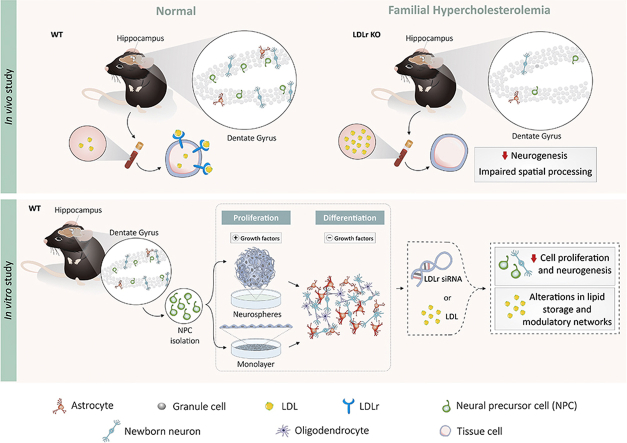
Abstract
Objective
In familial hypercholesterolemia (FH), mutations in the low-density lipoprotein (LDL) receptor (LDLr) gene result in increased plasma LDL cholesterol. Clinical and preclinical studies have revealed an association between FH and hippocampus-related memory and mood impairment. We here asked whether hippocampal pathology in FH might be a consequence of compromised adult hippocampal neurogenesis.
Methods
We evaluated hippocampus-dependent behavior and neurogenesis in adult C57BL/6JRj and LDLr−/− mice. We investigated the effects of elevated cholesterol and the function of LDLr in neural precursor cells (NPC) isolated from adult C57BL/6JRj mice in vitro.
Results
Behavioral tests revealed that adult LDLr−/− mice showed reduced performance in a dentate gyrus (DG)-dependent metric change task. This phenotype was accompanied by a reduction in cell proliferation and adult neurogenesis in the DG of LDLr−/− mice, suggesting a potential direct impact of LDLr mutation on NPC. Exposure of NPC to LDL as well as LDLr gene knockdown reduced proliferation and disrupted transcriptional activity of genes involved in endogenous cholesterol synthesis and metabolism. The LDL treatment also induced an increase in intracellular lipid storage. Functional analysis of differentially expressed genes revealed parallel modulation of distinct regulatory networks upon LDL treatment and LDLr knockdown.
Conclusions
Together, these results suggest that high LDL levels and a loss of LDLr function, which are characteristic to individuals with FH, might contribute to a disease-related impairment in adult hippocampal neurogenesis and, consequently, cognitive functions.
Keywords: Familial hypercholesterolemia, Adult neurogenesis, LDLr, Dentate gyrus, Neural precursor cell, Cholesterol
Graphical abstract
LDL receptor and cholesterol metabolism regulate adult hippocampal neurogenesis. The LDLr −/− mice show impaired hippocampal related behavior and adult neurogenesis. In vitro exposure of neural precursor cells (NPC) to LDL and LDLr knock-down reduces cell proliferation modulating distinct regulatory networks. Additionally, LDL exposure induces increase in lipid storage and impaired neuronal differentiation.
Highlights
-
•
The LDLr −/− mice show impaired hippocampal related behaviour and adult neurogenesis.
-
•
In vitro exposure of NPC to LDL and LDLr knock-down reduces cell proliferation.
-
•
LDL exposure induces lipid storage in NPC.
-
•
In vitro LDL and LDLr knock-down in NPC modulates distinct regulatory networks.
1. Introduction
Hypercholesterolemia is an important risk factor for the development of neurodegenerative diseases [1], [2]. Particularly, altered cholesterol metabolism is considered a critical factor in the pathogenesis of Alzheimer's disease [3], [4]. Compared to individuals with the sporadic form, those with familial hypercholesterolemia (FH) present a higher incidence of mild cognitive impairment in later life [5]. Ariza and coworkers [6] reported that even young FH subjects already showed neuropsychological deficits.
FH is caused by inherited genetic abnormalities, predominantly in the low-density lipoprotein (LDL) receptor (LDLr) gene, resulting in an ineffective metabolism of LDL particles. Defective uptake of LDL by the liver leads to elevated plasma LDL cholesterol from birth and development of premature atherosclerosis and cardiovascular disease [7]. Despite the increasing number of clinical and preclinical evidence of FH association with cognitive impairment, it remains unclear whether the chronic exposure to high circulating cholesterol levels or the dysfunction of the LDLr as such contribute to the altered central nervous system (CNS) function. Cholesterol-carrying lipoproteins, such as LDL, cannot readily cross the blood–brain barrier (BBB). Instead, the majority of cholesterol in the brain is synthesized de novo within the brain tissue [8]. However, recent data suggest that hypercholesterolemia might weaken BBB function, disrupting cholesterol balance between the brain and the periphery, and this could favor pathological processes in the CNS [9], [10], [11]. The decrease in the LDLr activity in FH individuals might also bear consequences on neuronal development and function. Neurons in the adult brain take up ApoE-cholesterol complexes, produced and released by astrocytes, via endocytosis through the LRP1 and LDLr receptors [12]. Besides the cholesterol uptake, however, the physiological and pathological roles of LDLr in the brain remain unclear. Although LDLr−/− mice have normal brain morphology, they exhibit impairment in learning and memory and a depressive-like phenotype [13], [14], [15], [16], [17], [18], [19]. Together, these findings suggest that FH is accompanied by hippocampal dysfunction that is reflected at the onset of cognitive deficit.
