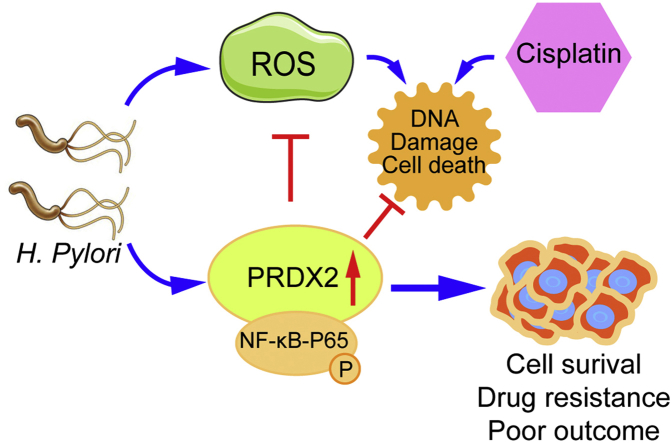
Background
Helicobacter pylori (H. pylori) infection is the main risk factor for gastric cancer. The role of antioxidant enzyme peroxiredoxin 2 (PRDX2) in gastric tumorigenesis remains unknown. In vitro (AGS and SNU-1 cell lines) and in vivo mouse models were utilized to investigate the role of PRDX2 in response to H. pylori infection (7.13, J166 or PMSS1 strain). We detected high levels of PRDX2 expression in gastric cancer tissues. Gastric cancer patients with high expression levels of PRDX2 had significantly worse overall and progression-free survival than those with low levels. H. pylori infection induced activation of NF-κB with increased expression of PRDX2, in in vitro and in vivo models. The knockdown of PRDX2 led to an increase in the levels of reactive oxygen species (ROS), oxidative DNA damage, and double-strand DNA breaks, in response to H. pylori infection, as measured by H2DCFDA, 8-oxoguanine, and p-H2AXγ assays. Luciferase reporter and ChIP assays confirmed the presence of a putative binding site of NF-κB-p65 on PRDX2 promoter region. The inhibition of PRDX2 significantly sensitized AGS and SNU-1 cells to cisplatin treatment. Our data suggest that the future development of therapeutic approaches targeting PRDX2 may be useful in the treatment of gastric cancer.
Keywords: NF-κB, Infection, Antioxidant response, Oxidative DNA damage, Apoptosis
Graphical abstract
Highlights
-
•
Infection with H. pylori induces PRDX2 expression, protecting gastric mucosa against reactive oxygen species, oxidative DNA damage and DNA double-strand breaks.
-
•
PRDX2 is a novel transcription downstream target of NF-kB-P65 in gastric mucosa.
-
•
Overexpression of PRDX2 in gastric cancer cells promotes resistance to cisplatin treatment and is associated with poor clinical outcome in gastric cancer patients.
Abbreviations
- PRDX2
Peroxiredoxin 2
- ROS
Reactive oxygen species
- H. pylori
Helicobacter pylori
- WB
Western blot
- qRT-PCR
Quantitative real-time PCR
- ChIP
Chromatin Immunoprecipitation
- CDDP
Cisplatin
- GC
Gastric cancer
- Redox
Reduction–oxidation reaction
- FBS
fetal bovine serum
- DAPI
4′, 6-diamidino-2-phenylindole
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- GEO
Gene Expression Omnibus
- EGA
European Genome-phenome Archive
- TCGA
The Cancer Genome Atlas
- H2DCFDA
2′, 7′-dichlorodihydrofluorescein diacetate
1. Background
Gastric cancer (GC) is the 4th most common cancer and 3rd leading cause of cancer-related deaths worldwide [1,2]. Infection with Helicobacter pylori (H. pylori) is one of the most established risk factors for the development of gastric cancer [[3], [4], [5], [6]]. H. pylori infection affects approximately 4.4 billion people, defining it as one of the most common infections worldwide [7]. Once infection with H. pylori is acquired, the gastric epithelium is the main interface of the host and the bacteria. Colonization of H. pylori infection in the gastric mucosa initiates chronic inflammatory response, leading to the development of gastritis that can progress in a multi-step gastric tumorigenesis cascade, known as Correa's cascade [[8], [9], [10]]. This chronic pro-inflammatory environment is associated with an increase in the levels of reactive oxygen species (ROS), oxidative DNA damage with activation of oncogenic signaling pathways involved in gastric carcinogenesis [[8], [9], [10], [11]].
Peroxiredoxin 2 (PRDX2) is a typical 2-Cys antioxidant enzyme belonging to the peroxiredoxin family and plays an important role in scavenging H2O2 and ROS levels, therefore protecting cells from oxidative stress [12]. Under normal physiological conditions, the levels of ROS are tightly controlled to maintain essential biological functions and normal cellular homeostasis [13]. Interruption of the physiological balance between oxidation and reduction (redox) leads to excessive accumulation of ROS with accumulation of DNA damage [14,15]. The expression of PRDX2 protein is quite abundant in mammalian cells and plays a critical role in keeping the redox balance and prolonging cell life span. A reduced PRDX protein is oxidized by ROS and H2O2 with the formation of oxidized PRDX, a key step in eliminating ROS levels with the thiol subunit of the cysteine residues [12,16]. Among all PRDXs, PRDX2 is one of the most efficient ROS and H2O2 scavenger proteins, protecting cells from oxidative stress, as compared with other PRDX family members [17]. PRDX2 could function as a suppressor or enhancer of tumorigenesis in a context- and cell-dependent manner, subject to etiological factors, cancer type, and stage of tumor progression [16,18]. As a tumor suppressor, decreased expression of PRDX2 promotes the proliferation and migration in melanoma and is associated with EMT and activation of β-catenin signaling [19]. On the other hand, overexpression of PRDX2 correlated with cancer progression in several malignancies, including cancers of the colon, cervix, lung and prostate [[20], [21], [22], [23], [24]].
The role of PRDX2 in gastric cancer, as it relates to H. pylori infection, remains largely unknown. In this study, we investigated the expression pattern and function of PRDX2 in response to H. pylori infection in gastric cancer.
2. Methods
2.1. Cell culture and reagents
Human gastric cancer cell lines AGS and SNU-1 were obtained from American Tissue Culture Collection (ATCC, Manassas, VA). SNU-1 was cultured in RPMI with 10% fetal bovine serum (FBS, Invitrogen Life Technologies, Carlsbad, California, USA) and 1% penicillin/streptomycin (GIBCO, Invitrogen Life Technologies). AGS cells were cultured in F12 with 10% fetal bovine serum (FBS, Invitrogen Life Technologies) and 1% penicillin/streptomycin (GIBCO, Invitrogen Life Technologies). Recombinant human TNF-α (No. 300-01A, PeproTech, New Jersey, USA) and Bay 11–7082 (No. S2913, Selleckchem, Houston, Texas, USA) were purchased.
2.2. Human samples
The de-identified human tissue samples used for qRT-PCR came from 40 gastric cancer and 40 normal gastric tissues. Samples were collected from the archives of Pathology and the National Cancer Institute Cooperative Human Tissue Network (CHTN). De-identified human tissues for immunohistochemistry were obtained from the Department of Gastric Surgery at the First Affiliated Hospital of Nanjing Medical University. Written informed consent was obtained from all patients. All tissue samples were obtained, coded, and de-identified in accordance with the Institutional Review Board-approved protocols.
2.3. H. pylori culture and infection
Wild-type CagA positive H. pylori 7.13, J166 and the rodent-adapted PMSS1 strain were a kind gift from Dr. Richard Peek, Jr. (Vanderbilt University Medical Center). Briefly, H. pylori strains were cultured in trypticase soy agar with 5% sheep blood (BD Biosciences, Bedford, Massachusetts, USA) for 3 days and transferred to a new agar plate [25]. The strains were then cultured in Brucella broth with 10% serum and 10 μg/mL vancomycin at 37 °C and placed in an incubator with 5% CO2 overnight. The H. pylori bacteria was added over gastric cells, for a co-culture, at a multiplicity of infection of 100:1 and cells were harvested at different time points.
