Abstract
Intellectual disability (ID) is a non-specific phenotype present in a genetically heterogeneous group of disorders. The genetic cause of ID remains elusive in the majority of patients due to this extreme heterogeneity. Whole exome sequencing technology has been applied to identify pathogenic gene variants responsible for ID. The present report described a 1.7-year-old female patient who had severe ID with the specific features of delayed motor development, language disorders and abnormal facial features. Exome analysis identified a novel pathogenic variant of the SETD5 gene [c.2025_2026delAG (p.Gly676Valfs*2)]. The variant was a frameshift mutation, causing termination of the protein in advance. These findings indicated that this mutation of the SETD5 gene may be a genetic cause for ID. The present study aimed to provide a meaningful exploration of ID and the identification of clinical core genetic pedigrees.
Keywords: intellectual disability, SETD5 gene, whole exome sequencing, developmental delay
Introduction
Intellectual disability (ID) is defined as substantial impairment of cognitive and adaptive functions that commonly occur in childhood, and has an estimated prevalence of 1.5~2.0% (1). Most reports conclude that severe ID (an intellectual quotient of <50) has a prevalence of 0.3~0.4%, while the estimated prevalence of mild ID differs between various studies (1). Related studies have shown that most affected patients exhibited development delay, speech impediment, feeding difficulties, gastrointestinal or abdominal wall defects and scoliosis (2,3). In addition, affected patients exhibited certain facial features, including abnormal ears, unusually shaped nose and micrognathia. To date, the application of next generation sequencing technology has allowed the rapid identification of new gene mutations that lead to ID (4). The search for de novo mutations through a trio approach is an efficient strategy for the identification of the genetic cause of ID in most cases (5,6). Grozeva et al (2) examined 996 individuals with ID by a targeted next-generation sequencing approach and discovered 7 de novo mutations of the SET domain-containing 5 (SETD5) gene. The clinical phenotype of ID was markedly similar to 3p deletion syndrome or 3p25 microdeletion syndrome, for which SETD5 is believed to be the strongest candidate gene (7,8). Kuechler et al (3) further extended the spectrum of ID-associated SETD5 mutation by reporting 2 de novo intragenic variants in two patients. These results suggested that SETD5 may play a role in the mammalian developmental process.
In this report, we present an ID patient with a de novo frameshift mutation in exon 15 of SETD5 that to the best of our knowledge has never been reported before. We give a detailed description of the clinical features of the patient to aid a more comprehensive understanding of the disease.
Case report
Patient
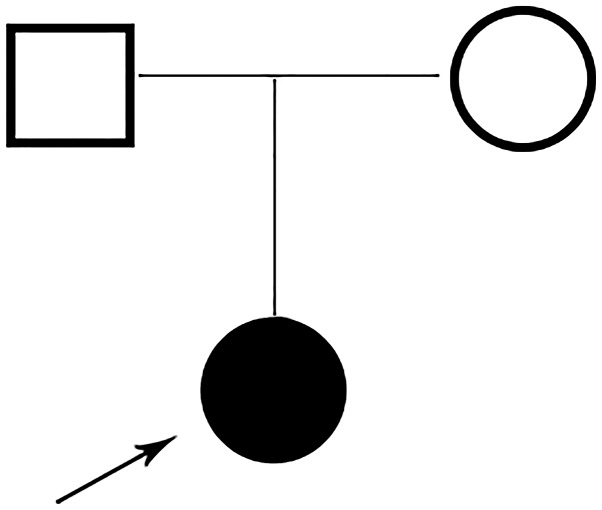
A Chinese family with a child was recruited in this study (Fig. 1). The proband was a 1.7-year-old girl who was born to a non-consanguineous 32-year mother and 31-year father. There was no family history of exposure to teratogens. The proband was born by Caesarean section at 40+5 weeks because of a fetal heartbeat of 68 times per minute, and Apgar scores were normal. Birthweight was 2,800 g and length 48 cm. Birth occipito-frontal circumference (OFC) was not available. The left eyelid showed slight droop not long after birth. The child's motor development was delayed, especially gross motor functions. She was unable crawl at 21 months. At the time of presentation (2 year 5 months), her height was 82 cm with weight (10 kg) and OFC (46 cm), within normal centile. She was not able to walk without support or speak fluently. Facial signs comprised prominent forehead, depressed nasal bridge, abnormal nose, thin upper lip and prominent philtrum. Moreover, the ears tended to be large and low set. The eyes were long, narrow and exhibited unilateral mild ptosis. Both hands were able to hold items and muscle tension was normal. At presentation, her anterior fontanelle had not been closed. Brain magnetic resonance imaging (MRI) showed high signal in bilateral parietal white matter area about T2W1 and fluid attenuated inversion recovery (FLAIR); in addition to a widened ventricle, outside the brain interval (Fig. 2). Routine chromosome analysis showed a normal female 46, XX karyotype. Screening for metabolic disorders and electromyography examinations detected no pathological results. The clinical phenotype of the parents was normal.