Adult hippocampal neurogenesis, the process that leads to the addition of new granule neurons in the dentate gyrus (DG), is believed to contribute to hippocampal functions such as cognition and emotional behavior [20], [21]. The new neurons originate from a pool of stem cells, located in the subgranular zone of the DG, that proliferate and give rise to precursor cells, which can potentially mature into functional glia or neurons past a number of defined stages [22]. There is a rising interest in how lipid metabolism can influence adult neural progenitors. Recent studies manipulated key components of fatty acids and cholesterol metabolism and found that, besides their requirement for new membrane production upon cell proliferation and differentiation, their availability can influence cell energetic states; moreover, they might act as signaling entities in adult neural precursor cells (NPC) [23]. Mulder and colleagues [13] have demonstrated that 14 months old LDLr−/− mice had reduced numbers of proliferating cells and synaptic connections in the DG compared to wild type controls. However, it remained unclear whether hippocampal cell proliferation and neurogenesis would be affected in younger animals, which already show cognitive impairment.
To address this question, we designed a set of experiments to explore if adult hippocampal neurogenesis might be involved in cognitive dysfunction associated with FH. We also investigated whether exposure to LDL and LDLr knock-down have a direct influence on cell proliferation and differentiation of hippocampal precursor cells in vitro.
2. Materials and methods
2.1. In vivo study
2.1.1. Animals
The progenitors of low-density lipoprotein receptor knockout mice (LDLr−/−; B6.129S7- Ldlrtm1Her/J) were obtained from Universidade Estadual de Campinas (UNICAMP, Sao Paulo, Brazil), originally purchased from Jackson Laboratory (JAX stock #002207; [24]) and bred at the same conditions as wild type C57BL/6J mice (JAX stock #000664) in our own breeding colony. Animals were maintained in groups of four to five animals per cage, under controlled temperature (23 ± 1 °C) and 12 h light cycle, with free access to food and water.
The LDLr−/− mice display constitutive increased plasma cholesterol levels (2–3 times higher) in standard feeding conditions [24], as used in this study (NUVILAB, Nuvital, Paraná, Brazil). All efforts were made to minimize the number and suffering of animals. The procedures used in the present study complied with the guidelines on animal care of the UFSC Ethics Committee on the Use of animals (PP00948), which follows the Principles of Laboratory Animal Care from National Institutes of Health.
2.1.2. BrdU injections
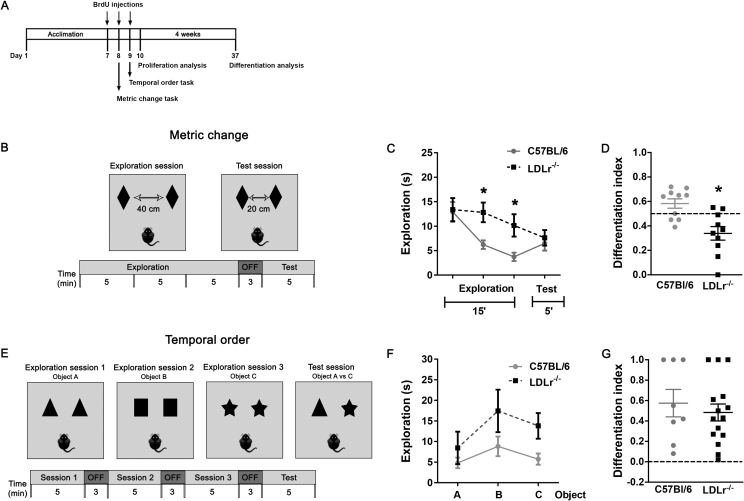
We used 5- bromo-2′-deoxyuridine (BrdU) to evaluate cell proliferation and overall neurogenesis. BrdU is a thymidine analogue that is incorporated into the DNA double-helix during the S-phase of the cell cycle, and thus marks actively proliferating cells [25]. C57BL/6J and LDLr−/− male mice at 3 months of age (n = 7/group) received i.p. injections of BrdU (100 mg/kg, in 0.1 M PBS, pH = 7.2; Sigma–Aldrich) every 12 h for 3 consecutive days. The same mice were then submitted to behavioral testing on the hippocampal sub-region dependent behavioral tasks and were euthanized 1 or 28 days later (Figure 1A) to assay proliferation or survival of new cells, respectively.
Figure 1.
