Abstract
Hypoxia is widely considered as a limiting factor in vertebrate embryonic development, which requires adequate oxygen delivery for efficient energy metabolism, while nowadays some researches have revealed that hypoxia can induce stem cells so as to improve embryonic development. Erythroid differentiation is the oxygen delivery method employed by vertebrates at the very early step of embryo development, however, the mechanism how erythroid progenitor cell was triggered into mature erythrocyte is still not clear. In this study, after detecting the upregulation of vgll4b in response to oxygen levels, we generated vgll4b mutant zebrafish using CRISPR/Cas9, and verified the resulting impaired heme and dysfunctional erythroid terminal differentiation phenotype. Neither the vgll4b-deficient nor the γ-secretase inhibitor IX (DAPT)-adapted zebrafish were able to mediate HIF1α-induced heme generation. In addition, we showed that vgll4b mutant zebrafish were associated with an impaired erythroid phenotype, induced by the downregulation of alas2, which could be rescued by irf2bp2 depletion. Further mechanistic studies revealed that zebrafish VGLL4 sequesters IRF2BP2, thereby inhibiting its repression of alas2 expression and heme biosynthesis. These processes occur primarily via the VGLL4 TDU1 and IRF2BP2 ring finger domains. Our study also indicates that VGLL4 is a key player in the mediation of NOTCH1-dependent HIF1α-regulated erythropoiesis and can be sensitively regulated by oxygen concentrations. On the other hand, VGLL4 is a pivotal regulator of heme biosynthesis and erythroid terminal differentiation, which collectively improve oxygen metabolism.
Keywords: Oxygen, HIF1α, VGLL4, IRF2BP2, Erythroid differentiation
1. Introduction
Aerobionts, organisms that require oxygen to survive, have evolved various mechanisms for absorbing oxygen from their environment in order to avoid continuous hypoxia, erythropoiesis is one of the most indispensable mechanisms widespread in vertebrates [1,2]. Hemoglobinogenesis, which occurs during terminal erythroid differentiation, involves the acquisition of heme, iron and globin chains via the upregulation of alas2, tfr1 and other related genes [3]. This process is regulated by a complex of repressive proteins that bind to the promoters of developmentally-regulated erythroid genes [4]. However, the signals required to trigger the dissociation of this regulatory complex remain unknown.
During development, the embryo is subjected to low oxygen concentrations, before peripheral blood circulation is established [5,6]. As a member of the hypoxia inducible factors (HIF) family, HIF1α responds to cellular oxygen concentrations with high levels of sensitivity [7]. Under hypoxic conditions (1–5% oxygen), HIF1α is stabilized in the cytoplasm and translocated to the nucleus to bind the promoter regions of various genes, triggering downstream transcriptional events [[8], [9], [10], [11]]. Hif1α has been reported to be involved in hematopoietic stem cells (HSC) formation of the aorta-gonad-mesonephros (AGM) region [7,12], as well as the expansion of hematopoietic stem and progenitor cells (HSPCs) via the hypothalamic-pituitary-adrenal (HPA)-central glucocorticoid (GR) axis [13]. In addition, hif1α has been shown to promote alas2 expression, although the exact mechanism is yet to be elucidated [14].
As an excellent model [15], zebrafish erythropoiesis comprises two sequential waves [16]: Primitive erythropoiesis begins in the posterior lateral mesoderm (PLM) and subsequently the intermediate cell mass (ICM), which contains gata1a+ proerythroblasts and erythroblasts [17] and generates all erythrocytes in the circulatory system until 4 days post-fertilization (dpf) [18]. From 24 h post-fertilization (hpf), gata1a+ primitive erythroid cells differentiate into mature erythrocytes, which express alas2 [19]. During definitive erythropoiesis in zebrafish, HSCs from the ventral arterial wall (VDA) at approximately 26 hpf [20], then moved into caudal hematopoietic tissue (CHT), thymus, and finally the pronephros from 4 dpf to adulthood [21], which can differentiate into erythrocytes.
Vestigial Like Family Member 4 (VGLL4) is a transcription cofactor which have no DNA binding domain and exert their biological functions through interaction with various transcription factors via their TONDU (TDU) domains [[22], [23], [24]]. VGLL4 contains two TDU domains, which exhibit different preferences for interaction with proteins on certain occasions [25]. For example, VGLL4 antagonizes TEA domain transcription factor/Yes associated protein (TEAD/YAP) signaling during cardiac growth largely via TDU1 [26], while delays malignant progression in breast cancer by interacting with TEA Domain Transcription Factor 1 (TEAD1) predominantly via TDU2 [27]. Among three paralogs of zebrafish vgll4, vgll4b showed highest similarity with human vgll4 [28].
In this study, we observed the rapid upregulation of vgll4b by mild low oxygen concentrations at the hemoglobin accumulation phase of zebrafish embryonic development, and showed that this can be mimicked by increased hif1α expression. Furthermore, VGLL4 regulates Interferon Regulatory Factor 2 Binding Protein 2 (IRF2BP2) transcriptional activity by protein sequestration, thus ensuring effective oxygen delivery in erythroid terminal differentiation.
2. Materials and methods
2.1. Zebrafish maintenance and the generation of mutants
The line of Tg(gata1a: dsRed) [29] was used in this study. After the generation of vgll4b mutant zebrafish was achieved using CRISPR/Cas9, the genotyping PCR primers were designed as follows: forward 5′-GGTGAACCAGCTGAAAGCTGTAACC-3’; reverse 5′-CTTTATCCGCCGAGATGTTGAAAG-3’.
The vgll4b-/- Tg(gata1a: dsRed) zebrafish maintenance was then established and used.
2.2. Plasmid construction
Zebrafish genes, and mutants, were amplified from reverse transcription products and inserted into the tagged PCS2+ vector with their respective primers. In order to generate the luciferase reporter, several 1–2 kb upstream frames of zebrafish vgll4b and alas2 were amplified by PCR and ligated into the Kpn 1 and Sac 1 sites of the PGL3+ basic vector (Promega). In order to construct key mutants and mutant sequence with the depletion that less than 40bp, we conducted a PCR using plasmids containing corresponding wild type (wt) genes as templates and primers that cover upstream and downstream regions of the deleted sites. The construction of mutants in which larger segments were deleted was generated using the same method as that for wild-type plasmid construction. All the primers used for plasmid construction are given in the Supplementary Tables 1–5.
2.3. Morpholinos and mRNA microinjection
Morpholinos (MOs) and mRNA microinjection are extremely valuable research tools used to study embryo development in vivo. While MOs mimic gene knock-down, mRNA microinjection simulates gene expression. The MOs were designed using Genetools (www.gene-tools.com) [30] and the sequences are as follows: Zebrafish irf2bp2a MO: 5′-ACGACATCGCTCTCTCTCGGGCGAA-3’; zebrafish tead1a MO: 5′-CATGGCAATGGATGTGATCTCAGA-3’; zebrafish hif1 MO: 5′-GGTAACAGTAGTACTCATG-3’. Full-length mRNA samples were all synthesized using an SP6 kit (Life Technologies) from plasmid products after endonuclease digestion. After preparation, microinjection of mRNA was conducted into the yolk sac site close to the single cell of embryos at the 1-cell stage (before 0.5 hpf) [31]. The concentrations of mRNA samples used for microinjection were approximately 40–120 ng/μl, depending on experimental conditions.
2.4. Histology
Histological samples were disposed for Hematoxylin and eosin (H & E) staining, and Prussian blue staining as described previously [32].
2.5. Flow cytometry and cell sorting
Peripheral blood was collected by cardiac puncture from two adult zebrafish lines, normal Tg(gata1a: dsRed) and vgll4b-/- Tg(gata1a: dsRed), following anesthesia with a solution of 0.1% tricaine (Sigma). Whole embryos were collected as well, then carefully dissected and maintained in 0.9 × phosphate-buffered saline (PBS) with 2% fetal bovine serum (FBS) on ice before being placed on a Falcon nylon cell strainer (Beckton Dickinson); gravity was then used to obtain a single-cell suspension. For gata1a+ erythrocyte sorting, at least 60 embryos per group were needed to separate 105 erythrocytes for further analysis, including cell staining, quantitative PCR and RNA-sequencing.
2.6. Cell staining
Sorted cells, or collected blood samples, were concentrated onto slides by centrifugation at a speed of 500 rpm for 3 min using a Cytospin column. These were then stained with Wright–Giemsa solution to observe erythrocyte modality.
2.7. Embryo staining
Embryos were fixed overnight before Sudan Black staining [33].
Embryos were stained with O-dianisidine according to a previous study [34]. In brief, the working concentration of o-dianisidine solution was 0.6 mg/mL and contained 0.01 M sodium acetate (pH 5), 40% ethanol and 0.8% hydrogen peroxide. Finally, fresh embryos were stained and gently shaken in the dark for 30 min at room temperature.
2.8. Embryo staining transmission electron microscopy (TEM)
Zebrafish embryos at 3dpf were collected and conducted using a previous method [35].
2.9. Extraction of DNA and RNA and quantitative PCR
DsRed+ cells were sorted from Tg(gata1a: DsRed) controls and Tg(gata1a:DsRed) vgll4b mutants at 72hpf. Total RNA was then extracted using TRIzol reagent (Life Technologies) and then reverse transcribed into cDNA using a Revert Aid First Strand cDNA Synthesis Kit (Thermo). Amplification products were synthesized using a SYBR Green Real-time PCR Master Mix (TOYOBO).
2.10. Whole mount in situ hybridization (WISH)
Whole-mount mRNA in situ hybridization was conducted as described previously [36]. Digoxigenin (DIG)-labeled RNA probes were transcribed with T7, T3 or SP6 polymerase (Ambion). Probes labeled with DIG (Roche) were detected using anti-digoxigenin Fab fragment antibody (Roche) with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) staining (Vector Laboratories).
2.11. RNA-Seq of zebrafish kidney marrow
Zebrafish kidney marrow were dissected from 6-month-old fish after anesthetized in ice, at least 15 fishes per group, then the kidney marrow were made into single-cell suspensions in 20 ml of 4 °C PBS and were resolved in TRIzol reagent (Life Technologies) after centrifuged and Remove the supernatant. Total RNA was extracted for RNA sequencing. RNA libraries were constructed using MGI Easy RNA Library Prep Set according to the manufacturer’s protocol. Libraries were sequenced using BGISEQ500 platforms.