Figure 1.
A family pedigree of two generations. The arrow indicates the proband.
Figure 2.
Brain MRI lesions of the proband. (A) In bilateral parietal white matter; (B) widened ventricle, cistern and sulci. MRI, magnetic resonance imaging.
Methods
Full informed consent was obtained from the guardian of the patient, after which peripheral blood samples were collected from the 3 family members, and genomic DNA was extracted from blood samples using a DNA Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. Whole exome sequencing was performed with trios including the proband and her parents using the Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, CA, USA). All sequencing fragments were aligned to the reference human genome (hg19) using Burrows-Wheeler Alignment (BWA) (9), and duplicated reads were removed. Single nucleotide variants (SNVs) and small indels were identified using the SAMtools (http://samtools.sourceforge.net/). For each sample, variants of indels and SNVs were called using the Genome Analysis Tool kit v3.2 (GATK) program (10). All the identified variations were annotated and classified by Variant Effect Predictor software (11). The variants were filtered to excluded those with <1% minor allele frequency in all the following databases: ExAC (http://exac.broadinstitute.org), 1,000 Genomes (http://www.1000genomes.org/), ESP6500 (http://evs.gs.washington.edu/EVS), and an in-house database of ~500 Polish exomes. Then, known disease-causing variants were screened based on ClinVar, OMIM and HGMD. Finally, the pathogenic variant was determined in combination with clinical manifestation.
The variant of interest was confirmed by Sanger sequencing of exon 15 of SETD5. DNA was amplified by polymerase chain reaction (forward primer: GACCAGTTCAGCCCAAAGAC, reverse primer: CAGCCTTTTGTGTCAAAGCA). The PCR products were identified by agarose gel electrophoresis and sequenced directly using an ABI3730 automated sequencer (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). SETD5 variants were annotated based on the transcript number NM_001080517.2.
Results
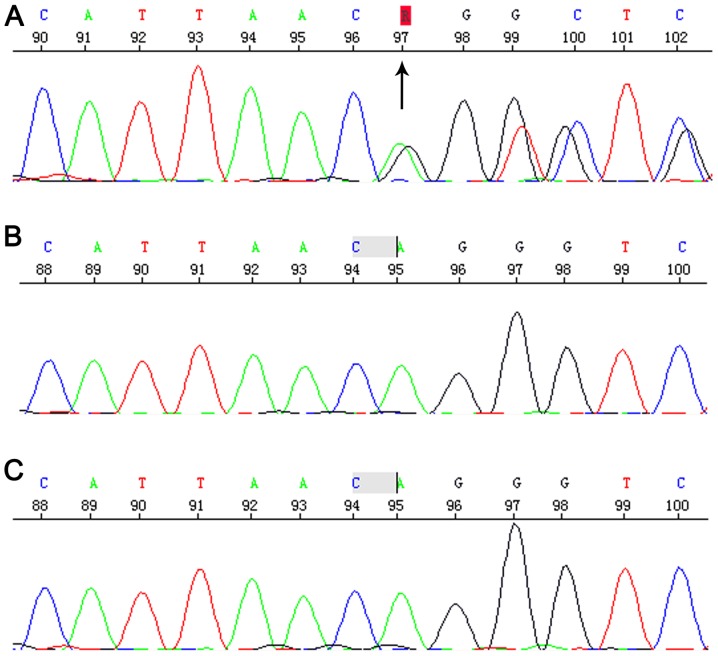
Sequencing revealed the Chr3:9489612-9489613 c.2025_2026del AG that located in 15 exon of SETD5 gene. This is a frameshift mutation, causing termination of the protein in advance (p.Gly676Vlfs*2). This variant had not previously reported in the literature and was absent from the ExAC and 1,000 Genomes databases. Parents tested negative for this particular variant, suggesting that this was a de novo mutation (Fig. 3) and autosomal dominant transmission of ID (MIM:615761). Therefore, this variant was considered as probably pathogenic in the proband.
Figure 3.
Sequencing diagram of the region verified by Sanger sequencing. (A) Patient with the variant; (B) father without the variant; (C) mother without the variant.
Discussion
ID patients can be characterized by specific features, including speech and motor deficits, growth retardation, cardiovascular and renal defects, and behavioral problems (2,12). These features can vary depending on the main underlying genetic cause. Previous papers have suggested that the ID caused by SETD5 de novo mutations might be part of a more complex clinical phenotype with variable expressivity. These observations are in conformity with the previously emphasized value of familial cases of newly recognized genetic syndromes in expanding knowledge of their phenotype (13).
The phenotypic features of individuals with the SETD5 de novo mutations in our family were similar to previously described patients, although there were some obvious differences (2). As to the reason for these differences, it should be noted that the previous studies reporting SETD5 mutations analyzed hundreds of ID patients for de novo variants (2,3).
