Abstract
Cannabidibutol (CBDB), a novel butyl analog of cannabidiol, was identified as impurity of commercial cannabidiol (CBD) extracted from hemp (for full data and results interpretation see “Analysis of impurities of cannabidiol from hemp. Isolation, characterization and synthesis of cannabidibutol, the novel cannabidiol butyl analog” Citti et al, 2019). The compound was isolated from a CBD sample and subject to a full characterization. First, a complete spectroscopic characterization was performed by Nuclear Magnetic Resonance (NMR): in particular, 1H-NMR, 13C-NMR, COSY, HSQC and HMBC, which were followed by UV absorption and circular dichroism (CD) spectra. In order to confirm the structural identity and stereochemistry of the compound, a stereoselective synthesis of the trans isomer (1R,6R) was carried out and all the chemical and spectroscopic properties were analyzed. The synthesized compound was characterized by NMR (1H-NMR, 13C-NMR, COSY, HSQC and HMBC), Infra-Red spectroscopy (IR), UV and CD absorption, matching the results obtained for the natural isolated compound. With the analytical standard in hand, a simple high-performance liquid chromatography method coupled to UV detection (HPLC-UV) was developed and validated in house in terms of linearity, accuracy, precision, dilution integrity and stability. The present data might be useful to any researcher or industry that may run into a very common impurity of CBD extracted from hemp, so it can be easily compared with their own experimental data.
Keywords: Cannabidiol-C4, Chemical and spectroscopic characterization of cannabidibutol, HPLC-UV method validation, NMR spectra, Circular dichroism, UV absorption
Specifications Table
| Subject | Pharmaceutical Science |
| Specific subject area | Identification of an impurity in a sample of the drug cannabidiol (CBD), by synthesis and spectroscopic characterization, development and validation of an analytical method for its qualitative and quantitative determination |
| Type of data | Table Figure |
| How data were acquired | NMR: DPX-600 Avance (Bruker) spectrometer (600.13 MHz for 1H NMR and 150.92 MHz for13C NMR) equipped with a CryoProbe BBO H&F 5 mm, and processed with TopSpin v4.0.6 (Bruker BioSpin 2018) IR: Perkin-Elmer Spectrum Two ATR-IR and processed with Spectrum 10™ software (PerkinElmer) UV and Circular dichroism (CD): Jasco J-1100 spectropolarimeter HPLC-UV: Agilent 1220 Infinity LC System (EZChrom software) |
| Data format | Raw and analyzed |
| Parameters for data collection | NMR spectra of compounds were acquired in CDCl3 at 99.96% of deuteration. CD and UV spectra were acquired in acetonitrile (ACN) using quartz cells with a 10 mm path length. Optical rotation was measured in ACN, using a 1 mL-100 mm cell-length. All the measurements were performed at 298 K. HPLC separation was performed with a Poroshell 120 C18 column, eluting with 0.1% formic acid in water and ACN. |
| Description of data collection | NMR spectra were recorded using standard Bruker pulse programs. IR spectra were acquired in the range 450–4000 cm−1. CD and UV spectra were acquired in the 400-200 nm range, using a 50 nm/min scanning speed. HPLC-UV conditions were set as follows: isocratic elution with 70% B for 10 minutes, then 95% B pumped for 5 min and re-equilibration of the column for 2 min, flow rate maintained constant at 0.5 mL/min. Ibuprofen (1 μg/mL) was used as internal standard. The UV trace was acquired at 228 nm. |
| Data source location | NMR, IR, CD, UV: City/Town/Region: Modena Country: Italy Latitude and longitude (and GPS coordinates): 44°37′54″N, 10°56′45″ E HPLC-UV: City/Town/Region: Lecce Country: Italy Latitude and longitude (and GPS coordinates): 40°20′11″N, 18°07′15″E |
| Data accessibility | Raw data are accessible at the following link: https://drive.google.com/open?id=1gEL6bo5_btm5Gxul2FFVnLxZKMLdIdN7 |
| Related research article | Author's name: Cinzia Citti, Pasquale Linciano, Flavio Forni, Maria Angela Vandelli, Giuseppe Gigli, Aldo Laganà, Giuseppe Cannazza Title: Analysis of impurities of cannabidiol from hemp. Isolation, characterization and synthesis of cannabidibutol, the novel cannabidiol butyl analog Journal: Journal of Pharmaceutical and Biomedical Analysis https://doi.org/10.1016/j.jpba.2019.06.049 |
Value of the Data
|
1. Data
Analysis of impurities of cannabidiol from hemp. Isolation, characterization and synthesis of cannabidibutol, the novel cannabidiol butyl analog [1]. The following figures refer to the chemical characterization of CBDB obtained by stereoselective synthesis and CBDB isolated from authentic CBD samples.