2.12. Cell transfection and luciferase reporter assay
HEK293T cells was cultured in DMEM (Gibco) with 10% FBS (Gibco). Plasmid samples were transfected into cells using Effectene Transfection Reagent (QIAGEN), and were then harvested after 40–48 h before protein extraction, or luciferase reporter assays conducted with the Dual Luciferase Reporter Assay Kit (Promega), as previously described [37].
2.13. Fluorescent analysis of heme levels [38]
The fluorescent heme assay was conducted using a protocol described previously [39].
2.14. Western blotting and co-immunoprecipitation assay (Co-IP)
Cell preparations were carried out as described previously [40]. Western blotting and co-immunoprecipitation assay were conducted using a previous methods [41]. Cells and tissues were degraded by RIPA lysis buffer (Beyotime) containing proteinase inhibitors (Roche). This mixture was then shaken on ice for 30 min and centrifuged at 15,000×g for 30–45 min until a clear precipitate appeared. The supernatant was then removed and a rabbit anti-HA antibody (Sigma) and protein G agarose beads (Sigma, 30 μL per sample) were added and shaken overnight at 4 °C. The next day, samples were washed three times in RIPA buffer and centrifuged at 6000×g for 1 min. The supernatant was then removed and mixed with sample buffer (Laemmli 2 × concentrate, Sigma). Proteins were then analyzed by immunoblotting using mouse anti-FLAG (Sigma).
2.15. Western blotting of wt and mutant VGLL4b proteins
Vgll4b sequences were amplified by PCR using cDNA from wt and vgll4b−/− zebrafish, respectively, and then inserted into FLAG-tagged PCS2+ vector to construct flag-tagged vgll4b plasmids. These plasmids were subsequently transfected into H293T cells and protein bands were visualized using an anti-FLAG antibody.
2.16. Statistical analysis
Data were expressed as mean ± standard error of the mean (SEM) [37]. Statistical significance was determined by a two-tailed unpaired t-test. For survival analysis, Kaplan–Meier survival curves were analyzed, statistical significances were calculated using GraphPad Prism 5.0. Quantitative data were collected from at least three independent experiments. For WISH and other embryo staining images, analysis involved at least 20–30 embryos. Positive areas of gene expression in every embryo involved in WISH analysis were selected and calculated as a proportion of the total captured embryo area using Photoshop_CS5.
3. Results
3.1. Vgll4b is upregulated in mild hypoxia during zebrafish embryonic differentiation
First, we conducted two experimental groups of zebrafish embryos, comprising normoxic condition as control and hypoxic condition (generated by MGC Anaero Pack incubation: 2.5 L, 5%–6% O2), result showed that vgll4b was up-regulated obviously in mild hypoxic condition (Supplementary Fig. 1A).
3.2. Vgll4b-deficient zebrafish mutant displayed an increased mortality rate
The zebrafish model of mutant vgll4b (vgll4b−/−) was successfully established in our laboratory using CRISPR/Cas9 (Supplementary Fig. 1B); eight nucleotides were deleted leading to a non-functional Sma1 restriction site (Supplementary Fig. 1D) and resulting in a truncated VGLL4b protein of 22.4kD without any TDU domain (Supplementary Figs. 1C and E).
Survival analysis indicated a higher mortality rate in this vgll4b mutant line at 48 hpf, implying embryo dysplasia, followed by sustained and consistently higher mortality rates during adulthood, which is different from the survival curve of a premature-aging phenotype [42] (Supplementary Fig. 1F).
3.3. Mutation of vgll4b led to abnormal erythrocyte in zebrafish
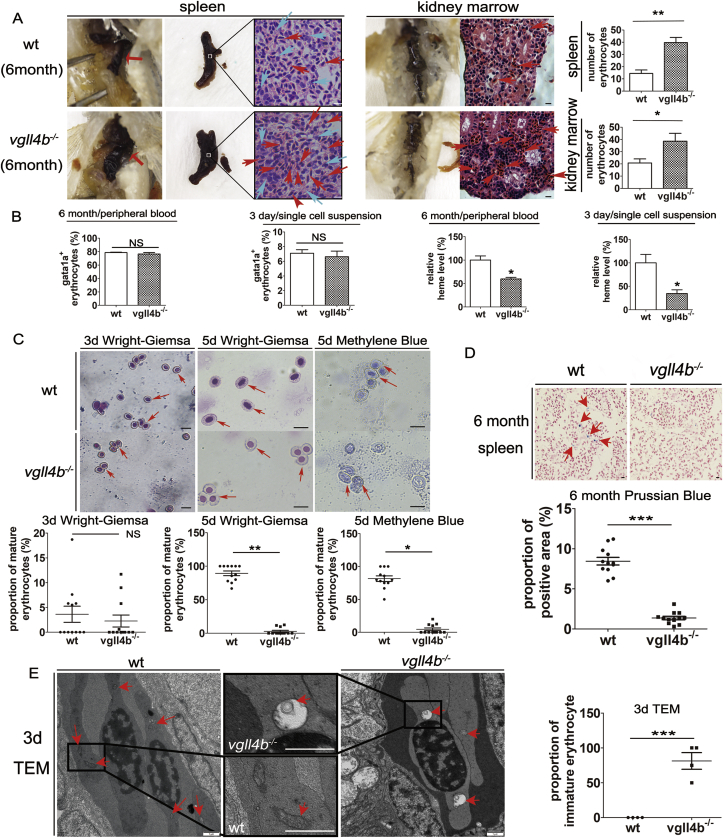
First, splenomegaly and increased irregular erythrocytes accumulation in vgll4b mutants implied increased clearance of abnormal erythrocytes; Kidney paramorphia and increased erythroid precursors implied compensatory generation of erythrocytes (Fig. 1A, red arrow). Flow cytometry analysis of both adult fish peripheral blood and embryonic single cell suspensions showed normal erythrocyte counts (Supplementary Figs. 2A and B and Fig. 1B) while reduced heme levels in vgll4b-/- zebrafish (Fig. 1B).
Fig. 1.
Gross anatomy and microscopic observation, analysis of erythrocyte count, morphology and function of vgll4−/−zebrafish. (A) Gross anatomy of adult mutant fish showing splenomegaly and kidney paramorphia compared with wild type (wt) fish. H&E staining showing that, in spleen, lymphocytes in the blood sinuses were normal in the mutant fish (blue arrow), while the peripheral space was filled with irregular erythrocytes, which were almost completely absent in wildtype animals (red arrow). Original magnification, × 600. Scale bar, 25 μm. In the mutant pronephros, the erythroid series was significantly advanced in control wild type samples (red arrow). Original magnification, × 400. Scale bar, 25 μm. Erythrocyte-counts were significantly increased in both the spleen and renal marrow of vgll4b-/- zebrafish. (B) Erythrocyte count comparisons between wt and vgll4b mutants in peripheral blood from 6-month-old adult zebrafish or single cell suspensions of 3 dpf embryos. Comparison of heme levels between wt and vgll4b mutants in peripheral blood samples from 6-month-old adult zebrafish or single cell suspensions of 3 dpf embryos (left and right, respectively). (C) Wright–Giemsa stain of 3 dpf embryos demonstrating slightly more immature erythrocytes in vgll4b mutants, with round, light colored nuclei. Wright–Giemsa stain of 5 dpf embryos, showing the pale cytoplasm and immature cell morphology compared with wild-type embryos. Methylene blue staining of mutant erythrocytes revealing that the cytoplasm contained hyperchromatic dots, resembling cytoplasmic DNA, while the nuclei were less compact, indicating an immature morphology compared with control cells. (D) Prussian Blue staining of the spleen showing few iron deposits in the tissue in vgll4b-/- zebrafish. (E) Transmission electron microscope image of erythrocytes isolated from control and vgll4b-/- zebrafish at 3 dpf. Erythrocyte was fusiform and Erythroid mitochondria were observed with normal membranes in WT, while in vgll4b mutants, erythrocyte was rounder shaped and immature, with swollen ruptured-membrane mitochondria. Erythrocyte-counts were calculated from three fields of view per fish, and six fish per group. All features are indicated by red arrows. Scale bar is 25 μm in optical micrograph and 1 μm in TEM micrograph. The statistical significance of differences between data points (mean ± s.e.m.) is denoted by *P < 0.05, **P < 0.01, and ***P<0.001; NS, no significant difference. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In vgll4b-/- embryos, Wright–Giemsa staining revealed significantly paler cytoplasm, methylene blue staining revealed that the cytoplasm of abnormal erythrocytes contained hyperchromatic cytoplasmic DNA thus indicating an immature morphology and dysplasia erythroid terminal differentiation (Fig. 1C); Prussian Blue staining of the spleen showed few iron deposits (Fig. 1D).
3.4. Mutation of vgll4b resulted in ruined mitochondria and stimulated mitophagy in zebrafish erythrocyte
Erythrocyte terminal differentiation involves the elimination of mitochondria [35]. which in zebrafish, begins at 3dpf, and is completed by 4 dpf [43,44]. In this study, transmission electron microscopy analysis showed that at 3 dpf, vgll4b mutants presented an immature erythrocyte phenotype with swollen membrane-ruptured mitochondria (Fig. 1E). Quantitative PCR analysis of the mitophagy-related gene, gata1a+ erythrocytes sorted at 3 dpf, revealed a slight increase in bnip3 expression in the mutant animals (Supplementary Fig. 2C), hinting at upregulated mitophagy [45].
Collectively, the data suggested that vgll4b-/- zebrafish generated abnormal and immature erythrocytes, leading to increased levels of compensatory erythroid cell turnover in an attempt to maintain erythroid cell numbers, albeit characterized by lower heme levels and abnormal mitochondria.