2.4. PRDX2 siRNA
PRDX2 siRNA was purchased from Ambion (AM51331, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and Santa Cruz (sc-40832, Santa Cruz, Dallas, Texas, USA). A concentration of 40 nM of siRNA into 1x LipoJet Transfection buffer with LipoJet reagent (SL100567, SignaGen, Rockville, Maryland, USA) was prepared for transfection. The knockdown of PRDX2 was verified using Western blot analysis.
2.5. Western blot
Proteins were extracted from cell lysates using standard methods. Proteins were separated by 10% or 12.5% running gel and stack gel electrophoresis, made of 40% acrylamide/bis solution, SDS, Tris and other components. Proteins were then transferred to the nitrocellulose membranes (Bio-rad, Hercules, California, USA), followed by blocking in 5% Bovine Fraction V (BSA, 9048-46-8, RPI (Research Products International), Mount Prospect, Illinois, USA) for 3h. After hybridization with primary antibodies at 4°C overnight, the membranes were washed and immunoblotted with secondary antibodies. Images were obtained by the Bio-Rad ChemiDoc XRS + System. Primary antibodies were as follows: PRDX2 (46855S, Cell Signaling Technology, Danvers, MA, USA), phospho–NF–κB-p65 (Ser536) (3033S, Cell Signaling Technology), NF-κB-p65 (D14E12) (8242s, Cell Signaling Technology), Phospho-Histone H2A.X (Ser139) (2577s, Cell Signaling Technology), Histone H2A.X (2595s, Cell Signaling Technology), Cleaved PARP (As214) (5625s, Cell Signaling Technology), PARP (46D11) (9532s, Cell Signaling Technology) and beta-Actin (A5441, Sigma-Aldrich, St. Louis, Missouri, USA).
2.6. Quantitative real-time PCR
Total RNA was extracted from cells and tumor tissues using TRIzol reagent. Integrity and quantity were assessed using the NanoDrop 2000 (Thermo Fisher Scientific). The RNA was reverse transcripted with high capacity cDNA Reverse Transcription kit (4368814, Applied Biosystems, Foster City, California, USA). Universal SYBR Green Master kit was used for quantitative PCR with a 1:5 dilution cDNA and PCR was performed on the Bio-Rad CFX Connect Real-time System (Bio-Rad). The Primers used were as follows: human PRDX2-F: 5′-CGTCTCGGTGGACTCTCAGT-3′, human PRDX2-R: 5′-TCAGACAAGCGTCTGGTCAC -3; mouse Prdx2-F: 5′- CACCTGGCGTGGATCAATACC-3′, mouse Prdx2-R: 5′- GACCCCTGTAAGCAATGCCC -3’; human HPRT1-F 5′-TTGGAAAGGGTGTTTATTCCTCA -3′, human HPRT1-R 5′-TCCAGCAGGTCAGCAAAGAA -3’; mouse Hprt1-F: 5′-TATGCCGAGGATTTGGAAAA-3′, mouse Hprt1-R: 5′- ACAGAGGGCCACAATGTGAT-3’; PMSS1-F: 5′- CGTCCGGCAATAGCTGCCATAGT-3′, PMSS1-R: 5′- GTAGGTCCTGCTACTGAAGCCTTA-3. Experiments were performed in triplicate and the ΔΔC(t) method was adopted for analysis.
2.7. Reactive oxygen species
Reactive oxygen species (ROS) were detected using H2DCFDA (Thermo Fisher Scientific) [26,27]. The H2DCFDA was dissolved in DMSO to achieve 1 mM as a stock concentration and diluted to 5 μM using PBS (phosphate buffered solution). After removing culture medium and washing with PBS, the cells were seeded in 24-well plates, incubated with dye at room temperature for 40 min in dark conditions. After removing the dye, cells were then covered in phenol red-free DMEM for 15 min at 37 °C in an incubator. Images were obtained with the All-in-One Fluorescence Microscope (BZ-X700) (Keyence, Itasca, IL, USA).
2.8. Luciferase reporter assay
To construct the PRDX2 promoter luciferase vector, we amplified three different promoter sequences of PRDX2 and inserted them into the luciferase reporter plasmid before the Kozak and luciferase sequences (P1: 1050 ——0 bp upstream of PRDX2 coding sequence, P2: 774 ——0 bp upstream of PRDX2 coding sequence, P3: 500 ——0 bp upstream of PRDX2 coding sequence), using VectorBuilder services (VectorBuilder Inc, Chicago, IL, USA). The NF-κB-p65 plasmid and the luciferase reporter plasmids were co-transfected into cells with empty vector as a control. The luciferase signal was detected using the luciferase reporter assay kit (Promega, Madison, Wisconsin, USA) and measured using BMG FLUOstar OPTIMA Microplate Reader (BMG LABTECH, Cary, North Carolina, USA).
2.9. Chromatin immunoprecipitation assay
The Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (17–10085, Millipore Sigma, Burlington, Massachusetts, USA) was used for the Chromatin Immunoprecipitation (ChIP) assay [25]. AGS cells cultured in 10-cm plates were fixed with 37% formaldehyde, followed by adding cell and nuclear lysis buffer. Chromatin fragments were sonicated on ice for 6 cycles (30s On and 30s Off at 40% amplitude). The shear cross-linked DNA was sonicated into 100–1000 bp for the best efficiency of pull-down. The immunoprecipitation antibody NF-kB-p65 (Ser536) (3033S, Cell Signaling Technology) and control antibody normal mouse IgG (12–371, Millipore Sigma), as well as protein A/G magnetic beads (CS204457, Millipore Sigma), were added into lysates and incubated at 4 °C overnight. Elution of the protein/DNA complexes was obtained after DNA purification using wash buffers and standard PCR. We designed three pairs of primers for potential NF-κB-p65 binding sites: PRDX2-1-CHIP-F: 5′- GATGGAGTCTTGCTGTGTGG -3′, PRDX2-1-CHIP-R: 5′- CATAGGGGAAAGGGGCAGAT -3’ (Primer 1: −1083 —— −921 bp); PRDX2-2-CHIP-F: 5′- CACACCTCACCGACCTCTTT -3′, PRDX2-2-CHIP-R: 5′- GAAGCTGTCACTCGGGGATA -3’ (Primer 1: −832 —— −594 bp); PRDX2-3-CHIP-F: 5′- TGCCCACACCCTCTCTTC -3′, PRDX2-3-CHIP-R: 5′- TTGCCCTACTTCTCCTGCTG -3’ (Primer 1: −587 —— −403 bp).
2.10. Immunofluorescence
5000 cells were seeded in each well of an 8-chamber slide. After transfection with PRDX2 and H. pylori infection, the cells were fixed with 4% paraformaldehyde solution for 45 min at 4 °C. After washing cells, the cells were incubated in the permeabilization solution for 3 min on ice. Each well was blocked using 100 μl non-immune goat serum (50062Z, Thermo Fisher Scientific) for 20 min in the dark. Cells were incubated with primary antibodies overnight, followed by incubation with secondary antibodies for 1 h. Slides were sealed with the cover glass after DAPI was added. Primary antibodies that were used: 8-Oxoguanine (MAB3560, Sigma Aldrich, St. Louis, MO, USA) (1:100) and Phospho-Histone H2A.X (Ser139) (2577s, Cell Signaling Technology) (1:250). Secondary antibodies used were: Alexa Fluor 568 goat anti-mouse IgG (1:400) and Alexa Fluor 488 goat anti-rabbit IgG (1:400). Cells were analyzed and imaged using the All-in-One Fluorescence Microscope (BZ-X700) (Keyence).