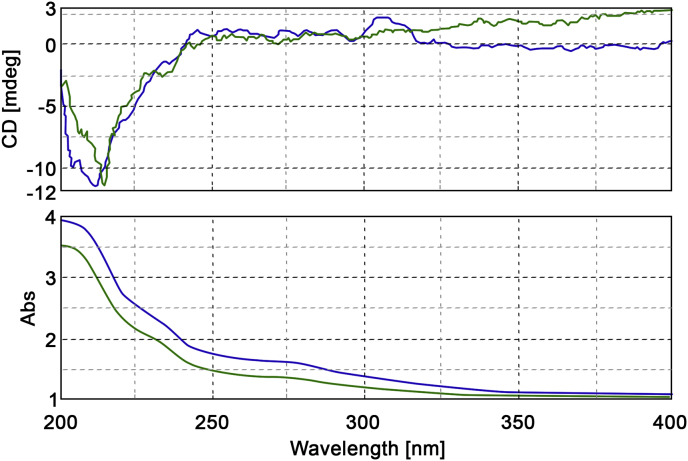
Fig. 12 reports the superimposition of the Circular Dichroism (CD) spectra of isolated (green) and synthesized (blue) CBDB. The solvent used id acetonitrile, the path length is 1 cm and the concentration loaded is 10 μg/mL.
Fig. 12.
Circular Dichroism (CD) of isolated (green) and synthesized (blue) CBDB. Solvent: acetonitrile, path length: 1 cm; concentration: 10 µg/mL.
The following figures and tables refer to the validation of the HPLC-UV method for the quantification of CBDV and CBDB in samples of CBD.
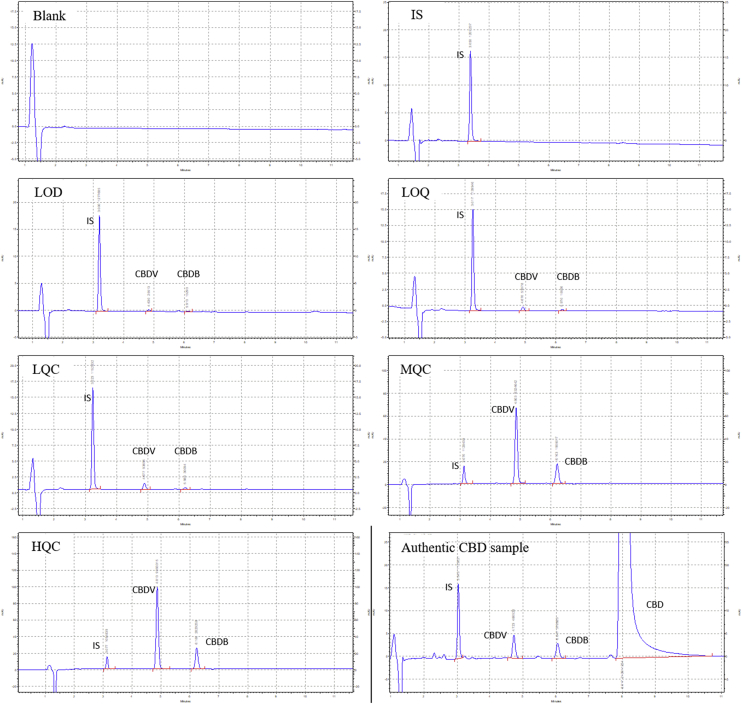
Fig. 13 reports the HPLC-UV chromatograms of a blank sample (acetonitrile), an internal standard (IS) working solution, a standard mixture of cannabidivarin (CBDV) and cannabidibutol (CBDB) in IS working solution at the limit of detection (LOD, 0.10 μg/mL for CBDV and 0.04 μg/mL for CBDB), a standard mixture at the lower limit of quantification (LLOQ, 0.28 μg/mL for CBDV and 0.12 μg/mL for CBDB), a standard mixture at three quality control (QC) levels, low (LQC, 0.56 μg/mL for CBDV and 0.24 μg/mL for CBDB), medium (MQC, 18.8 μg/mL for CBDV and 8.00 μg/mL for CBDB) and high (HQC, 45.1 μg/mL for CBDV and 19.2 μg/mL for CBDB), and an authentic cannabidiol (CBD) sample in IS working solution (the peak of CBD is not entirely visible as the chromatogram is zoomed in to highlight the impurities).
Fig. 13.
HPLC-UV chromatograms of a blank sample (acetonitrile), an internal standard (IS) working solution, a standard mixture of cannabidivarin (CBDV) and cannabidibutol (CBDB) in IS working solution at the limit of detection (LOD, 0.10 µg/mL for CBDV and 0.04 µg/mL for CBDB), a standard mixture at the lower limit of quantification (LLOQ, 0.28 µg/mL for CBDV and 0.12 µg/mL for CBDB), a standard mixture at three quality control (QC) levels, low (LQC, 0.56 µg/mL for CBDV and 0.24 µg/mL for CBDB), medium (MQC, 18.8 µg/mL for CBDV and 8.00 µg/mL for CBDB) and high (HQC, 45.1 µg/mL for CBDV and 19.2 µg/mL for CBDB), and an authentic cannabidiol (CBD) sample in IS working solution (the peak of CBD is not entirely visible as the chromatogram is zoomed in to highlight the impurities).
Table 1 reports the linearity parameters for CBDV and CBDB (slope, intercept, R2 weighted, and linear range). Values are expressed as mean ± standard error (n = 3).
Table 1.
Linearity parameters for CBDV and CBDB (slope, intercept and R2wheighted). Values are expressed as mean ± standard error (n = 3).
| Compound | Slope | Intercept | R2 (weighted) | Linear range (μg/mL) |
|---|---|---|---|---|
| CBDV | 0.2942000 ± 0.0018950 | −0.0011220 ± 0.0012680 | 0.9993 | 0.28–56.4 |
| CBDB | 0.2077000 ± 0.0016890 | 0.0006360 ± 0.0004843 | 0.9989 | 0.12–24.0 |
Table 2 reports the intra-day and inter-day accuracy and precision of CBDV and CBDB at four concentration levels (LLOQ, LCQ, MQC and HQC). Values are expressed as mean of three analyses for intra-day accuracy and precision and 15 analyses for inter-day accuracy and precision (n = 3 for 5 consecutive days).