Behavioral performance of C57BL/6J and LDLr−/− on metric change and temporal order tasks. (A) Experimental time-line for the animal behavioral experiments and BrdU administration. 3-months old C57BL/6J and LDLr−/− male mice were acclimated to the experimental apparatus for 7 days. Following this, at days 8 and 9, mice were evaluated on the metric change and temporal order tasks. In addition, starting at day 7, mice received BrdU injections (100 mg/kg i.p.) every 12 h for 3 consecutive days. 24 h after the last BrdU injection, one cohort of animals was sacrificed by transcardial perfusion, and their brains were processed for immunohistochemistry (proliferation analysis). A second cohort of animals was euthanized 4 weeks after BrdU injections (survival analysis). (B) In the metric change task, a mouse is allowed to explore the environment for 15 min with an identical pair of objects separated by 40 cm. Following the intersession period, in the 5-min test session, the mouse is allowed to explore the environment with objects repositioned 20 cm closer. (C) C57BL/6J and LDLr−/− mice explored progressively less during the 15-min habituation period reflecting familiarity with the environment. (D) Differentiation index score showing changes in object exploration between test session and the final 5 min of the habituation session. Index of 0.5 indicates equal exploration. Only C57BL/6J animals engaged in increased exploration during the test session, reflecting recognition of the metric change, while the LDLr−/− mice spent less time exploring objects during the exploration session, indicating lack of recognition of the metric change or continued habituation. (E) In the temporal order task, the mouse is introduced to 3 different pairs of identical objects and allowed to explore each pair for a period of 5 min. For the test session one of the objects from the third pair is exchanged for one of the objects from the first pair, and the mouse is allowed to freely explore for 5 min. (F) Animals exploration during the habituation sessions for object A, B and C. (G) Differentiation index of exploration between object A and object C indicates that most of C57BL/6J and LDLr−/− mice prefer to explore temporally distant object A. Zero indicates lack of preference. N = 8–15 per group; Results are expressed as means ± S.E.M; *p < 0.05, t-test. Statistical analysis for Figures 1–5 are available in Supplementary Table 1.
2.1.3. Hippocampal-dependent behavioral tasks
2.1.3.1. Behavioral apparatus and acclimation
The behavioral apparatus used for the metric and temporal ordering of visual objects consisted of a rectangular wooden box measuring 40 × 60 m × 50 cm high. Five sets of objects were used (one set for acclimation, one set for metric change, and three sets for temporal order). Objects included large Lego blocks, small glass bottles, and plastic toys and ranged in size between 2.5 and 5 cm at the base and between 5 and 15 cm tall. These objects were chosen as they are texturally and visually unique, and easily distinguishable for the mice. Between the habituation and test sessions, the mice were placed in a clean cage. The experimental box was wiped down with 70% ethanol after testing of each mouse to remove olfactory cues that could influence object exploration by subsequent mice. A video camera (Logitech, Newark, CA, USA) was mounted on the roof above the arena, and all test sessions were recorded using the ANY-maze software (Stoeling Co., Wooddale, IL, USA).
All animals (10–15 per group) underwent seven days of acclimation to the arena. On all days the animals were brought to the behavior room two hours prior to testing to allow them to acclimate to the room. On the first 3 days, animals were picked up by tails and placed in apparatus for 1 min with no objects present and then returned to their home cage. On the following 2 days, the animals were acclimated to the arena for 5 min with no objects present and then returned to their home cage. Finally, in the last 2 days of acclimation two objects were placed 40 cm apart in the center of the arena (these objects were randomly selected and were different from the ones used for either of the behavioral tests) and each animal was given a 10 min period to explore the arena and the objects before being placed back into its home cage. Following the 7 days of acclimation, the animals were subjected to the metric change task on day 8 and to the temporal order task on day 9 (Figure 1A).
2.1.3.2. Metric change task
The metric change task was conducted as initially described by Goodrich-Hunsaker [26], [27], [28] and composed of one habituation session and one test session. Mice were placed in the experimental box with two objects placed 40 cm apart. The mouse was allowed to freely explore the arena and stimulus objects for 15 min. After this habituation session, the mouse was removed to a clean cage for 3 min. During this intersession interval, the objects were moved closer to each other so that the objects were 20 cm apart. The mouse was then placed again in the experimental box and given 5 min to re-explore the objects during the test session. For the 15 min habituation session, total object exploration time (s) was calculated individually for the first, middle, and last 5 min time-blocks to facilitate comparison between the last 5 min of the exploration session and the 5 min test session.
When the time spent exploring objects through the three 5 min habituation periods did not decrease, it was taken as an indication that the animal did not habituate to the objects, and the mouse should be removed from the study. A decrease of less than 10% of the original exploration time is used as a criterion for elimination. In this study, all animals showed proper habituation behavior.
2.1.3.3. Temporal ordering task
The temporal order task was also conducted as initially described by Goodrich-Hunsaker [27], [29] and composed of 3 habituation sessions and 1 test session. Prior to the first habituation session, 2 identical copies of the first object (object A) were placed in the extremities of the experimental box, 10 cm from the end walls and centered between the long walls. For session 1, the mouse was placed in the center of the experimental box and given 5 min to freely explore the objects. Afterwards, the mouse was removed to a clean cage for 3 min. During this time, the first objects were replaced with 2 duplicates of a second object (object B). The procedure for the habituation session was then repeated once more, with 2 duplicates of a new object (object C). After session 3, the mouse was removed into a clean cage for 3 min and a copy of the first object and a copy of the third object (i.e., object A and object C) were placed in the box. The mouse was again allowed to explore the two objects during a 5 min test session. When a clear preference for one of the objects was detected during sessions 1–3 (i.e., difference in exploration time greater than 15 s), the animal was removed from the study (5 out of 13, for the C57BL/6J group and none for the LDLr−/- mice).
2.1.3.4. Behavioral analysis
A differentiation index for the metric spatial change task was calculated to facilitate the comparison between the test session and the last 5 min of the exploration session, as previously described [26]. Briefly, the index was calculated as: [(exploration time during the 5 min test session)/(exploration time during the 5 min test session + exploration time during the last 5 min of the exploration session)]. This constrained all the values between 0 and 1. With this index, increased exploration during the 5 min test session compared to the last 5 min of the habituation is reflected as a value > 0.5, while decreased exploration (or continued habituation) is reflected as a value < 0.5. Exploration during the temporal ordering test sessions was also converted into a differentiation index to constrain the values between −1 and 1. The index was calculated as follows: [(exploration of object A − exploration of object C)/(exploration of object A + exploration of object C)].