SETD5 is located on chromosome 3q3.25 and includes 49 exons, which code a 1,442 amino acid protein, SET domain-containing protein 5. The protein belongs to the SET methyltransferase family, which catalyzes methylation of histone H3 and H4 lysine residues (14). It is highly expressed in the cerebral cortex, the intestine and the eye (3). Related studies have confirmed that SET-containing-domain protein could possess histone H3-specific methyltransferase activity (15).
Although rare, patients with ID might exhibit large deletions (up to 12 Mb), small deletions, substitutions, or insertions (16). Our results indicated that a new de novo SETD5 gene variant [c.2025_2026 delAG (p.Gly676Vlfs*2)] as the genetic cause of ID in the patient under study, but her parents did not carry the mutation. Fortunately, related studies have shown that rare de novo loss of function (LoF) variants of SETD5 have recently been proposed to be a relatively frequent (0.7%) cause of ID (3). In addition, Rauch et al described a new one de novo nonsense mutation resulting in the deletion of 7 genes (5). Kuechler et al (3) revealed 2 de novo intragenic variants; one was a nonsense variant and the other an 81 bp deletion located across a splice-donor site. Szczałuba et al (17) prioritized a potential LoF mutation. Related studies have confirmed that ID could be caused by de novo variants in 45~55% of patients (5). These data demonstrated that mutations were commonly found in patients with ID. Surprisingly, these studies further showed that de novo mutations might be the main cause of most ID (18).
In conclusion, we described a novel variant in SETD5 gene as the putative genetic cause of a patient with ID. The findings may aid deeper understanding of this disease.
Acknowledgements
Not applicable.
Funding
This work was supported by the Key Project of Tianjin Health Care Professionals (grant no.16KG166), the National Natural Science Foundation of China (grant no.81771589), and the Program of Tianjin Science and Technology Plan (grant no. 18ZXDBSY00170).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YLF conceived and designed this study, and drafted the manuscript. RPZ performed the data analysis and interpretation. YZW and LRC performed the Sanger confirmation and analysis. YQZ and CQC participated in the design and coordination of this study in addition to revising and critiquing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Tianjin Children's Hospital. Informed consent for participation in the study or use of their medical data was obtained from all participants or their legal guardian.
Patient consent for publication
Informed consent was obtained from the guardian (parents), who agreed to join this study, and for the use of medical information and images for scientific research and publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Leonard H, Wen X. The epidemiology of mental retardation: Challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–134. doi: 10.1002/mrdd.10031. [DOI] [PubMed] [Google Scholar]
- 2.Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park SM, Canham N, Holder SE, Wilson M, et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet. 2014;94:618–624. doi: 10.1016/j.ajhg.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuechler A, Zink AM, Wieland T, Lüdecke HJ, Cremer K, Salviati L, Magini P, Najafi K, Zweier C, Czeschik JC, et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet. 2015;23:753–760. doi: 10.1038/ejhg.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 6.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 7.Shuib S, McMullan D, Rattenberry E, Barber RM, Rahman F, Zatyka M, Chapman C, Macdonald F, Latif F, Davison V, Maher ER. Microarray based analysis of 3p25-p26 deletions (3p-syndrome) Am J Med Genet A 149A. 2009:2099–2105. doi: 10.1002/ajmg.a.32824. [DOI] [PubMed] [Google Scholar]
- 8.Peltekova IT, Macdonald A, Armour CM. Microdeletion on 3p25 in a patient with features of 3p deletion syndrome. Am J Med Genet A 158A. 2012:2583–2586. doi: 10.1002/ajmg.a.35559. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey JC. Significance of case reports in the advancement of medical scientific knowledge. Am J Med Genet Part A. 2006;140:2131–2134. doi: 10.1002/ajmg.a.31449. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 15.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 16.Riess A, Grasshoff U, Schäferhoff K, Bonin M, Riess O, Horber V, Tzschach A. Interstitial 3p25.3-p26.1 deletion in a patient with intellectual disability. Am J Med Genet 158A. 2012:2587–2590. doi: 10.1002/ajmg.a.35562. [DOI] [PubMed] [Google Scholar]
- 17.Szczałuba K, Brzezinska M, Kot J, Rydzanicz M, Walczak A, Stawiński P, Werner B, Płoski R. SETD5 loss-of-function mutation as a likely cause of a familial syndromic intellectual disability with variable phenotypic expression. Am J Med Genet A. 2016;170:2322–2327. doi: 10.1002/ajmg.a.37832. [DOI] [PubMed] [Google Scholar]
- 18.Grozeva D, Carss K, Spasic-Boskovic O, Tejada MI, Gecz J, Shaw M, Corbett M, Haan E, Thompson E, Friend K, et al. Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutat. 2015;36:1197–1204. doi: 10.1002/humu.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.