Table 2.
Autosampler carryover calculated as percentage of the peak area of the analyte in a blank sample run after an HQC sample compared to the area of the analyte in the LLOQ sample.
| Compound | Carryover (%) |
|---|---|
| CBDV | 14.7 ± 3.8 |
| CBDB | 15.8 ± 1.4 |
| IS | <5 |
Table 3 shows the dilution integrity for CBDV and CBDB at three dilution factors (5, 10 and 20) calculated as accuracy and precision. Values are expressed as mean of five analyses (n = 5).
Table 3.
Intra-day and inter-day accuracy and precision of CBDV and CBDB at four concentration levels (LLOQ, LCQ, MQC and HQC). Values are expressed as mean of three analyses for intra-day accuracy and precision and 15 analyses for inter-day accuracy and preciusion (n = 3 for 5 consecutive days).
| CBDV |
CBDB |
||||
|---|---|---|---|---|---|
| Accuracy | Precision (CV) | Accuracy | Precision (CV) | ||
| Intra-day (n = 3) | LLOQ | 102.1 | 1.38 | 101.0 | 1.62 |
| LQC | 98.23 | 2.25 | 105.5 | 12.0 | |
| MQC | 101.1 | 0.98 | 100.3 | 2.57 | |
| HQC | 104.9 | 1.09 | 103.0 | 3.71 | |
| Inter-day (n = 15) | LLOQ | 104.3 | 2.76 | 100.7 | 3.62 |
| LQC | 102.0 | 2.25 | 91.67 | 9.14 | |
| MQC | 109.0 | 0.96 | 101.6 | 3.94 | |
| HQC | 104.1 | 1.07 | 102.0 | 2.37 | |
Table 4 relates to the stability data (bench-top and under refrigeration) for CBDV and CBDB calculated as mean of three analyses compared to nominal concentration of freshly prepared calibration curves.
Table 4.
Dilution integrity for CBDV and CBDB at three dilution factors (5, 10 and 20) calculated as accuracy and precision. Values are expressed as mean of five analyses (n = 5).
| Dilution factor | CBDV |
CBDB |
||
|---|---|---|---|---|
| Accuracy | CV | Accuracy | CV | |
| 5 | 98.91 | 0.74 | 95.67 | 0.96 |
| 10 | 96.72 | 1.25 | 93.47 | 0.48 |
| 20 | 99.53 | 2.05 | 96.00 | 2.27 |
Also, we have provided raw data files at this accessible link: https://drive.google.com/open?id=1gEL6bo5_btm5Gxul2FFVnLxZKMLdIdN7.
Where it is possible to find NMR FIDs for both synthetic and extracted cannabidibutol, HPLC-UV raw files with a blank sample, a sample at the LOD (limit of detection) concentration, a sample at the LOQ (limit of quantification) concentration, three samples at the QC levels (low, medium and high) and a commercial CBD sample. Moreover, we have provided a GraphPad file with linearity data and an Excel file with the calculations for the method validation.
2. Experimental design, materials, and methods
2.1. NMR analyses
One-dimensional 1H and 13C NMR and two-dimensional NMR (COSY, HSQC and HMBC) were acquired on a DPX-600 Avance (Bruker) spectrometer (600.13 MHz for 1H NMR and 150.92 MHz for 13C NMR). A 10 mg aliquot of synthetic CBDB and 1 mg aliquot of CBDB isolated from CBD were solubilized in 700 and 250 μL of CDCl3 (at 99.96% of deuteration) and placed in a 5 mm and 3 mm NMR tube, respectively. All NMR spectra were recorded at 298 K. All the NMR spectra were processed with TopSpin v4.0.6 (Bruker BioSpin 2018).