3.5. Expression of mutant vgll4b led to heme biosynthesis-related gene inhibition and alas2 downregulation in zebrafish erythropoiesis
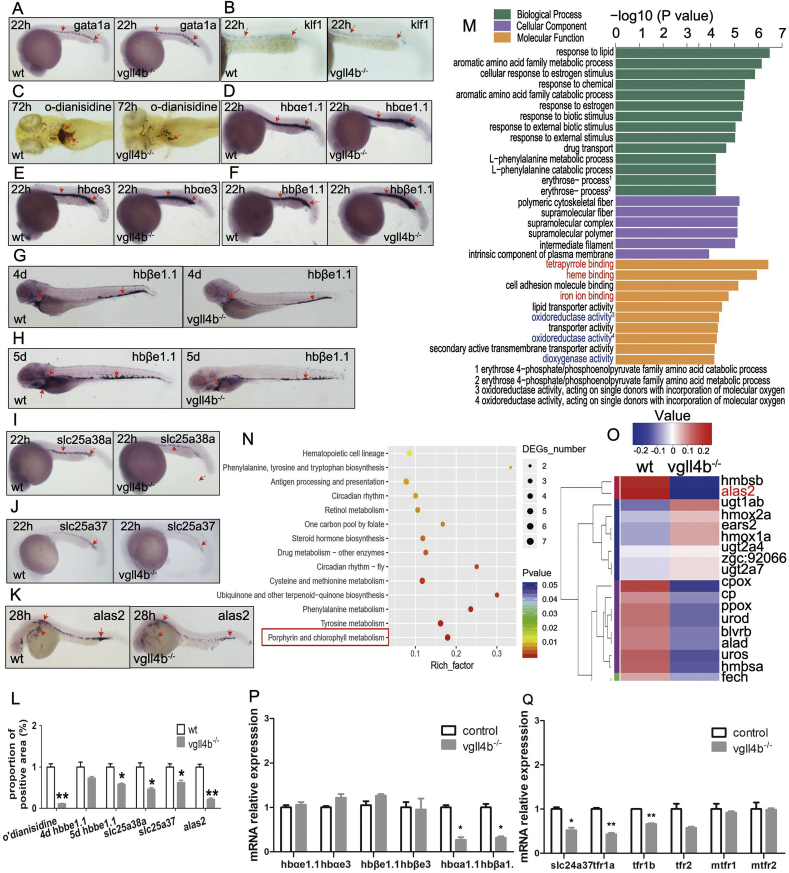
Firstly, HSC and lineage-specific markers in vgll4b mutants were examined by whole-mount in situ hybridization (WISH) to detect other hematopoietic lineage changes. We found that with the expression of the HSC marker c-myb, which was slightly elevated, the levels of all other assayed markers were unchanged (Supplementary Fig. 3).
We next investigated the effect of vgll4b depletion on erythropoiesis, globin chain expression, heme synthesis and hemoglobin maturation. Although we observed a significant reduction in o’dianisidine staining (P=0.008; Fig. 2C and L), the expression levels of the erythrocyte progenitor gene marker, gata1a [46], and the globin production-inducing gene, klf1 [47], remained unchanged (Fig. 2A and B), Expression levels of the major embryonic hemoglobin chain genes [48] varied little until 5 dpf, when hbβe1.1 was slightly downregulated (Fig. 2D–H, Supplementary Figs. 4A–C), suggesting a secondary phenomenon resulting from impaired heme levels [49]. However, we detected the clear downregulation of the genes involved in mitochonrial heme synthesis: slc25a38a[50], slc25a37 [51] and alas2 [52] (Fig. 2I–L).
Fig. 2.
Effect of mutant vgll4b on erythropoiesis, hemoglobin maturation, globin chain expression level and heme synthesis. (A–B) WISH analyses of gata1 (A) and klf1 (B) at 22 hpf, Red arrows indicate the main positions of the positive cells for each marker as follows. (C) O’dianisidine staining of mature hemoglobin at 72hpf showed impair in mutant embryos (P=0.008). (D–F) globin chain genes of hbαe1.1, hbαe3 and hbβe1.1 did not express abnormally at 22hpf; (G–H) expression of hbβe1.1 wasn’t affected until 4 to 5dpf (P=0.168 and 0.038, respectively); (I and J) WISH analyses of slc25a38a and slc25a37 expression showed statistic difference in vgll4b mutant embryos in control of wild-types at 22hpf (P=0.037 and 0.048, respectively). (K) WISH experiments on alas2 Expression at 24hpf in vgll4b mutants was significantly lower than in wild-types (P=0.009). (L) All positive results were gathered to control the difference between mutants and wild-type, the most significant difference was heme level and secondly alas2 expression, the mildest was hbβe1.1 expression level by WISH. (M) GO analysis detected vgll4b-mutation was significantly related with tetrapyrrole binding, heme binding and iron ion binding (in red); Oxidoreductase activity and dioxygenase activity (in blue) (ontology: molecular function). (N) KEGG pathway categories of differentially expressed genes (DEGs) in the pronephros of WT and vgll4b mutant adult fish. The porphyrin and chlorophyll metabolism pathway showed significant changes (in red frame). The vertical axis lists the most involved pathways while the horizontal axis indicates the richness factor. The richness factor is the proportion of the number of DEGs versus the total number of genes in each pathway. The point size shows the number of matched DEGs. The color represents the P value arising from Fisher’s exact test. (O) Hierarchical clustering analysis of DEGs related to the porphyrin and chlorophyll metabolism pathway; alas2 showed the most significant change (in red). The vertical axis lists the most DEGs in porphyrin and chlorophyll metabolism pathway while the colour band indicates the value of expression abundance after homogenization. (P) qPCR showing hemoglobin gene expression in the sorted peripheral blood from 18dpf embryos. (Q) qPCR showing transferrin-related gene expression in sorted peripheral blood from 18dpf embryos. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *p<0.05, **p<0.01, ***p<0.001. NS denotes no significant difference. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Expression of mutant vgll4b led to heme biosynthesis-related gene inhibition and alas2 downregulation in zebrafish erythropoiesis
To determine whether other potential erythropoiesis-related genes were altered, we conducted RNA-sequence analysis of adult zebrafish kidney marrow. The gene ontology (GO) analysis tool that detect vgll4b mutation in zebrafish was significantly associated with tetrapyrrole binding, heme binding and the redox reaction (Figure 2M). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of categories enriched for differentially-expressed genes (DEGs) indicated that the porphyrin pathway was strongly enriched (Fig. 2N). Furthermore, various genes related to heme biosynthesis were indeed downregulated, with alas2 exhibiting the greatest expression change (Fig. 2O and Supplementary Fig. 4J).
We next examined the expression of alas2 every 4 h from 16 to 32 hpf in vgll4b-/- zebrafish mutants (skipped 28hpf as tested in Fig. 2K). As early as 20 hpf, alas2 levels in the ICM region were markedly reduced, and declined further until 24 hpf (Supplementary Figs. 4D and E). From 28 to 32 hpf, alas2 expression became detectable in the peripheral blood circulation, the VDA, and the yolk sac, while total expression levels remained significantly lower in vgll4b mutants (Fig. 2K and Supplementary Fig. 4G). Alas2 expression is primarily regulated by the erythroid-specific transcription factors, GATA1 [52,53] and EPO/EPOR [54]. Since these factors were not decreased in the mutant fish (Fig. 2A and Supplementary Fig. 4H), the molecular mechanisms underlying alas2 downregulation remain to be determined. As zebrafish embryonic-to-adult globin chain switching occurs at around 10 dpf [48], qPCR analysis of the peripheral blood cells sorted at 18 dpf, showed a significant downregulation of adult hemoglobin and transferrin genes (Fig. 2P and Q, respectively).
Taken together, our results demonstrate that the presence of a dysfunctional vgll4b gene leads to impaired heme synthesis and hemoglobin generation, primarily via the suppression of alas2 and other related genes.
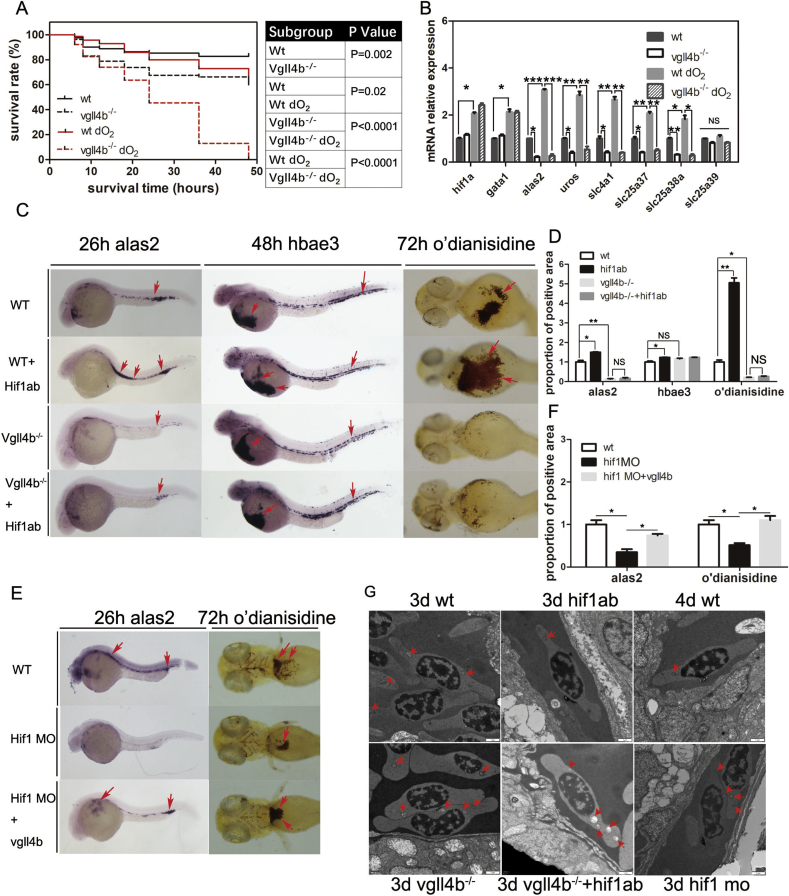
3.7. Vgll4b-depletion retarded hif1α-regulated hemoglobin maturation
The mitochondrial biosynthesis of heme is critical for oxygen transport and resistance to oxidative stress [29], In order to test the resistance to oxidative stress, we assessed the survival of the four experimental groups; wild-type (WT) and vgll4b mutant embryos, each under either normoxic or hypoxic conditions (using the MGC AnaeroPack® System, set to 2.5 L, 5%–6% O2). Gata1a+ erythrocytes from each group were collected at 3 dpf and RT-PCR was performed to evaluate the expression of genes associated with heme biosynthesis. The results showed that vgll4b mutants exhibited inferior survival relative to WT embryos under normoxic conditions, and that incubation under hypoxic conditions, exacerbated the difference in their prognoses (P=0.002 vs. P<0.001, respectively; Fig. 3A). Furthermore, genes, such as alas2, were upregulated in WT embryos in the hypoxia group, consistent with compensatory heme synthesis hypothesis, as observed previously in a mouse model [55]. In contrast, we detected the dramatic downregulation of alas2, uros, slc25a37, and slc25a38 expression in vgll4b mutants, along with normal levels of the slc25a38 homologs, slc25a39 [56], and slc25a39, which are ubiquitously expressed and not known to be involved in heme biosynthesis. Additionally, these genes were not upregulated during hypoxia (Fig. 3B).
