Abstract
Hydrogen peroxide (H2O2) is an essential second intracellular messenger. To reach its targets in the cytosol, H2O2 must cross a membrane, a feat that requires aquaporins (AQP) endowed with ‘peroxiporin’ activity (AQP3, AQP8, AQP9). Here, we exploit different organelle-targeted H2O2-sensitive probes to show that also AQP11 efficiently conduits H2O2. Unlike other peroxiporins, AQP11 is localized in the endoplasmic reticulum (ER), accumulating partly in mitochondrial-associated ER membranes (MAM). Its downregulation severely perturbs the flux of H2O2 through the ER, but not through the mitochondrial or plasma membranes. These properties make AQP11 a potential regulator of ER redox homeostasis and signaling.
Keywords: Aquaporins, Peroxiporins, Hydrogen peroxide, Membrane permeability, Endoplasmic reticulum, Redox homeostasis
Abbreviations: AQP, aquaporins; DTT, dithiothreitol; ER, endoplasmic reticulum; H2O2, hydrogen peroxide; MAM, mitochondria-associated ER membranes
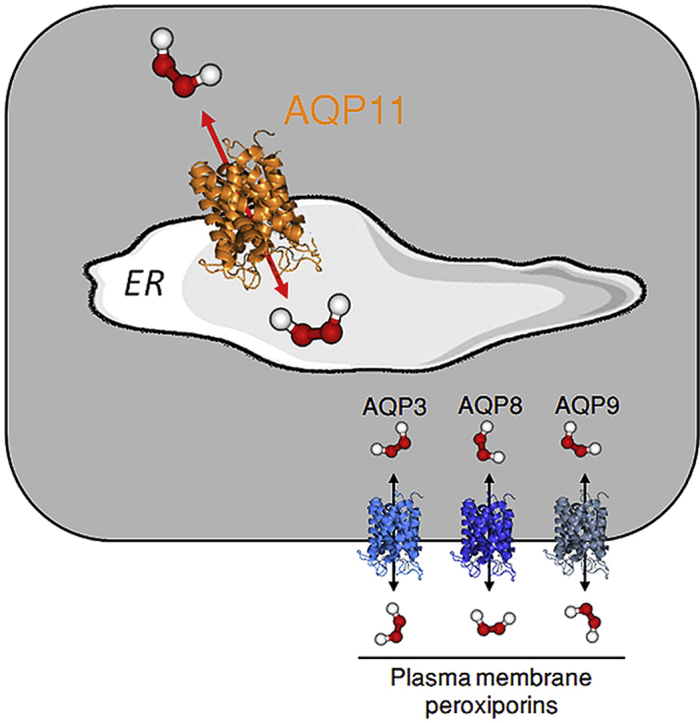
Graphical abstract
Highlights
-
•
AQP11 is an endoplasmic reticulum (ER)-resident peroxiporin.
-
•
AQP11 allows H2O2 fluxes across the ER membrane.
-
•
Its levels impact ER redox homeostasis.
1. Introduction
Aquaporins (AQP) are membrane channels widely distributed from archaea to mammals, that facilitate the passage of water and other solutes across biological membranes [1]. They share a common homotetrameric fold, each monomer containing a functional pore formed by six membrane-spanning α-helices connected by three extracellular (A, C, and E) and two intracellular (B and D) loops [2]. Two selectivity filters determine the type of molecules that can permeate each family member. The first one, composed by two opposing asparagine–proline–alanine (NPA) motifs that meet in the center of each channel, creates an electrostatic barrier for charged ions [3]. A second barrier near the extracellular entrance, the aromatic/arginine (ar/R) constriction site, limits instead the size of the solutes that can traverse the channel [2].
Based on their transporting capacities, the thirteen human isoforms were traditionally clustered in two main groups: orthodox AQP, involved in water permeability (AQP0, AQP1, AQP2, AQP4, AQP5, AQP6 and AQP8) and aquaglyceroporins (AQP3, AQP7, AQP9 and AQP10), that transport glycerol in addition to water [4]. With the discovery of two intracellular isoforms presenting little homology to conventional members, AQP11 and AQP12, a new category, named unorthodox or superaquaporins [5], was added. Selective permeability of this group is still poorly characterized, though some evidence has indicated that AQP11 can transport water [6] and glycerol [7]. However, this archetypical classification has been lately blurred, because some solutes, importantly H2O2, are transported both by orthodox (i.e. AQP8 [8]) and aquaglyceroporins (AQP3 [9] and AQP9 [10]). Since H2O2 modulates the activity of tyrosine phosphatases and kinases [11,12], those ‘peroxiporins’ may control key signaling circuits and have thus attracted much attention [13]. Indeed, transport of redox messengers through some of these channels is a regulated event [14,15], suggesting a possible role in topologically restrained signal amplification [16].
Remarkably, mice lacking AQP11, an isoform thought to accumulate primarily in the endoplasmic reticulum (ER), rapidly develop a lethal polycystic kidney-like disease [17] with marked signs of oxidative stress [18]. Considering that the ER is an important source of H2O2 and hub for signal integration [19], we investigated the localization, topology and function of AQP11. Our data demonstrate that AQP11 is an ER-resident peroxiporin capable of transporting H2O2 along concentration gradients.
