Graphical abstract
Keywords: Coumarin, Bergaptol O-methyltransferase, Rational design, Depigmentation disorder
Highlights
-
•
The PpBMT crystal structure identified key amino acids in bergaptol conversion.
-
•
Protein engineering was used to obtain PpBMT mutants with improved activity.
-
•
A high-activity mutant was used to produce bergapten for pharmacological analysis.
Abstract
Bergapten has long been used in combination with ultraviolet A irradiation to treat depigmentation disorder. However, extremely low bergapten contents in plants and difficulties in synthesizing bergapten have limited its application. Here, we developed an alternative bergapten-production method. We first determined the crystal structures of bergaptol O-methyltransferase from Peucedanum praeruptorum (PpBMT) and the ternary PpBMT–S-adenosyl-L-homocysteine (SAH)–bergaptol complex to identify key residues involved in bergaptol binding. Then, structure-based protein engineering was performed to obtain PpBMT mutants with improved catalytic activity towards bergaptol. Subsequently, a high-activity mutant was used to produce bergapten for pharmacological-activity analysis. Key PpBMT amino acids involved in bergaptol binding and substrate specificity were identified, such as Asp226, Asp246, Ser265, and Val320. Site-directed mutagenesis and biochemical analysis revealed that the V320I mutant efficiently transformed bergaptol to produce bergapten. Pharmacological-activity analysis indicated that bergapten positively affected hair pigmentation in mice and improved pigmentation levels in zebrafish embryos. This report provides the first description of the catalytic mechanism of coumarins-specific O-methyltransferase. The high-activity V320I mutant protein could be used in metabolic engineering to produce bergapten in order to treat depigmentation disorder. This structure–function study provides an alternative synthesis method and important advances for treating depigmentation disorders.
Introduction
Melanin is produced by melanocytes through a rate-limiting tyrosinase-catalysed reaction. Abnormal melanin accumulation often causes dermatological problems, such as age spots and vitiligo [1], [2]. Hence, the regulation of melanogenesis is a key method in treating depigmentation disorders, and numerous candidate agents targeting signalling pathways involved in melanin synthesis have been developed [3], [4], [5]. Of all compounds involved in treating depigmentation disorders, linear furocoumarins (such as psoralen and bergapten) are widely used in clinical trials (Fig. S1) [6], [7], [8]. However, their sources are mainly limited to plant extract and low abundance and season- or region-dependent sourcing limits their widespread application [6]. Using solvents for extraction or excavation raises environmental concerns [9], [10].
Metabolic engineering of microorganisms or plants shows promise for addressing these problems, and many efforts have succeeded [11], [12], [13]. Nevertheless, few reports have described metabolic engineering for coumarin production because their biosynthetic mechanisms are largely unsolved [14], [15]. Therefore, it is urgent to clarify the catalytic mechanisms of the proteins involved in coumarin biosynthesis to improve the catalytic activity to enhance the yield of target coumarins through metabolic engineering. In addition, proteins with high catalytic activity can also be used as candidate enzymes in synthetic biology to complete a target pathway [16]. Protein engineering is an industrially promising method for tailoring biocatalysts, and generating enzymes with good activity to produce target compounds is also desirable in microorganisms [17], [18], [19], [20]. However, the success of methods depends on accurate knowledge of the catalytic mechanisms and key amino acid residues mediating substrate binding.
To develop an alternative method for bergapten production and to generate a candidate protein for metabolic engineering, we previously cloned and functionally authenticated the bergaptol-specific O-methyltransferase (OMT) from P. praeruptorum (PpBMT) [21]. However, the activity of PpBMT needs to be improved [21]. Herein, the crystal structures of apo-PpBMT and a ternary PpBMT–SAH–bergaptol complex were first determined by X-ray diffraction with resolution of 2.0 and 2.2 Å, respectively. Then, computer-aided rational design was employed to improve the activity of PpBMT. A candidate mutant (V320I) with high catalytic efficiency was obtained for bergapten production. The produced bergapten positively affected mouse hair pigmentation and improve pigmentation in zebrafish embryos. The work provides a deep understanding of the substrate preferences and catalytic mechanism of PpBMT-mediated coumarin O-methylation, and also lays the foundation for metabolic engineering to increase the potential applications of coumarins.
Materials and methods
Protein expression and purification
PpBMT complementary DNA was ligated into the pGEX-6P-1 plasmid to generate pGEX-6P-PpBMT (Table S1) [21]. Subsequently, the recombinant plasmid was transformed into E. coli BL21 (DE3) for protein expression, according to our previous method [21]. The protein was attached to glutathione S-transferase-conjugated affinity resin and released overnight into lysis buffer via on-bead 3C protease. Finally, the protein was concentrated to 20 mg/ml for crystallization and other assays.