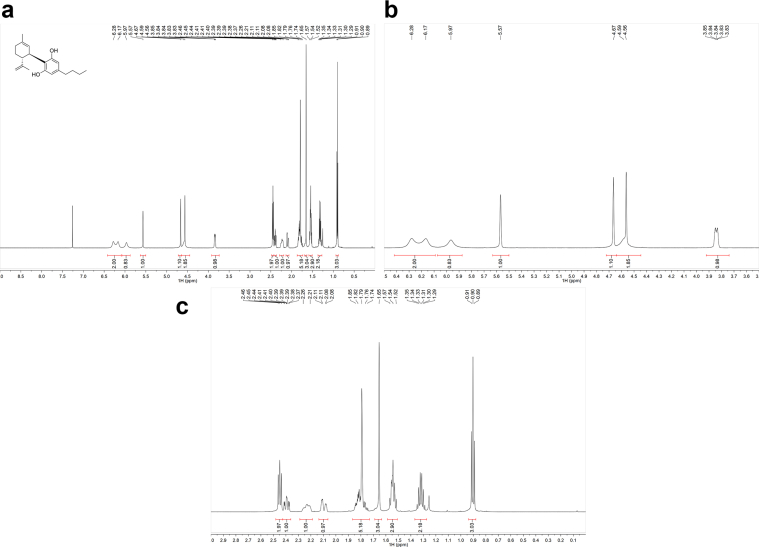
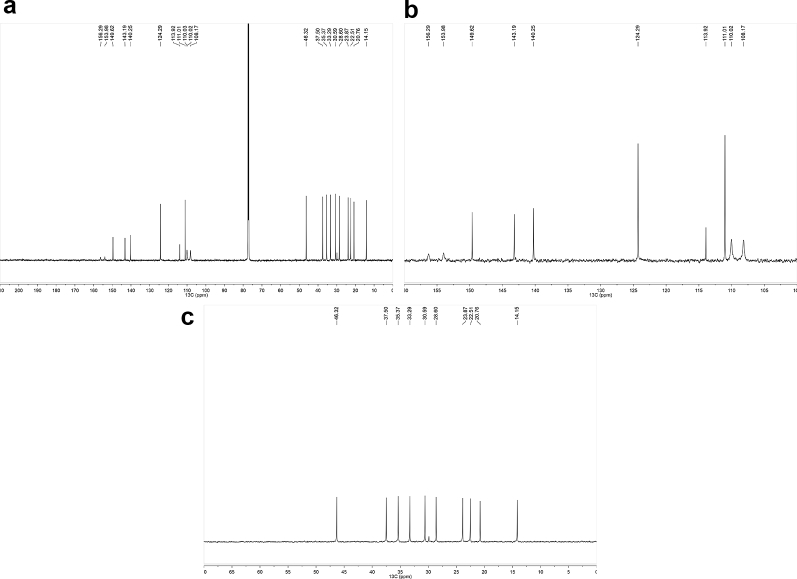
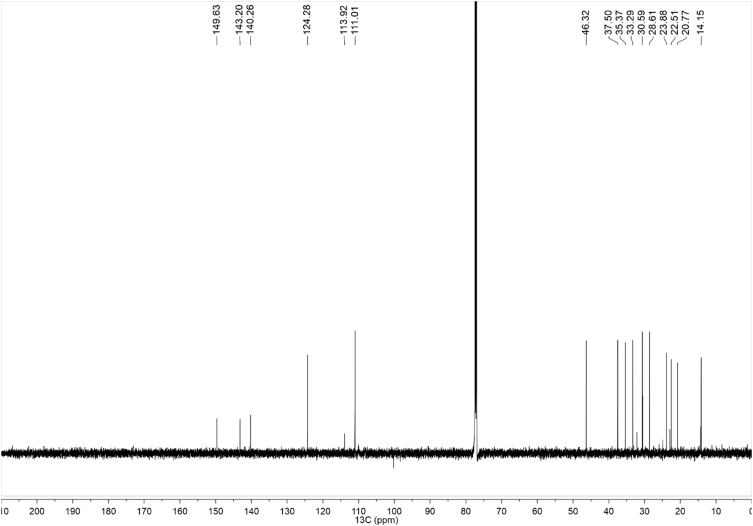
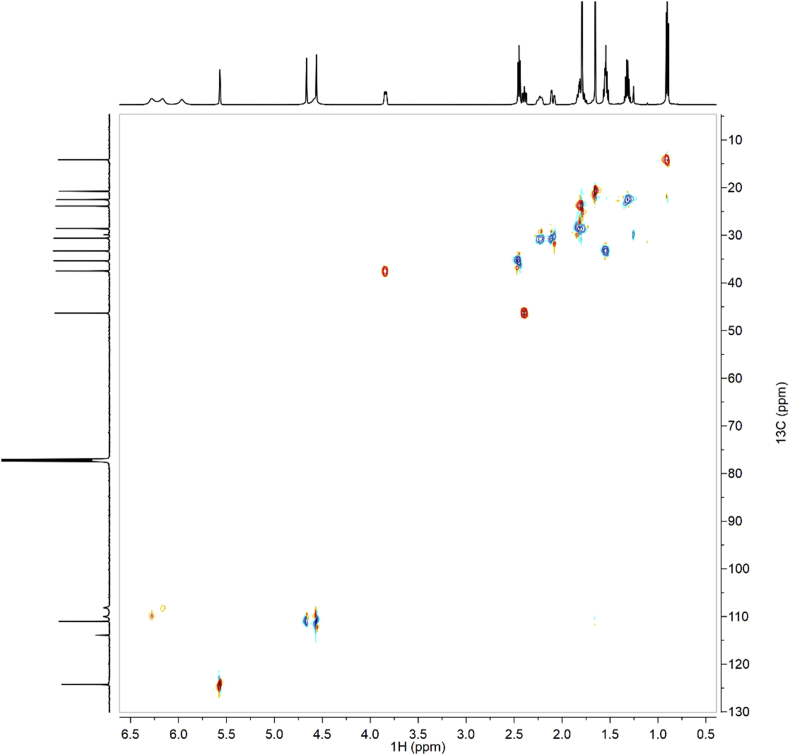
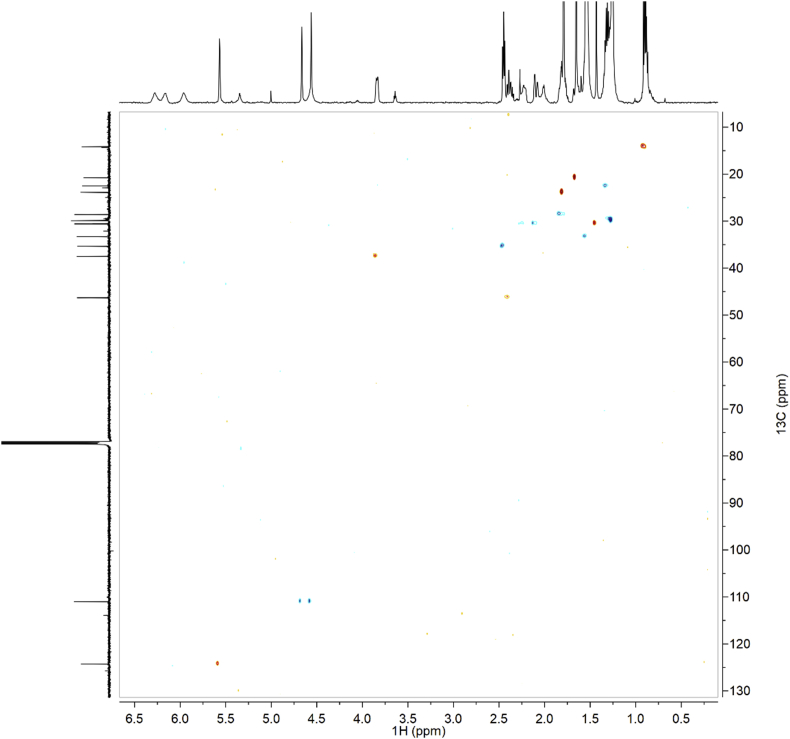
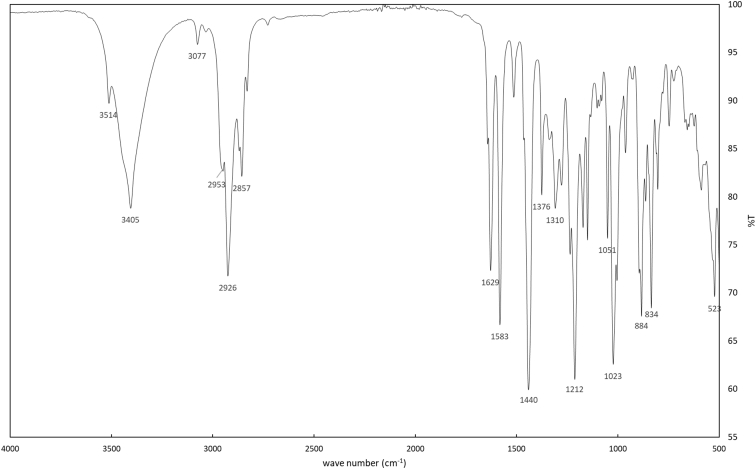
1H-NMR spectra (Fig. 1a–c for synthetic CBDB and Fig. 7 for isolated CBDB), were acquired with a spectral width of 13204.2 Hz, a relaxation delay of 5 s, a pulse width of 11.23 Hz and 16 number of transient. Proton chemical shifts were reported in parts per million (ppm, δ units) and referenced to the solvent residual peaks (CDCl3 δ = 7.26 ppm). Coupling constants are reported in Hertz (Hz). Splitting patterns are designed as s, singlet; d, doublet; t, triplet; q, quartet; dd, double doublet; m, multiplet; b, broad. 13C-NMR spectra (Fig. 2a–c for synthetic CBDB and Fig. 8 for isolated CBDB) were acquired with a spectral width of 33.3 kHz, a relaxation delay of 5 s, a pulse width of 10.00 Hz and 128 and 10240 number of transient for synthetic CBDB and extracted CBDB, respectively. Carbon chemical shifts were reported in parts per million (ppm, δ units) and referenced to the solvent residual peaks (CDCl3 δ = 77.20 ppm). The COSY, shown in Fig. 4, Fig. 10 for synthetic and