2. Results
2.1. AQP11 resides in the ER
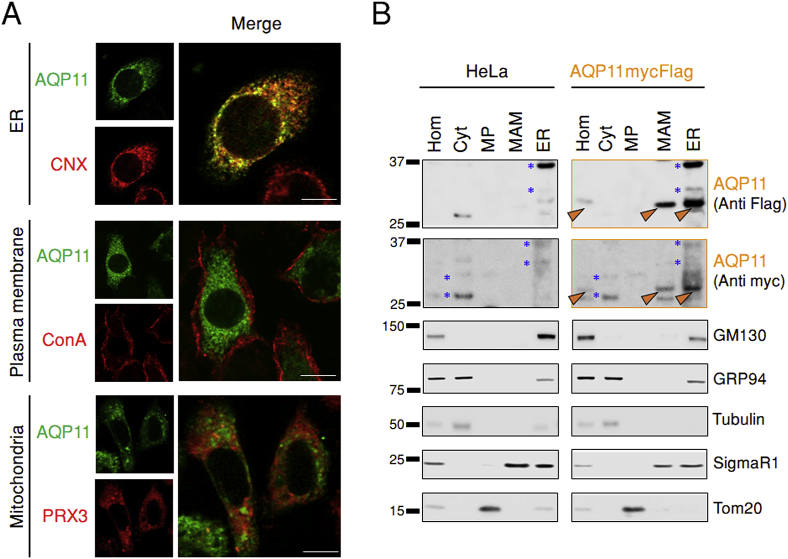
When expressed in different cell lines, chimeric human AQP11 accumulated in the ER, independently from the position and nature of appended tags (Fig. 1A and Fig. S1A). In all cases, subcellular localization was not related to protein expression levels (not shown), reducing the risk of artefactual mislocalization upon protein overexpression. HaloAQP11 largely co-localized with calnexin (CNX, Fig. 1A, upper panels) in the ER, but little if any was present in the plasma membrane (stained with ConA before cell permeabilization, middle panels) or in mitochondria (stained with antibodies to peroxiredoxin-3, PRX3, bottom panels). Analyses of the coincidence of the AQP11 signal for the entire cell volume with each organelle marker by the Pearson correlation coefficient [20], confirmed a high degree of co-distribution (>0.7) for the CNX-AQP11 pair and a low value (slightly over the >0.3 positive limit) for Prx3-AQP11 (Fig. S1B). The distinctive localization of AQP11 was further evident in cells co-expressing a Flag-tagged AQP11 and a plasma membrane-bound AQP such as a HaloAQP8 (Fig. S2A): a strong signal was observed for AQP8 at the cell perimeter, while AQP11 yielded the typical reticular staining of ER-resident molecules. Pixel intensity profiles of both AQP concurred this observation (Fig. S2B), ConA profile serving to delimit plasma membrane areas. That is, the vast majority of AQP11 occupied zones corresponding to the cytoplasm, whereas AQP8 profile showed a strong overlap with ConA profile and a minor signal at the cytoplasmic area, likely corresponding to molecules traversing from the ER on their way to the cell surface.
Fig. 1.
AQP11 resides in the ER.
A. HeLa transfectants expressing HaloAQP11 were co-stained with fluorescent Halo ligands, and with antibodies against calnexin (CNX) or peroxiredoxin 3 (PRX3), to decorate AQP11, the ER and mitochondria, respectively. To label glycoproteins on the plasma membrane, cells were stained with concanavalin A-FITC (ConA) in absence of cell permeabilization, and then fixed (middle panels). Scale bar = 10 μm.
B. HeLa cells expressing AQP11mycFlag were fractionated as previously described [22,23]. Aliquots were resolved electrophoretically and stripes of the blots decorated with the indicated antibodies. Hom, total post-nuclear homogenates. Cyt, cytosol. MP, pure mitochondrial fraction. MAM, mitochondria-associated membranes. ER, endoplasmic reticulum.
Even though AQP11 highly co-distributes with CNX, their respective localization pattern did not perfectly overlap, suggesting that the two proteins might reside in partially distinct subregions of the ER. In eukaryotic cells, subregions of the ER membrane are tethered to mitochondria [21]. These mitochondrial-associated membranes (MAM) are key signaling platforms that allow the exchange of redox signals between both organelles [19]. As we surmised that AQP11 functions as an ER-resident peroxiporin, we sought evidence to establish whether the channel is present at those sites. Following a validated protocol [22,23], we obtained fractions enriched for Sigma1 receptor [24], but devoid of markers of the cytosol, mitochondria, Golgi and ER (tubulin, Tom20, GM130 or GRP94, respectively. Fig. 1B), and hence likely corresponding to MAM. Accordingly, this fraction contained also IP3 receptors and VDAC [22,25]. Thus, part of AQP11 segregates with MAM. These biochemical data were in line with the results of the Pearson coefficient described above (Fig. S1B), in which a modest degree of co-distribution was observed between AQP11 and the mitochondrial marker Prx3.
2.2. AQP11 topology
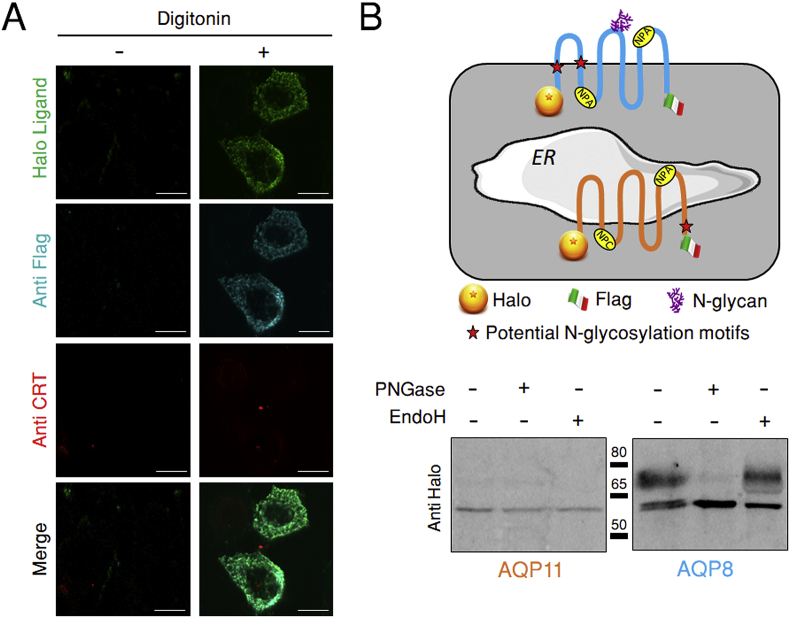
To determine whether AQP11 adopts the canonical AQP topology, we constructed a recombinant AQP11 with a Halo tag and a Flag tag at its extremities (HaloAQP11-Flag). If both its N- and C- termini were facing the cytosol, the tags should become accessible to antibodies or membrane-impermeant Halo ligands upon incubation with digitonin (Fig. S3), a detergent that permeabilizes the plasma membrane but not the membrane of the ER [26]. Clearly, both the non-permeant Halo ligand and an anti-Flag antibody produced a positive signal on digitonin-treated cells (Fig. 2A), indicating that AQP11 follows the regular AQP structural scheme, with both termini oriented to the cytosol. Calreticulin (CRT), an ER luminal protein, was not detected unless cells were treated with Triton X100 to permeabilize the ER (Fig. S3), confirming that digitonin did not hamper ER membrane integrity.
Fig. 2.
Both the N- and C- termini of AQP11 protrude into the cytosol.
A. HeLa transfectants expressing HaloAQP11-Flag were treated with or without digitonin to selectively solubilize the plasma membrane, and then incubated with a membrane-impermeant Halo ligand (green) or with antibodies specific for Flag (cyan) or calreticulin (CRT, red), a lumenal protein of the ER. See Supplementary Fig. S2 for further details on the protocol. Note that both the N- and C-termini of HaloAQP11-Flag face the cytosol, as schematized in panel B (top scheme). Scale bar = 10 μm.