Crystallization and structure determinations
To obtain the PpBMT–SAH–bergaptol (BGO) ternary complex, we mixed PpBMT, SAH, and BGO at a molar ratio of 1: 1.2: 1.2. The crystals were flash-frozen in liquid nitrogen for diffraction in the Shanghai Synchrotron Radiation Facility on beamline BL19U1. The dataset was first processed with the HKL-3000 program [22] and further processed with programs from the CCP4 suite [23]. The collected data and structural-refinement statistics are summarized in Table 1. The apo structure was solved by molecular replacement with chain A of Protein Data Bank (PDB) structure 1KYZ as a search model using the PHASER program, and the ternary-complex structure was solved using Autosol in PHENIX [24], [25]. The structure was manually and iteratively refined with PHENIX and COOT [25], [26]. All structural representations were prepared with PyMOL (1.7.4, http://www.pymol.org). The PpBMT structures were deposited in the PDB (www.rcsb.org/pdb/) under accession numbers 5XG6 and 5XOH.
Table 1.
Summary of crystallographyic data collection and refinement statistics for PpBMT-apo and ternary complex.
| 5XG6 (Native) | 5XOH (SeMet-SAD) | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P3121 |
| Cell dimensions□□ | ||
| a, b, c (Å) | 128.23, 76.11, 84 | 59.44, 59.44, 172.50 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90, 90, 120 |
| Resolution (Å) | 2.00 | 2.20 |
| Rmerge | 0.128 (0.362)* | 0.134 (0.76)* |
| I/σI | 19.9 (5.0)* | 29.8 (2.07)* |
| Completeness (%) | 98.7 (98.8)* | 99.8 (99.76)* |
| Redundancy | 12.1 (7.2)* | 9.8 (6.9)* |
| Refinement | ||
| Resolution (Å) | 49.03–2.00 | 33.06–2.20 |
| No. of reflections | 55,442 | 18,775 |
| Rwork/Rfree (%) | 17.1/19.9 | 19.7/24.9 |
| No. of atoms | ||
| Protein | 5406 | 2665 |
| Water | 540 | 59 |
| Ligand | 0 | 41 |
| Avg.B-factor | 32.38 | 54.23 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | 0.007 |
| Bond angles(°) | 1.27 | 1.03 |
| Ramanchandran plot | ||
| Favored region | 99 | 98.6 |
| Allowed region | 0.9 | 1.4 |
| Outlier region | 0.1 | 0 |
Highest resolution shell is shown in parenthesis.
Enzymatic activity and high-performance liquid chromatography (HPLC) analysis
All enzymatic-activity assays were performed in triplicate as described previously [21]. To investigate the protein stability, protein activities were determined after storage for 4 weeks at different temperatures (−80 °C, −20 °C, 4 °C, and 25 °C). Experiments were also conducted in different pH values (2.5–11.5), temperatures (15–60 °C), and with different ions (1 or 0.1 mM Fe2+, Fe3+, Ca2+, Mg2+, Zn2+, Cu2+, Mn2+, Co2+, Ag+, or Ni2+) [27]. HPLC analysis were conducted as described previously [21].
Bioinformatics analysis, docking, and computational and structure-based protein design
For molecular docking, we employed the Schrödinger (Schrödinger, Inc., New York, USA) and Molecular Operating Environment (Chemical Computing Group, Inc., Montreal, Canada) programs. The experiments were conducted after preparing a target protein with the protein-preparation wizard program in Schrödinger [28], [29], [30]. To improve the catalytic activity of BMT, the ‘calculate mutation energy (binding) protocol’ in Discovery Studio 4.1 was used to evaluate the effects of mutations on the bergaptol-binding affinity of BMT. The protocol performed combinatorial amino acid-scanning mutagenesis on selected amino acid residues neighbouring bergaptol by mutating them to other natural amino acids. The effect of each mutation on the binding affinity (mutation energy, ΔΔGmut) was calculated as the difference between the binding free energy (ΔΔGbind) in the mutant and wild-type proteins:
ΔΔGbind was defined as the difference between the free energy of the complex and that of the unbound state. All energy terms were calculated using CHARMm, and the electrostatic energy was calculated using a Generalized Born implicit-solvent model. The total energy was calculated as an empirical weighted sum of van der Waals (VDW) interactions, electrostatic interactions, an entropy contribution (-TSsc) related to side-chain mobility changes, and a non-polar, surface dependent, contribution to solvation energy. Candidate amino acids were selected for improving catalytic activity based on the index of a low mutation energy, which indicates a higher binding affinity. Total free-energy differences between the wild-type and mutated structures were calculated as a weighted sum of the VDW, electrostatic, entropic, and non-polar terms (Tables S2–S4). Finally, site-directed mutagenesis was conducted using a polymerase chain reaction-based method with KOD-plus-neo polymerase and primers shown in Table S1. The mutant plasmids were transformed into E. coli for protein expression, subsequent purification, and enzyme-activity analysis.