2.11. Cell viability assay
To detect cell survival after knockdown of PRDX2, we used 5 μM cisplatin (CDDP) to treat AGS and SNU-1 cells for 48 and 72 hours. For each time point, 100 μl of CellTiter-Glo Reagent (Promega, Madison, Wisconsin, USA) was added into each well of the 24-well plates and placed on a shaker at room temperature for 10 min. Next, 50 μl of media was added into 96-well plates. All experiments were performed in triplicate. Data was then obtained using BMG FLUOstar OPTIMA Microplate Reader (BMG LABTECH, Cary, North Carolina, USA).
2.12. In vivo experiment
All animal use protocols were approved by the Institutional Animal Care and Use committee of the University of Miami. C57BL/6 mice were purchased from Charles River Laboratories. Twenty mice were randomly assigned into control and PMSS1 groups with 10 mice per group. Mouse-adapted wild-type H. pylori strain PMSS1 (109 CFU/mouse) was introduced using orogastric gavage. The control group included mice inoculated with Brucella broth. Mice were euthanized at one week or two weeks. Glandular gastric tissues were obtained for Western blot and qRT-PCR analysis.
2.13. Colony formation
500 cells were seeded into 6-well plates and incubated at 37 °C and 5% CO2 for 2 weeks. Cells were fixed using ethanol and stained with 0.1% crystal violet. Images were obtained and colonies were counted.
2.14. Immunohistochemistry staining
Fifteen human gastric tissues and fifteen human normal adjacent tissues were included. Twelve male and three female patients (age 48 to 76) were diagnosed with gastric cancer with clinical stages from ⅠA to ⅡB. The gastric cancerous tissues were a mix of intestinal type (N = 9), diffuse type (N = 2) and mixed type (N = 4). These tissues were fixed in 4% paraformaldehyde and embedded in paraffin blocks. The tissues were embedded into paraffin as blocks for storage and cut into 5-μm-thick sections. For immunohistochemistry (IHC), 5-μm-thick sections were cut into glass slides. IHC procedure was performed following the standard protocol [25]. Sections were incubated with primary antibody at 4 °C overnight and then incubated with a secondary antibody at room temperature for 1 hour. After washing the slides, chromogen was applied and images were obtained with the All-in-One Fluorescence Microscope (BZ-X700) (Keyence). The primary PRDX2 antibody (LF-MA0144, Thermo Fisher Scientific) (1:250) was used. The scores of tissues were assessed by an independent pathologist with ImageJ [28,29]. The scores were automatically determined by imageJ and checked by the individual pathologist. The scores were presented as relative expression levels to adjacent non-cancerous tissues.
2.15. Statistical methods
Data was presented as mean ± SD in each experiment. The significant difference was considered as P < 0.05 in each experiment. One-way analysis of variance (ANOVA) and Student's t-test were applied.
3. Results
3.1. Overexpression of PRDX2 is a poor prognostic marker in gastric cancer
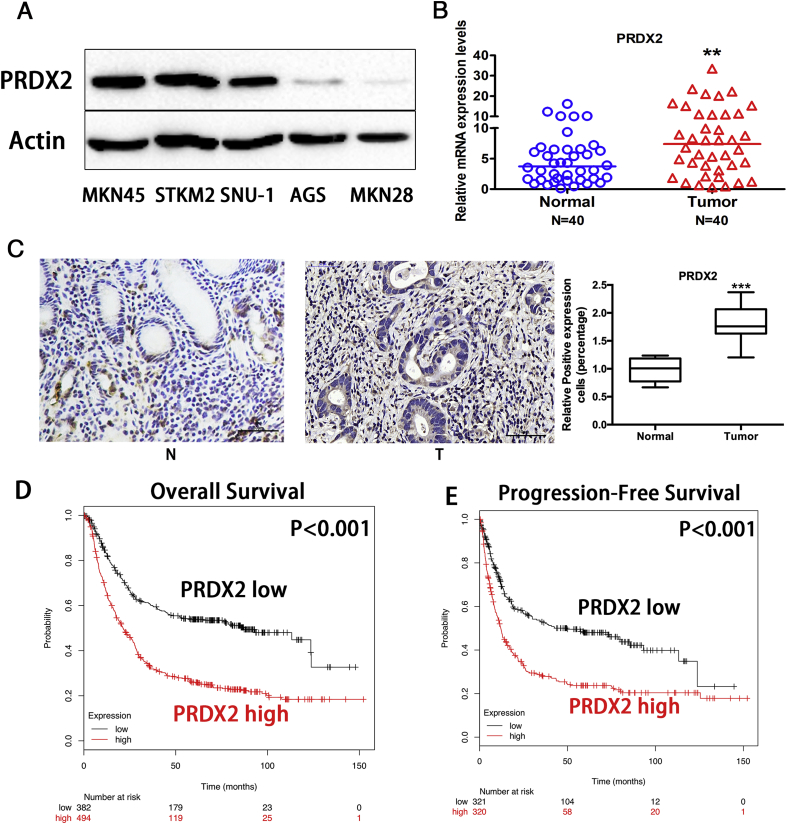
First, we detected PRDX2 protein expression levels in five gastric cancer cell lines. Western blot results indicated the protein expression level of PRDX2 was remarkably higher in MKN45, STKM2 and SUN-1 cells. Similar or lower expression of PRDX2 was observed in AGS and MKN28 cells. (Fig. 1A). To examine the PRDX2 expression levels in gastric cancer samples, qRT-PCR analysis in human gastric tissues samples demonstrated the PRDX2 mRNA level was significantly higher in the gastric cancer sample (N = 40) compared with adjacent normal gastric tissues (N = 40) (Fig. 1B, P < 0.05). However, we did not detect any significant correlation between PRDX2 mRNA expression level and TNM staging, possibly due to the limited number of cases. Furthermore, immunohistochemistry analysis was performed on human normal gastric (N = 15) and gastric cancer tissues (N = 15). Our data demonstrated that PRDX2 protein level is significantly higher in gastric cancer, as compared with normal gastric samples(Fig. 1C, P < 0.001). Similar to the mRNA expression results, we did not detect any significant association between PRDX2 protein expression level with histopathological or other clinical parameters, possibly because of the relative small sample size. To investigate the prognostic value of PRDX2 expression, overall survival (OS), progression-free survival (PFS) and PRDX2 mRNA expression levels were analyzed by KMplot (http://kmplot.com/analysis/index.php?p=service), downloaded from Gene Expression Omnibus (GEO), European Genome-phenome Archive (EGA) and The Cancer Genome Atlas (TCGA). The results revealed that patients with higher PRDX2 expression demonstrated significantly worse OS and PFS (N = 876, divided by median PRDX2 mRNA expression; Fig. 1D and E). These results indicate that PRDX2 is overexpressed in gastric cancer. Moreover, PRDX2 expression level is associated with poor survival and could serve as a valuable prognostic factor in gastric cancer.
Fig. 1.
PRDX2 is overexpressed in gastric cancer, predicting prognosis.
(A) Western blot analysis of PRDX2 and β-actin in gastric cancer cell lines. (B) qRT-PCR analysis of PRDX2 mRNA expression level in 40 normal and 40 gastric cancer tissue samples. (C) Immunohistochemistry staining of PRDX2 on human adjacent non-cancerous gastric and gastric cancer tissues. Scale Bar: 50 μm. (D) Kaplan-Meier analysis of overall survival (OS) in gastric cancer patients based on PRDX2 expression level. (E) Kaplan-Meier analysis of progression-free survival (PFS) in gastric cancer patients based on PRDX2 expression level. ***, P < 0.001.
