Abstract
Non-invasive measures of the response of individual patients to cancer therapeutics is an emerging strategy in precision medicine. Platelets offer a potential dynamic marker for metabolism and bioenergetic responses in individual patients since they have active glycolysis and mitochondrial oxidative phosphorylation and can be easily isolated from a small blood sample. We have recently shown how the bioenergetic-metabolite interactome can be defined in platelets isolated from human subjects by measuring metabolites and bioenergetics in the same sample. In the present study, we used a model system to assess test the hypothesis that this interactome is modified by xenobiotics using exposure to the anti-cancer drug doxorubicin (Dox) in individual donors. We found that unsupervised analysis of the metabolome showed clear differentiation between the control and Dox treated group. Dox treatment resulted in a concentration-dependent decrease in bioenergetic parameters with maximal respiration being most sensitive and this was associated with significant changes in over 166 features. A metabolome-wide association study of Dox was also conducted, and Dox was found to have associations with metabolites in the glycolytic and TCA cycle pathways. Lastly, network analysis showed the impact of Dox on the bioenergetic-metabolite interactome and revealed profound changes in the regulation of reserve capacity. Taken together, these data support the conclusion that platelets are a suitable platform to predict and monitor therapeutic efficacy as well as anticipate susceptibility to toxicity in the context of precision medicine.
Graphical abstract
Highlights
-
•
Chemotherapeutic drug doxorubicin dose-dependently inhibited bioenergetic parameters in platelets.
-
•
Doxorubicin induced metabolite alterations in multiple pathways that modulate cellular bioenergetics.
-
•
The interactome revealed novel interactions between bioenergetic parameters and metabolites in platelets.
-
•
The integration of platelet bioenergetics with the metabolome can be utilized for precision medicine strategies.
1. Introduction
Doxorubicin (Dox), a non-selective class I anthracycline, also known as Adriamycin, is a common chemotherapeutic used to treat a broad range of cancers including leukemia, multiple myeloma and cancers of the breast [[1], [2], [3]]. Dox was first isolated from Streptomyces peucetius in the 1960s and approved for medical use in the United States in 1974 [4]. Although several formulations of Dox are in use, the occurrence of dose limiting side effects remains relatively high [5]. The antineoplastic properties of Dox are attributed to its ability to intercalate into nucleic acids [6] leading to DNA/RNA damage and inhibition of macromolecule synthesis [7]. Dox crosses the cell membrane by passive diffusion [[8], [9], [10]] and accumulates in the nuclear compartment and the mitochondria [11]. In addition to passive diffusion the organic cation transporter SLC22A16, which is expressed in many cancers, also contributes to cellular uptake of Dox.
Pharmacokinetic studies of Dox show a rapid uptake by tissue following a single intravenous injection but a slow tissue elimination with a half-life of up to 48 h and a maximum serum concentration (Cmax) in the low micromolar range [7]. Doxorubicin undergoes three metabolic fates with 50% of the administered dose remaining as the parent molecule. Dox is primarily metabolized to doxorubicinol (Dox-ol) via a two-electron reduction mediated by carbonyl reductase [12,13]. A second metabolic route involves an enzymatically driven one-electron reduction of the quinone functional group by nitric oxide synthase, NADPH cytochrome P450 reductase and other oxidoreductases, leading to the formation of a semiquinone radical. Molecular oxygen (O2) can then oxidize the Dox semiquinone radical to form superoxide (O2˙-) and hydrogen peroxide (H2O2) [2,14] contributing to oxidative stress and macromolecule-damage. The final metabolic route of Dox involves the deglycosidation of the parent compound by microsomal reductases to form doxorubicinone (Dox-one), doxorubicinolone (Dox-olone), and 7-deoxy-doxorubicinolone (7-Dox-olone) [7,15,16]. This route accounts for a small fraction of Dox metabolism and has been linked to changes in human erythrocytes energy metabolism as seen by a shift in the pentose phosphate pathway and inhibition of antioxidant enzymes [17].
Although Dox is widely used as an anti-tumor agent, its use is associated with serious side effects ranging from cardiotoxicities to thrombocytopenia [5,[18], [19], [20]]. Dox mediates tumor cell death via inhibition of topoisomerase I and II and activation of apoptosis [6]. This mechanism also occurs in non-cancer cells, mediated by a p53-dependent signaling pathway [10], and leads to cardiomyopathy. Dox-induced cardiotoxicity has also been ascribed to mitochondrial cytochrome c release and permeability transition pore opening [21]. Longer exposures have been linked to decreased expression of mitochondrial proteins, and the redox signaling of the semiquinone exacerbates reactive oxygen species production further activating the apoptotic cascade [21,22]. Furthermore, Dox can form complexes with cardiolipin, a mitochondrial inner membrane phospholipid important for allosteric regulation of mitochondrial enzymes [2,23] and intercalation with mitochondrial DNA [11].
Dox-induced thrombocytopenia and destruction of mature platelets [6,18,21] suggests that they may be useful to monitor Dox effects on mitochondrial function. Platelets are enucleated cells derived from megakaryocytes and have a lifetime circulation of approximately 10 days in the blood [24,25]. Although platelets do not contain a nucleus, they do contain RNA transcripts and other RNAs acquired during circulation [26] and contain mitochondria which are essential for platelet function [27]. Interestingly, it is now becoming clear that platelets can serve as biomarkers for mitochondrial dysfunction and the metabolome is predictive of bioenergetic function [28,29]. Indeed, platelet bioenergetics have been shown to be potential biomarkers for the clinical severity of sickle cell disease, asthma, Alzheimer's and Parkinson's disease [[30], [31], [32]]. These findings raise the possibility that platelet bioenergetics can also serve to monitor the potential dose limiting toxicity of therapeutics, such as Dox, which have side effects mediated by their effects on metabolism.
In a recent study combining the mitochondrial stress test (MST) with high-resolution metabolomics, we found that subsets of metabolites, including fatty acids and xenobiotics correlated with mitochondrial parameters, establishing platelets as a platform to integrate bioenergetics and metabolism for analysis of mitochondrial function in precision medicine [29]. In the current study, we reasoned that the mitochondrial response to stress will be modulated by the activity of metabolic pathways prior to exposure to a therapeutic agent. If so, differences in the platelet metabolome between individuals could contribute to variability in the bioenergetic profiles of the intact platelet and therefore support use of platelet bioenergetic-metabolite interactome to predict and monitor therapeutic effects. This concept was addressed by measuring mitochondrial bioenergetic parameters among platelet donors in the presence and absence of Dox and integrating these data with non-targeted metabolomics. We found that the pre-treatment basal metabolome was correlated with maximal respiration, the response to Dox and its metabolism. These data provide the foundation for not only understanding Dox-mediated platelet toxicity but also contribute to precision medicine-based chemotherapies.
2. Materials and methods
2.1. Chemicals
All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. A mixture of internal standard stable isotopic chemicals [33,34] was purchased from Cambridge Isotope Laboratories, Inc. (Andover, Pennsylvania).
2.2. Platelet isolation
Platelet concentrates collected from individual donors were obtained from the University of Alabama at Birmingham blood bank and used between days 6–8 after collection. Platelet activation during preparation is suppressed by the inclusion of Prostaglandin I2 and any potential activation was assessed by microscopy [35]. In this study no platelet samples were excluded due to activation. Collection and use of these samples was approved by the University of Alabama at Birmingham Institutional Review Board (Protocol #X110718014). Platelets used for these studies were between day 6 and 8 after collection or freshly isolated as described previously [35,36]. In brief, platelets were pelleted by centrifuging at 1500 g for 10 min then washed with PBS containing prostaglandin I2 (1 μg/ml) and platelet number was determined by turbidimetry [37]. Prostaglandin I2 is included in the washing buffer to prevent activation during platelet preparation. The platelets for the experiments were isolated from 14 individual donors and data are reported for all patients for metabolomics and for 13 for cellular bioenergetics due to the failure of one assay. Platelet aggregation using the 96-well plate reader was measured as previously described [38].
2.3. Doxorubicin treatment
For both bioenergetics and metabolomics studies, Dox stocks (20 mM in MilliQ water) were diluted to a 10-fold working solution in XF DMEM assay media (pH adjusted to 7.3). All assays described below were performed in parallel with each other. Vehicle controls was XF assay media alone.
2.4. Platelet bioenergetics and mitochondrial function
The 96-well format Seahorse extracellular flux analyzer (Seahorse Bioscience, MA, USA) was used to measure bioenergetics [36]. Platelets were diluted to a concentration of 1 × 107 in 75 μl XF DMEM assay buffer (DMEM with 1 mM pyruvate, 5.5 mM d-glucose, 4 mM l-glutamine, pH 7.4) and were seeded onto Cell-Tak coated XF96 microplates and the mitochondrial stress test was performed as described [39]. The mitochondrial complex assay is performed using Plasma Membrane Permeabilizer (PMP) with injection of respiratory substrates with or without ADP [40].
2.5. High-Resolution Metabolomics (HRM)
For metabolic measurements, a protocol similar to the bioenergetic measurements was used [29], and untargeted metabolomics was performed using previously established HRM methods [[41], [42], [43], [44]]. Washed platelets were diluted to a concentration of 1 × 108/well in 0.75 ml DMEM assay buffer (DMEM with 1 mM pyruvate, 5.5 mM glucose, 4 mM glutamine, pH 7.4) and treated and were incubated with vehicle control or Dox (25 μM) for 3 h at 37 °C in a non-CO2 humidified incubator. Platelets were then washed with cold PBS and the proteins precipitated using acetonitrile (50 μl) containing a mixture of stable isotope-labeled internal standard [33,34]. Pooled platelets (3 × 108 platelets from 3 wells) in 150 μl of acetonitrile containing internal standard were incubated on ice for 10–15 min and precipitated proteins removed by centrifugation for 10 min at 13,000 rpm. The extraction protocol and the solvent are chosen to effectively clamp the metabolites which is verified by our previous study using oligomycin as a positive control [29]. Samples were randomized to minimize effects of instrumental drift during analysis, and 10 μL aliquots were analyzed with three technical replicates using reverse-phase C18 liquid chromatography (Targa C18 2.1 mm × 50 mm x 2.6 μm, Higgins Analytical) combined with a High-Field Q-Exactive mass spectrometer (Thermo Fisher). Mass spectral detection completed in negative mode electrospray ionization (ESI) at 120,000 (FHWM) resolution over a mass-to-charge ratio (m/z) range of 85–1250. A quality control pooled reference plasma sample (Q-Std3) was included at the beginning and end of each batch of 25 samples for quality control and quality assurance [45]. Raw data files were extracted using apLCMSv6.3.3 [46] with xMSanalyzerv2.0.7 [47], followed by batch correction with ComBat [48]. Uniquely detected ions consisted of m/z, retention time (RT) and ion abundance, are referred to as metabolic features.