Design of animal experiments and assessment of hair pigmentation
A wax/rosin mixture was applied to induce highly synchronized hair growth as described previously [31]. A model of vitiligo was generated with C57BL/6 mice by daily topical application of 2 mL of 2% hydroquinone (HQ) to shaved areas (2 × 2 cm) of the dorsal skin, for 12 days. To determine whether drugs could induce pigmentation, the mice were randomly divided into three groups: a control group (without treatment), a model (HQ treatment), and a bergapten group (bergapten delivered transdermally for 12 days). From days 3 to 15, the mice were treated with dermatology drugs for one hair-growth cycle. Topical HQ and bergapten were applied using oil-in-water emulsion creams that contained 61.3% water, 8.0% stearic acid, 8% white Vaseline, 7.0% glycerol, 6.0% octadecanol, 5.0% propylene glycol, 2.0% azone, 1.6% trolamine, 1.0% sodium dodecyl sulphate, and 0.1% ethylparaben. The HQ and bergapten concentrations were 2% and 1% (w/w), respectively. The mice were photographed with a digital camera (Canon, Japan) once daily after depilation to assess the hair cycle. Hematoxylin and eosin (HE) staining was used to determine the stage of hair pigmentation based on the morphology of the dermal papilla and sebaceous glands. In addition, the melanin granules in HE-stained tissue samples were visualised histochemically.
Immunofluorescence analysis
Immunofluorescence was performed as described below. Sections were dew axed, rehydrated, and immersed in citric acid buffer for antigen retrieval. After washing with 0.01 M potassium phosphate buffer (PBS), the specimens were treated with PBS containing Tween 20 (PBST) for 15 min at room temperature and then blocked for 1 h in blocking buffer (5% goat serum, 0.1% bovine serum albumin, and 0.1% Triton X-100). Thereafter, the specimens were incubated separately with each primary antibody at 4 °C for 24 h. After being washed with 0.01 M PBS, the specimens were incubated with a secondary antibody in the dark inside a cassette at 37 °C for 2 h. The specimens were washed with 0.01 M PBS and mounted using DAPI (Invitrogen, catalogue #D3571) and photographed with a DM400B fluorescence microscope (Leica, Wetzlar, Germany). The semi-quantitative data obtained using the immunofluorescence images for the control, HQ model, and bergapten groups are shown in Fig. S2.
Zebrafish maintenance and chemical treatments
Wild-type zebrafish were maintained according to the guides for the laboratory use of zebrafish in a circulating aquaculture system. Zebrafish embryos were incubated at 28.5 °C as described by Kimmel et al. [32]. For chemical treatments, bergapten was dissolved in dimethyl sulfoxide (DMSO) to prepare stock solutions and then diluted with fresh fish water to 10, 20, or 40 μM. The reagent 1-phenyl 2-thiourea (PTU; Sigma-Aldrich, St. Louis, MO, USA) was dissolved in water to prepare stock solutions and then diluted with fresh fish water to 200 μM for all treatments. The experimental design is shown in Fig. S3.
Cell culture, chemical treatments, and assays of melanin content and cellular tyrosinase (TYR) activity
The B16F10 melanoma cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (v/v) heat-inactivated foetal bovine serum. The cells were cultured in monolayers in a humidified incubator at 37 °C with 5% CO2. For chemical treatments, bergapten, psoralen, and the bergapten produced by the purified PpBMT V320I protein (synthesis) were dissolved in DMSO and then diluted in DMEM to a final concentration of 1, 10, or 100 μM. The melanin content and cellular TYR activity were measured as described previously [33], with minor modifications.
Statistical analysis and graph preparation
Unless indicated, three replicates were used to obtain the data, and the data are presented as the mean ± standard deviation (SD) for triplicate experiments. Statistical analysis of the results was performed using one-way analysis of variance with Tukey’s correction for multiple comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate the statistical significance.
Results and discussion
BMT as a good candidate for bergapten production
Owing to efficient separation/purification techniques and advances in pharmacological-activity research, new compounds are continually isolated and identified, and new pharmacological activities are frequently demonstrated [34]. However, low contents in medicinal plants have limited widespread applications of these compounds [9], [10]. Hence, improving their contents in plants or alternatively producing them in engineered microorganisms is rewarding. Previously, we cloned and functionally authenticated PpBMT [21]. Extreme stability, high solubility, and good temperature and pH tolerance made PpBMT an ideal enzyme for industrial bergapten production by metabolic engineering (Fig. S4) [35], [36]. However, the catalytic activity of the target protein needs to be improved because bergaptol could not be completely transformed by PpBMT, a phenomenon occurring with Ammi majus BMT (AmBMT) and Angelica dahurica BMT (AdBMT) as well [35], [37]. Structure-based rational design has helped improve the quality of enzymes [17], [18], [19], [20]. However, no corresponding structure involved in coumarin biosynthesis has been solved. To improve the enzymatic activity of PpBMT via protein engineering, the crystal structure of PpBMT needed to be solved.