B. The top panel shows the topology of AQP11 and AQP8 (orange and blue, respectively) as determined by immunofluorescence (Fig. 2A) and biochemical analyses (bottom panel). The absence of N-glycosylated AQP11 species in HeLa transfectants contrasts with the easily detected Endo-H resistant AQP8 glycoforms. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Analyzing protein glycosylation further confirmed that the C-terminus of AQP11 lies in the cytosol. Despite the presence of a potentially accessible N-glycosylation site in human AQP11 (NHT, residues 264–266, see scheme on Fig. 2B), neither endoglycosidase-H (Endo-H) nor peptide N-glycosidase F (PNGaseF) caused mobility shifts to the protein (Fig. 2B, lower panels and Fig.S4A), and a sharp band was present regardless of the enzyme applied. In contrast, human AQP8 yielded three bands, serving as control of glycosidase activities. The slower-migrating band contains glycoforms that are resistant to Endo-H and hence had traversed the Golgi complex. The slower of the two sharper bands is sensitive to both Endo-H and PNGaseF, and most likely corresponds to N-glycosylated molecules en route to the Golgi. The third band carries instead no N-glycans. Altogether, the above data suggest that whilst most AQP8 molecules travel through the secretory pathway and are processed before reaching the plasma membrane, AQP11 resides in the ER with its potential N-glycosylation site facing the cytosol.
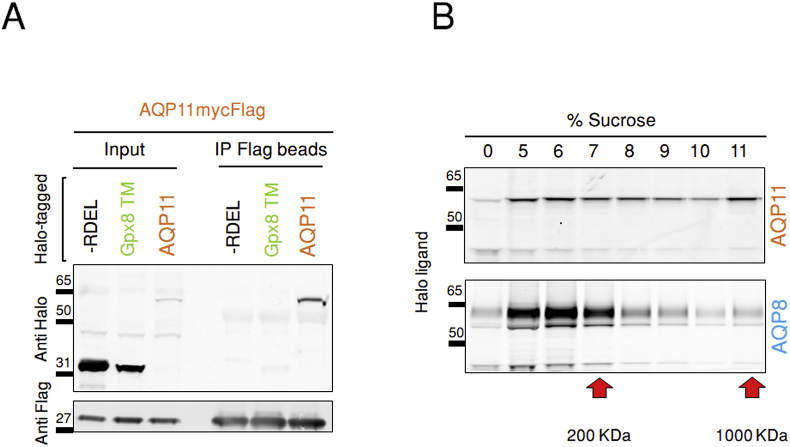
Next, we tested whether also AQP11 forms tetramers, as most other AQP do. To this end, AQP11mycFlag was coexpressed in HeLa cells with HaloAQP11 or, as specificity controls, with Halo proteins preceded by a signal sequence and extended at its C-terminus with an ER localization motif (-RDEL) or fused to the transmembrane domain of the ER protein Gpx8 (HaloGpx8 TM, see Ref. [22]). Clearly, anti-Flag antibodies precipitated virtually all HaloAQP11 (Fig. 3A). Neither Halo nor HaloGpx8 TM were instead coprecipitated, even if they were more abundant than HaloAQP11 in the cell lysates (compare the anti-Halo signals in the 3 left lanes, labeled as Input), indicating that mixed assemblies can be formed by differently tagged AQP11. To confirm and expand this notion, we fractionated lysates of HeLa transfectants expressing either HaloAQP11-Flag or HaloAQP8-Flag on discontinuous sucrose gradients. In Fig. 3B, the two red arrows indicate the migration of 7S and 19S purified immunoglobulins, and their respective molecular weights. Based on those settings, both AQP11 and AQP8 co-fractionated where molecules of around 200 kDa accumulate (Fig.3B and Fig.S4C), a size compatible with a tetrameric conformation. However, gel distribution profile suggests that AQP11 tends to form more high molecular weight complexes than AQP8.
Fig. 3.
AQP11 forms tetramers.
A. Aliquots of the lysates of HeLa co-transfectants expressing AQP11mycFlag and either Halo-RDEL, HaloGpx8 TM or HaloAQP11 proteins were immunoprecipitated using immobilized anti-Flag antibodies and analyzed by western blotting using anti-Halo or anti-Flag specific antibodies. IP, immunoprecipitation.
B. Lysates of HeLa cells expressing either HaloAQP11-Flag or HaloAQP8-Flag were separated through a discontinuous 5–
11% sucrose density gradient, and aliquots of each fraction subjected to reducing SDS-PAGE. The presence of the recombinant proteins was assessed by direct detection of bound fluorescent Halo ligands on the gels using a laser scanner imager.
2.3. AQP11 is a peroxiporin facilitating the transport of H2O2 across the ER membrane
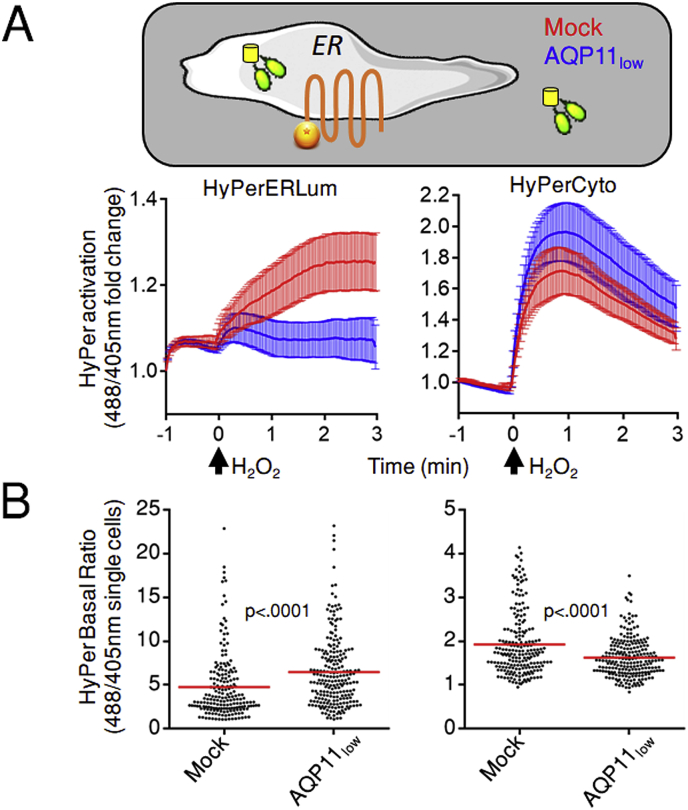
To analyze whether AQP11 can act as a peroxiporin, we engineered HeLa cells expressing HyPer sensors [27] in the ER lumen (HyPerERLum), cytosol (HyPerCyto) or in the mitochondrial matrix (HyPerMito). The HyPerERLum probe showed a higher level of activation at steady state (Fig. S5A), as reported previously [28]. Nonetheless, its oxidation upon exposure of cells to exogenous H2O2 was clearly detectable (Fig. 4). Lowering the basal oxidation state of HyPerERLum by pretreating cells with 5 mM DTT for 5 min allowed to better visualize the entry of H2O2 into the ER (green trace in Fig. S5A). However, as the results could be easily quantified also in the absence of DTT pretreatment, we performed our experiments without the addition of reducing agents.