2.6. Data processing and metabolic feature selection
Prior to data analysis, triplicate injections were averaged and only m/z features with at least 80% non-missing values in either of the groups and more than 40% non-missing values across all samples were retained. After filtering based on missing values, data were log2 transformed and quantile normalized [49]. Selection of differentially expressed m/z features was performed based on one-way repeated measures ANOVA, using the limma package in R [50]. For the metabolome-wide association study of Dox, metabolic features which were correlated with Dox were selected using linear regression analysis and transformed as described above. Benjamini-Hochberg false discovery method was used for multiple hypothesis testing corrections at a FDR<0.2 threshold [51]. Visualization of the data, which was based on similarity in expression, was performed using unsupervised two-way hierarchical clustering analysis (HCA) utilizing the hclust() function in R to determine the clustering pattern of selected m/z features and samples. Principal component analysis (PCA) was performed using the pca() function implemented in R package pcaMethods.
2.7. Pathway enrichment analysis
To evaluate metabolic alterations at a systems level metabolome-wide association analysis was performed for discriminatory metabolites at p < 0.05 and characterized for pathway enrichment using mummichog v1.0.10 software [52]. For this analysis, features differing at p < 0.05 were selected to protect against type 2 error, and permutation testing (p < 0.05) was used in pathway enrichment analysis to protect against type 1 error [53]. Pathways including minimum 4 matched metabolites in total size were selected and annotated using the criteria described below.
2.8. Metabolite annotation
Metabolic features were annotated using xMSannotator [54]; confidence scores for annotation by xMSannotator are derived from a multistage clustering algorithm. Identities of selected metabolites were confirmed by co-elution relative to authentic standards and ion dissociation mass spectrometry (Level 1 identification by criteria of Schymanski et al. [55]. Supplemental annotations were made based on high or medium confidence (≥4) with M-H adducts detected in the negative mode. Lower confidence annotations were made using KEGG, (Kyoto Encyclopedia of Genes and Genomes) [56]; HMDB (Human Metabolome Database) [57]; T3DB [58], and Lipid Maps [59] databases at 5 ppm tolerance.
2.9. xMWAS
Bioenergetic and HRM data from the same set of samples were integrated by using xMWAS based on the sparse partial least-squares (sPLS) regression method for data integration [60]. sPLS is a regression-based modeling approach which performs simultaneous variable selection and data integration, and is designed for problems where the sample size (n) is much smaller than the number of variables (p) and the variables are highly correlated [61]. In addition, xMWAS performs community detection using the multilevel community detection algorithm [62] to identify groups of nodes that are heavily connected with other nodes in the same community, but have sparse connections with the rest of the network. The input for xMWAS included the cellular bioenergetics (13 samples × 6 energetic parameters) and the metabolome (13 samples x 3240 metabolic features) for the vehicle control group and (13 samples × 6 energetic parameters) and the metabolome (13 samples x 3263 metabolic features) for the Dox treated group, which had been quantile normalized and log-transformed). Thresholds for determining significant associations met the correlation threshold criteria (|r| > 0.6) and p < 0.05 as determined by Student's t-test.
2.10. Bubble plots
Generation of the bubble plot, a visualization tool for metabolic pathways found to be associated with bioenergetic parameters, was performed using the R script corrplot() [63]. For these analyses, metabolic features which were previously found to be significant using the xMWAS correlation criteria, previously described in the xMWAS methods, were input into mummichog v1.0.10. Pathway analysis was performed for each bioenergetic parameter independently, and significant pathways were selected using the criteria previously described within the pathway enrichment methods. Both the size as well as the color of the bubble represent the pathway significance level based on the -Log10P value.
2.11. Statistical analysis
The data reported in the metabolomics analyses are derived from platelets isolated from 13 or 14 different donors. Each platelet group was comprised of 3–5 technical replicates, and the data is presented as mean ± SEM. Statistical significance was determined using either a T-TEST or ANOVA with Tukey's post hoc test for data with more than 2 groups, and p < 0.05 was considered significant. The linear correlation between multiple pairs of bioenergetic parameters were determined using the multivariate function of the JMP statistical program (JMP®, Version 13, SAS Institute Inc., Cary, NC). A correlations (r-values) table that summarizes the strength of the linear relationships between each pair of bioenergetic parameters and a table with corresponding p-values were generated to identify significant dependencies between parameters. Data for |r| ≥0.4 and p ≤ 0.01 were considered significant.
3. Results
3.1. Effect of Dox treatment on platelets bioenergetics
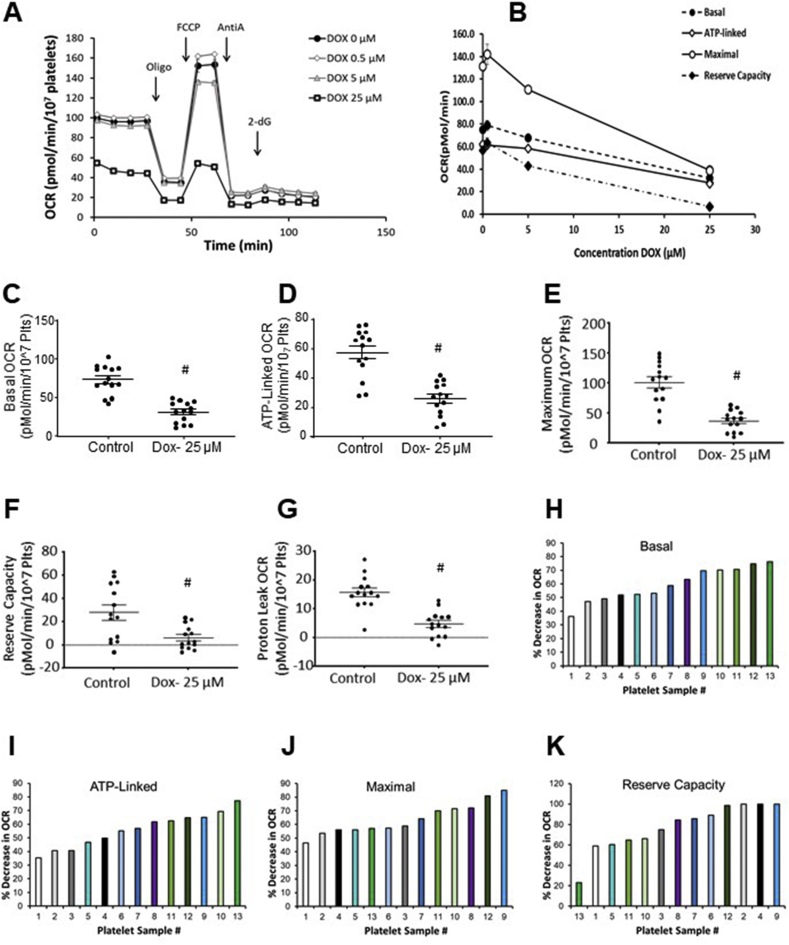
To assess the impact of Dox on bioenergetics platelets were pretreated for 3 h with increasing concentrations of Dox before the mitochondrial stress test (MST) was used to measure the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) [64]. A representative trace for a single individual is shown in Fig. 1A. The control platelets showed the expected response to oligomycin with a suppression of basal OCR which was reversed on addition of the uncoupler FCCP [64]. Antimycin inhibited OCR below post-oligomycin levels and 2-DG, an inhibitor of glycolysis, had no additional effect on OCR. Also shown in Fig. 1A and B is the concentration dependent effect of Dox (0–25 μM) on intact platelet bioenergetics. The MST parameters were calculated to characterize cellular bioenergetics [65] and showed (Fig. 1B) that the lowest concentrations of Dox suppressed reserve capacity and maximal respiration and an effect on ATP linked respiration was apparent at the 25 μM concentration. This concentration is a reasonable approximation for the higher range in human subjects treated with Dox which have a reported Cmax of 11–19 μM [66]. Platelet ECAR or platelet aggregation was not significantly changed by Dox treatment (results not shown).
Fig. 1.
Doxorubicin inhibits mitochondrial respiration. (A) Platelets from a representative donor are shown with and without incubation with Doxorubicin (Dox) (0–25 μM) for 3 h after which a mitochondrial stress test was performed by first measuring basal OCR, followed by sequential injection of oligomycin (Oligo) (1 μg/ml), FCCP (0.6 μM), antimycin A (AntiA) (10 μM) and 2-deoxyglucose (50 mM). (B) The Dox concentration dependent inhibition of Basal, ATP linked, Maximal and Reserve Capacity. Data is represented as the mean ± SEM for 5–6 technical replicates for this donor. Data from 13 individual donors are shown for (C) basal (basal OCR – AA OCR), (D) ATP-linked (AL) (basal OCR – oligomycin OCR), (E) maximal (FCCP OCR – AA OCR), (F) reserve capacity (RC) (FCCP OCR – basal OCR) (G) proton leak (PL) (oligomycin OCR – AA OCR), calculated and are expressed as mean ± SEM with each dot representing the average data from a single donor, n = 5–6 replicates for each parameter. (H–K) Decrease (% of pre-Dox value) for each donor for basal (H), ATP linked (I) Maximal (J) and Reserve Capacity (K) after treatment with 25 μM Dox.
The same protocol shown in Fig. 1 was used to determine the average response to Dox in platelets from 13 individual donors and is shown for the 25 μM Dox concentration in Fig. 1 (Panels C–G). All bioenergetic parameters were significantly decreased after 25 μM Dox treatment. We noted that both the pre and post-Dox treatment OCR values varied significantly between individuals. For example, the extent of Dox-dependent inhibition ranged from 30 to 70% decrease in basal OCR, 30–75% decrease in ATP-linked OCR, 50–80% decrease in maximal OCR and as little as a 20% decrease to complete loss of reserve capacity among the 13 individuals tested (Fig. 1H–K). Our previous studies suggested that the parameters from the mitochondrial stress test represent an integrated metabolic profile [67] and this can be demonstrated using a multi-variate analysis for the parameters from the MST in the presence or absence of 25 μM Dox (see Table 1 and Fig. 2).
Table 1.