Overall structure of PpBMT
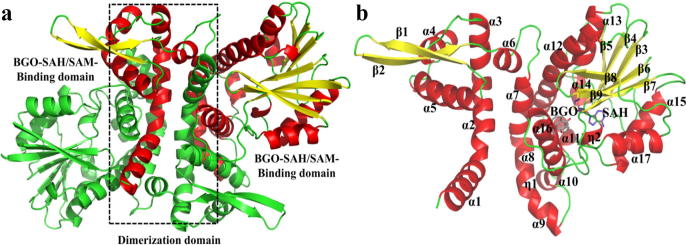
The crystal structures of apo-PpBMT and the ternary PpBMT–SAH–BGO complex were first determined by X-ray diffraction at a resolution of 2.0 and 2.2 Å, respectively (Fig. S5, Table 1). The PpBMT crystal structure contained 17 α-helices and 9 β-sheets and exhibited a central symmetric dimer. The N-terminal domain involved in dimerization contained 10 α-helices (α1–α9 and α16) and 2 β-sheets (β1 and β2), whereas the C-terminal catalytic domain contributes to substrate binding and enzyme activity with more β sheets (β3–β9) (Fig. 1a and Fig. 1b). Primary-sequence analysis and structural comparisons with other plant OMTs demonstrated that PpBMT is a type-I plant OMT resembling the Medicago sativa COMT (MsCOMT), which has a spacious and versatile binding pocket constructed with more flexible loops, although a rigid α14 helix and a four-end α15 helix was present in PpBMT (Figs. S6 and S7) [24]. However, the chemical structures of the OMT substrates differed to a large extent, which is determined by the shape-selectivity of the substrate pockets and surrounding amino acids (Fig. S8).
Fig. 1.
Overall PpBMT structure. The PpBMT dimer is shown with the monomers displayed in red or green. The SAH/BGO-binding site is located in the C-terminal region, and the domain involved in dimerization is located in the N-terminal region (a). The secondary structure of one monomer of the PpBMT–SAH–BGO complex is displayed in the figure with different colours (α-helixes, red; β-strands, yellow; loops, green), the SAH molecule in represented with purple sticks, and the BGO molecule is represented in grey (b).
SAH/SAM- and bergaptol-binding sites
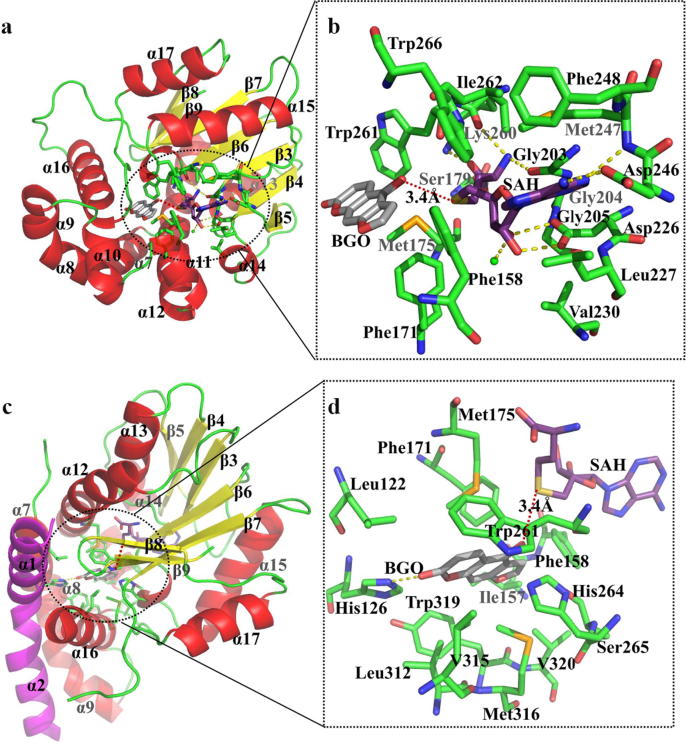
Thereafter, we analysed the SAH/SAM-binding site based on the overall structure of the BMT–SAH–BGO ternary complex (Fig. 2a, b). Most importantly, the carbonyl groups of Gly203 (motif I) and Lys260 (motif IV) hydrogen bonded with the terminal amino group of SAH, the hydroxyl group of Ser179 from α12 interacted with the homocysteine moiety of SAH, and the side chain of Asp226 (motif II) interacted with the SAH ribose hydroxyl groups (Fig. 2a, b). Considering that SAM is a common cofactor for all OMTs, which are known to possess a highly conserved SAM-binding region (Fig. S6) [24], the data discussed below are mainly focused on the residues relevant to substrate (bergaptol) specificity.
Fig. 2.
The binding sites of PpBMT. A cartoon diagram of the PpBMT in complex with SAH (a) and its local enlarged images (b) is shown. A cartoon diagram of PpBMT in complex with bergaptol (c) and its local enlarged images (d) is also shown. The secondary structures (α-helixes and β-sheets) and the important amino acid residues contributing atoms within 4 Å of SAH and BGO are represented with green sticks. SAH and BGO are shown in purple stick and grey, respectively. The yellow dotted line is a hydrogen bond and the red dotted line displays the distances between the 5-hydroxyl groups of bergaptol and SAH.