Fig. 4.
AQP11 is a peroxiporin.
A. Polyclonal HeLa cells stably expressing HyPer in the ER lumen (HyPerERLum) or in the cytosol (HyPerCyto) were treated with AQP11-specific or control small interfering RNAs (siRNAs) and the kinetics of probe activation upon exposure to 50 μM H2O2 plotted against time. The black arrow indicates when H2O2 was added. Data are shown as mean fold changes of the 488/405 nm ratio and corresponds to ≥5 experiments ± standard error of the mean (SEM). Note that AQP11 silencing inhibits HyPerERLum activation without interfering with the cytosolic sensor.
B. Measurement of the basal oxidation state of HyPerERLum and HyPerCyto. Each dot represents a single cell distributed in the graph according to its 488/405 nm ratio. Comparing the panels highlights that AQP11 silencing impacts in opposite ways the ER lumenal and cytosolic HyPer sensors.
Consistent with AQP11 being an ER-resident protein, silencing its expression (Fig. S5B) had different impact on HyPer sensors. While oxidation of HyPerERLum was severely inhibited (Fig. 4A, AQP11low, left graph and supplementary movies SM1 and SM2), AQP11 silencing had no significant effects on HyPerCyto (see Fig. 4A, right graph and SM3 and SM4) or HyPerMito (see Fig. S5C, SM5 and SM6) activation. Silencing AQP4 did not impair significantly HyPerERLum oxidation (Fig. S5D). Thus, in agreement with its subcellular distribution, knock-down of AQP11 yielded functional consequences only on the permeability of the ER membrane to H2O2.
Importantly, AQP11low cells consistently revealed a higher basal oxidation state of HyPerERLum when compared to mock cells (Fig. 4B, left graph). Therefore, a plausible explanation of the failure to detect HyPerERLum changes upon challenge with exogenous H2O2 could be that basal activation of the sensor is maximum in the more oxidizing environment of the ER. However, reproducible changes in the 488/405 ratio can be constantly recorded after addition of H2O2 also in AQP11low cells (Fig. S6A). These results suggest that a continuous efflux of the ER-produced H2O2 reaches the cytosol through the channel. Accordingly, HyPerCyto sensors were significantly less oxidized at steady state in AQP11low cells (Fig. 4B, right graph), whereas the HyPerMito probe showed minimal changes (Fig. S6B).
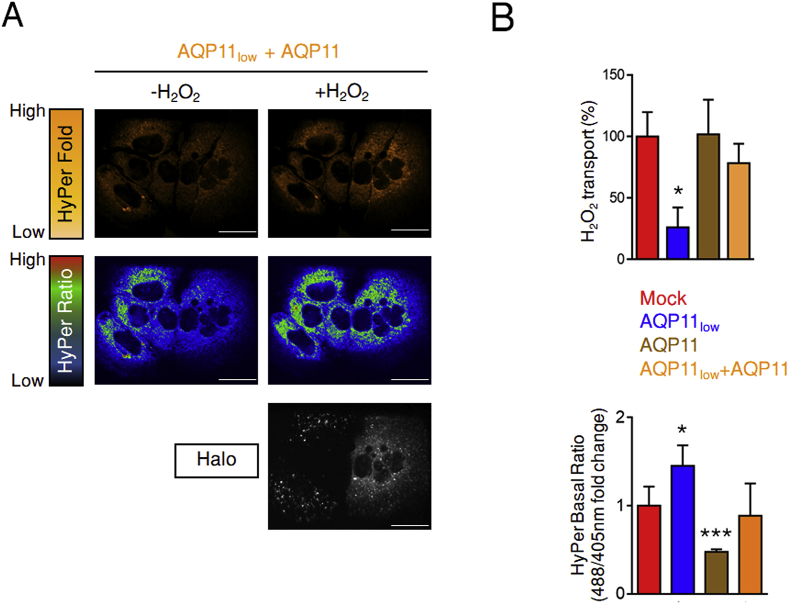
Unlike silencing, that impacts the majority of cells (Figs. S5B and S6A, lower panel), plasmid-driven transfection induces overexpression in half or less of cells. This allowed us to perform imaging experiments with internal controls, in that we could compare the responses of AQP11low cells with those that were reconstituted with a Halo-tagged chimeric variant engineered to be silencing-resistant (Fig. 5A). Clearly, expression of the transgene rescued H2O2 transport across the ER membrane, allowing the oxidation of the HyPerERLum probe (Fig. 5A, left panels, and 5B, upper graph). The higher basal ratio level of HyPerERLum in non-rescued AQP11low cells, evident also in this experiment (Fig. 5B, lower graph), and did not increase significantly upon exposure to H2O2 (Fig. 5A). Consistently with our previous results, in cells whose ER peroxiporin capacity had been rescued by HaloAQP11 expression, the HyPerERLum basal ratio returned to the levels seen in mock controls (Fig. 5B, lower panel). Cells expressing both transfected HaloAQP11 and the endogenous protein displayed the lowest basal ratios, suggesting that ER permeability to H2O2 is related to the number of channels expressed at its membrane.
Fig. 5.
AQP11 restores H2O2fluxes in silenced cells.HyPerERLum HeLa transfectants were silenced for AQP11 expression and then transfected with a silencing-resistant HaloAQP11.
A. Representative immunofluorescence analyses of one of the experiments averaged in panel B confirm the lower response to exogenous H2O2 (top panels, HyPerERLum fold changes, in orange) and higher basal oxidation of HyPer ER lumen (middle panels, basal ratio, in rainbow) in AQP11-silenced cells. Both features are rescued by re-expression of HaloAQP11, as monitored using fluorescent Halo ligands as shown in the lower panel (in white).