3.2. PRDX2 expression is induced by H. pylori infection in vitro and in vivo
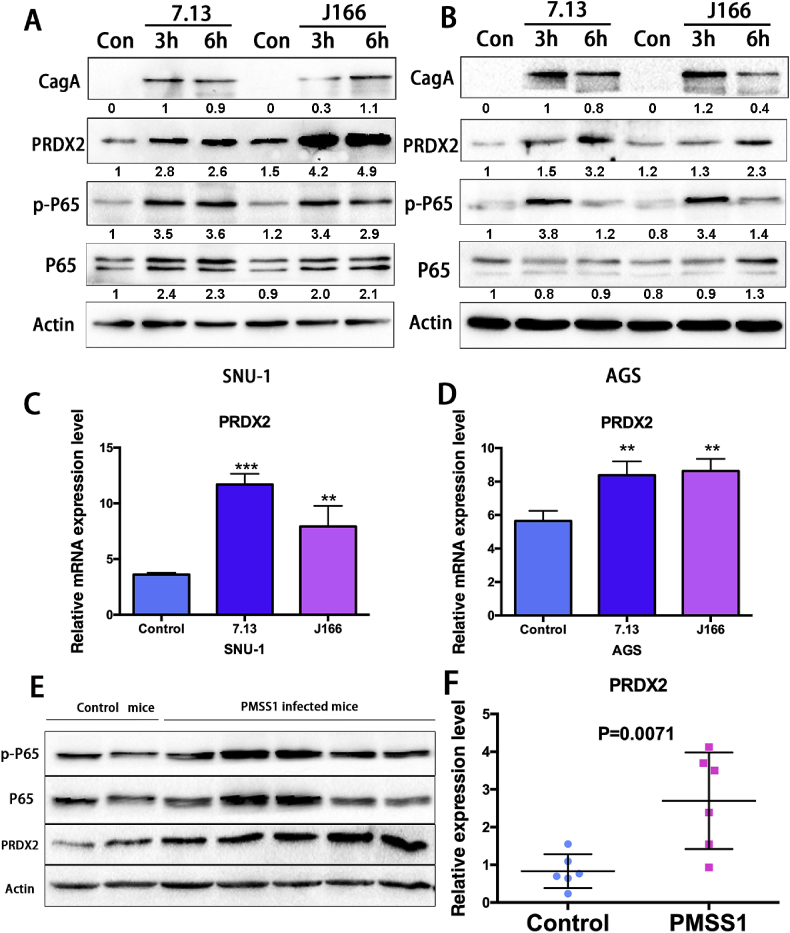
Since H. pylori infection is a major risk factor of gastric cancer, we tested whether PRDX2 expression in gastric cancer cells is related to H. pylori infection. AGS and SNU-1 gastric cancer cells, with relatively low endogenous PRDX2 expression level, were examined to investigate whether H. pylori infection induces PRDX2 expression in gastric cells. Two H. pylori strains 7.13 or J166 were co-cultured with AGS or SNU-1 cells for 3 or 6 h. Western blot analysis demonstrated that PRDX2 and phospho–NF–κB-p65 protein expression levels were induced with both 7.13 and J166 infections (3 h and 6 h) in SNU-1 and AGS cells. The expression of CagA was tested to confirm the H. pylori infection in gastric cancer cells (Fig. 2A and B). Consistently, qRT-PCR results indicated that PRDX2 mRNA levels were significantly increased after both H. pylori strain infections for 6 h in AGS and SNU-1 cells (Fig. 2C and D). H. pylori infection induced both mRNA and protein expression levels of PRDX2 in gastric cancer cells. We used rodent-adapted H. pylori strain PMSS1 to infect the wild-type (WT) C57BL/6 mice to confirm in vitro data. Protein and RNA samples were acquired from broth (control) or H. pylori infected (PMSS1) mice stomach tissues. Western blot data showed the PRDX2 protein expression level was remarkably induced at 1–2 weeks post PMSS1 infection, compared with control group (Fig. 2E). We also observed an increase of phospho–NF–κB-p65 in gastric cells after PMSS1 infection (Fig. 2E). qRT-PCR demonstrated successful infections of PMSS1 (Supplementary Fig. S1). We found that the protein expression of PRDX2 was significantly upregulated in tissues infected with PMSS1 for 1 or 2 weeks compared to control mice tissues. Next, qRT-PCR was performed to detect the PRDX2 mRNA level in control and PMSS1 infected mice gastric tissue samples. Our data revealed that PRDX2 mRNA level was significantly upregulated after PMSS1 infection (Fig. 2F). These results indicated that both PRDX2 mRNA and protein expression levels are induced by H. pylori infection in gastric cells in vitro and in vivo.
Fig. 2.
PRDX2 expression is induced by H. pylori infections in vitro and in vivo.
(A) and (B) Western blot data of CagA, PRDX2, p–NF–kB-p65, NF-kB-p65, and β-actin in AGS and SNU-1 cells infected with 7.13 (100 MOI), J166 (100 MOI) or PBS (Control) for 3 h and 6 h. (C) and (D) qRT-PCR analysis of PRDX2 mRNA expression levels in AGS and SNU-1 cells infected with 7.13 (100 MOI), J166 (100 MOI) or PBS (Control) for 6h. (E) Western blot analysis of PRDX2, p–NF–kB-p65, NF-kB-p65 and β-actin in control or PMSS1 infected mice for 1–2 weeks. (F) qRT-PCR analysis of PRDX2 mRNA expression level in control or PMSS1 infected mice 1–2 weeks. Quantification of Western blot data was performed using Image Lab software from Bio-Rad. *, P < 0.05, **, P < 0.01.
3.3. PRDX2 genetic knockdown induces reactive oxygen species (ROS) with or without H. pylori infection in gastric cancer cells
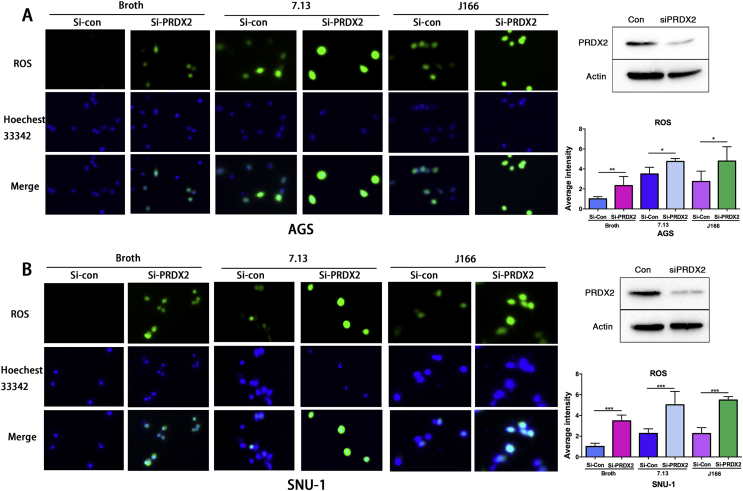
To test the role of PRDX2 in regulating ROS levels, we performed experiments with or without H. pylori infection. Immunofluorescence staining of 2′, 7′-dichlorodihydrofluorescein diacetate (H2DCFDA), an ROS indicator in cells [26,27], was performed in control or PRDX2 siRNA knockdown AGS and SNU-1 cells with or without H. pylori (7.13 or J166) infection (6 h). Our data indicated that PRDX2 knockdown significantly induced higher ROS levels without H. pylori infection, as compared with control (Fig. 3A and B, P < 0.05). In the meantime, both 7.13 and J166 H. pylori infections induced high ROS levels, as expected. PRDX2 knockdown with 7.13 or J166 infection showed significantly higher ROS levels (P < 0.05), compared with 7.13 or J166 infection alone in AGS or SNU-1 cells (Fig. 3A and B, P < 0.05). PRDX2 knockdown was confirmed using Western blot analysis (Fig. 3A and B right panels). These results indicated that knockdown of PRDX2 promoted a significant increase in ROS levels, especially noted with H. pylori infection, in gastric cancer cells.
Fig. 3.
Knockdown of PRDX2 enhances H. pylori-induced ROS level.