Platelet bioenergetic parameters with and without Dox treatment.
| Basal | AL | PL | Maximal | RC | NM | |
|---|---|---|---|---|---|---|
| Basal | ||||||
| AL | 0.9505* | |||||
| PL | 0.4892 | 0.1945 | ||||
| Maximal | 0.7315* | 0.7994* | 0.0748 | |||
| RC | 0.2584 | 0.3927 | −0.2737 | 0.8477* | ||
| NM |
0.5060 |
0.4800 |
0.2532 |
0.1407 |
−0.1967 |
1.0000 |
| Correlation between the platelet bioenergetic parameters following Dox (25 μM) treatment | ||||||
| Basal |
AL |
PL |
Maximal |
RC |
NM |
|
| Basal | ||||||
| AL | 0.9465* | |||||
| PL | 0.5790* | 0.2852 | ||||
| Maximal | 0.8108* | 0.7877* | 0.4258 | |||
| RC | 0.0719 | 0.1005 | −0.0281 | 0.642* | ||
| NM | 0.3779 | 0.3204 | 0.3031 | 0.0649 | −0.3818 | |
Table 1 provides the Pearson correlation coefficients (r values) calculated using the multivariate platform of JMP statistical program and shows the strength of the linear relationships between each pair of variables. * indicates p ≤ 0.05.
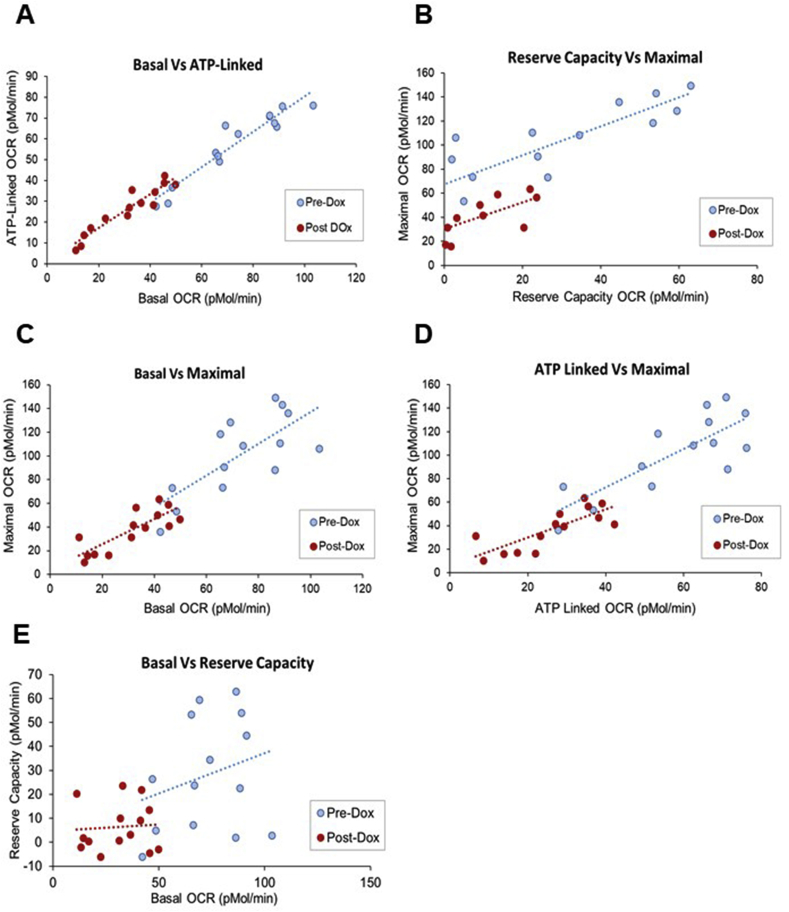
Fig. 2.
Relationships between bioenergetic parameters from the mitochondrial stress test (MST) with and without doxorubicin. Using the data shown in Fig. 1 a correlation analysis was performed using the JMP statistical program to assess the linear relationships between the bioenergetic parameters derived from the MST with and without 25 μM Dox. The bivariate plots of basal OCR and ATP-linked OCR (A), reserve capacity and maximal OCR (B), basal OCR and maximal OCR (C), ATP-linked OCR and maximal OCR (D) and basal OCR and reserve capacity (E) are shown. The r values, the measure of strength of linear relationships and results of the significance tests for these data are reported in Table 1.
In untreated control platelets significant relationships were evident between basal vs ATP linked respiration, basal vs. maximal, ATP linked vs maximal, and maximal vs reserve capacity (Fig. 2, Table 1). Basal vs reserve capacity OCR were not significantly related. Taken together these data confirm our previous findings that there is a wide normal range for platelet bioenergetics in healthy donors with several interesting features [29]. After Dox treatment, these relationships were largely preserved, but with a lower range of activities, as shown graphically in Fig. 2. Analysis of this data revealed that after Dox treatment the range of OCR for ATP linked and basal was narrowed (also evident from Fig. 1) consistent with a lower capacity to generate ATP. Interestingly significant relationships exist between the Dox metabolite and the % change in the bioenergetic parameters in individual platelet samples (Table 2). Dox and the Dox metabolite, Doxorubicinone) correlate positively with Basal and ATP-linked OCR whereas Reserve Capacity is negatively correlated with Dox levels. These data are consistent with a mechanism in which the Reserve Capacity is being used in response to the redox cycling of Dox. In addition, these data confirm our previous findings in other cell types that in response to oxidative stress, basal OCR increases with a corresponding decrease in Reserve Capacity [67,68]. Dox levels in the individual platelet samples ranged from 3.54 × 108 ± 2.06 × 108 to 1.43 × 109 ± 1.1 × 108 and its aglycone metabolite doxorubicinone from 2.71 × 107 ± 9.15 × 105 to 1.73 × 108 ± 3.83 × 107 which suggest that a significant variability exist between individuals in the ability to metabolize doxorubicin and is related to the susceptibility to mitochondrial damage.
Table 2.
Relationships between bioenergetic parameters in the individual platelet samples prior to Dox treatment and the Dox metabolites.
| Correlations | ||||||
|---|---|---|---|---|---|---|
| Basal | AL | PL | Max | Res Cap | NonMito | |
| Dox | 0.6504 | 0.698 | −0.0738 | 0.0518 | −0.6783 | 0.2246 |
| Doxorubicinone | 0.5413 | 0.5686 | −0.3219 | 0.3002 | −0.2234 | 0.2219 |
|
| ||||||
| Correlation Probability | ||||||
| Basal |
AL |
PL |
Max |
Res Cap |
NonMito |
|
| Dox | 0.022 | 0.0116 | 0.8197 | 0.873 | 0.0153 | 0.4828 |
| Doxorubicinone | 0.0692 | 0.0537 | 0.3076 | 0.3431 | 0.4851 | 0.4881 |
Table 2 demonstrates the relationship between bioenergetic parameters and the unchanged Dox parent compound or Dox metabolites, such as doxorubicinone. The Pearson correlation coefficients were calculated using the JMP statistical program. * indicates p ≤ 0.05.
3.2. The effect of Dox treatment on the platelet metabolome
Samples of platelets were prepared for untargeted metabolomics with and without exposure to 25 μM Dox for 3 h under the same conditions used for the bioenergetic measurements. 2831 features (distinct m/z values) were identified in both the control and Dox treated samples. Using the KEGG metabolic pathway mapping program a total of 48 pathway matches were found which included metabolites associated with platelet activation as well as fatty acid and arachidonic acid metabolism which are strongly associated with platelet function after Dox treatment (Supplementary Table 1).