2.1.4. Brain dissection and tissue processing for immunohistochemistry
Mice were deeply anesthetized with chloral hydrate (40%, i.p.) and perfused through the left cardiac ventricle with 0.9% saline solution, followed by 4% paraformaldehyde in 0.1 M phosphate buffered saline (PBS), pH 7.4. After perfusion, the brains were removed, post-fixed in the same fixative solution for 24 h at room temperature (RT) and immersed in a 30% sucrose solution in PBS at 4 °C. Serial coronal sections (30 μm) of hippocampi were obtained with a vibratome (Vibratome®, Series 1000, St. Louis, MO, USA) and stored in 0.1 M PBS at 4 °C.
To analyze cell proliferation and survival in the subgranular cell layer of the DG, a series of one-in-six free-floating sections were processed for detection of the BrdU and Ki67 immunoreactivity. Ki67 is a nuclear protein that is expressed during all active phases of the cell cycle, but is absent from cells at rest, and therefore used as an endogenous proliferative marker [30], [31]. Briefly, after DNA denaturation in 2 N HCl at 65 °C for 30 min and pre-incubation with 5% blocking solution (0.1M TBS: 0.15 NaCl, 0.1M Tris-HCl, pH 7.5, supplemented with 5% normal goat serum and 0.2% Triton X-100), sections were incubated for 48 h at 4 °C in mouse anti-BrdU primary antibody (1:50; Sigma) and rabbit polyclonal anti-Ki67 primary antibody (1:200; Vector Laboratories). The sections were then incubated with Alexa 564 donkey anti-mouse IgG secondary antibody (1:400; Jackson ImmunoResearch) and Alexa 488 donkey anti-rabbit IgG secondary antibody (1:400; Jackson ImmunoResearch) for 2 h at RT.
Differentiation was assessed by BrdU/neuronal nuclei (NeuN) double-labeling. Briefly, after DNA denaturation in 2 N HCl and blocking (as described before), the sections were incubated for 48 h at 4 °C in rat anti-BrdU (1:100; Bio-Rad) and mouse anti-NeuN (1:50; Chemicon) primary antibodies. The sections were then incubated with the secondary antibodies Alexa 488 donkey anti-rat IgG secondary antibody (1:400; Jackson ImmunoResearch) and Alexa 568 donkey anti-mouse IgG secondary antibody (1:400; Jackson ImmunoResearch) for 2 h at RT. All sections were mounted onto 2% gelatine-coated microscope slides, coverslipped with Fluor Mount (Sigma–Aldrich), and stored in the dark at 4 °C.
2.1.5. Morphological quantification
All morphological analyses were performed on coded slides, with the experimenter blind to the experimental group, using a Leica microscope (Leica DM5500 B). The total numbers of BrdU and Ki-67 immunopositive cells in the DG of the hippocampus were estimated by manually counting all positive cells present within two–three nuclear diameters below the granule cell layer (GCL). All sections containing the DG region of the hippocampus, from 1.34 mm posterior to Bregma to 3.52 mm posterior to Bregma [32], were used for the analysis. Thus, the results were expressed as the total number of labeled cells in the DG sub-region of the hippocampus by multiplying the average number of labeled cells/DG section by the total number of 30 μm thick-sections that contain the DG (estimated as 73 sections).
2.1.6. Phenotypic analysis
Newly-formed neurons in the DG of adult mice were analyzed in one-in-six series of sections by immunofluorescent double staining [33]. A maximum of 50 BrdU-labeled cells per mouse were randomly selected for analysis of co labeling with NeuN. Analysis was performed by confocal microscopy (Leica DMI6000 B) in sequential scanning mode to avoid cross-bleeding between channels. Double-labeling was confirmed by three-dimensional reconstruction of z-series covering the entire nucleus (or cell) of interest.
2.2. In vitro study
We used two complementary in vitro models to dissect direct roles of elevated LDL levels and LDLr downregulation in survival, proliferation and fate decisions of adult NPC: i) primary neurospheres, which provide a good read-out for effects of extrinsic factors (here serum LDL) on proliferative and differentiation potential of NPC isolated directly from the DG of wild type mice; and ii) adherent monolayers, in which cells remain more isolated and homogeneous, thus eliminating the heterogeneous nature of the neurospheres, since cells inside the neurosphere have the tendency to differentiate [34], [35], [36].
2.2.1. Animals
C57BL/6JRj mice were bred by Jackson Laboratory and purchased from Janvier, Germany. Animal handling and all procedures during the experiment were conducted in accordance with the applicable European and national regulations (Tierschutzgesetz) and were approved by the responsible authority (Landesdirektion Sachsen, TVA 24-9168.24-1/2013-16a). All animals were 8 weeks of age at the time of dissection. Animals were maintained on a 12 h light/dark cycle with food and water provided ad libitum.