(A) Left panel: the level of reactive oxygen species (ROS) was detected using H2DCFDA in control or PRDX2 siRNA knockdown AGS cells with or without H. pylori (7.13 or J166) infection for 6 h. Right upper panel: Western blot confirmed the knockdown of PRDX2 was efficient in AGS cells. Right lower panel: quantification data for A. (B), similar experiment in SUN-1 cells as in A. *, P < 0.05, **, P < 0.01, ***, P < 0.001.
3.4. PRDX2 abrogates H. pylori-induced DNA damage in gastric cancer cells
We investigated the role of PRDX2 in protecting gastric cancer cells from H. pylori induced oxidative stress. By using the same experiment settings as in Fig. 3, we measured DNA lesions with 8-Oxoguanine immunofluorescence staining. Immunostaining for 8-Oxoguanine demonstrated a significant increase in oxidative DNA damage in conditions of knockdown of PRDX2, most notable with H. pylori infection (P < 0.001) (Supplementary Fig. S2). PRDX2 knockdown was confirmed using Western blot analysis (Supplementary Fig. S2 right panels). These results indicated that PRDX2 protected gastric cancer cells from oxidative DNA damage induced by H. pylori infection.
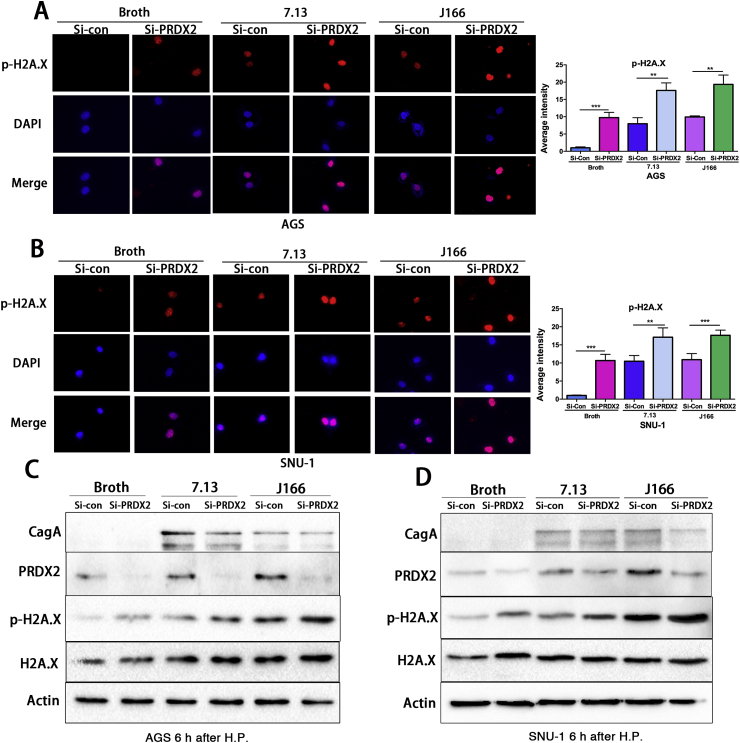
Because the occurrence of double-strand DNA breaks (DSB) is a natural progression of oxidative DNA damage lesions, if not repaired, we determined the levels of DSB in response to H. pylori and PRDX2 knockdown. p-H2A.X immunofluorescence staining in AGS or SNU-1 cells with or without H. pylori infection demonstrated a significant increase in DSB, following H. pylori infection and knockdown of PRDX2, as compared with controls (P < 0.01) (Fig. 4A and B, P < 0.01). Western blot analysis from these cells confirmed the increase in p-H2A.X protein expression levels after knockdown of PRDX2 (Fig. 4C & D) and similar results were found after treating cells with PRDX2 siRNA-2 (Supplementary Fig. S3). These data demonstrated that PRDX2 protects against the formation of double-strand DNA breaks in gastric cells.
Fig. 4.
Knockdown of PRDX2 increases the expression of p-H2A.X with or without H. pylori infection.
The immunofluorescence staining of p-H2A.X in control or PRDX2 siRNA knockdown AGS cells with or without H. pylori (7.13 or J166) infection for 6 h. Right upper panel: Western blot confirmed the knockdown of PRDX2 was efficient in AGS cells. Right lower panel: quantification data for A. (B), similar experiment in SUN-1 cells as in A. *, P < 0.05, **, P < 0.01, ***, P < 0.001. (C) Western blot analysis of CagA, PRDX2, p–NF–kB-p65, NF-kB-p65, p-H2A.X, H2A.X and β-actin in control or PRDX2 siRNA knockdown AGS cells with or without H. pylori (7.13 or J166) infection for 6 h. (D), similar result in SNU-1 cells as C. **, P < 0.01, ***, P < 0.001.
3.5. PRDX2 expression level is regulated by NF-κB signaling in gastric cancer cells
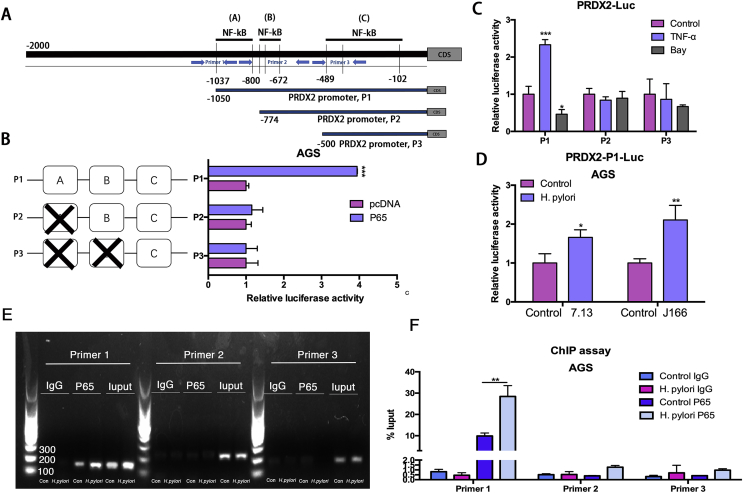
Several studies showed that H. pylori infection activates NF-κB signaling in gastric cancer cells [25,30]. Therefore, we investigated our hypothesis that NF-κB activation may promote PRDX2 expression to re-balance the oxidative stress and DNA damage induced by H. pylori infection. Western blot data demonstrated that NF-κB inhibitor Bay-11-7082 (Bay) treatment dramatically decreased the level of phospho–NF–κB-p65 and PRDX2. Meanwhile, H. pylori infection (7.13 or J166) induced NF-κB-p65 (Supplementary Fig. S4 A & B). Interestingly, Bay treatment abrogated the expression of PRDX2 induced by two H. pylori strains, indicating that H. pylori-induced PRDX2 expression is mediated by NF-κB signaling (Supplementary Fig. S4 A & B). To confirm our findings that PRDX2 expression was regulated by NF-κB pathway, we stimulated AGS and SNU-1 cells with TNF-α. Western blot data indicated that PRDX2 expression was induced after TNF-α treatment, as compared to control, suggesting that the NF-κB pathway played an important role in PRDX2 expression (Supplementary Figs. S4C and D). Next, to investigate if PRDX2 is a transcription target of NF-κB signaling in gastric cells, we used luciferase reporter assay containing predicted NF-κB binding sites on PRDX2 promoter. We analyzed 0–2000 bp upstream regions of PRDX2 transcription start site. Three promoter prediction websites were used to locate the putative NF-κB binding sites within the 2000 bp PRDX2 promoter region, including Promo (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), TFbinding (http://tfbind.hgc.jp) and Jaspar (http://jaspar.genereg.net). We found 12 predicted binding sites for NF-κB on PRDX2 promoter (Supplementary Fig. S4E). According to predictions, most predicted sites were located between the −1000 bp and +1bp of the promoter sequence. We designed three pGL3-based luciferase reporter constructs (P1–P3) of PRDX2 promoter regions containing potential binding areas (cluster A-C) (Fig. 5A). The luciferase reporter assay data revealed that NF-κB-p65 overexpression in AGS cells significantly increased PRDX2-P1 luciferase reporter activity, compared with empty vector control (Fig. 5B, P < 0.001). Of note, we did not detect increased luciferase reporter activity of P2 or P3, following similar conditions (Fig. 5B). Because our data demonstrated that the exogenous NF-κB-p65 could increase the luciferase activity, we confirmed these results using TNF-α to activate or Bay-11-7082 to inhibit NF-κB signaling. Our results indicated that TNF-α treatment significantly increased luciferase activity of cells transfected with PRDX2-P1 reporter, rather than P2 and P3 (Fig. 5C, P < 0.001). On the other hand, Bay-11-7082 treatment significantly decreased the luciferase activity in PRDX2-P1 reporter, but not P2 or P3 (Fig. 5C, P < 0.05). Next, we examined PRDX2-P1 reporter activity, following H. pylori infection. Our data demonstrated that H. pylori infection (7.13 and J166) induced significant increase of PRDX2-P1 reporter activity compared with control cells (Fig. 5D, P < 0.05). These results strongly suggest the presence of functional NF-κB binding sites on PRDX2 promoter, localized in cluster A of PRDX2-P1, that play an active role in gastric cells.