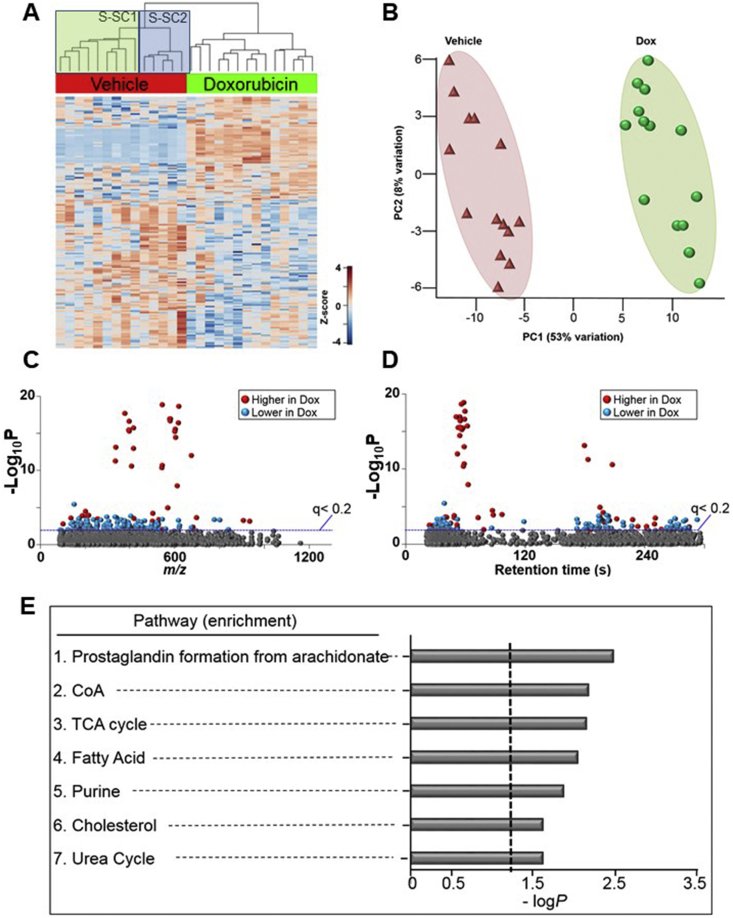
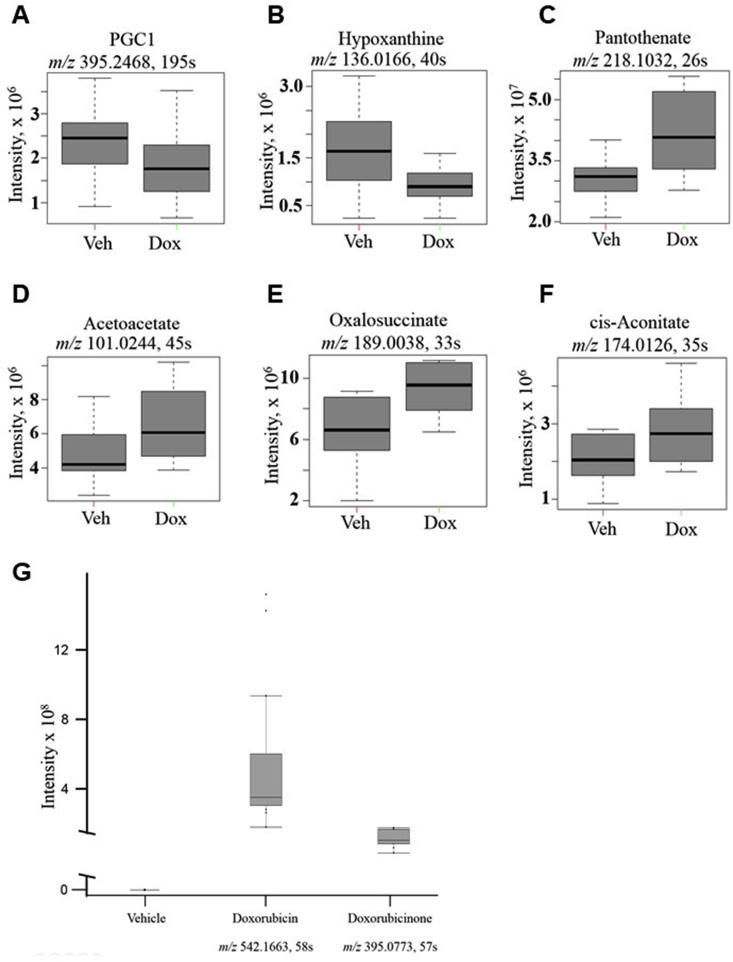
Next, we examined metabolic responses of the platelets to Dox treatment. ANOVA on the 2831 features revealed 166 features were changed after Dox treatment (p < 0.05 at FDR of 0.2) and are presented by hierarchical clustering analysis-heat map (Fig. 3A) and PCA plots (Fig. 3B). Manhattan plots, based upon RT, m/z and abundance of metabolites show that of the 166 metabolites, 68 metabolites were higher and 98 metabolites were lower in Dox-treated platelets compared to the control group (Fig. 3C,D). Supplementary Table 2 shows detailed information on 68 annotated metabolites. To examine the metabolic pathways altered by Dox treatment, pathway enrichment analysis was performed using mummichog. The results showed that metabolites from the prostaglandin formation from both arachidonate, as well as CoA, TCA cycle, fatty acid, purine, cholesterol, and urea cycle pathways were enriched by Dox treatment (Fig. 3E). The detailed information on metabolites associated with these pathways is provided in Supplementary Table 3. These data are consistent with inhibition of bioenergetic function by Dox in intact platelets. Selected metabolites from these pathways are shown in Fig. 4. For prostaglandin formation we observed that PGC1 (4A) was decreased, and similar observations were made for purine metabolite hypoxanthine (4B) which was also decreased. The CoA catabolism metabolite pantothenate (4C) was found to be increased after Dox treatment, which was also observed for acetoacetate (4D), and the TCA cycle metabolites, oxalosuccinate (4E) and cis-aconitate (4F). We also observed Dox (4G) and its metabolite doxorubicinone in the Dox treated group, showing that platelets metabolize Dox. We also found a number of metabolic features which were highly enriched in the Dox samples; however, many did not have known matches to public available databases, and are reported in Table 3.
Fig. 3.
Metabolic separation of features after treatment to Dox in human platelets. (A) Unsupervised hierarchal clustering heatmap indicate that intensity of 166 features which drive the separation between vehicle control and Dox treated groups. Subject sub clusters 1 and 2 are indicated in the vehicle group. (B) PCA plot showing separation of the vehicle control group (shown in green) and BaP exposed group (shown in red), through the 1st and 2nd principal components. (C) Type I Manhattan plot of m/z features plotted against the –Log10P value. Shown in gray are the 2831 features identified after filtering and normalization. 166 features were found to be different between the two groups using the criteria (p < 0.05, FDR < 0.2) as indicated by the blue dotted line. Shown in red were features identified to be increased after Dox exposure (68/166) and in blue features which were decreased (98/166). (D) Type II Manhattan plot using time plotted against –Log10P value. (E) Pathway enrichment analysis of stored human platelets after Dox exposure compared to vehicle control. A total of (7/119) enriched pathways were determined (Filled gray bars indicate significance and the cutoff (p < 0.05) is indicated by the dotted line). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Comparison of selected metabolites between the vehicle and doxorubicin groups. Representative metabolites were selected from mummichog pathway enrichment analysis. Data shown are mean and standard deviation for spectral intensities of Dox and vehicle groups. A. Prostaglandin C1 (PGC1; m/z 395.2468, 195s), B. Hypoxanthine (m/z 136.0166, 40s), C. Pantothenate (m/z 218.1032, 26s), D., Acetoacetate (m/z 101.0244, 45s), E. Oxalosuccinate (m/z 189.0038, 33s), and F. cis-Aconitate (m/z 174.0126, 36s). Metabolites of Dox metabolism were also annotated, which data shown as mean and standard deviation for spectral intensities of Dox and vehicle groups. G. Dox (m/z 542.1663, 58s), and Doxorubicinone (Doxone, m/z 395.0773, 57s).
Table 3.
Unidentified Metabolic Features which are highly enriched with Dox Treatment.
| No | m/z | RT (s) | Chemical_ID | Annotation Score | Formula | Name | Adduct | P value | Fold Change |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 542.1663 | 58 | C01661 | 4 | C27H29NO11 | Doxorubicin | M-H | 4.16E-11 | ∞ |
| 2 | 544.1723 | 58 | C01661 | 4 | C27H29NO11 | Doxorubicin | M-H [+2] | 1.37E-19 | ∞ |
| 3 | 377.067 | 59 | 5 | C21H14O7 | Doxorubicin Impurity 14 | M-H | 2.21E-18 | ∞ | |
| 4 | 395.0773 | 57 | 5 | C21H18O9 | Doxorubicinone | M-H2O–H | 3.52E-16 | ∞ | |
| 5 | 335.0547 | 183 | Unknown | Unknown | Unknown | Unknown | Unknown | 5.67E-12 | ∞ |
| 6 | 336.0594 | 179 | Unknown | Unknown | Unknown | Unknown | Unknown | 7.16E-14 | ∞ |
| 7 | 396.0808 | 59 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.56E-17 | ∞ |
| 8 | 397.0829 | 56 | Unknown | Unknown | Unknown | Unknown | Unknown | 5.36E-16 | ∞ |
| 9 | 407.3167 | 207 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.68E-11 | ∞ |
| 10 | 415.013 | 58 | Unknown | Unknown | Unknown | Unknown | Unknown | 1.13E-13 | ∞ |
| 11 | 416.0165 | 62 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.03E-16 | ∞ |
| 12 | 543.1697 | 59 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.01E-11 | ∞ |
| 13 | 578.1447 | 54 | Unknown | Unknown | Unknown | Unknown | Unknown | 1.12E-17 | ∞ |
| 14 | 579.1476 | 54 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.03E-17 | ∞ |
| 15 | 580.1391 | 50 | Unknown | Unknown | Unknown | Unknown | Unknown | 1.18E-17 | ∞ |
| 16 | 600.128 | 56 | Unknown | Unknown | Unknown | Unknown | Unknown | 5.53E-16 | ∞ |
| 17 | 601.1287 | 54 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.66E-16 | ∞ |
| 18 | 602.1213 | 53 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.97E-16 | ∞ |
| 19 | 603.1251 | 54 | Unknown | Unknown | Unknown | Unknown | Unknown | 3.45E-15 | ∞ |
| 20 | 610.1559 | 62 | Unknown | Unknown | Unknown | Unknown | Unknown | 1.14E-08 | ∞ |
| 21 | 616.1 | 54 | Unknown | Unknown | Unknown | Unknown | Unknown | 4.14E-17 | ∞ |
| 22 | 618.096 | 56 | Unknown | Unknown | Unknown | Unknown | Unknown | 2.26E-19 | ∞ |
| 23 | 674.0603 | 52 | Unknown | Unknown | Unknown | Unknown | Unknown | 9.92E-13 | ∞ |
Table 3 demonstrates differentially expressed metabolic features which were found to be highly enriched in the Dox treated group, but were not present in the Vehicle Control. ∞ indicates metabolites detected in Dox treated group but not in the baseline group (control).