isolated CBDB respectively, were recorded as a 1024 × 160 matrix with 2 transients per t1 increment and processed as a 1024 × 1024 matrix. The HSQC spectra, reported in Fig. 3, Fig. 9 for synthetic and isolated CBDB respectively, were collected as a 1024 × 256 matrix with 4 transients per t1 increment and processed as a 1024 × 1024 matrix, and the one-bond heteronuclear coupling value was set to 145 Hz. The HMBC spectra, shown in Fig. 5, Fig. 11 for synthetic and isolated CBDB respectively, were collected as a 2048 × 220 matrix with 8 transients per t1 increment and processed as a 2048 × 1024 matrix, and the long-range coupling value was set to 8 Hz. IR spectra, reported in Fig. 6 for synthetic CBDB, were recorded at 25 °C on a Perkin-Elmer Spectrum Two ATR-IR, scanning from 450 to 4000 cm−1, and processed with Spectrum 10™ software (PerkinElmer). Circular dichroism (CD) and UV spectra (Fig. 12) were acquired on a Jasco (Tokyo, Japan) J-1100 spectropolarimeter using a 50 nm/min scanning speed. Quartz cells with a 10 mm path length were employed to record spectra in the 400-200 nm range.
Fig. 1.
a: 1H-NMR of synthetic CBDB. b: 1H-NMR of synthetic CBDB from 3 to 5 ppm. c: 1H-NMR of synthetic CBDB from 0 to 3 ppm.