Fig. 5.
NF-κB-p65 binds directly to PRDX2 promoter region.
(A), Schematic diagram shows three luciferase reporters cover different DNA sequences of PRDX2 promoter region. Primers for ChiP assay are marked using 3 pairs of blue arrows. (B) Luciferase reporter assay analysis of three PRDX2 promoter luciferase reporters in AGS cells transfected with NF-kB-p65 or pcDNA. (C) Luciferase reporter assay analysis of three PRDX2 promoter luciferase reporters in AGS cells treated with 5 μM TNF-α (3 h), Bay-11-7082 11–7082 (6 h) before luciferase detection. (D) Luciferase reporter assay analysis of P1 in AGS cells infected with 100 MOI H. pylori for 3 h. (E) RT-PCR was performed in AGS cells after pull down NF-kB-p65 using three pairs of Primers to investigate the potential NF-kB-p65 binding sites in PRDX2 promoter region. (F) qRT-PCR analysis of DNA pulled down in E. *, P < 0.05, **, P < 0.01, ***, P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. NF-kB-p65 binds to PRDX2 gene promoter region
To further probe the direct NF-κB-p65 binding site to the PRDX2 promoter region within the natural chromatin context of gastric cancer cells, we designed three pairs of primers to detect the binding using chromatin immunoprecipitation (ChIP) assay. The amplification regions of the primers are shown in Fig. 5A and the nucleotide sequence of primers were shown in the methods section. Primer 1 contains the first predicted NF-κB-p65 binding site in cluster A mentioned in Fig. 5A. The second predicted NF-κB-p65 binding site in cluster A is included in the amplification sequence of Primer 2 along with all predicted binding sites of cluster B. Primer 3 targeted two predicted NF-κB binding sites in cluster C to confirm our findings of Fig. 5B to C. The DNA pull down by beads containing NF-κB-p65 antibody was utilized as a template in the RT-PCR reaction using the primers mentioned above. RT-PCR results revealed amplification of the pull downed DNA region using Primer 1 was significantly higher compared with Primer 2 or 3 (Fig. 5E and F, P < 0.01). Because H. pylori 7.13 strain induced higher expression of PRDX2 at 6 h (Fig. 2C &D), we performed ChIP assay in AGS with or without infection using this strain to test whether H. pylori infection regulates the binding of NF-κB-p65 to PRDX2 promoter region. RT-PCR results showed that binding of NF-κB-p65 to the PRDX2 promoter region was stronger with 7.13 infection for 6 h compared with control cells, indicating that the H. pylori infection promotes the binding of NF-κB-p65 to the PRDX2 promoter region amplified by Primer 1, rather than Primer 2 or 3 (Fig. 5E and F, P < 0.01). We also performed quantitative ChIP analysis using qRT-PCR from the same settings in Fig. 5E. Our data indicated that the pull downed DNA amplification with Primer 1 after H. pylori infection is significantly higher than without H. pylori infection (P < 0.01) (Fig. 5F). We did not detect induction or significant difference in the amplification products of Primers 2 or 3, with or without H. pylori infection, consistent with Fig. 5E. Overall, our data indicated that PRDX2 is regulated by transcriptional factor NF-κB-p65. Our data, for the first time, reveals that there are active NF-κB-p65 direct binding sites on the PRDX2 promoter region.
3.7. PRDX2 promotes cisplatin resistance in gastric cancer cells
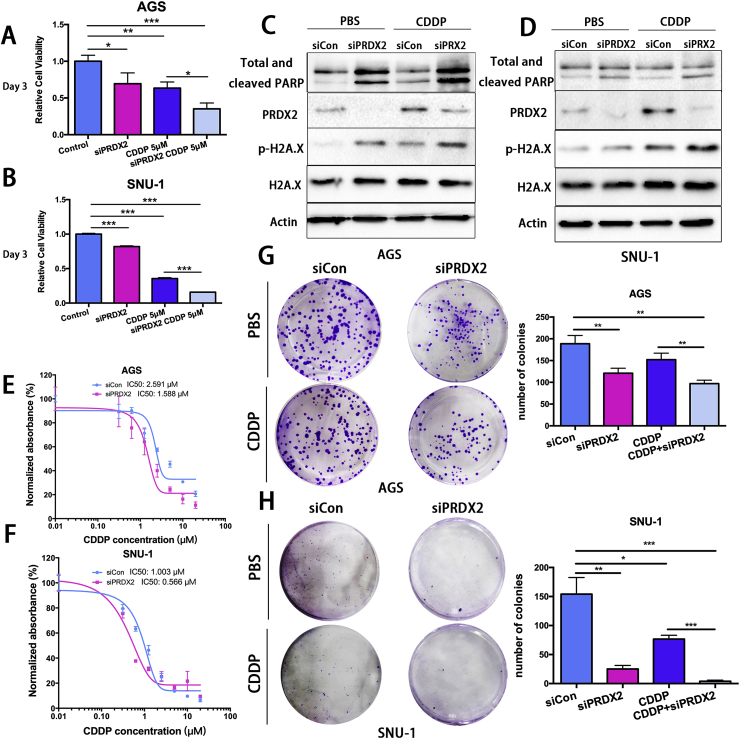
Our analysis of public databases demonstrated poor survival in patients with high levels of PRDX2. cisplatin (CDDP) treatment is a chemotherapeutic drug frequently used in the standard care of gastric cancer patients. We also found that PRDX2 protected gastric cancer cells from oxidative stress and DNA damage. Therefore, we postulated that PRDX2 may play a role in resistance to CDDP. Our results demonstrated that cell viability decreased significantly after knockdown of PRDX2, as compared with control (Fig. 6A and B, P < 0.05). CDDP treatment significantly diminished cell viability in cells with knockdown of PRDX2 (Fig. 6A and B, P < 0.05). The expression of cleaved PARP was strongly induced after knockdown of PRDX2 compared to control with or without CDDP treatment, indicating that knockdown of PRDX2 promotes cell apoptosis independently or induced by CDDP. The expression of p-H2A.X was also increased, similar to cleaved-PARP (Fig. 6C and D). Similar WB results were found after we transfected the other siRNA into cells with or without CDDP treatment and extracted proteins (Supplementary Figs. S5A and S5B). Next, we performed CellTiter-Glo assay in cells treated with control or PRDX2 siRNA. Results indicated that PRDX2 knockdown sensitized AGS and SNU-1 cells to CDDP treatment. The IC50 of CDDP in AGS cells decreased from 2.591 μM (Control) to 1.588 μM with PRDX2 siRNA knockdown. A Similar observation was obtained in SNU-1 cells (Fig. 6E and F). Colony formation assay confirmed the IC50 findings and demonstrated significantly less colonies following PRDX2 knockdown, as compared with control cells. Collectively, our results indicated a critical cell-protective role of PRDX2, suggesting a possible role of PRDX2 in promoting cisplatin resistance in gastric cancer cells.