Fig. 3.
Vgll4b mediates hif1α-regulated hemoglobin maturation. (A) Survival analysis of zebrafish subgroups (80 embryos per subgroup): WT in normoxia, vgll4b mutant in normoxia, WT in hypoxia, and vgll4b mutant in hypoxia. Hypoxia increased the difference in prognosis between WT and vgll4b mutant fish (P = 0.002 vs. P < 0.001). (B) Gene expression analysis in the four zebrafish subgroups described above (erythrocytes from 120 embryos per subgroup). The hif1α and gata1 genes were up-regulated in hypoxia, while mitochondria-related genes, including alas2 and slc25a38a, were up-regulated in WT animals under hypoxic conditions, while they were reduced in vgll4b mutants. (C and D) WISH analyses of alas2 and hbae3 expression and o’dianisidine staining in the embryonic subgroups: WT, hif1ab mRNA micro-injection, vgll4b mutant, and vgll4b mutant injected with Hif1ab mRNA. High levels of hif1ab were up-regulated both alas2 and hbae3 expression, and induced hematoglobin maturation, as demonstrated by o’dianisidine staining. Vgll4b depletion eliminated the alas2 expression and o’dianisidine staining induced by hif1ab, while it had no effect on hbae3 expression. (E and F) Hif1ab depletion by MO micro-injection reduced alas2 levels and o’dianisidine staining, which could be partially rescued by injection of mRNA encoding VGLL4b. (G) Erythroid morphology in subgroups by TEM, hif1ab overexpression group exhibited an accelerated erythroid maturation. Vgll4b−/−hif1ab overexpression group did not rescue the despaired phenotype in control of Vgll4b−/− group. Hif1 MO group exhibited similar and milder phenotype than vgll4b mutants (Fig. 3G). All features are indicated by red arrows. Scale bar is 1 μm in TEM micrograph. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *P<0.05; **P<0.01; ***P<0.001. NS denotes no significant difference. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Taking into account that: i) elevated hif1α levels can induce heme biosynthesis by upregulating alas2 expression [55]; and that ii) hif1ab (which shares a high level of similarity with human hif1α) is highly expressed in the great vessels of zebrafish between 24 hpf and 28 hpf [57]; we next set out to explore the possibility that HIF1α-induced erythroid compensation is mediated by VGLL4. Hif1ab mRNAs were injected into either WT control or vgll4b mutant embryos, and the expression levels of alas2, hbαe3 and a heme marker were quantified using o’dianisidine staining. In vgll4b mutant embryos, a clear drop in expression in alas2 expression levels and o’dianisidine staining occurred, even following hif1ab mRNA injection. In contrast, the expression levels of hbαe3 remained unaltered (Fig. 3C and D). Importantly, vgll4b mRNA rescued the impaired hemoglobin phenotype triggered by hif1a MO knockdown (Fig. 3E and F).
Based on our TEM analysis of erythroid morphology, hif1ab mRNA injection rendered the 3 dpf erythrocyte more fusiform-shaped and less mitochondria-like, similar as seen in WT erythrocyte at 4 dpf, implying accelerated erythroid terminal differentiation. However, hif1ab mRNA injection did not rescue this phenotype in vgll4b mutants. Erythrocyte receiving a Hif1a MO injection, exhibited a similar, albeit milder phenotype than vgll4b mutants (Fig. 3G). Taken together, our data suggest that HIF1α-induced heme biosynthesis is partly dependent on VGLL4.
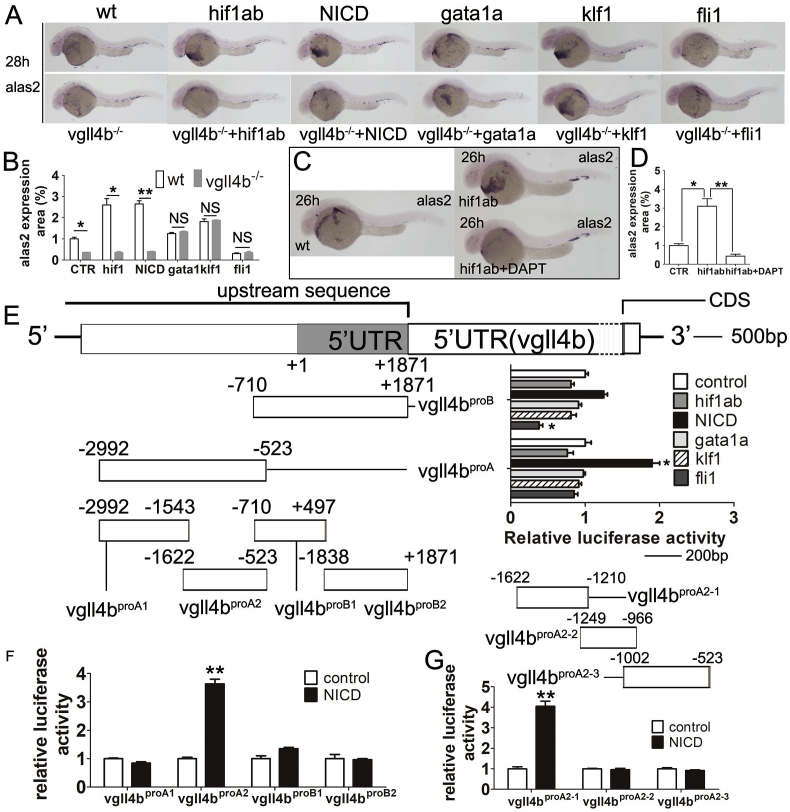
3.8. HIF1a -VGLL4 axis induced erythroid differentiation dependent on NOTCH1
We set out to find potential genes downstream hif1α. On condition that genes notch1 [58], gata1a [59], klf1 [60,61], fli1 [62] can all be regulated by hif1α and they have all been reported to affect hematopoiesis, we set to inject all the corresponding mRNAs to detect alas2 expression level, among them we overexpressed notch1 intracellular domain (NICD) in presentation of NOTCH1 [58]. As a result, only HIF1α and NICD led high expression of alas2, which was attenuated by vgll4b mutation (Fig. 4A and B). Furthermore, HIF1α could not stimulated alas2 expression any longer when wt zebrafish embryo were exposure to DAPT (GSI-IX, γ-secretase inhibitor IX) at 300 μM, implied an alas2-inducing axis of HIF1α-NOTCH1-VGLL4 (Fig. 4C).
Fig. 4.
Vgll4b mediates hif1α-regulated hemoglobin maturation based on notch1. (A and B) WISH analyses of alas2 expression in WT and vgll4b mutant zebrafish injected with mRNA of hif1ab, NICD, gata1a, klf1 and fli1, respectively. (C and D) WISH analyses of alas2 expression in zebrafish subgroups of WT, hif1ab mRNA injection and hif1ab injection with exposure of 300 μM. (E) The sketch map of vgll4b promoter segments construction. Two main segments on the upstream of vgll4b coding region were firstly constructed named vgll4bproA and vgll4bproB (−2992 to −523, −710 to +1870, respectively), vgll4bproA was up-regulate by the transfection of NICD, vgll4bproB was down-regulated by the transfection of FLI1 detected by luciferase assay. (F) Both vgll4bproA2 reached significant suppress effect on vgll4b expression by luciferase assay when transfected with NICD. (G) vgll4bproA2 was further divided and finally vgll4bproA2−1 reached the largest stimulative effect by luciferase assay. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *P<0.05; **P<0.01; ***P<0.001. NS denotes no significant difference.
To verify a potential target site of HIF1α or NOTCH1 to vgll4b, two successive 4863 bp segments of the sequences upstream of the vgll4b coding region were cloned: alas2proA (−2992 to −523) and alas2proB (−710 to +1871) (Fig. 4D). Luciferase reporter gene analysis results reflected that HIF1α did not make significant difference to vgll4b express, while an about 400 bp segment (−1622 to −1210) upstream of the vgll4b open reading frame seems to be a binding site of NOTCH1 and can mediated the stimulation of NICD to vgll4b express (Fig. 4E and G).
3.9. VGLL4b activation of heme biosynthesis is mainly dependent on its TDU1 domain and can be enhanced by mutation of the K225 acetylation site
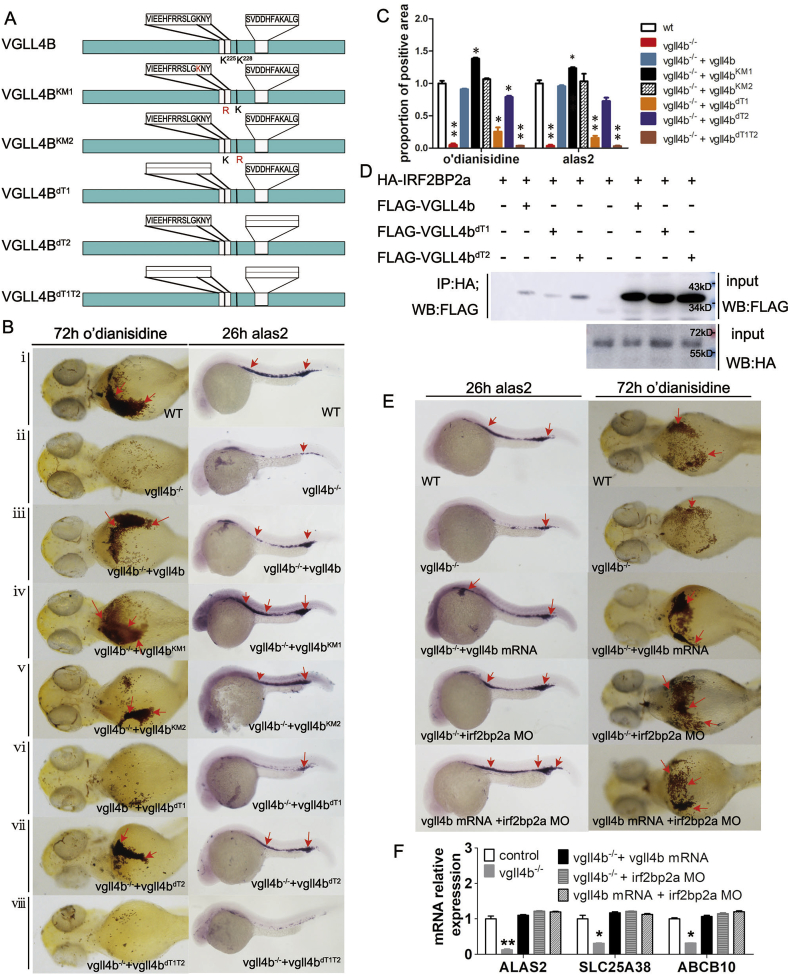
As VGLL4 is an ancillary transcription factor which can interact with several proteins via its two TDU domains [63], and has an acetylation site at K225 (equivalent to K221 in zebrafish VGLL4b) that is important for its impact on VGLL4-TEAD compound [26], five mutants of VGLL4b were constructed: VGLL4bKM1 (an A to R change at K221), VGLL4bKM2 (an A to R change of K224); VGLL4bdT1 (TDU1 domain deletion), VGLL4bdT2 (TDU2 domain deletion), and VGLL4bdT1T2 (deletion of both TDU domains) (Fig. 5A).