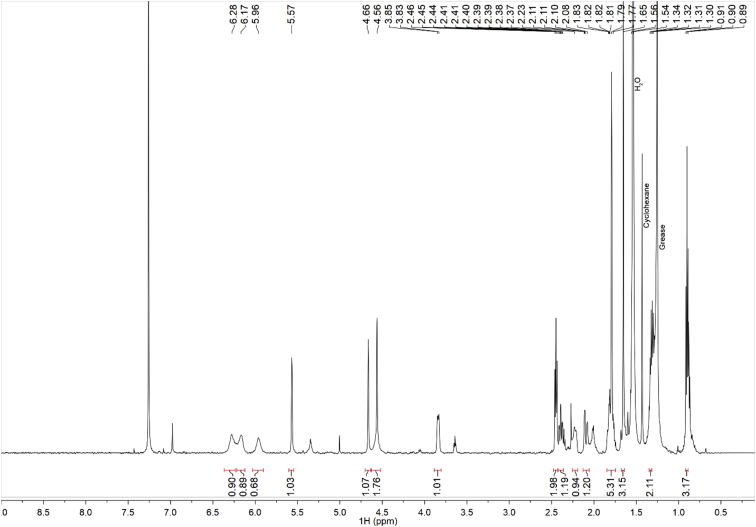
Fig. 7.
1H-NMR of isolated CBDB.
Fig. 2.
a: 13C-NMR of synthetic CBDB. b: 13C-NMR of synthetic CBDB from 100 to 160 ppm. c: 13C-NMR of synthetic CBDB from 0 to 70 ppm.
Fig. 8.
13C-NMR of isolated CBDB.
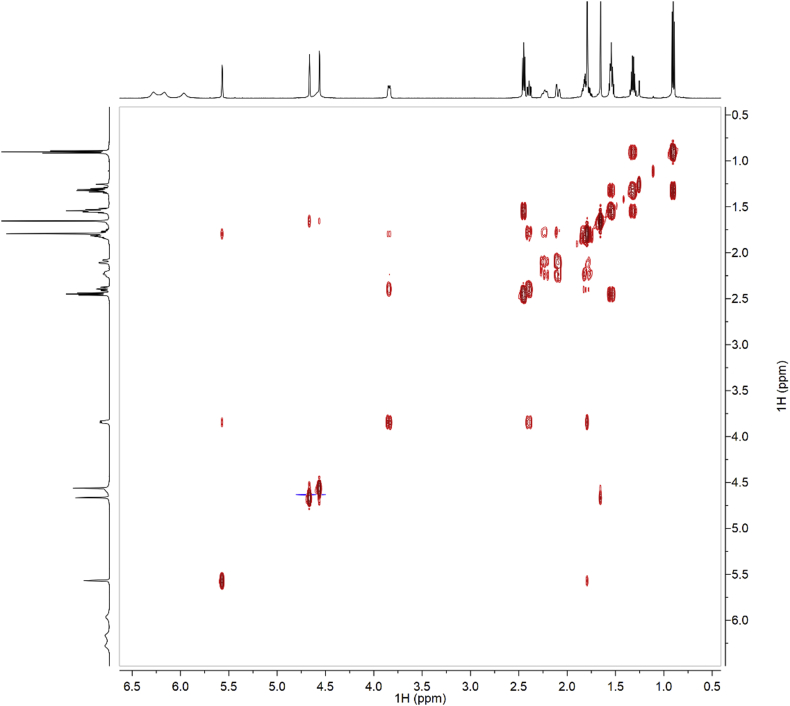
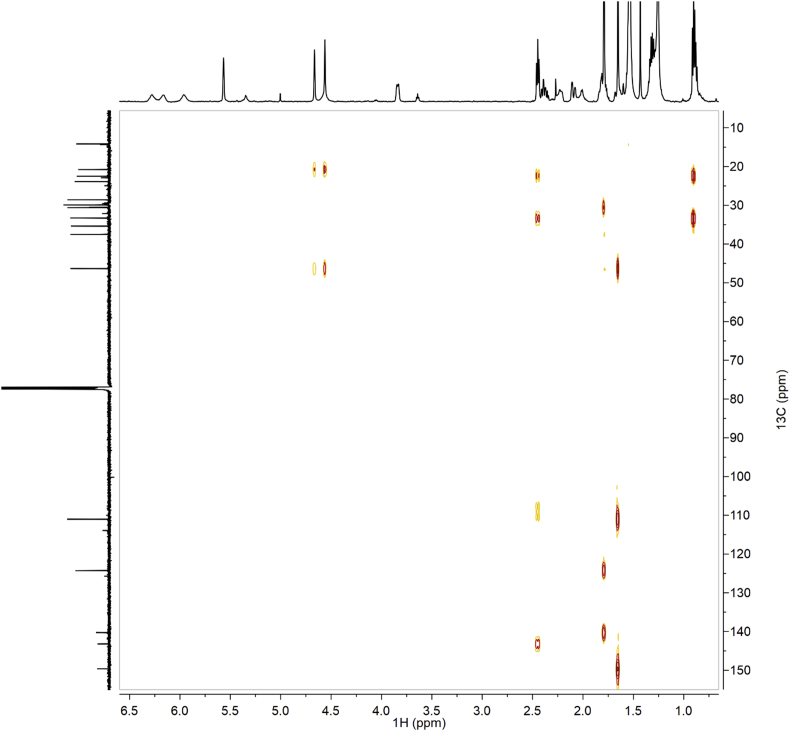
Fig. 4.
HSQC of synthetic CBDB.
Fig. 10.
HSQC of isolated CBDB.