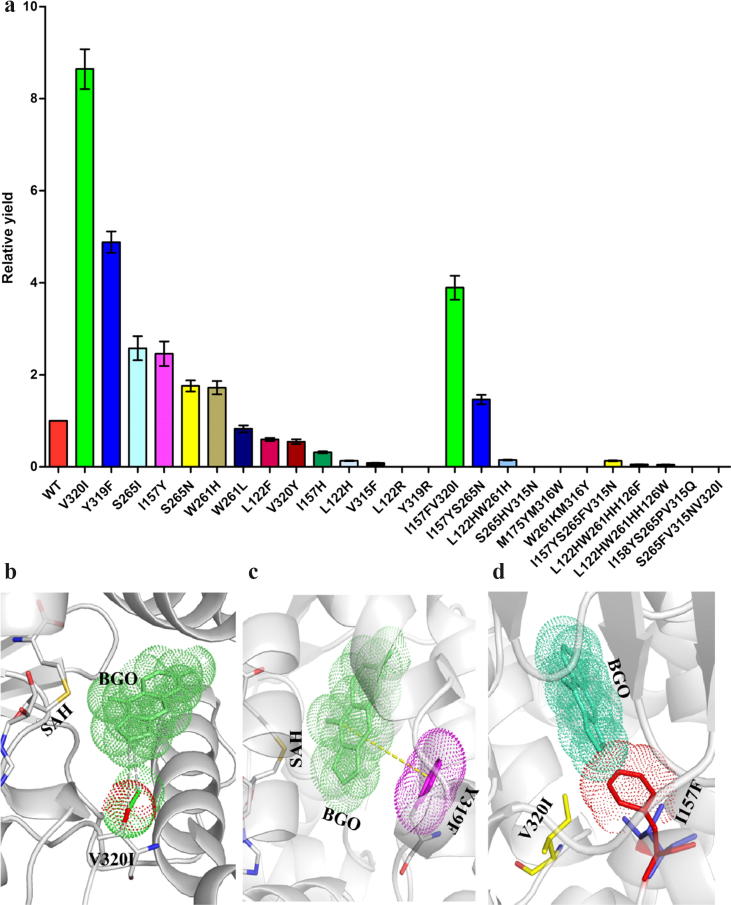
The bergaptol-binding site was surrounded by 8 α-helices. α1, α2 in one monomer and α7, α8, α10–12, α15 in another monomer interacted to establish a specific cavity for bergaptol entrance (Fig. 1a, b). The 5-hydroxyl group of bergaptol formed two hydrogen bonds with the imidazole nitrogen atom of His264 and the carbonyl oxygen of Trp261, respectively. Additionally, bergaptol was fixed by two hydrogen bonds with His126 to hold the 5-hydroxyl group close to the methyl group of SAM (Fig. 2c, 2d). In addition, aromatic amino acids (Phe158, Phe171, Trp261, and Tyr319) and aliphatic amino acids (Leu122, Ile157, Met175, Leu312, Val315, Met316, and Val320) were also involved in VDW forces and π–π interactions with bergaptol (Fig. 2d). In conclusion, the hydrogen bonds and hydrophobic effects constructed the active centre of PpBMT, which played a crucial role in its catalytic activity. When these important amino acids were mutated to arginine or alanine, nearly all mutants showed abolished activity, illuminating their significance in designing proteins with improved catalytic activity (Fig. 3a).
Fig. 3.
Catalytic activity of PpBMT and its mutant variants generated by computer-aided protein design. The data shown represent the mean ± SD of three replicates and the fold-changes relative to the control (a). The bergaptol-docking results with mutants V320I (b), Y319F (c) and I157FV320I (d) are shown.
Rational design, mutagenesis, and catalytic activities
Considering that PpBMT is highly temperature and pH tolerant, and has higher stability and solubility than other BMTs [21], [35], [36], we mainly focused on improving its catalytic activity based on identifying important amino acids. Because no high-throughput method has been developed for selecting a variant protein with improved activity, structure-based rational design was employed for protein design, as done previously [17], [18], [19], [20]. We first mutated 10 important amino acids using computer-aided protein design with Discovery Studio 4.1. By comparing the mutation energies (Table S2), amino acid mutations with the potential for improving protein activities, such as V320I, Y319F, S265I, I157Y, and L122R (with low mutation energies of −0.2, 0.02, −0.11, −0.26, and −0.41, respectively), were selected for site-specific mutagenesis and activity analysis. The catalytic activities of mutants such as V320I, Y319F, S265I, and I157Y increased significantly (Fig. 3a and S9). However, the activities of L122R, H126R, and I157R, among other mutants, decreased to some extent (Fig. 3a). We then employed site-specific mutagenesis to design double and triple mutants, based on the single mutants with improved activities and the mutation energies (Tables S3 and S4). Unfortunately, no candidate mutants with further improved activity were obtained. A stronger hydrophobic interaction was formed between Ile320 and bergaptol (Fig. 3b), which may account for the improved activity. In addition, the relatively longer side chain of isoleucine (compared to that of valine) may also enhance its performance. BGO was trapped in a hydrophobic pocket in wild type BMT, while Y319F mutant contributed a more hydrophobic side chain in the pocket and reduced its steric hindrance (Fig. 3c). Hence, Y319F mutant could also significantly enhance the affinity activity of BMT to bergaptol (Fig. 3a). This phenomenon can be proved by another mutation, Y319R. As expect, when Y319 was substituted by arginine (a longer polar side chain amino acid), the activity was totally abolished (Fig. 3a). While, when we further mutated Ile157 to Phe157 in V320I mutant (Fig. 3d), the π–π interaction and steric hindrance caused by Phe157 pushed the BGO away and resulted in a poor orientation for catalysis in the I157FV320I double mutant (Fig. 3a). Hence, V320I was ultimately selected as the candidate high-catalytic activity mutant.