Fig. 5.
Rescue assays using vgll4b mRNA in vgll4b mutant embryos indicating that the biological effect of VGLL4b on hemoglobin is IRF2BP2-dependent. (A) Schematics of wt vgll4b, two K mutant vgll4b isoforms, and three vgll4b mutants lacking TDU domains. (B) Treatment with normal or various mutant vgll4b mRNAs resulted in different rescue effects on reduced alas2 expression and hemoglobin maturation in vgll4b-/- embryos. Red arrows indicate the positions of cells positive for each marker. Microinjection of both WT vgll4b and vgll4bKM2 mRNA fully restored the defective phenotypes (ⅲ and ⅴ), while vgll4bKM1 led to a significant increase in o’dianisidine staining and in the area of alas2 expression (ⅳ). Vgll4bdT2 could partly rescue the impaired phenotypes (ⅶ), while the mutants, vgll4bdT1 and vgll4bdT1T2 completely failed to rescue the defects (ⅵ and ⅷ). (C) Statistical calculation of the proportional areas of marker expression in each subgroup. (D) Co-IP demonstrating the interaction between zebrafish VGLL4b and IRF2BP2a and that the interaction was TDU1-dependent. (E and F) Depletion of IRF2BP2a by MO injection rescued impaired erythroid phenotypes in vgll4b mutants and abrogated the biological effects of VGLL4, as determined by WISH assay, embryonic staining, and qPCR analyses. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *p<0. 05, **p<0.01, ***p<0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, VGLL4bKM1 had a superior rescue effect relative to the wild type VGLL4b, indicating enhanced protein-protein interaction caused by the K221 mutation, leading to more effective rescue of the erythroid dysfunction. As expected, VGLL4bKM2 achieved a similar rescue effect to the wild type protein. Furthermore, VGLL4bdT2 exhibited a better rescue effect than VGLL4bdT1 and VGLL4bdT1T2, implying that VGLL4b likely exerts its function in erythropoiesis by interacting with a protein that preferentially binds to the TDU1 domain, rather than TDU2 (Fig. 5B and C).
RT-PCR at 3 dpf showed similar results in genes of uros and slc25a38a (Supplementary Fig. 5E).
3.10. VGLL4b induces heme biosynthesis and erythrocyte terminal differentiation via the sequestration of IRF2BP2, thus inhibiting the IRF2BP2-mediated suppression of target genes
The VGLL4 TDU domains form bridges to interact with proteins such as TEAD1(26), IRF2BP2 [64] and Myocyte Enhancer Factor 2 (MEF2) [65]. Therefore, we injected Irf2bp2a, irf2bp2b, tead1a, tead3b, mef2ca and mef2cb mRNAs into zebrafish embryos. The results showed that the overexpression of both Irf2bp2a and irf2bp2b, suppressed hemoglobin generation (Supplementary Fig. 5A ⅰ, ⅳ and ⅴ), while the other four genes had no effect (Supplementary Fig. 5A, B and D). This result was further confirmed by whole-embryo RT-PCR analysis (Supplementary Fig. 5C).
We next confirmed a direct interaction between zebrafish VGLL4b and IRF2BP2a by co-immunoprecipitation (Co-IP) experiments conducted in H293T cells (Fig. 5D). Consistent with previous reports [66], in this study, this interaction appeared to be more dependent on zebrafish VGLL4b TDU1 (Fig. 5D).
Interestingly, irf2bp2a MO knockdown rescued the impaired hemoglobin generation in vgll4b-/- mutants. More importantly, the effects exerted by vgll4b overexpression in the mutants were also abolished by irf2bp2a MO (Fig. 5E and F). This surprising result shows that irf2bp2a knockdown negated vgll4b′s influence, meaning that without irf2bp2a, vgll4b has little effect on erythroid maturation. Similar results were observed in vgll4b-/-irf2bp2b-/- embryos (Supplementary Figs. 5F and G), suggesting that VGLL4b function in the context of hemoglobin maturation and alas2 expression, is dependent on IRF2BP2.
3.11. VGLL4b induced alas2 expression by reducing the IRF2BP2-mediated transcriptional suppression, and uncoupled the co-suppressive effects of IRF2BP2 and its partner ETO2
To verify whether the inhibition of hemoglobin generation in vgll4b mutants was mediated by IRF2BP2, we next examined the influence of irf2bp2 on alas2 expression and hemoglobin generation. Previous studies had identified an IRF2BP2 binding site in the alas2 gene promoter region using the MEL cell line [4]. To identify the IRF2BP2 target site in the zebrafish alas2 gene, two successive 5361 bp segments upstream of the alas2 coding region were cloned: alas2proA (−679 to +2433) and alas2proB (−2996 to −82) (Supplementary Fig. 6A). A series of constructs were then generated for use in luciferase reporter gene analysis, which showed that a 240 bp segment (−2700 to −2460) upstream of the alas2 open reading frame (ORF), referred to as alas2proB’, was a potential target site for IRF2BP2 (Supplementary Figs. 6B–C). This target site also contains several predicted binding sites for other erythroid terminal differentiation regulators [4,67].
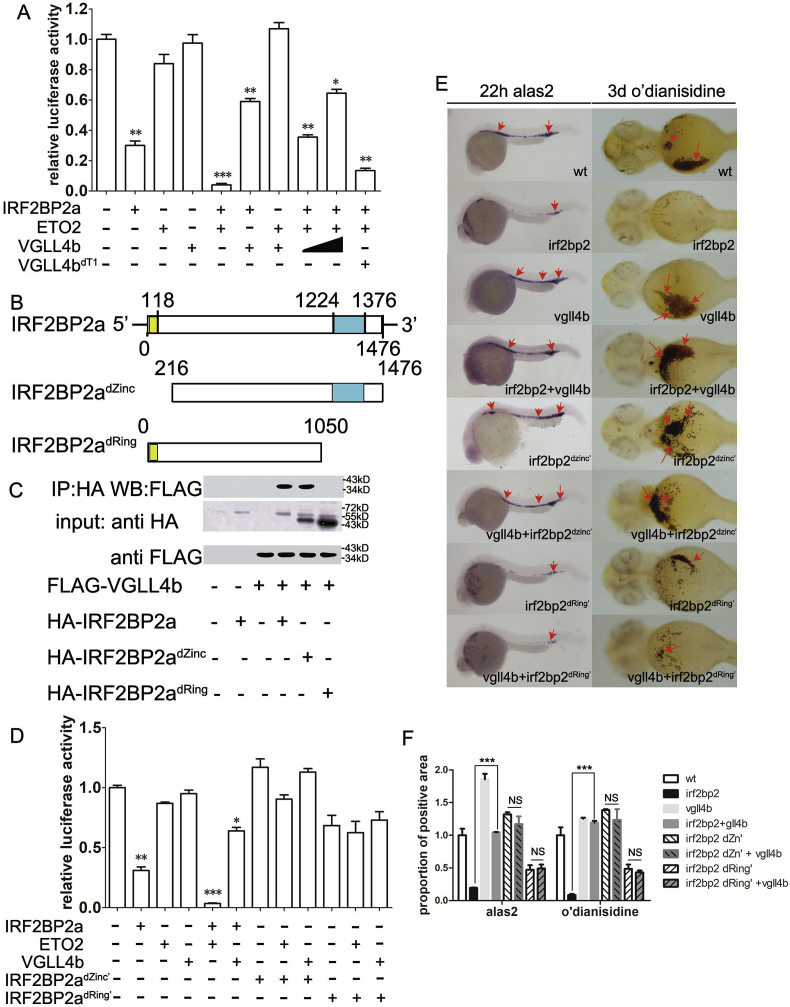
CBFA2/RUNX1 Partner Transcriptional Co-Repressor 3 (CBFA2T3, namely ETO2) is a transcriptional co-repressor involved in the coordination of erythroid cellular proliferation, differentiation, and maturation [4,68], which can interact with IRF2BP2 to co-suppress erythroid terminal differentiation [4]. We performed comprehensive luciferase assays to dissect the roles of IRF2BP2, ETO2 and VGLL4 in the regulation of alas2 expression (Fig. 6A). While VGLL4 or ETO2 alone did not exhibit any repressive effect, IRF2BP2 alone suppressed luciferase activity by approximately 30%, compared with control samples (P=0.004). Importantly, the co-expression of ETO2 further enhanced IRF2BP2-mediated transcriptional repression (P=0.001). However, this enhancement was counteracted by VGLL4b in a dose-dependent manner. VGLL4b only lost its ability to counteract IRF2BP2/ETO2 transcriptional repression activity following the deletion of its TDU1 domain (VGLL4bdT1), consistent with our previous in vivo rescue data (Fig. 5B and D).
Fig. 6.
Mechanism underlying VGLL4b interference with the suppressive effect of IRF2BP2 on erythroid terminal differentiation. (A) Luciferase assays demonstrating the effects of IRF2BP2a, ETO2 and VGLL4b in regulating alas2 expression. Neither ETO2 nor VGLL4b alone influenced alas2 expression; however, they induced and reduced suppression via IRF2BP2a. (B) Construction of the IRF2BP2a mutants, IRF2BP2adZinc’ and IRF2BP2adRing’. (C) Co-IP assay demonstrating that interaction of VGLL4b and IRF2BP2a depends on the ring finger domain and its surrounding region. (D) Luciferase assays using IRF2BP2a mutants. Neither IRF2BP2adZinc’ nor IRF2BP2adRing’ influenced the effects of ETO2 or VGLL4b on alas2 expression. (E and F) Biological effects of IRF2BP2a mutants on heme biosynthesis suppression and VGLL4 function. IRF2BP2adZinc’ was unable to repress heme biosynthesis, while IRF2BP2adRing’ was partially effective. Both mutants prevented the rescue effect of VGLL4 on heme biosynthesis. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *p<0.05, **p<0.01, ***p<0.001. NS denotes no significant difference.