B. The efficiency of the entry of exogenous H2O2 (50 μM, top graph) and the basal oxidation state of the HyPer ER lumenal probe (bottom panel) were measured as in Fig. 4, in control or AQP11low cells, before or after rescuing. Results are represented as percentage of H2O2 transported 1.5 min after of H2O2 addition relative to controls (Mock), and as the fold change in the basal 488/405 nm ratio (bottom). Average on ≥3 experiments ± SEM.
(For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
The results shown herein demonstrate that AQP11 is a peroxiporin residing in the ER. This conclusion is largely based on the use of organelle-targeted, ratiometric sensors, that undergo a fluorescent shift upon binding H2O2. When expressed in the ER lumen, the HyPer probe is partly oxidized in basal conditions and hence not ideal to detect H2O2 fluxes, particularly in secretory cells [28]. To circumvent this obstacle, we selected a HeLa subline in which the basal activation state was low enough to allow swift changes upon increased H2O2 fluxes. Thus, in all circumstances, we could detect a rise in the oxidized fraction of the probe upon exposure to exogenous H2O2, which was severely inhibited by silencing AQP11 but not AQP4. In accordance, expression of AQP11 rescued permeability of the ER membrane to H2O2 in AQP11low cells. These findings suggest that AQP11 is a key H2O2 channel in the ER membrane, a function that might be assisted partly by AQP8 molecules in transit to the plasma membrane [8].
Why the need of an ER-resident peroxiporin? The localization of AQP11 in the ER highlights yet another evolutionary pressure, that of allowing exchange of signals and solutes to-from selected organelles. Oxidative folding in the ER is a source of H2O2, a byproduct of Ero1 pathway [29,30]. Exiting the ER via AQP11, H2O2 may act as an indirect indicator of the fitness of the organelle and –as most secretory proteins contain disulfide bonds-of the prowess of protein biogenesis. Furthermore, considering the wide distribution and rapid rate of reaction of peroxidases [31], specific interactions with signaling molecules are likely to occur preferentially in the vicinity of H2O2 channels [16]. Therefore, the existence of a dedicated ER membrane peroxiporin opens yet another possibility for tuning or localize cellular signaling circuits in restrained areas of the cytosol or vicinal organelles. Indeed, a likely relevant target of AQP11-derived H2O2 may well be PTP1B, a redox-modulated phosphatase anchored to the cytoplasmic face of the ER [32]. In principle, this function could be also played by other peroxiporins as they transit through the ER en route to the membrane. Thus, additional features likely make AQP11 a better suited ER-resident peroxiporin. In this connection, AQP11, AQP12 and most ER-resident plant AQPs (SIPs [33,34]), share NPC and NPT motifs in place of the characteristic NPA-NPA family signature motif. They might allow for transport of other solutes or permit a different channel regulation from plasma membrane isoforms, the cysteine possibly acting as a redox sensor.
The mechanisms of ER retention are presently unknown. A di-lysine motif is present at the C-terminus of human AQP11 (-NKKE) as well as in some SIPs [35]. However, neither its deletion nor extending the C-terminus with different tags allowed AQP11 to reach the plasma membrane (not shown). A possible explanation might be the shortness of AQP11 transmembrane regions, some of which are predicted to be < 20 residues while being ≥21 in AQP8 (TMHMM and TMPred online analysis tools). Finally, previous studies exploiting biotinylation techniques detected some AQP11 at the plasma membrane [36,37]. Likewise, some cell types secrete KDEL bearing ER-resident proteins [38]. Why and under which circumstances ER retention mechanisms are selectively weakened is presently unclear. Surely, the mechanisms that regulate the localization and function of AQP11 in the ER -especially in subregions that establish contact with other organelles-deserve further studies.
4. Materials and methods
4.1. Cell culture and generation of HeLa polyclonal stable cell lines
HeLa, CHO, COS-7 and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) + GlutaMAXTM-I medium (Gibco) supplemented with 10% fetal bovine serum (FBS; EuroClone) and 5 mg/ml penicillin-streptomycin (Lonza).
HeLa transfectants stably expressing HyPer in the ER (HyPerERLum), in the cytosol (HyPerCyto), or in the mitochondrial matrix (HyPerMito) were generated by PEI transfection and then selected with 0.5 mg/ml of G418 (Sigma).
4.2. Plasmids, small interfering RNAs, and transfection procedures
Plasmids for expression of HyPerCyto, HyPerMito and HyPerERLum were gifts of Drs. V.Belousov and M.Geistz. The plasmids to express HaloAQP8 and HaloAQP8-Flag were generated as previously described [8,14], while the AQP11mycFlag construct was obtained from Origene (NM_173039) and used to build the proteins HaloAQP11-Flag and HaloAQP11. Briefly, the insert containing AQP11 fused to myc and Flag was excised from the original vector pCMV6-Entry and cloned into the pHTN-HaloTag® plasmid (Promega), to generate the HaloAQP11-Flag construct. Then, and using the later as template, a stop codon was introduced between AQP11 and the C-terminal tags in order to exclude them, with the primers: 5′-AAGGAATAGCGTACGCGGCCGC-3’ (forward) and 5′-GCGGCCGCGTACGCTATTCCTT-3’ (reverse). Lastly, three consecutive rounds of site-directed mutagenesis were performed on HaloAQP11 to obtain the silencing-resistant version bearing 5 mutations in the region that pairs the 21 bp target sequence of the AQP11 siRNA. The primers used were as follows: first round, 5′-GGAGCTTCGCCTGTAAGAATCCC-3’ (forward) and 5′-GGGATTCTTACAGGCGAAGCTCC-3’ (reverse); second round 5′-CAGCGAGAGGAGTTTTGCCTGTAAG-3’ (forward) and 5′-CTTACAGGCAAAACTCCTCTCGCTG-3’ (reverse); and third round 5′-GCCTGTAAAAACCCCATCCGAGTC-3’ (forward) and 5′-GACTCGGATGGGGTTTTTACAGGC-3’ (reverse). All constructs and mutations were validated by sequencing.
Reagents to silence AQP11 (5′-GAGCUUCGCUUGCAAGAAU-3′) and AQP4 (5′-GAUCAGCAUCGCCAAGUCU-3′), and an unrelated control (Block-itTM) were purchased from Ambion (Life Technologies), and its efficiency was monitored by real time RT-PCR [8].