Producing bergapten using the V320I mutant to treat depigmentation disorders
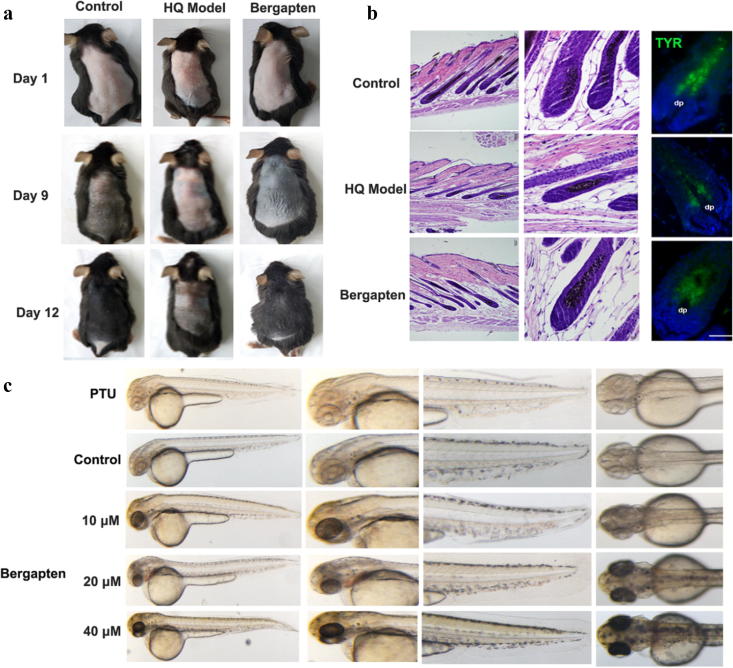
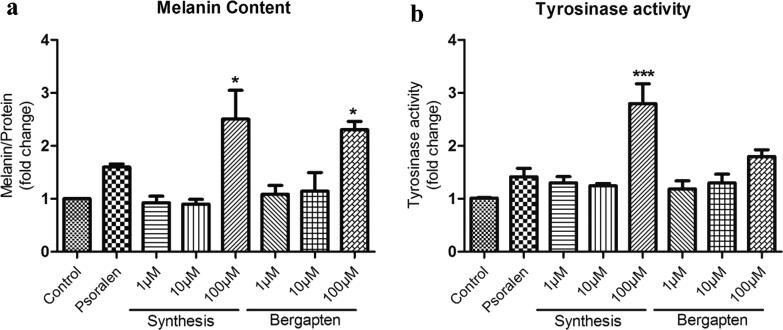
Considering the low bergapten content in medicinal plants, producing the target compound through plant-metabolic engineering or microbial-cell factories is potentially a useful alternative method. Hence, we used both engineered microorganisms with a V320I mutation and the purified V320I protein to produce bergapten. However, whether bergapten produced by microorganisms or the purified V320I protein has the same function as plant-based bergapten remains unsolved. Because bergapten has been used in clinical trials for treating depigmentation disorders for a long time, we tested bergapten for depigmentation disorder treatment in animals [6]. On days 9 and 12 after depilation and treatment, HQ-induced vitiligo mice showed obvious whitening of their dorsal skin (Fig. 4a). In contrast, the control and bergapten-treatment groups showed progressive darkening of the dorsal coat. In addition, we harvested skin samples for HE staining, and morphological observations revealed fewer histochemically identified melanin granules in HQ mice. In addition, the number of follicular melanin granules in the bergapten-treated group increased to reverse the dorsal whitening. These results suggest that bergapten positively affected hair pigmentation (Fig. 4b). However, whether bergapten rescues the effects on pigmentation by exerting positive effects on melanocyte survival remains undetermined. TYR could be considered a melanocyte marker as melanocytes can be labelled an anti-TYR protein antibody conjugated with fluorescein isothiocyanate, isomer I; thus, an immunofluorescence experiment was conducted [38]. The HQ mouse model clearly showed fewer TYR-labelled melanocytes in the hair follicles, demonstrating that the number of melanocytes decreased following HQ treatment (Fig. 4b and S2). However, the melanocyte loss was reversed after bergapten treatment. The experiment also showed that bergapten increased TYR protein expression. HQ acts by inhibiting the enzymatic oxidation of tyrosinase and phenoloxidase and is also a strong oxidant that rapidly converts to the melanocyte-toxic substances p-benzoquinone and hydroxybenzoquinone, both of which cause skin-melanocyte loss and then depigmentation [39]. To test whether bergapten could relieve depigmentation induced by different causes, a copper chelator (PTU) was used as a moulding agent to induce depigmentation. A relatively low concentration of bergapten increased pigmentation in zebrafish embryos (Fig. 4c). Bergapten dose-dependently increased the melanin content and tyrosinase activity in B16F10 melanoma cells, and an extremely significant effect was observed at a concentration of 100 μM (Fig. 5).
Fig. 4.
Macroscopic observations of pigmentation responses after bergapten treatment. The areas showing significant colour changes in mouse dorsal skin spanned from the neck to the tail (a, n = 10). A representative area of each group on day 12 after depilation with most hair follicles (b, n = 3). The original magnification was 100× (left) or 400× (right). n = 10 in each group. A representative therapeutic effect of bergapten on depigmentation caused by PTU is shown (c, n = 30).