2.2.2. Culture of adherent adult hippocampal precursor cells
Primary mouse dentate gyrus NPC were isolated and the adherent monolayer cultures prepared as previously reported [34], [35]. Cells were maintained in neurobasal growth medium (Gibco, Life Technologies) supplemented with 2% B27 (Invitrogen), 1X GlutaMAX (Life Technologies) and 50 units/ml penicillin/streptomycin (Life Technologies), 20 ng/mL human basic fibroblast growth factor (bFGF; PeproTech, Rocky Hill, NJ), and 20 ng/mL epidermal growth factor (EGF; PeproTech). Every 2 days, 75% of the culture medium was replaced with fresh medium. Precursor cells were passaged at approximately 80% confluence by treating with Accutase (PAA Laboratories, Linz, Austria) for 2 min, and reseeded in PDL/laminin-coated dishes at a density of 1 × 104 cells per cm2.
For the time-course differentiation analysis of gene expression, NPC were seeded in PDL/laminin-coated 60 × 15mm Petri dishes. Differentiation was induced by growth factor withdrawal whereby cells growing under standard proliferation conditions (20 ng/ml each of bFGF and EGF) were switched to medium without any growth factors [34]. Cells harvested before removal of growth factors were considered time 0, while the following dishes were harvested at 24–96 h after growth factor withdrawal.
2.2.3. Gene silencing
To knockdown the LDLr gene expression, adherent NPC were transfected with a pool of four different small interfering RNAs (siRNAs) targeting mouse LDLR (ON-TARGETplus SMARTpool, Cat. No. L-048572-00-0005, Dharmacon). Cells were cultured for 48 h in an antibiotic-free medium before the addition of 50 nM siRNA in culture medium containing 0.001% transfection agent (DharmaFECT, Dharmacon). After 24 h, the transfection media was replaced by regular culture media and cells were incubated for further 24 h before harvesting, fixation or cell viability assay.
2.2.4. LDL and 27-HC treatment
Adherent cells were cultured in 6 or 24-well plates in growth medium for 2 days followed by 2 additional days in the presence of human LDL (0, 25, 50, 100 or 200 μg/mL) or 27-hydroxycholesterol (27-HC; 0, 1, 5 or 10 μM; Santa Cruz). LDL was purchased from Sigma or prepared as described previously [37]. For neurospheres assays, the freshly obtained cells were plated into 48 wells of a 96-well plate in the presence of LDL (0, 25, 50, 100 or 200 μg/mL) and grown as described above.
2.2.5. Immunocytochemistry
To assess cell proliferation, adherent monolayers were cultured on coverslips coated with PDL/laminin in 24-well plates. After treatments, adherent cells were incubated with BrdU (10 μM) for 2 h, then fixed with 4% PFA in 0.1 M PBS for 10 min at RT. For BrdU staining, cells were washed with PBS, and DNA was denatured with 1N HCl for 30 min at 37 °C. For all stainings, cells were blocked for 1 h in blocking solution containing 10% donkey serum and 0.2% Triton X-100 in PBS, followed by an incubation with a primary (rat monoclonal anti-BrdU; 1:500; AbD Serotec); and secondary (Cy3; 1:1000 Jackson ImmunoResearch) antibodies prepared in blocking solution for 2 and 1 h, respectively. The nuclei were labeled with Hoechst and coverslips were mounted onto glass slides for Zeiss Apotome microscope imaging.
To assess apoptosis induction the number of pyknotic nuclei, average nucleus size and integrated density (fluorescence intensity normalized by cell size) were quantified per image using the ImageJ software.
2.2.6. CFSE cell proliferation assay
The influence of treatments on proliferation kinetics of NPC was estimated by cell division tracing with the fluorescein compound CFSE (Carboxyfluorescein succinimidyl ester). Before seeding, 1 × 106 cells were loaded with 10 μM CFSE for 10 min at 37 °C. The addition of media at 4 °C for 5 min stopped the reaction. After washing and resuspension, cells were seeded in 6-well plates, grown for 2 days and treated with LDL (100 μg/mL) or transfected with siRNAs for 48 h as described above. Finally, cells were washed, harvested and fixed in 2% PFA for 20 min at RT. Cell suspension was analyzed in BD FACSCanto II flow cytometer (BD Biosciences).
2.2.7. Quantitative RT-PCR
Total RNA was prepared using the Qiagen RNeasy kit (Qiagen, Hilden, Germany). The adherent NPC cultures were washed once with PBS and then harvested by on-plate lysis with RLT buffer. Lysate was collected using a cell scraper and RNA extracted following the manufacturer's protocol, including the optional DNAse step. The concentration and the purity of RNA were analyzed using a NanoDrop 1000 spectrophotometer (Thermo Scientific). For reverse transcription, 1 μg of RNA was used. The cDNA was prepared using Superscript II kit (Invitrogen) using oligo(dT) primers following the manufacturer's protocol. A cDNA negative reaction was also set up without addition of reverse transcriptase enzyme to ensure no genomic DNA contamination.
Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) for LRP1, LDLR, CYP27A1, CYP43A1, FDFT1, and HMGCR was performed using a SYBR Green PCR Mix (Qiagen) following the manufacturer's protocol. Template cDNA was prepared as above and each reaction contained cDNA from 0.1 μg total RNA. Thermal cycling and fluorescence detection were carried on a CFX96 Real Time PCR detection system (Bio-Rad, California, USA). Quantification was done using the ΔΔCt method after normalizing to the TATA box binding protein (TBP) housekeeping gene. Primer sequences for LRP1 were 5′- TTCTCCTGCTGCTTCTGGTG- 3′ and 5′- TCGGGCTCTCCACCTTCATA-3′; for LDLR 5′- GGAGCAGCCACATGGTATGA- 3′ and 5′- ATGTTCTTCAGCCGCCAGTT- 3′; for CYP27A1 5′- GGGTATCTGGCTACCTGCAC-3′ and 5′- CTCCTGGATCTCTGGGCTCT- 3′; CYP46A1 5′- GGTGGATGAGGTTGTCGGTT and 5′- TCAAGGTCTCCTCCTCCAGC- 3′; FDFT1 5′- AACATGCCTGCCGTCAAAG- 3′ and 5′- GGAGTAGTGGCTTCGGGAGA- 3′; HMGCR 5′- AAGCCAGTGGTCCCACAAAT- 3′ and 5′- TTGCATGCTCCTTGAACACC- 3′ and TBP 5′- CCAGAACAACAGCCTTCCAC- 3′ and 5′ - GGAGTAAGTCCTGTGCCGT- 3′.
2.2.8. Lipid droplets staining
Treated and control cells grown on glass coverslips in 24-well plates were fixed in 4% PFA in 0.1 M PBS for 10 min at RT, washed with PBS and incubated with a solution of 0.1 μg/mL LD540 (generously donated by Dr. Uenal Coskun) in PBS for 15 min [38]. Afterwards, the nuclei were labeled with Hoechst, coverslips were washed three times with PBS and mounted with fluorescence mounting medium without glycerol (Dako). Images were acquired with an SD spinning disk confocal microscope (Carl Zeiss), excitation at 530 nm. The number of lipid droplets were quantified per image, and normalized by cell number on the field, using the ImageJ software.
2.2.9. Fluorescent LDL internalization assay
48 h after treatment, adherent cells plated on PDL/laminin-coated coverslips in 24-well plates were washed twice with media and incubated for 45 min at 37 °C. The first LDL pulse (10 μg/mL; BODIPY™ FL LDL, Invitrogen) was given 60 min before fixation. After 10 min of incubation with LDL-Bodipy, wells were washed twice with media and returned to the incubator until the second LDL pulse. LDL-DiI (10 μg/mL; DiI LDL, Invitrogen) was given 20 min before fixation. After 10 min of incubation wells were washed twice with media and kept at 37 °C/5% CO2 until the end of the protocol (fixation). Cells were washed with cold PBS and fixed in 4% PFA for 10 min at RT. The coverslips were washed three times with PBS and mounted with fluorescence mounting medium. Images were acquired with a SD spinning disk confocal microscope (Carl Zeiss).
2.2.10. Microarray processing
Cells grown in 6-well plates were treated with 100 μg/mL LDL or saline as a control or transfected with LDLr or non-target siRNAs. Total RNA from three replicate cultures for each condition was extracted as described above. For each sample, 100 ng total RNA was used to synthetize Cy-3 labeled target cRNA with the Low Input Quick Amp Labeling Kit (Agilent Technologies, Germany), followed by hybridization to SurePrint G3 Mouse GE 8 × 60K one-colour microarrays (AMADID 028005; Agilent Technologies). Raw signal intensities were obtained with Feature Extraction software (protocol 028005_D_F_20150604; Agilent Technologies). The data were further analyzed in R using the limma package, which fits to each gene a linear model with treatment as a factor and computes moderated t-statistics applying the empirical Bayes approach [39]. Raw intensities were background corrected (subtraction method) and normalized between arrays applying the cyclic loess normalization. Only probes which were at least 10% brighter than the negative control (the 95th percentile of negative probes intensities for each array) on at least 3 arrays were retained. Replicated probes were averaged. A 10% FDR (false discovery rate) cut-off and an absolute value of fold change of 1.5 were used to identify differentially expressed genes. Functional enrichment analysis of canonical pathways and potential upstream regulators was performed with Ingenuity Pathway Analysis software (IPA; Qiagen) at default settings [40]. Array data are available online at the Gene Expression Omnibus repository(www.ncbi.nlm.nih.gov/geo/; accession: GSE136798).
2.3. Statistics analysis
Data were analyzed using GraphPad Prism and R. The statistical analyses were carried out using unpaired two-tailed Student's t-test, one-way analysis of variance (ANOVA) or ANOVA with repeated measures when appropriate. Post hoc comparisons were performed using the Dunnett's test. Data are presented as means ± standard error of the mean (SEM). A p value < 0.05 was considered to be statistically significant. Full results of statistical tests are available in the Supplementary Table 1.
3. Results
3.1. LDLr−/− mice perform poorly in the DG-dependent metric change task
It has been demonstrated that LDLr−/− mice perform poorly in a spatial memory task, suggesting a hippocampus related cognitive impairment [14]. In this study, seeking a DG-specific contribution to the spatial memory deficit in the LDLr−/− mice, we used the metric spatial change task (Figure 1A) to examine spatial pattern processing [26], [27]. During the task, mice are allowed to explore a pair of objects placed 40 cm apart in a 15 min long habituation session (Figure 1B). In the test session the same objects are replaced only 20 cm apart, and the mice are allowed to explore for 5 min. An increased interest to re-explore the objects in the test session indicates that the mice were able to notice the metric novelty [27], [41]. The habituation of mice during the 15 min session was evaluated by comparing the time that mice spent exploring the objects in three 5 min periods. Successful habituation was observed in both groups, as evidenced by a decrease in object exploration over the training session [one-way ANOVA with repeated measures; C57BL/6J: F (9, 18) = 2.94, p = 0.025; LDLr−/−: F (9, 18) = 4.19, p = 0.047; Figure 1C]. Interestingly, there is difference between the exploration time by C56BL/6J and LDLr−/− mice at the time points of 10 and 15 min (two-tailed t-test t (18) = 3.023, p = 0.0073 and t (18) = 2,635, respectively). However, during the 5 min test after repositioning of the objects, wild type mice, but not LDLr−/− mice, showed an increase in object exploration compared to the final 5 min of the habituation session [Figure 1D; t-test; t (18) = 3.62, p < 0.01].