3.3. Baseline metabolic pathways associated with subsequent response to Dox
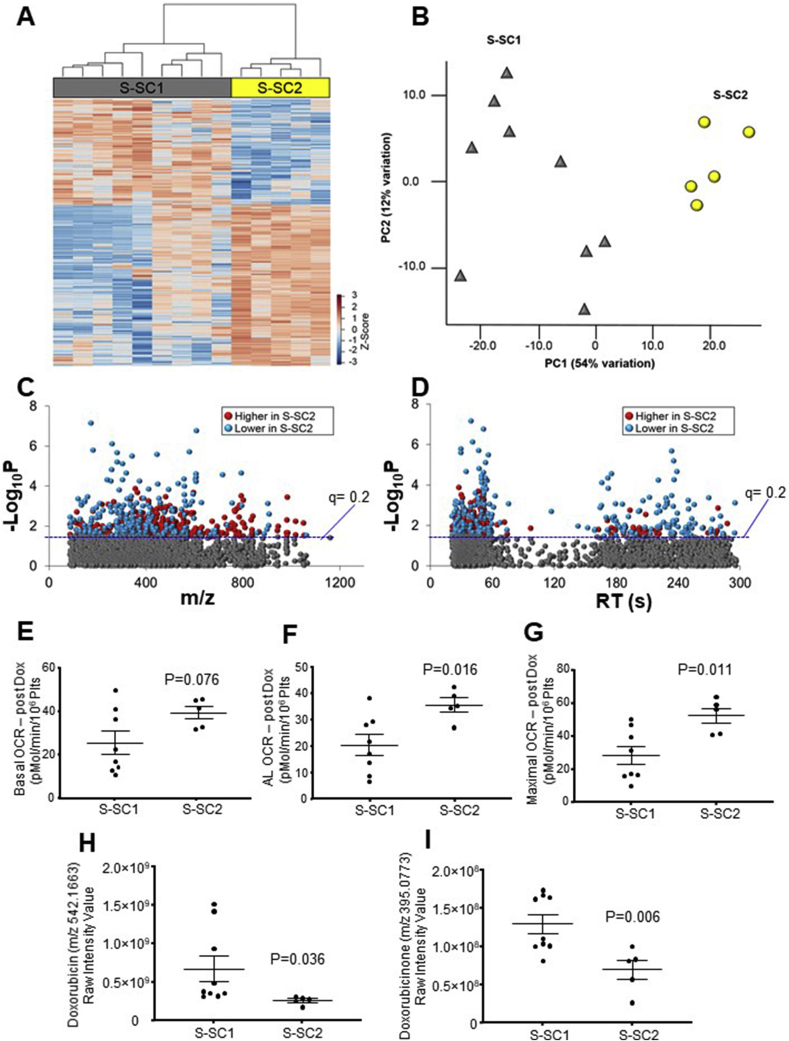
Based on previous evidence for sub-clustering of metabolic profiles in untreated platelets [29], we hypothesized that the metabolic response of platelets to Dox could be influenced by the baseline metabolic status. In the vehicle control heat map in Fig 3A the dendrogram reveals 2 main sub-clusters (S-SC1 and S-SC2) which we hypothesized would respond differently to Dox. To test this, we used these subject sub-clusters in the vehicle group and used it to segregate the Dox total metabolome data between the 2 groups. An ANOVA analysis was then used to identify metabolites which were significantly different between SC. ANOVA of the respective metabolic data following Dox treatment showed that 567 features differed between S-SC1 and S-SC2 groups as shown in the HCA-heat map (Fig. 5A). PCA for these metabolites showed complete separation of the sub-clusters, with 66% of variation associated with PC1 and PC2 (Fig. 5B). Manhattan plots showed that of the 567 metabolites, 342 metabolites were lower and 225 metabolites were higher in Dox-treated platelets of S-SC1 compared to the Dox treated platelets of S-SC2 (Fig. 5C; D). Annotation and details of these features are provided in Supplementary Table 4. Further examination of the magnitude of effect of Dox on MST bioenergetic parameters in sub-clusters 1 and 2 showed that platelets from sub-cluster 2 showed significantly less effect of Dox on basal, ATP and maximal OCR (Fig. 5 E-F). The results also showed differences in abundance between the groups for Dox and doxorubicinone, with characteristics suggesting that sub-cluster 2 had considerably greater Dox elimination rate (or lower uptake rate) than sub-cluster 1and lower toxicity (Fig. 5H; I).
Fig. 5.
Metabolic separation of individuals in Subject Sub-clusters 1 and 2. A. Unsupervised hierarchal clustering heatmap indicate that intensity of 567 features drive the separation between subject sub-clusters 1 and 2 (S-SC1 and S-SC2). B. PCA plot showing separation of S-SC1 (shown in gray), and S-SC2 (shown in yellow) through the 1st (54% variation) and 2nd (12% variation) principal components. C. Type I Manhattan plot of m/z features plotted against the –Log10P value. Shown in gray are the 3247 features identified after filtering and normalization. 567 features were found to be different between the two groups using the criteria (p < 0.05, FDR < 0.2) as indicated by the blue dotted line. Shown in red were features identified to be increased in S-SC2 (225/567) while features which were lower are shown in blue (342/567). D. Type II Manhattan plot using time plotted against –Log10P value. The more polar features elute earlier than the more non-polar metabolites which elute at a later time due to the methods and column described above in the methods. n = 9 for S-SC1 and n = 5 for S-SC2. Comparison of the bioenergetic parameters (basal (E), ATP-Linked (F) and maximal OCR (G)) between the sub clusters S-SC1 and S-SC2. H. Dox detected in the platelet samples using metabolomics analysis. I. Doxrubicinone, a major metabolite present in the platelet samples following Dox treatment as measured by mass specrometry. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Data-driven integrative network analysis of MST and metabolic responses using xMWAS
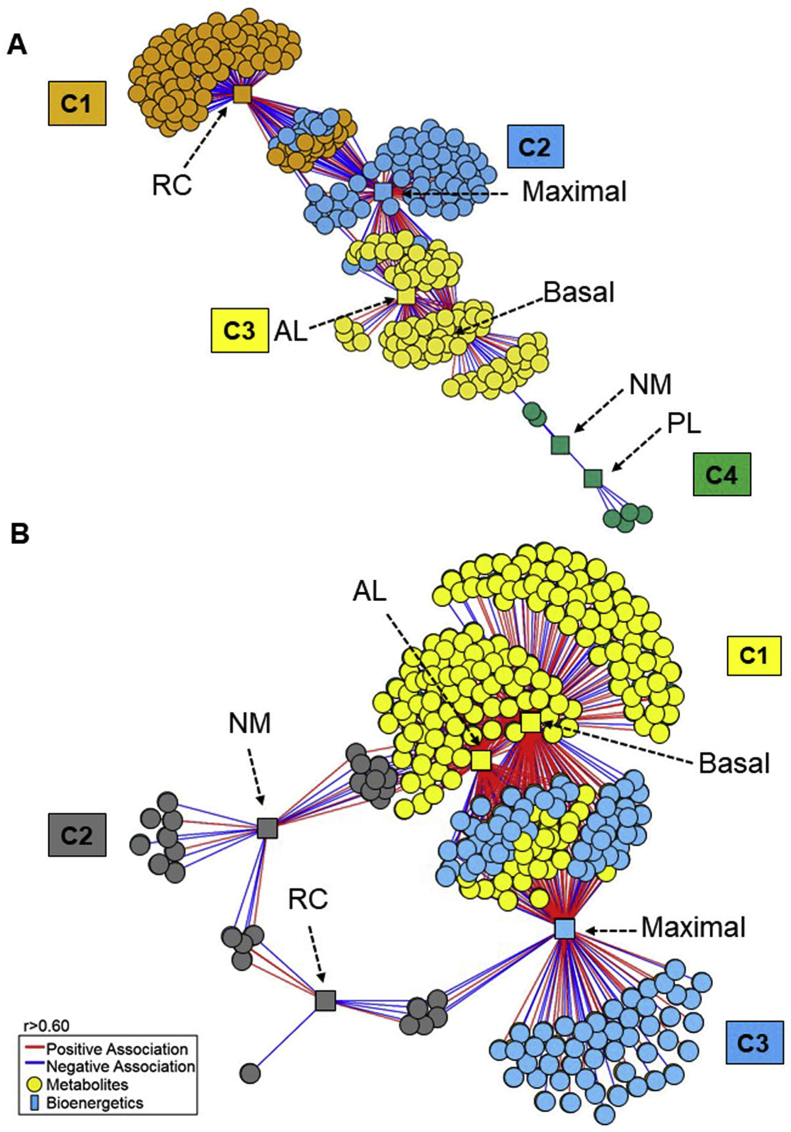
To gain an understanding of the differences in respiration-linked metabolic changes that occur in response to Dox, we used data-driven analysis with xMWAS. The associations between bioenergetics and the metabolome for vehicle-treated platelets (Fig. 6A) and Dox-treated platelets (Fig. 6B) revealed profound differences in metabolic organization. In this representation, the vehicle analysis revealed 4 communities, which had closely related interactions and were designated by different colors. Community 1 (orange) had 117 metabolic features associated with reserve capacity, while community 2 (blue) had 143 metabolic features associated with maximal respiration. Community 3 (yellow) had 112 feature associations with both basal and ATP-linked respiration. Community 4 (green) had 7 features which were associated with non-mitochondrial respiration as well as proton leak (Fig. 6A). In comparison, the Dox network had only 3 communities; communities 1 (green) and 3 (orange) had the highest number of associations. Basal and ATP-linked OCR had 239 metabolic associations and maximal respiration had 138 metabolic features. Community 2 (blue) associated with both non-mitochondrial and reserve capacity, had an increased number of 27 metabolic features compared to control (Fig. 6B). A complete listing of the positive and significant relationships between metabolites and bioenergetic parameters for both conditions is provided in Supplementary Table 5 a; b.
Fig. 6.
Association of the baseline mitochondrial bioenergetics with Vehicle Control and Dox treated metabolome. A. xMWAS network of the vehicle treated platelets revealed 4 metabolic communities. Community 1 (orange) has features predominantly associated with reserve capacity, while community 2 (blue) has metabolic features associated with maximal respiration. Community 3 (yellow) has features associated with both basal and ATP-linked respiration. Community 4 (green) has features are associated with non-mitochondrial respiration as well as proton leak B. xMWAS network of the Dox treated platelet metabolome and bioenergetics revealed 3 metabolic communities. Community 1 (yellow) associated with basal and ATP linked OCR. Community 2 (gray) associated with both non-mitochondrial and reserve capacity. Community 3 (blue) associated with maximal respiration. (|r| > 0.6 at p < 0.05). Molecular features are depicted as circles and bioenergetic parameters as squares. Red lines indicate positive associations, and blue lines indicate negative associations. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
For both vehicle and Dox-treated platelets, clusters associated with basal and maximal respiration were most central in the network structures (see Fig. 6A, C3 and C2; Fig. 6B, C1 and C3, respectively). Among the most obvious differences between vehicle control and Dox-treated network structures was the loss of the large cluster of metabolites associated with reserve capacity following treatment with Dox (Compare Cluster 1 in Fig. 6A with Cluster 2 in Fig. 6B). In the vehicle treated group (Fig. 6A), the metabolites that are correlating with non-mitochondrial (C4) do not correlate with Reserve Capacity whereas the Dox treatment generates stronger relationships (both positive and negative) between non-mitochondrial and Reserve Capacity (Fig. 6B). The strength of the interactions, the closeness and commonality of the metabolites in influencing non-mitochondrial and Reserve Capacity places them under one community which suggests a mechanistic association between the two bioenergetic parameters during stress. Although no direct correlation exist between non-mitochondrial and ATP linked in the vehicle treated group (Fig. 6A), the Dox treatments induces a realignment of the interactome map where common metabolites correlating with non-mitochondrial and ATP linked (Fig. 6B) exist. Another difference was that Dox treatment induced a greater number of metabolic associations with basal and maximal respiration. Proton-leak and non-mitochondria-linked respiration had few metabolic associations following either vehicle or Dox-treatment.