Fig. 5.
Effects of bergapten on melanin contents and tyrosinase activities. The extracellular and intracellular melanin levels were determined by measuring the absorbance at 405 nm and normalized to the total protein content (a). TYR activities were measured in terms of L-DOPA oxidation using lysates obtained from B16-F10 cells after bergapten treatment (b). The data shown are expressed as the mean ± SD (n = 3). The data were analysed by one-way analysis of variance followed by Tukey’s post-hoc test. *p < 0.1 and ***p < 0.001 vs. the control group.
Conclusions
In summary, using a structure-based protein-engineering approach, we developed an alternative method for producing rare valuable products from medicinal plants. We determined the first crystal structure of a coumarin-specific BMT. The well-displayed atomic structure may favour a deeper understanding of the substrate preferences and catalytic mechanism of O-methylation in coumarins. Using structure-based rational design, a candidate mutant with improved activity in bergapten production was obtained. The mutant protein provides a good candidate for further metabolic engineering to produce bergapten for use in treating depigmentation disorders. The preliminary pharmacological activity of bergapten was also estimated both in animals and cells, which may be helpful for subsequent study of the mechanisms of furocoumarins in depigmentation disorder treatment.
Compliance with Ethics Requirements
All Institutional and National Guidelines for the care and use of animals (fisheries) were followed.
Declaration of Competing Interest
The authors have no conflict of interest to declare.
Acknowledgments
This project was funded by the China Postdoctoral Science Foundation (2016M601922, 2018T110577), the Natural Science Fund in Jiangsu Province (BK20170736), the National Natural Science Foundation of China (81430092, 81703637, 31970596), and the Open Project of State Key Laboratory of Natural Medicines (SKLNMKF201708). This research was also supported by the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R63) and the 111 Project from the Ministry of Education of China and the State Administration of Foreign Export Affairs of China (B18056). We thank the staff of the BL19U1 beamline of the National Facility for Protein Science at the Shanghai Synchrotron Radiation Facility for assistance during data collection.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2019.10.003.
Contributor Information
Zhixiong Zeng, Email: zengzx@mail.hzau.edu.cn.
Lingyi Kong, Email: cpu_lykong@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Pillaiyar T., Namasivayam V., Manickam M., Jung S.H. Inhibitors of melanogenesis: an updated review. J Med Chem. 2018;61:7395–7418. doi: 10.1021/acs.jmedchem.7b00967. [DOI] [PubMed] [Google Scholar]
- 2.Lai X., Wichers H.J., Soler-Lopez M., Dijkstra B.W. Structure of human Tyrosinase related protein 1 reveals a binuclear zinc active site important for Melanogenesis. Angew Chem Int Ed. 2017;56:9812–9815. doi: 10.1002/anie.201704616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu P.Y., You Y.J., Liu Y.J., Hou C.W., Wu C.S., Wen K.C. Sesamol inhibited Melanogenesis by regulating melanin-related signal transduction in B16F10 cells. Int J Mol Sci. 2018;19:1108. doi: 10.3390/ijms19041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hałdys K., Goldeman W., Jewgiński M., Wolińska E., Anger N., Rossowska J. Inhibitory properties of aromatic Thiosemicarbazones on mushroom Tyrosinase: synthesis, kinetic studies, molecular docking and effectiveness in melanogenesis inhibition. Bioorg Chem. 2018;81:577–586. doi: 10.1016/j.bioorg.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Cheng M.-C., Lee T.-H., Chu Y.-T., Syu L.-L., Hsu S.-J., Cheng C.-H. Melanogenesis inhibitors from the Rhizoma of Ligusticum Sinense in B16–F10 melanoma cells in vitro and Zebrafish in vivo. Int J Mol Sci. 2018;19:3994. doi: 10.3390/ijms19123994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mcneely W., Kl D.C.N. 5-methoxypsoralen: a review of its effects in psoriasis and vitiligo. Drugs. 1998;56:667–690. doi: 10.2165/00003495-199856040-00015. [DOI] [PubMed] [Google Scholar]
- 7.Da S.V., Kawano D.F., Carvalho I., Da C.E., De F.O., Da S.C. Psoralen and bergapten: in silico metabolism and toxicophoric analysis of drugs used to treat vitiligo. Int J Mol Sci. 2009;12(3):378. doi: 10.18433/j3w01d. [DOI] [PubMed] [Google Scholar]
- 8.Bae J.M., Jung H.M., Hong B.Y., Lee J.H., Choi W.J., Kim G.M. Phototherapy for vitiligo: a systematic review and meta-analysis. JAMA Dermatol. 2017;153:666–674. doi: 10.1001/jamadermatol.2017.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Z., Luo J., Wang J., Kong L. Separation of minor coumarins from Peucedanum praeruptorum using HSCCC and preparative HPLC guided by HPLC/MS. Sep Purif Technol. 2010;75:132–137. [Google Scholar]
- 10.Ling-Yi K., Yi L., Zhi-Da M., Xian L., Ting-Ru Z. Coumarins from Peucedanum praeruptorum. Phytochemistry. 1996;41:1423–1426. [Google Scholar]