Fig. 6.
Knockdown of PRDX2 sensitized GC cells to Cisplatin treatments.
(A) and (B) ATP-Glo cell viability assay in AGS or SNU-1 cells with control or PRDX2 siRNA knockdown with or without CDDP treatment. (C) and (D) Western blot analysis of PARP, cleaved-PARP, PRDX2, P–
H2A.X, H2A.X and β-actin in AGS or SNU-1 cells as in A and B. (E) and (F) CDDP IC50 was calculated in AGS and SNU-1 cells. (G) and (H) The clonogenic formation was performed in AGS and SNU-1 control or PRDX2 siRNA knockdown cells. A total of 500 cells were seeded in each well in a 6-well plate. *, P < 0.05, **, P < 0.01, ***, P < 0.001.
4. Discussion
Gastric cancer is the third most common cause of cancer-related deaths [1,2,31]. Our study demonstrated poor clinical outcome for patients with high levels of PRDX2. Recent studies showed that PRDX2 was involved in scavenging H2O2 with important cellular physiological functions, including cell growth, differentiation, and survival [16,32]. H. pylori is the main risk factor for gastric cancer, affecting 4.4 billion people world-wide [[3], [4], [5], [6], [7]]. Several earlier studies have shown that H. pylori infection increases the levels of reactive oxygen species, oxidative stress. and DNA damage [7,8]. However, accumulation of uncontrolled levels of DNA damage is lethal and not compatible with physiological cellular functions and viability [[13], [14], [15]]. In this context, gastric cells must develop protective mechanisms to counteract H. pylori-induced oxidative damage. We discovered that PRDX2 overexpression in gastric cancer cells is induced by H. pylori infection. We have shown that H. pylori-mediated induction of PRDX2, in vitro and in vivo, protected gastric cells from H. pylori-induced oxidative stress and DNA damage; a step that could be critical for survival of gastric cells in response to H. pylori infection.
H. pylori infection is known to create a pro-inflammatory environment with activation of NF-ĸB signaling that also promotes gastric tumorigenesis. Activation of NF-ĸB by H. pylori infection induces expression of several cytokines/chemokines, growth factors, anti-apoptotic factors, angiogenesis regulators and metalloproteinases to promote gastric carcinogenesis [25,33,34]. Our results indicated that H. pylori induced PRDX2 at mRNA levels and this expression was dependent on the activation of NF-ĸB signaling in gastric cancer cells. Bioinformatics analysis predicted multiple potential binding sites of NF-ĸB on PRDX2 promoter. We have taken a systematic approach to analyzing these potential binding sites. Our results indicated that PRDX2 promoter contains NF-ĸB binding sites, responsive to H. pylori- and TNFα-mediated activation of NF-ĸB. In fact, ChIP assay confirmed the luciferase reporter findings, demonstrating the presence of novel NF-ĸB binding sites on the PRDX2 promoter. However, we can not exclude the presence of other regulatory mechanisms that mediate PRDX2 induction. PRDX2 is also a direct target of HIF and its expression is induced by prolonged hypoxia in hypoxic HeLa cells [35]. PRDX2 overexpression in colorectal cancer is regulated by miR-200b-3p [36]. Although, we have not examined this regulatory mechanism in the context of H. pylori and inflammation, we demonstrated a novel regulatory mechanism mediating PRDX2 expression in gastric cells, highly relevant to H. pylori infection and inflammation, the main risk factors for gastric cancer.
Chemotherapeutic regimens containing platinum analogs, such as cisplatin, remain a standard therapeutic approach in the treatment of gastric cancer. Treatment with cisplatin induces high levels of reactive oxygen species and DNA damage that mediate cancer cell death [37,38]. We, therefore, investigated the role of PRDX2 in cisplatin resistance in gastric cancer. Our data indicated that PRDX2 protected against cisplatin-induced cell death. PRDX2 could have functions not limited to its antioxidant capacity. Lv et al. recently reported that PRDX2 is associated with colorectal cancer invasion, metastasis and chemotherapeutic resistance [36]. In addition, PRDX2 expression is positively correlated with activation of AKT in colon cancer cells, promoting cancer cell resistance to 5-Fluorouracil treatment [21]. Our results can partially explain the observed correlation between high levels of PRDX2 with poor clinical outcome. In this context, PRDX2 may be a key player of chemo/radio resistance in gastric cancer.
In conclusion, we discovered that PRDX2 is induced by H. pylori infection, protecting gastric cells from ROS, oxidative DNA damage, and DNA double-strand breaks. We detected and validated PRDX2 as a novel transcription target of NF-kB in gastric cancer. A high level of PRDX2 is a poor prognostic marker in gastric cancer associated with resistance to cisplatin. The future development of therapeutic strategies that target PRDX2 may be useful in the treatment of gastric cancer.
Ethical approval and consent to participate
The research presented here has been performed in accordance with the Declaration of Helsinki and has been approved by the ethics committee of the University of Miami, University of Vanderbilt and the First Affiliated Hospital of Nanjing Medical University. The patients were informed about the sample's collection and signed informed consents were obtained. All human samples used in this study were de-identified.
Consent for publication
All authors of this article have directly participated in the planning and drafting and all authors listed have read and approved the final version including details and images.
Conflicts of interest
The authors declare that they have no competing interests.
Funding
Research reported in this publication was supported by a Research Career Scientist award (1IK6BX003787) and merit award (I01BX001179) from the U.S. Department of Veterans Affairs (W. El-Rifai), and the following grants from the U.S. National Institutes of Health: R01CA93999 and Sylvester Comprehensive Cancer Center (P30CA240139). This work was partially supported by the National Natural Science Foundation of China (81572362); the National Natural Science Foundation Project of International Cooperation (NSFC-NIH, 81361120398); the Primary Research & Development Plan of Jiangsu Province (BE2016786). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health, University of Miami, or Nanjing Medical University.
Authors’ contributions
WER and ZX conceived and supervised the project. SW performed the key experiments and drafting of the manuscript. ZC provided support and troubleshooting of experiments and molecular techniques. SZ provided support for H. pylori culture, ChIP and luciferase assay. HL provided support with immunofluorescence and ChIP assay. DP provided support with immunohistochemistry and ROS level detection. MS was the major contributor of mouse experiments. HN provided support for quantitative PCR experiments. RP, Jr. Provided H. pylori strains for this study, HX assisted in drafting of the manuscript and organizing data. AZ assisted in reviewing the experiments and manuscript draft. All authors read and approved the final manuscript.
Acknowledgments
This manuscript has been read and approved by all the authors for publication and has not been submitted or under consideration for publication elsewhere.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101319.
Contributor Information
Zekuan Xu, Email: xuzekuan@njmu.edu.cn.
