Abstract
Terbinafine resistant Indian ITS genotype VIII T. mentagrophytes strain was identified by partial sequencing of the squalene epoxidase gene using DNA isolated from infected scales. This method allowed the rapid identification of single point mutations within the squalene epoxidase gene, even before a fungal culture was obtained. Terbinafine resistance was indicated by the amino acid position switch Phe397Leu based on a single point mutation of the codon changing it from TTC to CTC.
Keywords: Terbinafine resistance, Squalene epoxidase, Trichophyton mentagrophytes, Indian ITS genotype VIII
1. Introduction
Trichophyton mentagrophytes isolates or formerly designated as zoophilic T. interdigitale species replaced T. rubrum infections as the most often found type of skin diseases in India [[1], [2], [3]]. High proportion of this Indian fungal isolates showed resistance against terbinafine, some also against fluconazole, itraconazole or voriconazole [3].
To distinguish between the different sub genotypes of T. mentagrophytes and Trichophyton interdigitale, it was necessary to sequence amplified DNA fragments, for example the internal transcribed spacer region (ITS) of the ribosomal DNA unit [1,2]. Although this method allowed the identification of Indian ITS genotype VIII T. mentagrophytes, it was not adequate to differentiate between terbinafine-sensitive or resistant isolates of this sub population [3]. Single point mutation variants of squalene epoxidase gene were found within the Indian T. mentagrophytes population [3], similar to squalene epoxidase point mutations of other Trichophyton species [4,5] responsible for high resistance against terbinafine. Partial sequencing of amplified squalene epoxidase fragments was necessary to determine the most important mutation sites associated with the c-terminal part of the coding sequence.
2. Case
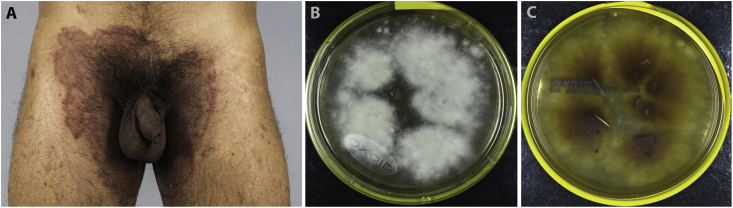
In March 2019 a 26-year old male Indian patient presented at the clinic (day 0) with livid, extensive centrally scaly plaques featuring an erythematous edge and 1 mm large pustules at the lower abdomen as well as bilateral inguinal reaching to gluteal (Fig. 1A). Tinea corporis was suspected and infection with T. mentagrophytes/interdigitale was confirmed by routine diagnostics using realtime PCR at day +3.
Fig. 1.
The male patient showed extensive formation of a Tinea cruris on day 0 (A). Strain morphology after cultivation on Dermasel agar on day +21 (B, C).
For subtype differentiation and resistance determination, the squalene epoxidase gene fragment was directly amplified from scale DNA of sample UKJ 594_19 at day +4. The automated tissue lysis DNA extraction protocol (Qiagen, Qiacube) was used to obtain DNA from scales of the patient. To amplify the squalene epoxidase gene fragments from scale DNA, the primer pair TmSQLEF5 5′-TGGGGCCTGGAGCTTATAGATG-3′ and TmSQLER4 5′-GATGACCCTGCAGGCAGTAAG-3′ was used, yielding a fragment of 844 bp (GenBank Acc. No. MN068043). Three weeks later at day +21, after growing a culture from sample UKJ 594_19 (Fig. 1B and C), DNA was also isolated from the obtained strain UKJ 594_19 as a control. For DNA preparation derived from fungal culture, primer TmSQLEF4 5′-AACGGCTTTGCGAATGGCTCC-3′ instead of TmSQLEF5 was used (GenBank Acc. No. MN068042). Each PCR reaction contained 25 ng DNA (for fungal culture or lower amounts if scale DNA was used), 200 μM dNTPs, 2.5 mM MgCl2, 0.5 μM of each forward and backward primer 80 mM Tris HCl pH 9.4, 100 mM (NH4)2SO4, 0.1% (w/v) Tween 20, and 1 U Taq polymerase (Bio Budget Technologies GmbH, Germany Krefeld) in a total volume of 20 μl. PCR started with pre-treatment of 5 min at 95 °C, 35 cycles of 30 sec at 95 °C, 30 sec at 54 °C, 1 min at 72 °C and post-treatment of 4 min at 72 °C. Additionally, ITS region was determined as described [2] using primer pair LSU266 and V9G [6,7] and fungal DNA obtained from the culture (GenBank Acc. No. MN064822).