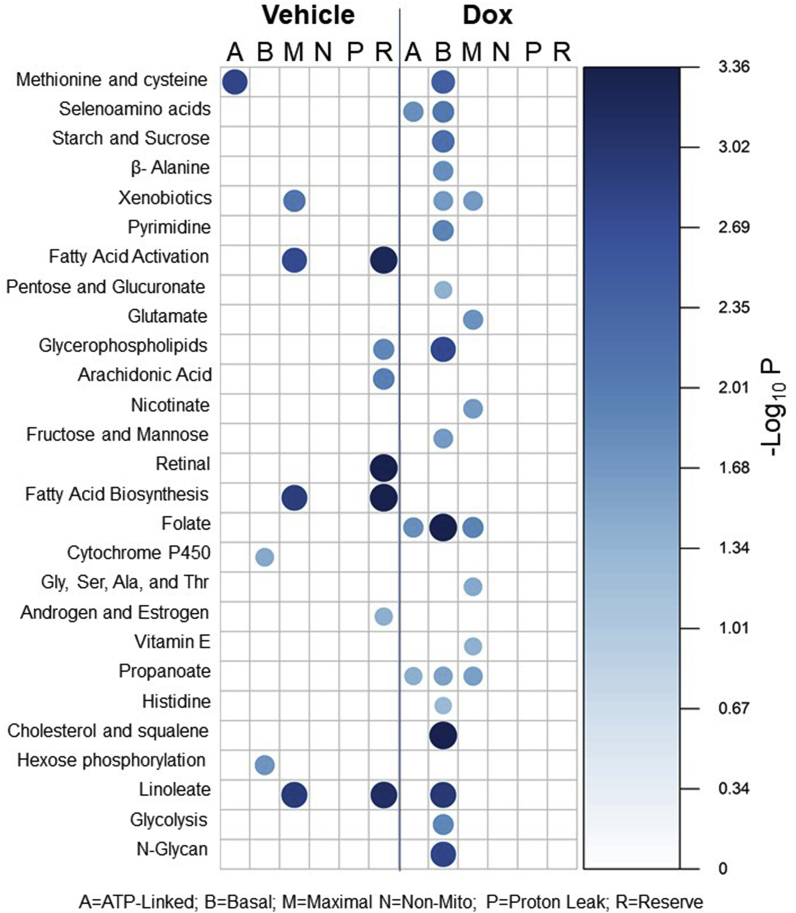
Because of the differences in the community network structure before and after Dox treatment, we sought to determine what metabolic pathways were associated with the mitochondrial bioenergetic parameters. Using pathway enrichment analysis for each bioenergetic parameter independently, we determined that a total of twenty-seven metabolic pathways were significantly associated with the bioenergetic parameters for both vehicle control and Dox treated networks. We then utilized bubble plots to visualize the results for the respective treatment conditions (Fig. 7). Both the size and color of each bubble represents pathway significance, based on the -Log10P values, between the metabolic pathways (left vertical-axis) and the respective bioenergetic parameter (horizontal-axis).
Fig. 7.
Metabolic pathways associated with mitochondrial bioenergetics in vehicle control and Dox treated samples. Bubble plot showing metabolic pathways associated with mitochondrial bioenergetics before and after Dox Treatment. Pathway analysis was completed using mummichog on each bioenergetic parameter independently. The size and color of the bubbles represents the pathway significance level based on -Log10P. Metabolic pathway analysis was performed using metabolic features associated with bioenergetics at |r|>0.6 and p < 0.05 using mummichog. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We determined that 11 of the 27 metabolic pathways identified were in the vehicle treated bioenergetics, and 7 of those pathways were associated with Reserve Capacity. These pathways included fatty acid, linoleate, glycerophospholipid, arachidonic acid, retinal, and sex-steroid metabolic pathways. We also found that ATP-linked respiration was linked with methionine and cysteine metabolism, while basal respiration was associated with cytochrome P450 and hexose metabolism. Maximal respiration was also associated with fatty acid, and linoleate metabolism as well as xenobiotic metabolism. These results show that fatty acid metabolism is highly associated with bioenergetics, which is supported by previous findings by our group in other studies using human platelets [69].
Treatment with Dox caused metabolic perturbation as many of the metabolic associations found in the vehicle treatment were lost. We identified that 20 of the 27 total metabolic pathways were associated with Dox treated bioenergetic conditions, and that 16 of these 20 were associated with basal respiration. These pathways included amino acids, selenoamino acids, sugars, xenobiotics, pyrimidine, pentose and glucoronate, glycerophospholipid, folate, propanoate, cholesterol and squalene, linoleate, N-glycans, and lastly glycolysis. Selenoamino acid, folate, and propanoate were also found to be associated with ATP-linked respiration. Xenobiotic, folate and propanoate metabolism were also found to be associated with maximal respiration; however, unique metabolic pathways for maximal included glutamate and glycine amino acids, nicotinate, and vitamin E metabolism.
4. Discussion
Metabolomics is emerging as an important tool for defining how individuals in human populations differ between each other and as a basis to predict outcomes for pathological processes or the response to treatments. For example, studies of the plasma metabolome in sepsis patients have identified lactate, alanine and lipid metabolites as markers related to clinical outcome [70,71]. Plasma metabolites can originate from different cells and organs, however, and this limits ability to relate metabolites in plasma to defined changes in bioenergetics or metabolism. In this respect, platelets offer an interesting platform because they provide a metabolically active cell fragment subject to the systemic stressors associated with pathology or environmental exposure in which both bioenergetics and metabolomics can be measured [28,29,64,72,73]. Furthermore, since platelets are without a nucleus, they have a limited capacity for repair and accumulate damage. Consequently, platelets can serve as a real-time sensor of metabolic stress.
The potential of platelets to act as a biomarker for disease or an indicator of the susceptibility to disease has been well recognized [30,73,74]. Indeed, recent data showed that in non-human primates brain glucose metabolism correlates with platelet energetics in the same individuals [74]. These findings are critically important in identifying biomarkers for the progression of Alzheimer's and other age-related diseases. Another important idea is that metabolic status sampled at any given time is determined by the interactions with our genetic profile and environment/diet. The pre-programmed metabolic plasticity can then impact the susceptibility to developing diseases afflicting the developing world such as diabetes [75]. Interestingly, the variation in basal bioenergetic parameters and the metabolome is a representation of an individual's variation in metabolic health and is defined by the bioenergetic-metabolite-interactome [29]. These concepts raises the important question of whether the healthy metabolome prior to exposure to a pathological or environmental stressor can be used to predict the severity of the response.
In the present study we addressed this question using a model system in platelets from healthy donors exposed to the widely used anti-cancer drug Dox. The baseline bioenergetic parameters from the MST showed the expected variation among individual donors (Fig. 1, Fig. 2). Interestingly, several of the parameters from the MST are well correlated among a population of healthy donors. This is important and supports our previous proposal that the MST represents an integrated metabolic program designed to meet both the resting and increased demands on the cell during normal physiology [29,67]. For example, we have shown that reserve capacity is essential in combatting the effects of oxidative stress [65,68,76]. The limits of the plasticity may then be an important factor in the capacity of an individual to combat stress and contribute to the variable outcomes in a population exposed to a pathogen, pathological processes or environmental stressors. It then follows that if we can assess this, we may be able to use metabolomics and bioenergetics as translational biomarkers for predicting the onset and progression of pathologies such as Alzheimer's disease or the dose limiting therapeutic levels for individual patients for drugs such as Dox. In this study we applied a widely used cancer chemotherapeutic to assess the impact on the bioenergetic-metabolite interactome in platelets and determine if individual responses could be predicted by basal metabolism.
The first series of experiments (Fig. 1, Fig. 2) show that Dox suppresses bioenergetics in platelets and this varies between individuals. The bioenergetic program in platelets can be revealed by a multi-variate statistical analysis as shown in Fig. 2. This analysis reveals that the capacity to generate ATP is retained however the dynamic range is suppressed by Dox and this is associated with a loss of Reserve Capacity. Overall these data show that platelets from different individuals respond differently to Dox exposure albeit through similar mechanisms which decrease the metabolic plasticity of the platelet. Next we tested the hypothesis that this individual variation will be related to the basal metabolome. This concept is based on our recent finding that the basal metabolome is a robust predictor of the platelet's response to metabolic stress [29].
The untargeted metabolomics of platelets included approximately 3000 mass spectral features which populated 48 major metabolic pathways expected to be present in platelets (Supplementary Table 1). Treatment of the platelets with Dox resulted in a profound inhibition of bioenergetic parameters (Fig. 1, Fig. 2) and enrichment of pathways related to metabolism (Fig. 3). Many of the pathways affected by Dox relate to platelet function (Fig. 3E). Not surprisingly, among the differentially expressed features some of Dox the metabolites were identified (Fig. 4 G-I). These data show that Dox is taken up and metabolized by platelets and that intra-platelet levels are strongly related to the suppression of bioenergetic function which in turn is related to the metabolism in the Dox pre-treated individual donors. We also found a number of metabolic features which were highly correlated with Dox-metabolites which could not be identified, highlighting the possibility that novel Dox-related metabolites, are being generated (see Table 3). This would need to be defined in a separate series of experiments using labeled Dox. These results also show that individuals metabolized Dox differently (Fig. 5 H, I), which could suggest differential activity of the enzymes, such as carbonyl reductase and glycosidase, which are responsible for detoxification of Dox [77,78]. Further studies analyzing the levels and activities of these enzymes in predicting drug-response to Dox may also be a useful approach in the future. Importantly, a lower level of Dox and Doxorubicinone was associated with an improved bioenergetic response.
Using the multi-omic approach by integrating mitochondrial function with the metabolome allows for broad determination of off-therapeutic effects of drugs. Using this approach allows us to determine which metabolic pathways contribute to mitochondrial function, and how those pathways are perturbed as a result of therapeutic intervention. In this instance of Dox treatment, changes in metabolic pathways linked to detoxification, such as the pentose and glucoronate, methionine and cysteine as well as xenobiotic metabolism were found to be associated with basal respiration, but either were not found to be associated at all, or were associated with other parameters under control conditions. Glycerophospholipids, which is involved in intermediary metabolism, was found to be associated with reserve capacity under vehicle treatment but changed to basal after treatment with Dox. Adaptive energetic changes to maintain platelet survival could explain the observation that the association of glycolysis with basal respiration were observed only after Dox treatment and not within the control.