For silencing experiments, 8 × 104 HeLa cells were grown overnight in six-well plates and transfected with 30 pmol of siRNA, using RNAiMAX lipofectamin (Invitrogen) according to the manufacturer's instructions. Silencing was validated by RT-PCR. To transiently express HaloAQP11, HaloAQP11-Flag or HaloAQP8, cell lines were transfected using JetPei (Polyplus) and cultured for 48 h before immunofluorescence or biochemical analyses. For experiments in which AQP11 expression was reconstituted using the silencing-resistant HaloAQP11 version, cells were first silenced for 24 h, then transfected with the correspondent plasmid, and cultured for further 24 h before imaging.
4.3. Imaging HyPer oxidation
To perform confocal live imaging experiments, 8 × 104 HyPerERLum-expressing HeLa cells were silenced and/or transfected on glass coverslips. To identify HaloAQP11-expressing cells, 2 nM HaloTag® TMRDirect (Promega) was added 24 h after transfection. After further 24 h of culture with the fluorescent ligand, cells on coverslips were equilibrated in Ringer buffer (RB: 140 mM NaCl, 2 mM CaCl2, 1 mM MgSO4, 1.5 mM K2HPO4, 10 mM Glucose, pH 7.3) for 10 min at room temperature (RT) before addition of 50 μM H2O2. Confocal images were collected every 2sec for 1-to-4min by dual excitation with a 488-nm argon and 405-nm violet diode lasers. We used an Ultraview confocal laser scanning microscope equipped with either an EC Plan - Neofluar 20X (NA 0.45) Dry (Carl Zeiss) or a 40X oil immersion lens (PerkinElmer). The 488/405-nm ratios were calculated by ImageJ software for ≥20 cells, averaged, and showed as ratio or mean fold change of ratio plotted against time ± SEM. To facilitate quantification and statistical analyses of some experiments, we averaged the data obtained at 1.5min after H2O2 addition and represented them as the percentage of H2O2 transport relative to the corresponding untreated cells. From 3 to 6 experiments were conducted for each condition. The same procedure was followed for HyPerCyto- (n = 6 experiments) and HyPerMito-expressing cells (n = 2 experiments). At least 15 cells per experiment were averaged and plotted.
4.4. Co-localization studies
To assess the localization of AQP11 using the HaloAQP11 or the AQP11mycFlag recombinant proteins, the indicated cell lines were plated on 13 mm coverslips and transfected as described above. The Halo-tagged version of AQP11 was labeled using 2 nM of the permeable HaloTag® TMRDirect, added 24 h after transfection, while the AQP11mycFlag version was detected using mouse anti-Flag antibodies (Sigma) 48 h after transfection. To stain glycoproteins at the plasma membrane, cells were incubated with Concanavalin A-FITC (Sigma) for 1 h at 4 °C before fixation with 4%PFA for 20 min at RT. Selective plasma membrane permeabilization was performed with digitonin and validated as described previously [26]. Permeabilization of the ER membrane was with 0.1% Triton X100. Rabbit anti-Calnexin and anti-Peroxiredoxin-3 were from Enzo Life Sciences and Thermo Scientific, respectively. Suitable anti-mouse or anti-rabbit secondary antibodies Alexa Fluor 488, Alexa Fluor 546 and Alexa Fluor 647 were from Molecular Probes. Fluorescent images were acquired either by using a Leica TCS SP8 SMD FLIM microscope equipped with a 63X oil lens (HC PL APO CS 2 63X (NA 1.4)) or an Ultraview confocal laser scanning microscope (PerkinElmer) with a Plan - APOCHROMAT 63X (NA 1.4) Oil lens. Images were processed using LASX imaging software from Leica or ImageJ. Pearson Correlation Coefficient was calculated through all the image z-stack by using the ImageJ plugin Coloc2.
4.5. Subcellular fractionation
After homogenization of the cells, crude mitochondrial and microsomal fractions were separated by classical differential centrifugation. Subsequently, the crude mitochondrial fraction was resuspended in isolation buffer (250 mM mannitol, 5 mM Hepes, 0.5 mM EGTA, pH 7.4) and further separated on a 30% Percoll gradient to obtain low-density (denoted as MAM) and high-density (denoted as pure mitochondria, MP) fractions as described in Wieckowski et al., 2009. Aliquots of the fractions were collected and analyzed by Western blot after SDS-PAGE was performed on an equal quantity of proteins using the antibodies mouse anti-Flag, mouse anti-β−Tubulin and rabbit anti-Sigma R1 from Sigma, mouse anti-myc and rabbit anti-TOM20 from SantaCruz Biotechnology, mouse anti-GM130 from BD Biosciences and rabbit anti-GRP94 from Enzo Life Sciences.
4.6. AQP11 end-tail analyses
To determine the orientation of the end-tails of AQP11, HeLa cells were plated on 13 mm coverslips and transfected for 48 h with the double-tagged construct HaloAQP11-Flag. A scheme of the protocol followed is depicted in Fig. S3. Briefly, the plasma membrane was permeabilized with 40 μg/ml of Digitonin (Sigma) and cells incubated for 1 h at 4 °C with 2 nM of an impermeant fluorescent Halo ligand (HaloTag® TMR Ligand, Promega). Subsequently, the Flag tag was detected with a mouse anti-Flag antibody (Sigma) decorated by using the fluorescently-labeled secondaries of Molecular Probes, as described above. Cells were then fixed and treated or not with the stronger detergent Triton X100. Finally, the ER lumen was stained using an anti-calreticulin antibody (Sigma).
4.7. Deglycosylation assays
HeLa transfectants transiently expressing HaloAQP11-Flag were grown in p100 plates for 48 h, and then washed once with ice-cold PBS and scraped in RIPA buffer (0.1% SDS, 1% NP40, 150 mM NaCl, 50 mM Tris, pH 7.4) supplemented with freshly added protease inhibitors, 10 mM NEM, 0.4 mM Na3VO4, and 10 mM NaF. Whole HeLa cell lysates were centrifuged at 15,000 rpm, 15 min at 4 °C, and 30 μg aliquots of the post-nuclear fractions incubated either with EndoH or PNGase F enzymes from New England Biolabs for 1 h at 37 °C. Resulting deglycosylated protein mixtures were used in standard reducing electrophoresis, followed by western blotting using a rabbit anti-Halo antibody (Promega). Images were acquired using a Typhoon FLA-9000 (GE HealthCare) and processed with ImageJ. The experiments were repeated twice and gave similar results.