Wael El-Rifai, Email: wxe45@miami.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics. CA Cancer J. Clin. 2012;65(2):87–108. doi: 10.3322/caac.21262. 2015. [DOI] [PubMed] [Google Scholar]
- 3.Amieva M., Peek R.M., Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham D.Y. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148(4):719–731.e3. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y.C., Chiang T.H., Chou C.K., Tu Y.K., Liao W.C., Wu M.S. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic Review and meta-analysis. Gastroenterology. 2016;150(5):1113–11124 e5. doi: 10.1053/j.gastro.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Wei J., Noto J.M., Zaika E., Romero-Gallo J., Piazuelo M.B., Schneider B. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut. 2015;64(7):1040–1048. doi: 10.1136/gutjnl-2014-307295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savoldi A., Carrara E., Graham D.Y., Conti M., Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic Review and meta-analysis in World Health organization regions. Gastroenterology. 2018;155(5):1372–1382 e17. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gobert A.P., Wilson K.T. Polyamine- and NADPH-dependent generation of ROS during Helicobacter pylori infection: a blessing in disguise. Free Radic. Biol. Med. 2017;105:16–27. doi: 10.1016/j.freeradbiomed.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson A.J., Menheniott T.R., O'Connor L., Walduck A.K., Fox J.G., Kawakami K. Helicobacter pylori infection promotes methylation and silencing of trefoil factor 2, leading to gastric tumor development in mice and humans. Gastroenterology. 2010;139(6):2005–2017. doi: 10.1053/j.gastro.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng A.S., Li M.S., Kang W., Cheng V.Y., Chou J.L., Lau S.S. Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology. 2013;144(1):122–133 e9. doi: 10.1053/j.gastro.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 11.den Hartog G., Chattopadhyay R., Ablack A., Hall E.H., Butcher L.D., Bhattacharyya A. Regulation of Rac1 and reactive oxygen species production in response to infection of gastrointestinal epithelia. PLoS Pathog. 2016;12(1) doi: 10.1371/journal.ppat.1005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Franceschi L., Bertoldi M., De Falco L., Santos Franco S., Ronzoni L., Turrini F. Oxidative stress modulates heme synthesis and induces peroxiredoxin-2 as a novel cytoprotective response in beta-thalassemic erythropoiesis. Haematologica. 2011;96(11):1595–1604. doi: 10.3324/haematol.2011.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez S., Talens-Visconti R., Rius-Perez S., Finamor I., Sastre J. Redox signaling in the gastrointestinal tract. Free Radic. Biol. Med. 2017;104:75–103. doi: 10.1016/j.freeradbiomed.2016.12.048. [DOI] [PubMed] [Google Scholar]
- 14.Xie C., Yi J., Lu J., Nie M., Huang M., Rong J. N-acetylcysteine reduces ROS-mediated oxidative DNA damage and PI3K/akt pathway activation induced by Helicobacter pylori infection. Oxid. Med. Cell Longev. 2018;2018:1874985. doi: 10.1155/2018/1874985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bera R., Chiu M.C., Huang Y.J., Liang D.C., Lee Y.S., Shih L.Y. Genetic and epigenetic perturbations by DNMT3A-r882 mutants impaired apoptosis through augmentation of PRDX2 in myeloid Leukemia cells. Neoplasia. 2018;20(11):1106–1120. doi: 10.1016/j.neo.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolussi A., D'Inzeo S., Capalbo C., Giannini G., Coppa A. The role of peroxiredoxins in cancer. Mol. Clin. Oncol. 2017;6(2):139–153. doi: 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzano S., Checconi P., Hanschmann E.M., Lillig C.H., Bowler L.D., Chan P. Linkage of inflammation and oxidative stress via release of glutathionylated peroxiredoxin-2, which acts as a danger signal. Proc. Natl. Acad. Sci. U. S. A. 2014;111(33):12157–12162. doi: 10.1073/pnas.1401712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei X.G., Zhu J.H., Cheng W.H., Bao Y., Ho Y.S., Reddi A.R. Paradoxical roles of antioxidant enzymes: basic mechanisms and Health implications. Physiol. Rev. 2016;96(1):307–364. doi: 10.1152/physrev.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuta J., Nobeyama Y., Umebayashi Y., Otsuka F., Kikuchi K., Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66(12):6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- 20.Lu W., Fu Z., Wang H., Feng J., Wei J., Guo J. Peroxiredoxin 2 knockdown by RNA interference inhibits the growth of colorectal cancer cells by downregulating Wnt/beta-catenin signaling. Cancer Lett. 2014;343(2):190–199. doi: 10.1016/j.canlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Xu J., Zhang S., Wang R., Wu X., Zeng L., Fu Z. Knockdown of PRDX2 sensitizes colon cancer cells to 5-FU by suppressing the PI3K/AKT signaling pathway. Biosci. Rep. 2017;37(3) doi: 10.1042/BSR20160447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomnytska M.I., Becker S., Bodin I., Olsson A., Hellman K., Hellstrom A.C. Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: diagnostic and prognostic value. Br. J. Canc. 2011;104(1):110–119. doi: 10.1038/sj.bjc.6605992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ummanni R., Duscharla D., Barett C., Venz S., Schlomm T., Heinzer H. Prostate cancer-associated autoantibodies in serum against tumor-associated antigens as potential new biomarkers. J. Proteom. 2015;119:218–229. doi: 10.1016/j.jprot.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Stresing V., Baltziskueta E., Rubio N., Blanco J., Arriba M.C., Valls J. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32(6):724–735. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S., Soutto M., Chen Z., Peng D., Romero-Gallo J., Krishna U.S. Helicobacter pylori-induced cell death is counteracted by NF-kappaB-mediated transcription of DARPP-32. Gut. 2017;66(5):761–762. doi: 10.1136/gutjnl-2016-312141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hrabe J.E., O'Leary B.R., Fath M.A., Rodman S.N., Button A.M., Domann F.E. Disruption of thioredoxin metabolism enhances the toxicity of transforming growth factor beta-activated kinase 1 (TAK1) inhibition in KRAS-mutated colon cancer cells. Redox Biol. 2015;5:319–327. doi: 10.1016/j.redox.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciag A.E., Holland R.J., Robert Cheng Y.S., Rodriguez L.G., Saavedra J.E., Anderson L.M. Nitric oxide-releasing prodrug triggers cancer cell death through deregulation of cellular redox balance. Redox Biol. 2013;1:115–124. doi: 10.1016/j.redox.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorter R.P., Stephenson J., Nutma E., Anink J., de Jonge J.C., Baron W. Rapidly progressive amyotrophic lateral sclerosis is associated with microglial reactivity and small heat shock protein expression in reactive astrocytes. Neuropathol. Appl. Neurobiol. 2018;45:459–475. doi: 10.1111/nan.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimm M., Krimmel M., Polligkeit J., Alexander D., Munz A., Kluba S. ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur. J. Cancer. 2012;48(17):3186–3197. doi: 10.1016/j.ejca.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 30.Maubach G., Sokolova O., Wolfien M., Rothkotter H.J., Naumann M. Ca2+/calmodulin-dependent kinase II contributes to inhibitor of nuclear factor-kappa B kinase complex activation in Helicobacter pylori infection. Int. J. Cancer. 2013;133(6):1507–1512. doi: 10.1002/ijc.28148. [DOI] [PubMed] [Google Scholar]
- 31.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 32.Wu F., Tian F., Zeng W., Liu X., Fan J., Lin Y. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 2017;8(6) doi: 10.1038/cddis.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb A., Yang X.D., Tsang Y.H., Li J.D., Higashi H., Hatakeyama M. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10(11):1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartung M.L., Gruber D.C., Koch K.N., Gruter L., Rehrauer H., Tegtmeyer N. H. pylori-induced DNA strand breaks are introduced by nucleotide excision repair endonucleases and promote NF-kappaB target Gene expression. Cell Rep. 2015;13(1):70–79. doi: 10.1016/j.celrep.2015.08.074. [DOI] [PubMed] [Google Scholar]
- 35.Luo W., Chen I., Chen Y., Alkam D., Wang Y., Semenza G.L. PRDX2 and PRDX4 are negative regulators of hypoxia-inducible factors under conditions of prolonged hypoxia. Oncotarget. 2016;7(6):6379–6397. doi: 10.18632/oncotarget.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv Z., Wei J., You W., Wang R., Shang J., Xiong Y. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. J. Transl. Med. 2017;15(1):257. doi: 10.1186/s12967-017-1357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yimit A., Adebali O., Sancar A., Jiang Y. Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Nat. Commun. 2019;10(1):309. doi: 10.1038/s41467-019-08290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J.H., Zhang P., Chen W.D., Li D.D., Wu X.Q., Deng R. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy. 2015;11(2):239–252. doi: 10.1080/15548627.2015.1009767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.