The data from Fig. 1, Fig. 2 show that the basal metabolome and bioenergetics vary between individuals reflecting the metabolic programs. This is important in predicting the response to stress since it suggests that in utilizing metabolic plasticity to meet physiological demand the capacity of a protective pathway maybe different between one individual compared to another. For example, if mitochondrial function is inhibited by Dox then the extent to which fuel switching can occur will determine the overall toxicity of the same dose of compound. If one individual has a lower level of the enzymes needed to utilize fatty acids as a fuel than another then the impact of a toxic dose of Dox will be more severe. This can be revealed by the bioenergetic-metabolite-interactome maps shown in Fig. 6. From this data it is clear that the bioenergetic-metabolite interactome is profoundly altered by Dox, which is compounded by donor variation. Fig. 7 is a different representation of the data shown in Fig. 6 highlighting the importance of different metabolic pathways in the platelet basal state and in the presence of Dox. For example, fatty acid activation is much less prominent in the Dox-treated group resulting in glycolysis emerging as a new feature. This type of analysis is agnostic and reveals connections which are not easily placed within existing mechanistic paradigms and reveals new avenues for research. For example, why folate and cholesterol metabolism become more prominent after Dox treatment is not immediately obvious and will require further study. Importantly, many of the energetic pathways which have been found to be critical for maintaining Reserve Capacity are being redirected towards basal respiration, ultimately leading to platelet survival. Future studies performing time course analysis can determine the flux of these pathways after treatment with Dox to determine adaptive versus causal metabolic paradigm shifts.
Taken together, these data builds upon the concept of how the metabolome in the platelets is integrated with bioenergetics and how platelets are viable as metabolic sensors in the context of environmental exposure and determination of severity of the toxic response among individuals. This study also has significant implications for precision medicine, in particular related to pathological conditions such as cancer and the utility of integrated approaches as exemplified by the bioenergetic-metabolite interactome [79].
Acknowledgment
The authors would like to acknowledge ViLinh Tran and Ken Liu for their technical expertise with the mass spectrometer.
This work was supported by UAB-UCSD O'Brien Center P30 DK079337 (VDU), UAB Blue Sky program (VDU) and UAB Nathan Shock Center P30 AG050886 (VDU), NIEHS R01 ES023485 (DJ, YMG), R21 ES025632 (DJ, YMG), P30 ES019776 (DJ) and U2C ES026560 (DJ), and NIH S10 OD018006 (DJ). Platelet bioenergetics assays were funded by the UAB Mitochondrial Medicine Endowment Fund with contributions from Agilent and the Mitochondrial Foundation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101311.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.P F.G.E., Müller S.C. New agents in intravesical chemotherapy of superficial bladder cancer. Scand. J. Urol. Nephrol. 2005;39:108–116. doi: 10.1080/00365590510007676. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho C., Santos R.X., Cardoso S., Correia S., Oliveira P.J., Santos M.S., Moreira P.I. Doxorubicin: the good, the bad and the ugly effect. Curr. Med. Chem. 2009;16:3267–3285. doi: 10.2174/092986709788803312. [DOI] [PubMed] [Google Scholar]
- 3.Weiss R.B., Sarosy G., Clagett-Carr K., Russo M., Leyland-Jones B. Anthracycline analogs: the past, present, and future. Cancer Chemother. Pharmacol. 1986;18:185–197. doi: 10.1007/BF00273384. [DOI] [PubMed] [Google Scholar]
- 4.Arcamone F., Cassinelli G., Fantini G., Grein A., Orezzi P., Pol C., Spalla C. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol. Bioeng. 1969;11:1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee K., Zhang J., Honbo N., Karliner J.S. Doxorubicin cardiomyopathy. Cardiology. 2010;115:155–162. doi: 10.1159/000265166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doroshow J.H., Synold T.W., Somlo G., Akman S.A., Gajewski E. Oxidative DNA base modifications in peripheral blood mononuclear cells of patients treated with high-dose infusional doxorubicin. Blood. 2018;97 doi: 10.1182/blood.v97.9.2839. [DOI] [PubMed] [Google Scholar]
- 7.Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 8.Rook M., Lely A.T., Kramer A.B., van Goor H., Navis G. Individual differences in renal ACE activity in healthy rats predict susceptibility to adriamycin-induced renal damage. Nephrol. Dial. Transplant. 2005;20:59–64. doi: 10.1093/ndt/gfh579. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Wang Y.P., Tay Y.C., Harris D.C. Progressive adriamycin nephropathy in mice: sequence of histologic and immunohistochemical events. Kidney Int. 2000;58:1797–1804. doi: 10.1046/j.1523-1755.2000.00342.x. [DOI] [PubMed] [Google Scholar]
- 10.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 11.Ashley N., Poulton J. Mitochondrial DNA is a direct target of anti-cancer anthracycline drugs. Biochem. Biophys. Res. Commun. 2009;378:450–455. doi: 10.1016/j.bbrc.2008.11.059. [DOI] [PubMed] [Google Scholar]
- 12.Misiti F., Giardina B., Mordente A., Clementi M.E. The secondary alcohol and aglycone metabolites of doxorubicin alter metabolism of human erythrocytes. BDroaxzoilriaunb iJcoinu rmnael taobf oMlietedsi caanl da nedry Bthiorolocgyitcea el nReersgeya rmch. 2003;36:1643–1651. doi: 10.1590/s0100-879x2003001200005. [DOI] [PubMed] [Google Scholar]
- 13.Mordente A., Meucci E., Martorana G.E., Giardina B., Minotti G. Human heart cytosolic reductases and anthracycline cardiotoxicity. IUBMB Life. 2001;52:83–88. doi: 10.1080/15216540252774829. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H., Sarkar S., Scott L., Danelisen I., Trush M., Jia Z., Li Y.R. Doxorubicin redox biology: redox cycling, topoisomerase inhibition, and oxidative stress. Reactive Oxygen Species. 2016:189–198. doi: 10.20455/ros.2016.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraier D., Frigerio E., Pianezzola E., Strolin Benedetti M., Cassidy J., Vasey P. A sensitive procedure for the quantitation of free and N-(2-hydroxypropyl) methacrylamide polymer-bound doxorubicin (PK1) and some of its metabolites, 13-dihydrodoxorubicin, 13-dihydrodoxorubicinone and doxorubicinone, in human plasma and urine by reversed-phase HPLC with fluorimetric detection. J. Pharm. Biomed. Anal. 1995;13:625–633. doi: 10.1016/0731-7085(95)01301-z. [DOI] [PubMed] [Google Scholar]
- 16.Beijnen J.H., Meenhorst P.L., van Gijn R., Fromme M., Rosing H., Underberg W.J. HPLC determination of doxorubicin, doxorubicinol and four aglycone metabolites in plasma of AIDS patients. J. Pharm. Biomed. Anal. 1991;9:995–1002. doi: 10.1016/0731-7085(91)80036-9. [DOI] [PubMed] [Google Scholar]
- 17.Misiti F., Giardina B., Mordente A., Clementi M.E. The secondary alcohol and aglycone metabolites of doxorubicin alter metabolism of human erythrocytes. Braz. J. Med. Biol. Res. 2003;36:1643–1651. doi: 10.1590/s0100-879x2003001200005. [DOI] [PubMed] [Google Scholar]
- 18.Kim E.J., Lim K.M., Kim K.Y., Bae O.N., Noh J.Y., Chung S.M., Shin S., Yun Y.P., Chung J.H. Doxorubicin-induced platelet cytotoxicity: a new contributory factor for doxorubicin-mediated thrombocytopenia. J. Thromb. Haemost. 2009;7:1172–1183. doi: 10.1111/j.1538-7836.2009.03477.x. [DOI] [PubMed] [Google Scholar]
- 19.Wallace K.B. Dox induced cardiac mitochondrionopathy. Pharmacol. Toxicol. 2003;93 doi: 10.1034/j.1600-0773.2003.930301.x. [DOI] [PubMed] [Google Scholar]
- 20.Minotti G., Ronchi R., Salvatorelli E., Menna P., Cairo G. Doxorubicin irreversibly inactivates iron regulatory proteins 1 and 2 in cardiomyocytes: evidence for distinct metabolic pathways and implications for iron-mediated cardiotoxicity of antitumor therapy. Cancer Res. 2001;61:8422–8428. [PubMed] [Google Scholar]
- 21.Wang Z., Wang J., Xie R., Liu R., Lu Y. Mitochondria-derived reactive oxygen species play an important role in Doxorubicin-induced platelet apoptosis. Int. J. Mol. Sci. 2015;16:11087–11100. doi: 10.3390/ijms160511087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung L.K., Wang T.T. Differential effects of chemotherapeutic agents on the Bcl-2/Bax apoptosis pathway in human breast cancer cell line MCF-7. Breast Canc. Res. Treat. 1999;55:73–83. doi: 10.1023/a:1006190802590. [DOI] [PubMed] [Google Scholar]
- 23.Goormaghtigh E., Huart P., Praet M., Brasseur R., Ruysschaert J.M. Structure of the adriamycin-cardiolipin complex. Role in mitochondrial toxicity. Biophys. Chem. 1990;35:247–257. doi: 10.1016/0301-4622(90)80012-v. [DOI] [PubMed] [Google Scholar]
- 24.Hanson S.R., Slichter S.J. Platelet kinetics in patients with bone marrow hypoplasia: evidence for a fixed platelet requirement. Blood. 1985;66:1105–1109. [PubMed] [Google Scholar]
- [25.Rumbaut R.E., Thiagarajan P. 2010. Platelet-Vessel Wall Interactions in Hemostasis and Thrombosis. Platelet-Vessel Wall Interactions in Hemostasis and Thrombosis. San Rafael (CA) [PubMed] [Google Scholar]
- 26.Sol N., Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. 2017;36:263–272. doi: 10.1007/s10555-017-9674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi S., Chacko B., Kramer P.A., Sawada H., Johnson M.S., Zhi D., Marques M.B., Darley-Usmar V.M. Defining the effects of storage on platelet bioenergetics: the role of increased proton leak. Biochim. Biophys. Acta. 2015;1852:2525–2534. doi: 10.1016/j.bbadis.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zharikov S., Shiva S. Platelet mitochondrial function: from regulation of thrombosis to biomarker of disease. Biochem. Soc. Trans. 2013;41:118–123. doi: 10.1042/BST20120327. [DOI] [PubMed] [Google Scholar]