3.2. Performance in the temporal ordering task is not affected by the absence of LDLr
The next day, we tested the mice in the temporal ordering task (Figure 1A), which is primarily dependent on the intact CA region of the hippocampus [42], [43]. In this task, mice are presented three different pairs of identical objects in three consecutive sample sessions (Figure 1E). In the test trial, mice are exposed to one object from the first session and one object from the last session. Intact animals show preference for the object more distant in time. To exclude object preference during acquisition sessions, for each genotype we performed a one-way ANOVA of the object exploration time with the session as a factor. We did not detect an effect of the session for the C57BL/6J [F (2, 36) = 1.46, p = 0.25] or the LDLr−/− mice [F (2, 42) = 1.96, p = 0.15], suggesting that all mice similarly explored different objects across sessions 1–3. To determine group differences in temporal ordering, the differentiation scores of the exploration time of each of the objects during the test session were calculated (Figure 1G). The LDLr−/− group showed similar preference towards temporally distant object as the wild type group [t-test; t (21) = 0.61, p = 0.55].
3.3. Hippocampal cell proliferation and adult neurogenesis are decreased in LDLr−/− mice
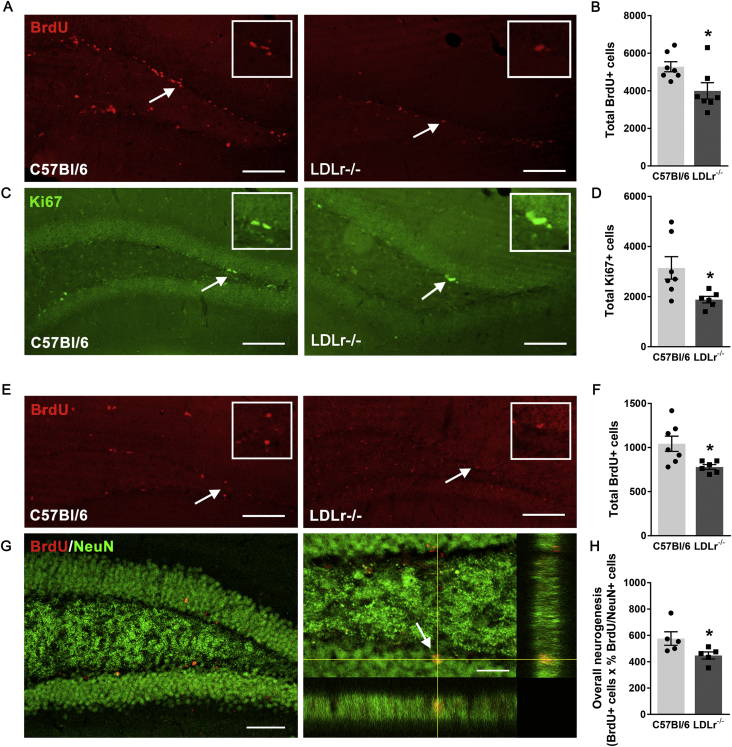
To assess the impact of the LDLr knock-out on adult hippocampal neurogenesis, we first compared the rates of cellular proliferation between 3 months old C57BL/6J and LDLr−/− mice. Both groups of animals were injected with the thymidine analogue BrdU and euthanized 24 h later (Figure 1A). As shown in Figure 2B, the LDLr−/− mice had reduced numbers of BrdU labeled cells compared to wild-type mice [t-test; t (12) = 2.51, p = 0.0273], which indicates lower numbers of proliferating precursor cells. We corroborated this finding using immunohistochemistry against an endogenous antigen Ki-67 present on a nuclear protein that is expressed during all active phases of the cell cycle but is absent from cells in G0 [44]. This marker also indicated that cell proliferation was reduced in the LDLr−/− mice when compared to the wild type group [t-test; t (11) = 2.52; p = 0.028; Figure 2D].
Figure 2.
Cell proliferation and differentiation in the granular cell layer of the DG. (A–D) C57BL/6J and LDLr−/− mice were sacrificed 24 h after the last BrdU injection by transcardial perfusion and their brains were processed for immunohistochemistry for BrdU (A) and an endogenous cell proliferation marker Ki-67 (D). LDLr−/− mice showed a reduced number of both BrdU (B) and Ki-67 (D) immunopositive cells. (E, F) Immunolabeling for BrdU-positive cells present in the DG 28 days after the last BrdU administration (E) reveals lower survival of new cells in the LDLr−/− mice (F). (G). The newborn neurons were identified by the BrdU/NeuN co-labeling and confocal microscopy. (H) An estimation of overall neurogenesis based on the total number of BrdU-positive cells that survived the 28-day period multiplied by the proportion of BrdU-positive cells that acquired a mature neuronal phenotype indicates a significant decrease in the overall number of new neurons of LDLr−/− mice. White arrows indicate either BrdU, Ki67 or BrdU/NeuN immunopositive cells in the DG of C57BL/6J and LDLr−/− mice. Scale bars = 100 μm (A–G) and 20 μm (G in BrdU/NeuN insert). Data are presented as means ± SEM. N = 7 per group. *p < 0.05, t-test.