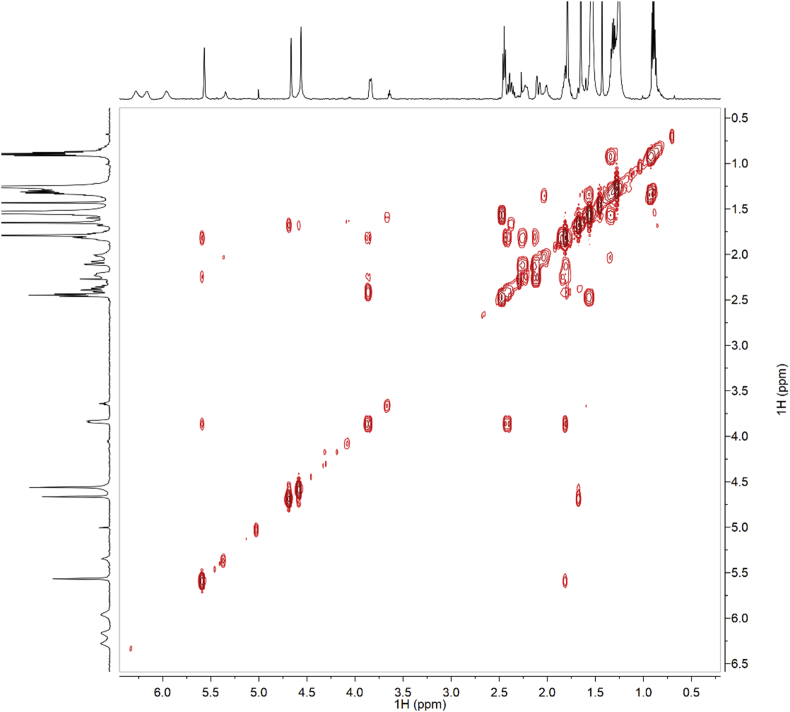
Fig. 3.
COSY of synthetic CBDB.
Fig. 9.
COSY of isolated CBDB.
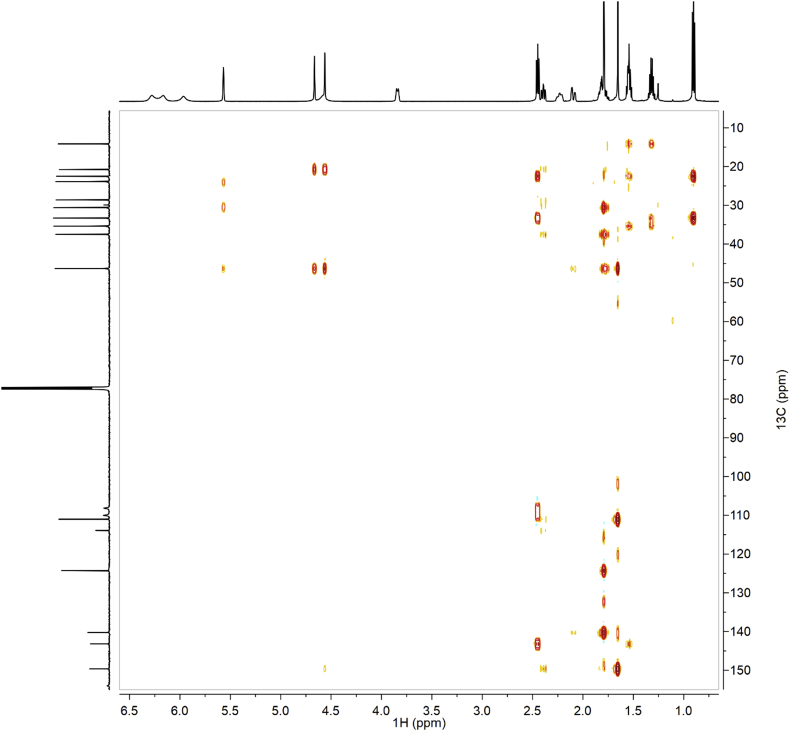
Fig. 5.
HMBC of synthetic CBDB.
Fig. 11.
HMBC of isolated CBDB.
Fig. 6.
IR of synthetic CBDB.
2.2. HPLC-UV method validation
The chromatograms were acquired with the software EZChrom on an Agilent 1220 Infinity LC System (Waldbronn, Germany), consisting of a vacuum degasser, a binary pump, a manual injector, a column compartment and a UV detector. The separation of the analytes was performed with a Poroshell 120 C18 column (Poroshell 120 SB-C18, 3.0 × 150 mm, 2.7 μm, Agilent, Milan, Italy) eluting a mobile phase composed of 0.1% formic acid in both (A) water and (B) acetonitrile (ACN). The chromatographic conditions were set as follows: isocratic elution with 70% B for 10 minutes, then 95% B pumped for 5 min and re-equilibration of the column for 2 min for a total run time of 17 min. The flow rate was maintained constant at 0.5 mL/min. The loading loop capacity was 6 μL. The loop was washed before each run first with 50 μL of ethanol 96% then with 50 μL of mobile phase. The UV trace was acquired at 228 nm. The analytes peaks were manually integrated using the EZChrom software (Agilent Technologies), which was employed also for controlling the online analysis.