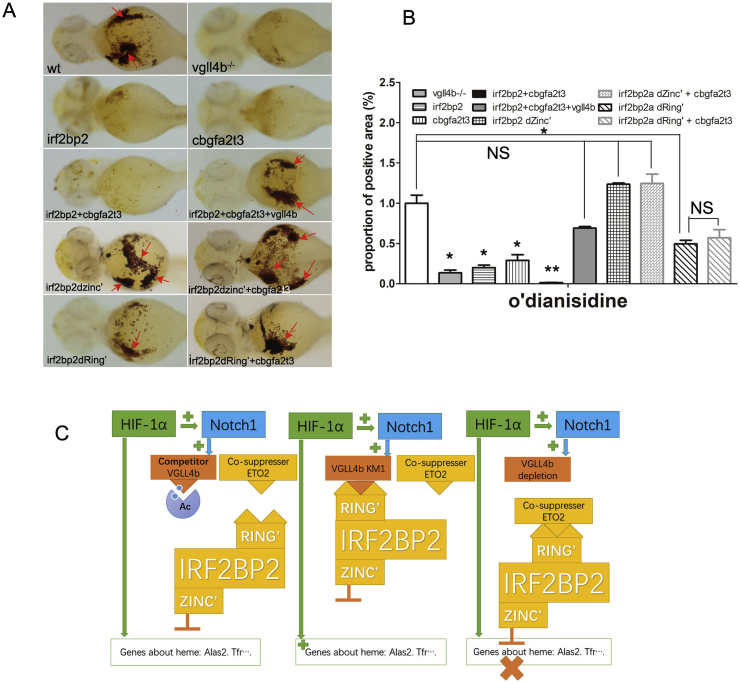
IRF2BP2 contains a potential DNA-binding zinc finger motif and a protein-protein interaction ring finger motif [64,66,69]; Therefore, we constructed two mutants with each of these domains deleted: IRF2BP2adZinc’ and IRF2BP2adRing’, respectively (Fig. 6B). Co-IP analysis showed that IRF2BP2 interacted with VGLL4b via amino acids 1050–1476, and the deletion of the zinc finger did not interfere with this interaction (Fig. 6C). Furthermore, we found that in the luciferase assays, IRF2BP2adZinc’ could no longer repress alas2 expression, negating both the synergistic effect of ETO2 and the rescue function of VGLL4b, simultaneously. Interestingly, while IRF2BP2adRing’ still repressed alas2 expression to some extent, it completely lost either the ability to synergize with ETO2 or be rescued by VGLL4b. These results indicate that, by coordinating zebrafish alas2 expression, both the zinc finger and the ring finger domains of IRF2BP2 are indispensable. For productive interactions with either ETO2 or VGLL4b, ring finger domain of IRF2BP2 is indispensable. Thus, the inhibitory function of VGLL4b, mediated via IRF2BP2, depends on both its zinc finger and ring finger (Fig. 6D). Furthermore, the in vivo experiments yielded similar results, with the exception that high cbgfa2t3 (the gene encoding ETO2) levels suppressed heme biosynthesis feasibly via its endogenous partners (Fig. 6E and F).
4. Discussion
Low oxygen density is typical during the early stage of embryonic development [5,6]. On sensing hypoxic conditions (1–5% oxygen). HIF1α is a sensitive oxygen responsor [[8], [9], [10], [11]] and has been widely reported to be essential for HSC formation in the AGM [7,12] and expand HSPCs [13]. However, persistent hypoxia is detrimental to embryonic development [70,71] and biological evolution [72,73], and can in some cases result in serious disease or death [74]. Thus, erythropoiesis represents a considerably more effective method for oxygen consumption. To our knowledge, this study is the first to delineate the oxygen sensing pathway used in erythropoiesis terminal differentiation to improve oxygen consumption, and thus serves to expand our knowledge on the relationship between erythroid differentiation and the environment.
Heme synthesis [29] is indispensable for the erythroid differentiation and oxygen delivering, and regulates the expression of the adult α- and β-globin genes to ensure balanced synthesis [3]. Dysfunction of ALAS2 in erythroblasts within the hematopoietic organs of erythroblasts, typically results in impaired heme biosynthesis, excessive iron accumulation and oxidative stress-induced cell damage, as a consequence of inadequate PPIX levels, relative to the available iron. Thus patients with ALAS2 dysfunction invariably exhibit hypochromic, microcytic anemia [75] and iron deposition in the cytoplasm [16,76], observations which are largely consistent with the vgll4b mutant zebrafish phenotype [77]. However, vgll4b mutant zebrafish exhibit normal erythrocyte counts without iron deposition, which could be potentially explained by impaired iron absorption mechanisms [78,79] (Figure 2Q). As a result, decreased iron absorption reduces oxidative stress-induced cell damage, thereby not altering erythrocyte counts.
The vgll4b mutant embryos exhibited normal β embryonic globin chain expression until 4dpf, while further qPCR analysis showed that the adult globin genes were significantly down-regulated in vgll4b mutants (Fig. 2P), thus confirming the impaired adult globin levels, reported in vgll4b mutants [4].
It has been reported that the VGLL4-TEAD1 interaction can be enhanced by the mutation of the K225 acetylation site in VGLL4 TDU1 domain during mouse cardiac growth [26]. TDU1 is crucial for the interaction of human VGLL4 with human IRF2BP2, as demonstrated in C2C12 mouse myoblasts [64]. On deletion of both TDUs, VGLL4 is still able to interact with IRF2BP2 in HEK293T cells [66]. Furthermore, the TDU domains were shown not to be required for the interaction of VGLL4 with IRF2BP2 and the regulation of PD-L1 in human lung cancer A549 cells [66]. In this study, however, the interaction between zebrafish VGLL4b and IRF2BP2a is dependent on the TDU1 domain. The mutation at the TDU1 acetylation site, enhanced the rescue effect exerted by VGLL4b (Fig. 5B iv). Consequently, TDU1 deletion significantly weakened: i) the VGLL4b/IRF2BP2a interaction (Fig. 5D); ii) the ability of VGLL4b to rescue alas2 expression (Fig. 6A); and iii) the ability of VGLL4b to rescue zebrafish heme biosynthesis (Fig. 5B vi). Collectively, these results assign an important role to the TDU1 domain of VGLL4 in the context of heme biosynthesis. Interestingly, TDU1 appears to have different roles in different mouse cells and is important in zebrafish in terms of promoting the IRF2BP2/VGLL4b interaction. However, while TDU1 appears critically important for the IRF2BP2/VGLL4b interaction in zebrafish and humanized animal models, its function may be redundant in this regard, in human cells. Whether the IRF2BP2/VGLL4b protein interaction is compensated for in more advanced species like humans, or whether this interaction is highly dependent on the species-specific environmental conditions, deserves further explored.
VGLL4 is reported to stabilize and enhance the suppressive effect of IRF2BP2 on IRF2 expression, in the regulation of PD-L1 expression in malignant cells [66]. In this study, we set out to test vgll4b mutant zebrafish model, and surprisingly detected the potentially IRF2BP2-mediated downregulation of various genes [4], implying that the inhibitory effect of IRF2BP2 was enhanced by vgll4b depletion. This seemingly opposing result observed in zebrafish erythrocytes, compared to tumor cell lines, implies that the VGLL4 and IRF2BP2 relationship may be delicately fine-tuned, depending on the environmental setting, with further research required on the relationship between the degradation and bioeffect of IRF2BP2.
Furthermore, since VGLL4 is also essential for heart growth [26] and valve development [80], it makes sense that VGLL4 may be regulated by oxygen level in other tissues. Thus, it is probable that VGLL4 can trigger the oxygen-delivery system development in both the cardiomyocytes and erythrocyte. The involvement of VGLL4 in oxygen sensing, delivery and organ development merits further exploration.
Additionally, other NOTCH family members may be involved in regulating oxygen-dependent organ development processes. For example, NOTCH2 regulates cortical neurogenesis and bone metabolism [81,82], while, NOTCH3 plays a critical role in esophageal squamous cell differentiation and angiogenesis [83,84].
In conclusion, this study shows that vgll4b is up-regulated at low oxygen concentration and can mediate NOTCH1-dependent HIF1α-induced heme biosynthesis. VGLL4b induces alas2 expression by reducing the suppressive effects of IRF2BP2 and its binding partner ETO2. As a result, erythroid maturation is improved through enhanced oxygen metabolism (Fig. 7). Hypoxia is considered as a limiting factor in embryonic development. Our results reveal that oxygen actually acts as a diffusible inducing factor for embryonic erythropoiesis under aerobic conditions. Based on published work, describing how the spatial distribution of oxygen or HIF1α plays a crucial role in local organ development [58,85], we have highlighted a loop that exists between oxygen distribution, molecular modulation and the development of oxygen delivery systems. Thus, the precise regulation of oxygen provision and its sensory downstream receptors are critical factors in embryonic development at spatial and temporal levels.
Fig. 7.
Bioeffects of IRF2BP2 mutants in synergistic effect with ETO2. (A) Bioeffects of IRF2BP2a mutants on heme biosynthesis suppression in addition to ETO2 detected by o’dianisidine stain. (B) Staining positive area compare among subgroups of IRF2BP2 mutants, depletion of either Zinc Finger or Ring Finger can abolish suppress effect of ETO2. (C) Model for the regulation of heme biosynthesis by alas2 et al., regulated by HIF1α, VGLL4 and IRF2BP2. HIF1α induced alas2 expression dependent on NOTCH1 and VGLL4. VGLL4 induced alas2 expression by reducing the IRF2BP2 transcription suppression and uncoupling the co-suppressor effects of IRF2BP2 and its partner ETO2. Data are shown as the mean ± SEM of at least 15 to 30 embryos in each subgroup. *p<0. 05, **p<0.01, ***p<0.001. NS denotes no significant difference.
Declarations
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Conflicts of interest
The authors declare that they have no competing financial interests.
Authors’ contributions
JZ contributed to the conception and design of the study. YW, XL and BX performed the experiments and analyzed the data. HY and YZ assisted with the experiments. JZ and YW wrote the paper. All authors read and approved the final manuscript.
Funding
This research was supported by National Natural Science Foundation of China (No.81770311).
Acknowledgments
We thank Yi Chen, Yi Jin and Zheng Ruan for their excellent technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101313.
Contributor Information
Yiqin Wang, Email: wyqforstudy@163.com.
Xiaohui Liu, Email: liuxiaohuiuestc@126.com.
