, et al. . Detection of circulating miRNA levels in schizophrenia. Am J Psychiatry. 2015;172(11):1141–1147. [DOI] [PubMed] [Google Scholar]
- 17. Zhang J, Li S, Li L, et al. . Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fries GR, Quevedo J. Exosomal microRNAs as potential biomarkers in neuropsychiatric disorders. Methods Mol Biol. 2018;1733:79–85. [DOI] [PubMed] [Google Scholar]
- 19. Alexander M, Hu R, Runtsch MC, et al. . Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bang C, Batkai S, Dangwal S, et al. . Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124(5):2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu D, Tahara H. The role of exosomes and microRNAs in senescence and aging. Adv Drug Deliv Rev. 2013;65(3):368–375. [DOI] [PubMed] [Google Scholar]
- 22. Tang MK, Wong AS. Exosomes: emerging biomarkers and targets for ovarian cancer. Cancer Lett. 2015;367(1):26–33. [DOI] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z, Joyce BT, Kresovich JK, et al. . Blood pressure and expression of microRNAs in whole blood. PLoS One. 2017;12(12):e0173550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao K, Liang G, Sun X, Guan le L. Comparative miRNAome analysis revealed different miRNA expression profiles in bovine sera and exosomes. BMC Genomics. 2016;17(1):630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee ST, Chu K, Jung KH, et al. . miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–277. [DOI] [PubMed] [Google Scholar]
- 27. Camkurt MA, Karababa F, Erdal ME, et al. . Investigation of dysregulation of several microRNAs in peripheral blood of schizophrenia patients. Clin Psychopharmacol Neurosci. 2016;14(3):256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai CY, Yu SL, Hsieh MH, et al. . MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6(6):e21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi W, Du J, Qi Y, et al. . Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res. 2012;46(2):198–204. [DOI] [PubMed] [Google Scholar]
- 30. Song HT, Sun XY, Zhang L, et al. . A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J Psychiatr Res. 2014;54:134–140. [DOI] [PubMed] [Google Scholar]
- 31. Sun XY, Lu J, Zhang L, et al. . Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci. 2015;22(3):570–574. [DOI] [PubMed] [Google Scholar]
- 32. Wu S, Zhang R, Nie F, et al. . MicroRNA-137 inhibits EFNB2 expression affected by a genetic variant and is expressed aberrantly in peripheral blood of schizophrenia patients. EBioMedicine. 2016;12:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thome J, Foley P, Riederer P. Neurotrophic factors and the maldevelopmental hypothesis of schizophrenic psychoses. Review article. J Neural Transm (Vienna). 1998;105(1):85–100. [DOI] [PubMed] [Google Scholar]
- 34. Angelucci F, Brenè S, Mathé AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10(4):345–352. [DOI] [PubMed] [Google Scholar]
- 35. Fernandes BS, Steiner J, Berk M, et al. . Peripheral brain-derived neurotrophic factor in schizophrenia and the role of antipsychotics: meta-analysis and implications. Mol Psychiatry. 2015;20(9):1108–1119. [DOI] [PubMed] [Google Scholar]
- 36. Green MJ, Matheson SL, Shepherd A, Weickert CS, Carr VJ. Brain-derived neurotrophic factor levels in schizophrenia: a systematic review with meta-analysis. Mol Psychiatry. 2011;16(9):960–972. [DOI] [PubMed] [Google Scholar]
- 37. Tran DT, Ten Hagen KG. Mucin-type O-glycosylation during development. J Biol Chem. 2013;288(10):6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62(3):1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hornig T, Haas C, Sturm L, Fiebich B, Tebartz van Elst L. Increased blood-reelin-levels in first episode schizophrenia. PLoS One. 2015;10(8):e0134671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCarthy NS, Melton PE, Ward SV, et al. . Exome array analysis suggests an increased variant burden in families with schizophrenia. Schizophr Res. 2017;185:9–16. [DOI] [PubMed] [Google Scholar]
- 41. Qu M, Lin Q, Huang L, et al. . Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J Control Release. 2018;287:156–166. [DOI] [PubMed] [Google Scholar]
- 42. Ridder K, Keller S, Dams M, et al. . Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12(6):e1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.