ITS and squalene epoxidase sequence data identified the sub genotype VIII T. mentagrophytes in sample UKJ 594_19 and a single point mutation at position 397 of the squalene epoxidase amino acid sequence replacing codon TTC encoding phenylalanine by codon CTC for leucine in this strain. The single point mutation of squalene epoxidase gene leads to high terbinafine resistance [3]. Consequently, it was decided to change the therapy regime of the patient from terbinafine to itraconazole at representation in the clinic on day +16. In addition, the patient was advised to apply ciclopiroxolamine-containing cream locally.
3. Discussion
The recent upsurge of T. mentagrophytes ITS genotype VIII [1,2] infections in India led to the replacement of T. rubrum as the most frequent human pathogen in this country by the zoophilic dermatophyte. The squalene epoxidase DNA fragments of UKJ 594_19 showed 100% identical sequences to other Indian mutant strains with MICs of above 35 μg/mL for terbinafine [3] and identical types of point mutations. The new Indian population of T. mentagrophytes strains with a high ratio of terbinafine resistance is a challenge for fast clinical diagnostic is in order to initiate adequate therapy. In future, it is not sufficient to determine sub genotypes of species of dermatomycetes, but also to analyse their resistance genes.
Ethical Form
Please note that this journal requires full disclosure of all sources of funding and potential conflicts of interest. The journal also requires a declaration that the author(s) have obtained written and signed consent to publish the case report photographs from the patient or legal guardian(s).
Conflict of interest
There is no conflict of interest. The study was funded by the University Hospital Jena.
Acknowledgements
None.
References
- 1.Nenoff P., Verma S.B., Uhrlaß S., Burmester A., Gräser Y. A clarion call for preventing taxonomical errors of dermatophytes using the example of the novel Trichophyton mentagrophytes genotype VIII uniformly isolated in the Indian epidemic of superficial dermatophytosis. Mycoses. 2019;62:6–10. doi: 10.1111/myc.12848. [DOI] [PubMed] [Google Scholar]
- 2.Nenoff P., Verma S.B., Vasani R., Burmester A., Hipler U.-C., Wittig F. The current Indian epidemic of superficial dermatophytosis due to Trichophyton mentagrophytes - a molecular study. Mycoses. 2019;62:336–356. doi: 10.1111/myc.12878. [DOI] [PubMed] [Google Scholar]
- 3.Singh A., Masih A., Khurana A., Singh P.K., Hagen M. Gupta F. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses. 2018;61:477‐484. doi: 10.1111/myc.12772. [DOI] [PubMed] [Google Scholar]
- 4.Rudramurthy S.M., Shankarnarayan S.A., Dogra S., Shaw D., Mushtaq K., Paul R.A. Mutation in the squalene epoxidase gene of Trichophyton interdigitale and Trichophyton rubrum associated with allylamine resistance. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02522-17. e02522-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada T., Maeda M., Alshahni M.M., Tanaka R., Yaguchi T., Bontems O. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob. Agents Chemother. 2017;61:e00115–e00117. doi: 10.1128/AAC.00115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Hoog G.S., van den Ende A.H.G.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 7.Heidemann S., Monod M., Gräser Y. Signature polymorphisms in the internal transcribedspacer region relevant for the differentiation of zoophilic and anthropophilic strains of Trichophyton interdigitale and other species of T. mentagrophytes sensu lato. Br. J. Dermatol. 2010;162:282–295. doi: 10.1111/j.1365-2133.2009.09494.x. [DOI] [PubMed] [Google Scholar]