Table 1 summarizes the experimental data obtained for testing linearity between the concentrations of CBDV and CBDB and the UV signal. Calibration curves were constructed at six non-zero calibration levels 0.28, 1.41, 2.82, 9.40, 28.2 and 56.4 μg/mL for CBDV, 0.12, 0.60, 1.20, 4.00, 12.0 and 24.0 μg/mL for CBDB, and 1.00 μg/mL for IS. Peak area ratios of analyte-to-IS (IS, ibuprofen 1 μg/mL) were plotted vs actual concentrations. Calibration curve was built at the beginning of each validation day of five consecutive days (n = 5). A linear correlation was assumed if the coefficient of determination (R2) was greater than 0.998 using weighed regression method (1/x2). The back calculated concentrations should be within 15% of the nominal concentrations, and within 20% of the lower limit of quantification (LLOQ).
Limit of detection (LOD) and limit of quantification (LOQ). Limit of detection (LOD) was estimated based on a 3:1 signal-to-noise (S/N) ratio. Standard stock solutions of the analytes were appropriately diluted at the levels of their respective estimated LOD values. The LOD values were then calculated as three times the standard deviation (SD) obtained by repeatedly analyzed standards (n = 5). Lower limit of quantification (LLOQ) was estimated based on a 10:1 S/N ratio and calculated as ten times the SD of repeatedly analyzed standards. The upper limit of quantification (ULOQ) was set at 10% above the highest concentration of the analytes in a concentrated sample of CBD (10 mg/mL).
Table 2 reports the results of the evaluation of the autosampler carryover. Autosampler carryover was evaluated by running two blank samples after a calibration standard at the ULOQ and after a high concentration sample (CBD 10 mg/mL). The carryover should not be greater than 20% of the LOQ for the analytes and 5% for IS.
Table 3 reports the data of accuracy and precision. The precision and accuracy were evaluated at four levels, LLOQ (0.28 μg/mL for CBDV and 0.12 μg/mL for CBDB), LQC (0.56 μg/mL for CBDV and 0.24 μg/mL for CBDB), MQC (18.8 μg/mL for CBDV and 8.00 μg/mL for CBDB), and HQC (45.1 μg/mL for CBDV and 19.2 μg/mL for CBDB). Each sample was analyzed in triplicate within a single day to determine the intra-day precision and accuracy. The replicate analyses were repeated on freshly prepared standard solutions for five successive days (n = 15) to determine the inter-day precision and accuracy. The precision was expressed as coefficient of variation (CV), and the accuracy was expressed as the percentage of mean calculated compared to nominal concentration.
Table 4 shows the results of the dilution integrity experiments. Dilution integrity was carried out using a spiked standard solution of the analytes prepared by diluting standard stock solutions to a final concentration that is three times that of the ULOQ (170.4 μg/mL for CBDV and 72.00 μg/mL for CBDB). Dilution integrity was demonstrated by diluting the spiking solution in IS to 1/5, 1/10 and 1/20 of its original concentration. Five replicates per dilution factor were run. The concentrations were calculated by applying the dilution factor 5, 10 and 20 against freshly prepared calibration curve. Dilution integrity is ensured as long as precision and accuracy are ≤15% and ±15% respectively.
Table 5 summarizes the stability tests. The short-term stability of the standard analytes was determined for LQC and HQC samples for 24 h at room temperature and under refrigeration (2–8 °C). Compounds were considered stable if the mean concentration (n = 3 for each sample) was within ±15% of the nominal concentration.
Table 5.
Stability data (bench-top and under refrigeration) for CBDV and CBDB calculated as mean of three analyses compared to nominal concentration of freshly prepared calibration curves.
| Stability | QC level | CBDV | CBDB |
|---|---|---|---|
| Bench-top (25 °C, 24 h) | LQC | 96.72 | 103.1 |
| HQC | 104.5 | 101.5 | |
| Refrigeration (2–8 °C, 24 h) | LQC | 103.2 | 101.2 |
| HQC | 102.7 | 100.6 |
Acknowledgments
This work was supported by UNIHEMP research project “Use of iNdustrIal Hemp biomass for Energy and new biocheMicals Production” (ARS01_00668) funded by Fondo Europeo di Sviluppo Regionale (FESR) (within the PON R&I 2017–2020 – Axis 2 – Action II – OS 1.b). Grant decree UNIHEMP prot. n. 2016 of 27/07/2018; CUP B76C18000520005 −COR 571,294.
Also, this research work has been partly funded by the project “Development of a cannabis based galenical preparation” FONDO DI ATENEO PER LA RICERCA ANNO 2017 - FAR2017, Italy (A.006@FAR2017DIP@05FA-CANNAZZA_FAR2017-(.20) CUP E53C17000720005).
The authors would like to thank Boris Bañas of CBDepot (Prague, Czech Republic), RSM S.p.a. (Montale, Italy), Linnea SA (Riazzino, Switzerland), Crystal Hemp (Lugano, Switzerland), Farmacia Sant’Elia (Corato, Italy) and Farmacia Tundo Dr. Alfredo (Alliste, Italy) for providing authentic CBD samples.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Citti C., Linciano P., Forni F., Vandelli M.A., Gigli G., Laganà A., Cannazza G. Analysis of impurities of cannabidiol from hemp. Isolation, characterization and synthesis of cannabidibutol, the novel cannabidiol butyl analog. J. Pharm. Biomed. Anal. 2019;175:112752. doi: 10.1016/j.jpba.2019.06.049. [DOI] [PubMed] [Google Scholar]