- 29.Chacko B.K., Smith M.R., Johnson M.S., Benavides G., Culp M.L., Pilli J., Shiva S., Uppal K., Go Y.M., Jones D.P., Darley-Usmar V.M. Mitochondria in precision medicine; linking bioenergetics and metabolomics in platelets. Redox Biol. 2019;22:101165. doi: 10.1016/j.redox.2019.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W., Cardenes N., Corey C., Erzurum S.C., Shiva S. Platelets from asthmatic individuals show less reliance on glycolysis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardenes N., Corey C., Geary L., Jain S., Zharikov S., Barge S., Novelli E.M., Shiva S. Platelet bioenergetic screen in sickle cell patients reveals mitochondrial complex V inhibition, which contributes to platelet activation. Blood. 2014;123:2864–2872. doi: 10.1182/blood-2013-09-529420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins H.M., Koppel S.J., Bothwell R., Mahnken J., Burns J.M., Swerdlow R.H. Platelet cytochrome oxidase and citrate synthase activities in APOE epsilon4 carrier and non-carrier Alzheimer's disease patients. Redox Biol. 2017;12:828–832. doi: 10.1016/j.redox.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Go Y.M., Kim C.W., Walker D.I., Kang D.W., Kumar S., Orr M., Uppal K., Quyyumi A.A., Jo H., Jones D.P. Disturbed flow induces systemic changes in metabolites in mouse plasma: a metabolomics study using ApoE(-)/(-) mice with partial carotid ligation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;308:R62–R72. doi: 10.1152/ajpregu.00278.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soltow Q.A., Strobel F.H., Mansfield K.G., Wachtman L., Park Y., Jones D.P. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chacko B.K., Kramer P.A., Ravi S., Johnson M.S., Hardy R.W., Ballinger S.W., Darley-Usmar V.M. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Laboratory investigation; a journal of technical methods and pathology. 2013;93:690–700. doi: 10.1038/labinvest.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer P.A., Chacko B.K., Ravi S., Johnson M.S., Mitchell T., Darley-Usmar V.M. Bioenergetics and the oxidative burst: protocols for the isolation and evaluation of human leukocytes and platelets. J. Vis. Exp. 2014 doi: 10.3791/51301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walkowiak B., Kesy A., Michalec L. Microplate reader--a convenient tool in studies of blood coagulation. Thromb. Res. 1997;87:95–103. doi: 10.1016/s0049-3848(97)00108-4. [DOI] [PubMed] [Google Scholar]
- 38.Bednar B., Condra C., Gould R.J., Connolly T.M. Platelet aggregation monitored in a 96 well microplate reader is useful for evaluation of platelet agonists and antagonists. Thromb. Res. 1995;77:453–463. doi: 10.1016/0049-3848(95)93881-y. [DOI] [PubMed] [Google Scholar]
- 39.Chacko B.K., Kramer P.A., Ravi S., Johnson M.S., Hardy R.W., Ballinger S.W., Darley-Usmar V.M. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Laboratory investigation; a journal of technical methods and pathology. 2013;93:690–700. doi: 10.1038/labinvest.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Go Y.M., Uppal K., Walker D.I., Tran V., Dury L., Strobel F.H., Baubichon-Cortay H., Pennell K.D., Roede J.R., Jones D.P. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol. Biol. 2014;1198:43–73. doi: 10.1007/978-1-4939-1258-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Go Y.M., Fernandes J., Hu X., Uppal K., Jones D.P. Mitochondrial network responses in oxidative physiology and disease. Free Radic. Biol. Med. 2018;116:31–40. doi: 10.1016/j.freeradbiomed.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X., Chandler J.D., Park S., Liu K., Fernandes J., Orr M., Smith M.R., Ma C., Kang S.M., Uppal K., Jones D.P., Go Y.M. Low-dose cadmium disrupts mitochondrial citric acid cycle and lipid metabolism in mouse lung. Free Radic. Biol. Med. 2018;131:209–217. doi: 10.1016/j.freeradbiomed.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker D., Pennell K., Jones D. 2018. The Mitochondrial Exposome; pp. 613–637. [Google Scholar]
- 45.Go Y.M., Walker D.I., Liang Y., Uppal K., Soltow Q.A., Tran V., Strobel F., Quyyumi A.A., Ziegler T.R., Pennell K.D., Miller G.W., Jones D.P. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci. 2015;148:531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alves G., Yu Y.K. Improving peptide identification sensitivity in shotgun proteomics by stratification of search space. J. Proteome Res. 2013;12:2571–2581. doi: 10.1021/pr301139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uppal K., Soltow Q.A., Strobel F.H., Pittard W.S., Gernert K.M., Yu T., Jones D.P. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinf. 2013;14(15) doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 49.Patel R.M., Roback J.D., Uppal K., Yu T., Jones D.P., Josephson C.D. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion. 2015;55:544–552. doi: 10.1111/trf.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth G.K., Michaud J., Scott H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 51.Benjamin Y., Hockberg Y. Controlling the false discovery rate: a practical and poweful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 52.Li S., Park Y., Duraisingham S., Strobel F.H., Khan N., Soltow Q.A., Jones D.P., Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uppal K., Walker D.I., Liu K., Li S., Go Y.M., Jones D.P. Computational metabolomics: a framework for the million metabolome. Chem. Res. Toxicol. 2016;29:1956–1975. doi: 10.1021/acs.chemrestox.6b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uppal K., Walker D.I., Jones D.P. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017;89:1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schymanski E.L., Jeon J., Gulde R., Fenner K., Ruff M., Singer H.P., Hollender J. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 2014;48:2097–2098. doi: 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 56.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S., Fung C., Nikolai L., Lewis M., Coutouly M.A., Forsythe I., Tang P., Shrivastava S., Jeroncic K., Stothard P., Amegbey G., Block D., Hau D.D., Wagner J., Miniaci J., Clements M., Gebremedhin M., Guo N., Zhang Y., Duggan G.E., Macinnis G.D., Weljie A.M., Dowlatabadi R., Bamforth F., Clive D., Greiner R., Li L., Marrie T., Sykes B.D., Vogel H.J., Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart D., Arndt D., Pon A., Sajed T., Guo A.C., Djoumbou Y., Knox C., Wilson M., Liang Y., Grant J., Liu Y., Goldansaz S.A., Rappaport S.M. T3DB: the toxic exposome database. Nucleic Acids Res. 2015;43:D928–D934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fahy E., Sud M., Cotter D., Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uppal K., Ma C., Go Y.M., Jones D.P., Wren J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. 2018;34:701–702. doi: 10.1093/bioinformatics/btx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chun H., Keles S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J. R. Stat. Ser. Soc. B Stat. Methodol. 2010;72:3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008;2008:P10008. [Google Scholar]
- 63.Wei T., S V. 2017. R Package "corrplot": Visualization of a Correlation Matrix.https://github.com/taiyun/corrplot Version 0.84. [Google Scholar]
- 64.Ravi S., Chacko B., Sawada H., Kramer P.A., Johnson M.S., Benavides G.A., O'Donnell V., Marques M.B., Darley-Usmar V.M. Metabolic plasticity in resting and thrombin activated platelets. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L.Y., Chatham J.C., Hill B.G., Zhang J.H., Landar A., Darley-Usmar V.M. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mross K., Maessen P., van der Vijgh W.J., Gall H., Boven E., Pinedo H.M. Pharmacokinetics and metabolism of epidoxorubicin and doxorubicin in humans. J. Clin. Oncol. 1988;6:517–526. doi: 10.1200/JCO.1988.6.3.517. [DOI] [PubMed] [Google Scholar]
- 67.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell'Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dranka B.P., Hill B.G., Darley-Usmar V.M. Mitochondrial reserve capacity in endothelial cells: the impact of nitric oxide and reactive oxygen species. Free Radic. Biol. Med. 2010;48:905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slatter D.A., Aldrovandi M., O'Connor A., Allen S.M., Brasher C.J., Murphy R.C., Mecklemann S., Ravi S., Darley-Usmar V., O'Donnell V.B. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metabol. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evangelatos N., Bauer P., Reumann M., Satyamoorthy K., Lehrach H., Brand A. Metabolomics in sepsis and its impact on public health. Public Health Genomics. 2017;20:274–285. doi: 10.1159/000486362. [DOI] [PubMed] [Google Scholar]
- 71.Cambiaghi A., Pinto B.B., Brunelli L., Falcetta F., Aletti F., Bendjelid K., Pastorelli R., Ferrario M. Characterization of a metabolomic profile associated with responsiveness to therapy in the acute phase of septic shock. Sci. Rep. 2017;7:9748. doi: 10.1038/s41598-017-09619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gardinassi L.G., Arevalo-Herrera M., Herrera S., Cordy R.J., Tran V., Smith M.R., Johnson M.S., Chacko B., Liu K.H., Darley-Usmar V.M., Go Y.M., Ma H.C., Jones D.P., Galinski M.R., Li S. Integrative metabolomics and transcriptomics signatures of clinical tolerance to Plasmodium vivax reveal activation of innate cell immunity and T cell signaling. Redox Biol. 2018;17:158–170. doi: 10.1016/j.redox.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kramer P.A., Ravi S., Chacko B., Johnson M.S., Darley-Usmar V.M. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol. 2014;2:206–210. doi: 10.1016/j.redox.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tyrrell D.J., Bharadwaj M.S., Jorgensen M.J., Register T.C., Shively C., Andrews R.N., Neth B., Dirk Keene C., Mintz A., Craft S., Molina A.J.A. Blood-based bioenergetic profiling reflects differences in brain bioenergetics and metabolism. Oxid Med Cell Longev. 2017;2017:7317251. doi: 10.1155/2017/7317251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rolo A.P., Palmeira C.M. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol. Appl. Pharmacol. 2006;212:167–178. doi: 10.1016/j.taap.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 76.Chacko B.K., Zhi D., Darley-Usmar V.M., Mitchell T. The Bioenergetic Health Index is a sensitive measure of oxidative stress in human monocytes. Redox Biol. 2016;8:43–50. doi: 10.1016/j.redox.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kassner N., Huse K., Martin H.J., Godtel-Armbrust U., Metzger A., Meineke I., Brockmoller J., Klein K., Zanger U.M., Maser E., Wojnowski L. Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab. Dispos. 2008;36:2113–2120. doi: 10.1124/dmd.108.022251. [DOI] [PubMed] [Google Scholar]
- 78.Gutierrez P.L., Gee M.V., Bachur N.R. Kinetics of anthracycline antibiotic free radical formation and reductive glycosidase activity. Arch. Biochem. Biophys. 1983;223:68–75. doi: 10.1016/0003-9861(83)90572-6. [DOI] [PubMed] [Google Scholar]
- 79.Hill B.G., Shiva Sruti, Ballinger Scott, Zhang Jianhua, Darley-Usmar, Victor M. Bioenergetics and translational metabolism: implications for genetics, physiology and precision medicine. Biol. Chem. 2019 doi: 10.1515/hsz-2019-0268. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.