Baoshu Xie, Email: clairtree@163.com.
Hao Yuan, Email: hyuan@sibs.ac.cn.
Yiyue Zhang, Email: mczhangyy@scut.edu.cn.
Jun Zhu, Email: zhuj1966@yahoo.com, jun.zhu@paris7.jussieu.fr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wu R.S.S. Hypoxia: from molecular responses to ecosystem responses. Mar. Pollut. Bull. 2002;45:35–45. doi: 10.1016/s0025-326x(02)00061-9. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G.L. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology. 2009;24:97–106. doi: 10.1152/physiol.00045.2008. [DOI] [PubMed] [Google Scholar]
- 3.Chiabrando D., Mercurio S., Tolosano E. Heme and erythropoieis: more than a structural role. Haematologica. 2014;99:973–983. doi: 10.3324/haematol.2013.091991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadhouders R., Cico A., Stephen T., Thongjuea S., Kolovos P., Baymaz H.I., Yu X., Demmers J., Bezstarosti K., Maas A. Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat. Commun. 2015;6 doi: 10.1038/ncomms9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steptoe P.C., Edwards R.G., Purdy J.M. Human blastocysts grown in culture. Nature. 1971;229:132–133. doi: 10.1038/229132a0. [DOI] [PubMed] [Google Scholar]
- 6.Gore A.V., Pillay L.M., Galanternik M.V., Weinstein B.M. vol. 7. Wiley Interdisciplinary Reviews-Developmental Biology; 2018. (The Zebrafish: A Fintastic Model for Hematopoietic Development and Disease). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imanirad P., Solaimani Kartalaei P., Crisan M., Vink C., Yamada-Inagawa T., de Pater E., Kurek D., Kaimakis P., van der Linden R., Speck N. HIF1alpha is a regulator of hematopoietic progenitor and stem cell development in hypoxic sites of the mouse embryo. Stem Cell Res. 2014;12:24–35. doi: 10.1016/j.scr.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danet G.H., Pan Y., Luongo J.L., Bonnet D.A., Simon M.C. Expansion of human SCID-repopulating cells under hypoxic conditions. J. Clin. Investig. 2003;112:126–135. doi: 10.1172/JCI17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith B., Johnson R.S., Simon M.C. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sitkovsky M., Lukashev D. Regulation of immune cells by local. tissue oxygen tension: hif1 alpha and adenosine receptors. Nat. Rev. Immunol. 2005;5:712–721. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin A., Cornelissen P.W.A., Kirchmaier B.C., Mokry M., Iich E., Nirmala E., Liang K.-H., Vegh A.M.D., Scholman K.T., Koerkamp M.J.G. Genome-wide analysis reveals NRP1 as a direct HIF1 alpha-E2F7 target in the regulation of motorneuron guidance in vivo. Nucleic Acids Res. 2016;44:3549–3566. doi: 10.1093/nar/gkv1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris J.M., Esain V., Frechette G.M., Harris L.J., Cox A.G., Cortes M., Garnaas M.K., Carroll K.J., Cutting C.C., Khan T. Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood. 2013;121:2483–2493. doi: 10.1182/blood-2012-12-471201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwan W., Cortes M., Frost I., Esain V., Theodore L.N., Liu S.Y., Budrow N., Goessling W., North T.E. The central nervous system regulates embryonic HSPC production via stress-responsive glucocorticoid receptor signaling. Cell Stem Cell. 2016;19:370–382. doi: 10.1016/j.stem.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon D., Pastore Y.D., Divoky V., Liu E., Mlodnicka A.E., Rainey K., Ponka P., Semenza G.L., Schumacher A., Prchal J.T. Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development. J. Biol. Chem. 2006;281:25703–25711. doi: 10.1074/jbc.M602329200. [DOI] [PubMed] [Google Scholar]
- 15.Carradice D., Lieschke G.J. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkeaw K., Sugiyama D. Zebrafish erythropoiesis and the utility of fish as models of anemia. Stem Cell Res. Ther. 2012;3 doi: 10.1186/scrt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galloway J.L., Wingert R.A., Thisse C., Thisse B., Zon L.I. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev. Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Willett C.E., Cortes A., Zuasti A., Zapata A.G. vol. 214. 1999. pp. 323–336. (Early Hematopoiesis and Developing Lymphoid Organs in the Zebrafish. Developmental Dynamics : an Official Publication of the American Association of Anatomists). [DOI] [PubMed] [Google Scholar]
- 19.Long Q., Meng A., Wang H., Jessen J.R., Farrell M.J., Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 20.Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y., Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kissa K., Murayama E., Zapata A., Cortes A., Perret E., Machu C., Herbomel P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 22.Maeda T., Gupta M.P., Stewart A.F.R. TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem. Biophys. Res. Commun. 2002;294:791–797. doi: 10.1016/S0006-291X(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T., Chapman D.L., Stewart A.F.R. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 2002;277:48889–48898. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 24.Karasseva N., Tsika G., Ji J., Zhang A.J., Mao X.Q., Tsika R. Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol. Cell. Biol. 2003;23:5143–5164. doi: 10.1128/MCB.23.15.5143-5164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faucheux C., Naye F., Treguer K., Fedou S., Thiebaud P., Theze N. Vestigial like gene family expression in Xenopus: common and divergent features with other vertebrates. Int. J. Dev. Biol. 2010;54:1375–1382. doi: 10.1387/ijdb.103080cf. [DOI] [PubMed] [Google Scholar]
- 26.Lin Z., Guo H., Cao Y., Zohrabian S., Zhou P., Ma Q., VanDusen N., Guo Y., Zhang J., Stevens S.M. Acetylation of VGLL4 regulates hippo-YAP signaling and postnatal cardiac growth. Dev. Cell. 2016;39:466–479. doi: 10.1016/j.devcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Shen H., Withers H.G., Yang N., Denson K.E., Mussell A.L., Truskinovsky A., Fan Q., Gelman I.H., Frangou C. VGLL4 selectively represses YAP-dependent gene induction and tumorigenic phenotypes in breast cancer. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-06227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue C., Wang H.H., Zhu J., Zhou J. The expression patterns of vestigial like family member 4 genes in zebrafish embryogenesis. Gene Expr. Patterns : GEP. 2018;28:34–41. doi: 10.1016/j.gep.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Mercurio S., Petrillo S., Chiabrando D., Bassi Z.I., Gays D., Camporeale A., Vacaru A., Miniscalco B., Valperga G., Silengo L. The heme exporter Flvcr1 regulates expansion and differentiation of committed erythroid progenitors by controlling intracellular heme accumulation. Haematologica. 2015;100:720–729. doi: 10.3324/haematol.2014.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brautigam L., Zhang J., Dreij K., Spahiu L., Holmgren A., Abe H., Tew K.D., Townsend D.M., Kelner M.J., Morgenstern R. MGST1, a GSH transferase/peroxidase essential for development and hematopoietic stem cell differentiation. Redox Biology. 2018;17:171–179. doi: 10.1016/j.redox.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shokhina A.G., Kostyuk A.I., Ermakova Y.G., Panova A.S., Staroverov D.B., Egorov E.S., Baranov M.S., van Belle G.J., Katschinski D.M., Belousov V.V. Red fluorescent redox-sensitive biosensor Grx1-roCherry. Redox Biology. 2019;21 doi: 10.1016/j.redox.2018.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung I.H., Chung Y.Y., Jung D.E., Kim Y.J., Kim do H., Kim K.S., Park S.W. Impaired lymphocytes development and xenotransplantation of gastrointestinal tumor cells in prkdc-null SCID zebrafish model. Neoplasia. 2016;18:468–479. doi: 10.1016/j.neo.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Guyader D., Redd M.J., Colucci-Guyon E., Murayama E., Kissa K., Briolat V., Mordelet E., Zapata A., Shinomiya H., Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 34.Devireddy L.R., Hart D.O., Goetz D.H., Green M.R. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell. 2010;141:1006–1017. doi: 10.1016/j.cell.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betin V.M., Singleton B.K., Parsons S.F., Anstee D.J., Lane J.D. Autophagy facilitates organelle clearance during differentiation of human erythroblasts: evidence for a role for ATG4 paralogs during autophagosome maturation. Autophagy. 2013;9:881–893. doi: 10.4161/auto.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chitramuthu B.P., Bennett H.P. High resolution whole mount in situ hybridization within zebrafish embryos to study gene expression and function. J. Vis. Exp. : J. Vis. Exp. 2013 doi: 10.3791/50644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Xie F., Chu F., Zhang Z., Yang B., Dai T., Gao L., Wang L., Ling L., Jia J. YAP antagonizes innate antiviral immunity and is targeted for lysosomal degradation through IKK epsilon-mediated phosphorylation. Nat. Immunol. 2017;18 doi: 10.1038/ni.3744. 733-+ [DOI] [PubMed] [Google Scholar]
- 38.Lian J., Chen J., Wang K., Zhao L., Meng P., Yang L., Wei J., Ma N., Xu J., Zhang W. Alas1 is essential for neutrophil maturation in zebrafish. Haematologica. 2018;103:1785–1795. doi: 10.3324/haematol.2018.194316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin H., Huang Z., Chi Y., Wu M., Zhou R., Zhao L., Xu J., Zhen F., Lan Y., Li L. c-Myb acts in parallel and cooperatively with Cebp1 to regulate neutrophil maturation in zebrafish. Blood. 2016;128:415–426. doi: 10.1182/blood-2015-12-686147. [DOI] [PubMed] [Google Scholar]
- 40.Wang L., Fu C., Fan H., Du T., Dong M., Chen Y., Jin Y., Zhou Y., Deng M., Gu A. miR-34b regulates multiciliogenesis during organ formation in zebrafish. Development. 2013;140:2755–2764. doi: 10.1242/dev.092825. [DOI] [PubMed] [Google Scholar]