4.8. Immunoprecipitation
HeLa transfectants transiently expressing AQP11mycFlag and either Halo-RDEL, HaloGpx8 TM [22], or HaloAQP11 were grown and lysed as above. Postnuclear fractions were quantified and 1 mg of total protein incubated o/n at 4 °C with commercial anti-Flag crosslinked beads (Sigma). Next, beads were washed three times with a buffer containing 0.25% NP40 150 mM NaCl 10 mM Tris pH 7.4 and boiled in standard protein sample buffer. Aliquots of the postnuclear lysates were run with those immunoprecipitated samples in reducing SDS-PAGE, followed for Western blot with rabbit anti-Halo (Promega) and mouse anti-Flag antibodies (Sigma). Images were acquired using a Typhoon FLA-9000 (GE HealthCare) and processed with ImageJ.
4.9. Sucrose density gradients
HeLa cells were transfected for 48 h with vectors to express either HaloAQP11-Flag or HaloAQP8-Flag and then washed once with ice-cold PBS and scraped in RIPA buffer (0.1% SDS, 1% NP40, 150 mM NaCl, 50 mM Tris, pH 7.4) supplemented with freshly added protease inhibitors, 10 mM NEM, 0.4 mM Na3VO4, and 10 mM NaF. To label both recombinant proteins 30 nM HaloTag® TMRDirect (Promega) was added 24 h after transfection. Postnuclear lysates were loaded on top of a discontinuous 5–11% sucrose gradient and centrifugated at 32700 rpm 3 h at 4 °C in a Beckman Coulter OPTIMA L90K ultracentrifuge equipped with a Beckman Coulter SW55Ti rotor. Aliquots of each phase were precipitated using a 10% solution of trichloroacetic acid (Sigma) and resuspended in standard reducing protein sample buffer. Samples were run in 3–8% precast gradient gels (NovexTM, Life Technologies) and directly scanned using a Typhoon FLA-9000 (GE HealthCare).
4.10. Statistical analyses
Statistics were calculated either by using the two-sample t-test for correlated samples or the one-way ANOVA method for multiple samples. When using the latter, Tukey's HSD post hoc test was also applied to find out which groups were significantly different from others. In all cases, statistical significance was defined as p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
Funding
This work was supported in part through grants from the Associazione Italiana Ricerca sul Cancro (IG 2016-18824 to R.S.), the Fondazione Cariplo (2015-0591 to R.S.), and the Telethon (GGP15059 to R.S.). A.R. was supported by local funds from the University of Ferrara and the Italian Ministry of Health (GR-2016-02364602). Italian Association for Cancer Research (AIRC, IG-18624), Telethon (GGP11139B), and local funds from the University of Ferrara to P.P.
Author contributions
I.M.-F. and R.S. designed the strategy of the study. S.B. and I.S. performed most IF experiments. M.G. and I.S. H2O2 transport and RT-PCR assays and A.R. and P.P. were responsible for subcellular fractionations. I.M.-F. performed biochemical and the basal ratio analyses. I.S. also performed the sucrose gradients and calculated the Pearson Correlation Coefficient. All authors contributed in interpreting the data. I.S., R.S. and I.M.-F. wrote the manuscript.
Conflicts of interest
The authors declare no competing interests.
Data and materials availability
Most data needed to evaluate the conclusions in the study are present in the text, figures and supplementary materials. All raw data and additional details concerning protocols and reagents may be obtained upon request.
Acknowledgments
In addition to all members of our laboratory and our Division, we thank P. Panina, L. Rampoldi, A. Rubartelli and E. van Anken (San Raffaele Scientific Institute, Milan, Italy); G.P. Bienert and M.D. Bienert (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany) and E. Hidalgo (Universidad Pompeu-Fabra, Barcelona, Spain) for useful suggestions, discussions and constructive criticisms. Paolo Pinton and Alessandro Rimessi are grateful to Camilla degli Scrovegni for continuous support.
We are indebted with Drs. V. Belousov (Institute of Bioorganic Chemistry, Moscow, RU), E. Margittai and M. Geistz (Semmelweis University, Budapest, HU) for providing plasmids and reagents. Because of space limitations, as well as restriction of cited references, we apologize to all those colleagues and researchers in the field whose work is not directly cited here.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101326.
Contributor Information
Roberto Sitia, Email: sitia.roberto@hsr.it.
Iria Medraño-Fernandez, Email: medranofernandez.iria@hsr.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
H2O2 uptake of HyPerERLum-expressing mock cells
H2O2 uptake of HyPerERLum-expressing AQP11low cells
H2O2 uptake of HyPerCyto-expressing mock cells
H2O2 uptake of HyPerCyto-expressing AQP11low cells
H2O2 uptake of HyPerMito-expressing mock cells
H2O2 uptake of HyPerMito-expressing AQP11low cells
References
- 1.Finn R.N., Cerda J. Evolution and functional diversity of aquaporins. Biol. Bull. 2015;229(1):6–23. doi: 10.1086/BBLv229n1p6. [DOI] [PubMed] [Google Scholar]
- 2.Tornroth-Horsefield S., Hedfalk K., Fischer G., Lindkvist-Petersson K., Neutze R. Structural insights into eukaryotic aquaporin regulation. FEBS Lett. 2010;584(12):2580–2588. doi: 10.1016/j.febslet.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J.B. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407(6804):599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 4.Verkman A.S. Aquaporins at a glance. J. Cell Sci. 2011;124(13):2107–2112. doi: 10.1242/jcs.079467. [DOI] [PubMed] [Google Scholar]
- 5.Ishibashi K., Tanaka Y., Morishita Y. The role of mammalian superaquaporins inside the cell. Biochim. Biophys. Acta. 2014;1840(5):1507–1512. doi: 10.1016/j.bbagen.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Yakata K., Tani K., Fujiyoshi Y. Water permeability and characterization of aquaporin-11. J. Struct. Biol. 2011;174(2):315–320. doi: 10.1016/j.jsb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Madeira A., Fernandez-Veledo S., Camps M., Zorzano A., Moura T.F., Ceperuelo-Mallafre V. Human aquaporin-11 is a water and glycerol channel and localizes in the vicinity of lipid droplets in human adipocytes. Obesity. 2014;22(9):2010–2017. doi: 10.1002/oby.20792. [DOI] [PubMed] [Google Scholar]