- 11.Choi K.R., Jang W.D., Yang D., Cho J.S., Park D., Lee S.Y. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019;37:817–837. doi: 10.1016/j.tibtech.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Dastmalchi M., Chen X., Hagel J.M., Chang L., Chen R., Ramasamy S. Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat Chem Biol. 2019;15:384–390. doi: 10.1038/s41589-019-0247-0. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen J. Cell factory engineering for improved production of natural products. Nat Prod Rep. 2019;36:1233–1236. doi: 10.1039/c9np00005d. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Jian X., Wu j., Huang W., Huang C., Luo J., Kong L. Elucidation of the biosynthesis pathway and heterologous construction of a sustainable route for producing umbelliferone. J Biol Eng. 2019;13:44. doi: 10.1186/s13036-019-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y., Liu T., Luo J., Zhang Q., Xu S., Han C. Integration of a decrescent transcriptome and metabolomics dataset of Peucedanum praeruptorum to investigate the CYP450 and MDR genes involved in coumarins biosynthesis and transport. Front Plant Sci. 2015;6:996. doi: 10.3389/fpls.2015.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng W. Defining and redefining its role in biology: Synthetic biology as an emerging field at the interface of engineering and biology. PeerJ Preprints. 2016;4:e2634v2631. [Google Scholar]
- 17.Bhuiy M.W., Liu C.J. Engineering monolignol 4-O-methyltransferases to modulate lignin biosynthesis. J. Bio. Chem. 2010;285:277–285. doi: 10.1074/jbc.M109.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K., Bhuiya M.W., Pazo J.R., Miao Y., Kim H., Ralph J. An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell. 2012;24:3135–3152. doi: 10.1105/tpc.112.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternke M., Tripp K.W., Barrick D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. P Natl Acad Sci USA. 2019;116:11275. doi: 10.1073/pnas.1816707116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W., Yao J., Meng J., Han W., Tao Y., Chen Y. Promiscuous enzymatic activity-aided multiple-pathway network design for metabolic flux rearrangement in hydroxytyrosol biosynthesis. Nat Commun. 2019;10:960. doi: 10.1038/s41467-019-08781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y., Wang N., Zeng Z., Xu S., Huang C., Wang W. Cloning, functional characterization, and catalytic mechanism of a Bergaptol O-Methyltransferase from Peucedanum praeruptorum Dunn. Front Plant Sci. 2016;7:72. doi: 10.3389/fpls.2016.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 23.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zubieta C., Kota P., Ferrer J.L., Dixon R.A., Noel J.P. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002;14:1265–1277. doi: 10.1105/tpc.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y., Wang N., Sui Z., Huang C., Zeng Z., Kong L. The molecular and structural basis of O-methylation reaction in Coumarin Biosynthesis in Peucedanum praeruptorum Dunn. Int J Mol Sci. 2019;20:1533. doi: 10.3390/ijms20071533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aid Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 29.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 30.Sherman W., Beard H.S., Farid R. Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des. 2006;67:83–84. doi: 10.1111/j.1747-0285.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 31.Muller-Rover S., Handjiski B., van der Veen C., Eichmuller S., Foitzik K., McKay I.A. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 32.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 33.Lee M.H., Lin Y.P., Hsu F.L., Zhan G.R., Yen K.Y. Bioactive constituents of Spatholobus suberectus in regulating tyrosinase-related proteins and mRNA in HEMn cells. Phytochemistry. 2006;67(12):1262–1270. doi: 10.1016/j.phytochem.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Menezes J.C., Diederich M. Translational role of natural coumarins and their derivatives as anticancer agents. Future Med Chem. 2019;11:1057–1082. doi: 10.4155/fmc-2018-0375. [DOI] [PubMed] [Google Scholar]
- 35.Hehmann M., Lukačin R., Ekiert H., Matern U. Furanocoumarin biosynthesis in Ammi majus L. Eur J Biochem. 2004;271:932–940. doi: 10.1111/j.1432-1033.2004.03995.x. [DOI] [PubMed] [Google Scholar]
- 36.Lo S.C., Chung P.E., Wang C.S. Molecular cloning and functional analysis of bergaptol-O-methyltransferase from Angelica dahurica (Bai Zhi) and using it to efficiently produce bergapten in E. coli. Bot Stud. 2012;53:197–206. [Google Scholar]
- 37.Ishikawa A., Kuma T., Sasaki H., Sasaki N., Ozeki Y., Kobayashi N. Constitutive expression of bergaptol O-methyltransferase in Glehnia littoralis cell cultures. Plant Cell Rep. 2009;28:257–265. doi: 10.1007/s00299-008-0631-9. [DOI] [PubMed] [Google Scholar]
- 38.Seberg H.E., Van Otterloo E., Loftus S.K., Liu H., Bonde G., Sompallae R. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet. 2017;13:e1006636. doi: 10.1371/journal.pgen.1006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osei M., Ali M., Owusu A., Baiden F. Skin-lightening practices among female high school students in Ghana. Public Health. 2018;155:81–87. doi: 10.1016/j.puhe.2017.11.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.