- 41.Yang R.M., Tao J., Zhan M., Yuan H., Wang H.H., Chen S.J., Chen Z., de The H., Zhou J., Guo Y. TAMM41 is required for heart valve differentiation via regulation of PINK-PARK2 dependent mitophagy. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carneiro M.C., Henriques C.M., Nabais J., Ferreira T., Carvalho T., Ferreira M.G. Short telomeres in key tissues initiate local and systemic aging in zebrafish. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davuluri G., Song P., Liu Z., Wald D., Sakaguchi T.F., Green M.R., Devireddy L. Inactivation of 3-hydroxybutyrate dehydrogenase 2 delays zebrafish erythroid maturation by conferring premature mitophagy. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1460–E1469. doi: 10.1073/pnas.1600077113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pase L., Layton J.E., Kloosterman W.P., Carradice D., Waterhouse P.M., Lieschke G.J. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweers R.L., Zhang J., Randall M.S., Loyd M.R., Li W., Dorsey F.C., Kundu M., Opferman J.T., Cleveland J.L., Miller J.L. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishikawa K., Kobayashi M., Masumi A., Lyons S.E., Weinstein B.M., Liu P.P., Yamamoto M. Self-association of Gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol. Cell. Biol. 2003;23:8295–8305. doi: 10.1128/MCB.23.22.8295-8305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oates A.C., Pratt S.J., Vail B., Yan Y., Ho R.K., Johnson S.L., Postlethwait J.H., Zon L.I. The zebrafish klf gene family. Blood. 2001;98:1792–1801. doi: 10.1182/blood.v98.6.1792. [DOI] [PubMed] [Google Scholar]
- 48.Ganis J.J., Hsia N., Trompouki E., de Jong J.L.O., DiBiase A., Lambert J.S., Jia Z., Sabo P.J., Weaver M., Sandstrom R. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 2012;366:185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crielaard B.J., Rivella S. beta-Thalassemia and Polycythemia vera: targeting chronic stress erythropoiesis. Int. J. Biochem. Cell Biol. 2014;51:89–92. doi: 10.1016/j.biocel.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Murray J.P., Prykhozhij S.V., Dufay J.N., Steele S.L., Gaston D., Nasrallah G.K., Coombs A.J., Liwski R.S., Fernandez C.V., Berman J.N. Glycine and folate ameliorate models of congenital sideroblastic anemia. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasrallah G.K., Younes N.N., Baji M.H., Shraim A.M., Mustafa I. Zebrafish larvae as a model to demonstrate secondary iron overload. Eur. J. Haematol. 2018;100:536–543. doi: 10.1111/ejh.13035. [DOI] [PubMed] [Google Scholar]
- 52.Kaneko K., Furuyama K., Fujiwara T., Kobayashi R., Ishida H., Harigae H., Shibahara S. Identification of a novel erythroid-specific enhancer for the ALAS2 gene and its loss-of-function mutation which is associated with congenital sideroblastic anemia. Haematologica. 2014;99:252–261. doi: 10.3324/haematol.2013.085449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Surinya K.H., Cox T.C., May B.K. Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. J. Biol. Chem. 1998;273:16798–16809. doi: 10.1074/jbc.273.27.16798. [DOI] [PubMed] [Google Scholar]
- 54.Paffett-Lugassy N., Hsia N., Fraenkel P.G., Paw B., Leshinsky I., Barut B., Bahary N., Caro J., Handin R., Zon L.I. Functional conservation of erythropoietin signaling in zebrafish. Blood. 2007;110:2718–2726. doi: 10.1182/blood-2006-04-016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon D., Pastore Y.D., Divoky V., Liu E., Mlodnicka A.E., Rainey K., Ponka P., Semenza G.L., Schumacher A., Prchal J.T. Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development. J. Biol. Chem. 2006;281:25703–25711. doi: 10.1074/jbc.M602329200. [DOI] [PubMed] [Google Scholar]
- 56.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Asp. Med. 2013;34:465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Rojas D.A., Perez-Munizaga D.A., Centanin L., Antonelli M., Wappner P., Allende M.L., Reyes A.E. Cloning of hif-1alpha and hif-2alpha and mRNA expression pattern during development in zebrafish. Gene Expr. Patterns. 2007;7:339–345. doi: 10.1016/j.modgep.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Gerri C., Marass M., Rossi A., Stainier D.Y.R. Hif-1alpha and Hif-2alpha regulate hemogenic endothelium and hematopoietic stem cell formation in zebrafish. Blood. 2018;131:963–973. doi: 10.1182/blood-2017-07-797795. [DOI] [PubMed] [Google Scholar]
- 59.Zhang F.-L., Shen G.-M., Liu X.-L., Wang F., Zhao Y.-Z., Zhang J.-W. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. J. Cell Mol. Med. 2012;16:1889–1899. doi: 10.1111/j.1582-4934.2011.01484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tallack M.R., Whitington T., Yuen W.S., Wainwright E.N., Keys J.R., Gardiner B.B., Nourbakhsh E., Cloonan N., Grimmond S.M., Bailey T.L. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 2010;20:1052–1063. doi: 10.1101/gr.106575.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palis J. Primitive and definitive erythropoiesis in mammals. Front. Physiol. 2014;5 doi: 10.3389/fphys.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aryee D.N.T., Niedan S., Kauer M., Schwentner R., Bennani-Baiti I.M., Ban J., Muehlbacher K., Kreppel M., Walker R.L., Meltzer P. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of ewing's sarcoma cells in vitro. Cancer Res. 2010;70:4015–4023. doi: 10.1158/0008-5472.CAN-09-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiao S., Li C., Hao Q., Miao H., Zhang L., Li L., Zhou Z. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat. Commun. 2017;8 doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teng A.C.T., Kuraitis D., Deeke S.A., Ahmadi A., Dugan S.G., Cheng B.L.M., Crowson M.G., Burgon P.G., Suuronen E.J., Chen H.-H. IRF2BP2 is a skeletal and cardiac muscle-enriched ischemia-inducible activator of VEGFA expression. FASEB J. 2010;24:4825–4834. doi: 10.1096/fj.10-167049. [DOI] [PubMed] [Google Scholar]
- 65.Barrionuevo M.-G., Aybar M.J., Tribulo C. Two different vestigial like 4 genes are differentially expressed during Xenopus laevis development. Int. J. Dev. Biol. 2014;58:369–377. doi: 10.1387/ijdb.130353ct. [DOI] [PubMed] [Google Scholar]
- 66.Wu A., Wu Q., Deng Y., Liu Y., Lu J., Liu L., Li X., Liao C., Zhao B., Song H. Loss of VGLL4 suppresses tumor PD-L1 expression and immune evasion. EMBO J. 2018;38(1) doi: 10.15252/embj.201899506. E99506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saleque S., Cameron S., Orkin S.H. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002;16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goardon N., Lambert J.A., Rodriguez P., Nissaire P., Herblot S., Thibault P., Dumenil D., Strouboulis J., Romeo P.H., Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koeppel M., van Heeringen S.J., Smeenk L., Navis A.C., Janssen-Megens E.M., Lohrum M. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 2009;37:322–335. doi: 10.1093/nar/gkn940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timme-Laragy A.R., Hahn M.E., Hansen J.M., Rastogi A., Roy M.A. Redox stress and signaling during vertebrate embryonic development: regulation and responses. Semin. Cell Dev. Biol. 2018;80:17–28. doi: 10.1016/j.semcdb.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Souders C.L., Liang X., Wang X., Ector N., Zhao Y.H., Martyniuk C.J. High-throughput assessment of oxidative respiration in fish embryos: advancing adverse outcome pathways for mitochondrial dysfunction. Aquat. Toxicol. 2018;199:162–173. doi: 10.1016/j.aquatox.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 72.Ducsay C.A., Goyal R., Pearce W.J., Wilson S., Hu X.Q., Zhang L. Gestational hypoxia and developmental plasticity. Physiol. Rev. 2018;98:1241–1334. doi: 10.1152/physrev.00043.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Falkowski P.G., Katz M.E., Milligan A.J., Fennel K., Cramer B.S., Aubry M.P., Berner R.A., Novacek M.J., Zapol W.M. The rise of oxygen over the past 205 million years and the evolution of large placental mammals. Science. 2005;309:2202–2204. doi: 10.1126/science.1116047. [DOI] [PubMed] [Google Scholar]
- 74.Burton G.J., Fowden A.L., Thornburg K.L. Placental origins of chronic disease. Physiol. Rev. 2016;96:1509–1565. doi: 10.1152/physrev.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Camaschella C. Hereditary sideroblastic anemias: pathophysiology, diagnosis, and treatment. Semin. Hematol. 2009;46:371–377. doi: 10.1053/j.seminhematol.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 76.Nakajima O., Takahashi S., Harigae H., Furuyama K., Hayashi N., Sassa S., Yamamoto M. Heme deficiency in erythroid lineage causes differentiation arrest and cytoplasmic iron overload. EMBO J. 1999;18:6282–6289. doi: 10.1093/emboj/18.22.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brownlie A., Donovan A., Pratt S.J., Paw B.H., Oates A.C., Brugnara C., Witkowska H.E., Sassa S., Zon L.I. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat. Genet. 1998;20:244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- 78.Pantopoulos K., Porwal S.K., Tartakoe A., Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richardson D.R., Lane D.J., Becker E.M., Huang M.L., Whitnall M., Suryo Rahmanto Y., Sheftel A.D., Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu W M.X., Xu J., Heumüller A.W., Fei Z., Feng X., Wang X., Liu K., Li J., Cui G., Peng G., Ji H., Li J., Jing N., Song H., Lin Z., Zhao Y., Wang Z., Zhou B., Zhang L. VGLL4 plays a critical role in heart valve development and homeostasis. PLoS Genet. 2019;15(2) doi: 10.1371/journal.pgen.1007977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Isidor B., Lindenbaum P., Pichon O., Bezieau S., Dina C., Jacquemont S., Martin-Coignard D., Thauvin-Robinet C., Le Merrer M., Mandel J.L. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 2011;43:306–308. doi: 10.1038/ng.778. [DOI] [PubMed] [Google Scholar]
- 82.Fiddes I.T., Lodewijk G.A., Mooring M., Bosworth C.M., Ewing A.D., Mantalas G.L., Novak A.M., van den Bout A., Bishara A., Rosenkrantz J.L. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369. doi: 10.1016/j.cell.2018.03.051. e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu H., Zhang W., Kennard S., Caldwell R.B., Lilly B. Notch3 is critical for proper angiogenesis and mural cell investment. Circ. Res. 2010;107 doi: 10.1161/CIRCRESAHA.110.218271. 860-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ohashi S., Natsuizaka M., Yashiro-Ohtani Y., Kalman R.A., Nakagawa M., Wu L., Klein-Szanto A.J., Herlyn M., Diehl J.A., Katz J.P. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–2123. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weits D.A., Kunkowska A.B., Kamps N.C.W., Portz K.M.S., Packbier N.K., Venza Z.N., Gaillochet C., Lohmann J.U., Pedersen O., van Dongen J.T. An apical hypoxic niche sets the pace of shoot meristem activity. Nature. 2019;569 doi: 10.1038/s41586-019-1203-6. 714-+ [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].