- 8.Bertolotti M., Bestetti S., Garcia-Manteiga J.M., Medraño-Fernandez I., Dal Mas A., Malosio M.L. Tyrosine kinase signal modulation: a matter of H2O2 membrane permeability? Antioxidants Redox Signal. 2013;19(13):1447–1451. doi: 10.1089/ars.2013.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller E.W., Dickinson B.C., Chang C.J. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. U. S. A. 2010;107(36):15681–15686. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe S., Moniaga C.S., Nielsen S., Hara-Chikuma M. Aquaporin-9 facilitates membrane transport of hydrogen peroxide in mammalian cells. Biochem. Biophys. Res. Commun. 2016;471(1):191–197. doi: 10.1016/j.bbrc.2016.01.153. [DOI] [PubMed] [Google Scholar]
- 11.Rhee S.G., Bae Y.S., Lee S.R., Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE. 2000;2000(53):pe1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 12.Chiarugi P. PTPs versus PTKs: the redox side of the coin. Free Radic. Res. 2005;39(4):353–364. doi: 10.1080/10715760400027987. [DOI] [PubMed] [Google Scholar]
- 13.Bienert G.P., Medraño-Fernandez I., Sitia R. Regulation of H2O2 transport across cell membranes. In: HMBKTJ Vissers C.M., editor. Hydrogen Peroxide Metabolism in Health and Disease. Oxidative Stress and Disease. 44. CRC Press; Boca Raton: 2018. pp. 365–386. [Google Scholar]
- 14.Medraño-Fernandez I., Bestetti S., Bertolotti M., Bienert G.P., Bottino C., Laforenza U. Stress regulates aquaporin-8 permeability to impact cell growth and survival. Antioxidants Redox Signal. 2016;24(18):1031–1044. doi: 10.1089/ars.2016.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bestetti S., Medraño-Fernandez I., Galli M., Ghitti M., Bienert G.P., Musco G. A persulfidation-based mechanism controls aquaporin-8 conductance. Sci Adv. 2018;4(5) doi: 10.1126/sciadv.aar5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordzieke D.E., Medraño-Fernandez I. The plasma membrane: a platform for intra- and intercellular redox signaling. Antioxidants-Basel. 2018;7(11) doi: 10.3390/antiox7110168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morishita Y., Matsuzaki T., Hara-chikuma M., Andoo A., Shimono M., Matsuki A. Disruption of aquaporin-11 produces polycystic kidneys following vacuolization of the proximal tubule. Mol. Cell. Biol. 2005;25(17):7770–7779. doi: 10.1128/MCB.25.17.7770-7779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino Y., Sonoda H., Nishimura R., Mori K., Ishibashi K., Ikeda M. Involvement of the NADPH oxidase 2 pathway in renal oxidative stress in Aqp11 (-/-) mice. Biochem Biophys Rep. 2019;17:169–176. doi: 10.1016/j.bbrep.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoboue E.D., Sitia R., Simmen T. Redox crosstalk at endoplasmic reticulum (ER) membrane contact sites (MCS) uses toxic waste to deliver messages. Cell Death Dis. 2018;9(3):331. doi: 10.1038/s41419-017-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300(4):C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Vliet A.R., Verfaillie T., Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochim. Biophys. Acta. 2014;1843(10):2253–2262. doi: 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Yoboue E.D., Rimessi A., Anelli T., Pinton P., Sitia R. Regulation of calcium fluxes by GPX8, a type-II transmembrane peroxidase enriched at the mitochondria-associated endoplasmic reticulum membrane. Antioxidants Redox Signal. 2017;27(9):583–595. doi: 10.1089/ars.2016.6866. [DOI] [PubMed] [Google Scholar]
- 23.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009;4(11):1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi T., Su T.P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131(3):596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 25.Anelli T., Bergamelli L., Margittai E., Rimessi A., Fagioli C., Malgaroli A. Ero1alpha regulates Ca(2+) fluxes at the endoplasmic reticulum-mitochondria interface (MAM) Antioxidants Redox Signal. 2012;16(10):1077–1087. doi: 10.1089/ars.2011.4004. [DOI] [PubMed] [Google Scholar]
- 26.Molteni S.N., Fassio A., Ciriolo M.R., Filomeni G., Pasqualetto E., Fagioli C. Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J. Biol. Chem. 2004;279(31):32667–32673. doi: 10.1074/jbc.M404992200. [DOI] [PubMed] [Google Scholar]
- 27.Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3(4):281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 28.Malinouski M., Zhou Y., Belousov V.V., Hatfield D.L., Gladyshev V.N. Hydrogen peroxide probes directed to different cellular compartments. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20(22):6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu B.P., Weissman J.S. The FAD- and O-2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 2002;10(5):983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 31.Winterbourn C.C. Biological production, detection, and fate of hydrogen peroxide. Antioxidants Redox Signal. 2018;29(6):541–551. doi: 10.1089/ars.2017.7425. [DOI] [PubMed] [Google Scholar]
- 32.Tonks N.K. PTP1B: from the sidelines to the front lines! FEBS Lett. 2003;546(1):140–148. doi: 10.1016/s0014-5793(03)00603-3. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa F., Suga S., Uemura T., Sato M.H., Maeshima M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 2005;579(25):5814–5820. doi: 10.1016/j.febslet.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 34.Park W., Scheffler B.E., Bauer P.J., Campbell B.T. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2010;10:142. doi: 10.1186/1471-2229-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeshima M., Ishikawa F. ER membrane aquaporins in plants. Pflüg. Arch. 2008;456(4):709–716. doi: 10.1007/s00424-007-0363-7. [DOI] [PubMed] [Google Scholar]
- 36.Gorelick D.A., Praetorius J., Tsunenari T., Nielsen S., Agre P. Aquaporin-11: a channel protein lacking apparent transport function expressed in brain. BMC Biochem. 2006;7:14. doi: 10.1186/1471-2091-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda M., Andoo A., Shimono M., Takamatsu N., Taki A., Muta K. The NPC motif of aquaporin-11, unlike the NPA motif of known aquaporins, is essential for full expression of molecular function. J. Biol. Chem. 2011;286(5):3342–3350. doi: 10.1074/jbc.M110.180968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sannino S., Anelli T., Cortini M., Masui S., Degano M., Fagioli C. Progressive quality control of secretory proteins in the early secretory compartment by ERp44. J. Cell Sci. 2014;127(Pt 19):4260–4269. doi: 10.1242/jcs.153239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
H2O2 uptake of HyPerERLum-expressing mock cells
H2O2 uptake of HyPerERLum-expressing AQP11low cells
H2O2 uptake of HyPerCyto-expressing mock cells
H2O2 uptake of HyPerCyto-expressing AQP11low cells
H2O2 uptake of HyPerMito-expressing mock cells
H2O2 uptake of HyPerMito-expressing AQP11low cells