Next, we assayed the number and neuronal differentiation of newly generated cells that survived 4 weeks after incorporation of BrdU to estimate net neurogenesis (Figure 1A). We observed a decline in the total number of 4 weeks old BrdU-labeled cells in the LDLr−/− animals (Figure 2F; t-test; t (12) = 2.71, p = 0.02). The proportion of new neurons among the cells that incorporated BrdU was determined using the mature neuronal marker NeuN and confocal microscopy (Figure 2G). The net neurogenesis, estimated by multiplying the number of BrdU-positive cells that survived the 28 day period by the proportion of BrdU-positive cells that co-expressed NeuN, was also significantly reduced in the hypercholesterolemic mice (Figure 2H; one-tailed t-test; t (8) = 2.22, p = 0.028).
3.4. LDL treatment and LDLr downregulation affect the proliferation of adult hippocampal precursor cells in vitro
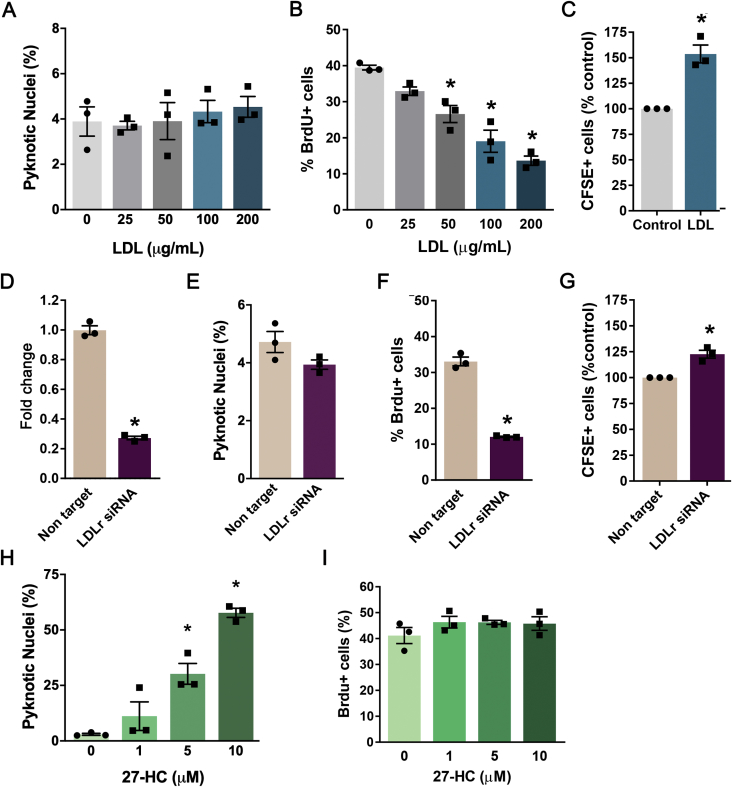
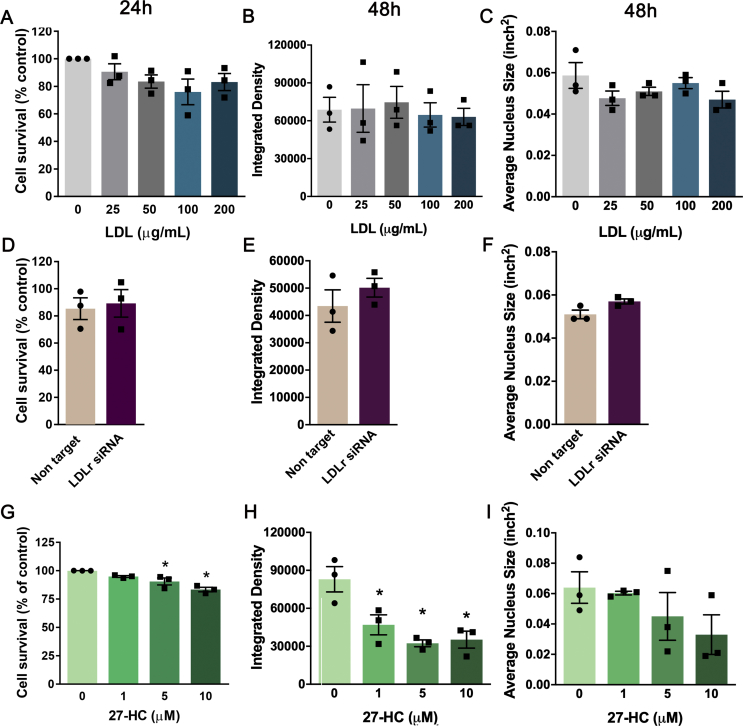
Varying concentrations of LDL did not affect cell viability of adherent hippocampal NPC after 24 h [one-way ANOVA; F (4,10) = 2.27; p = 0.13; Fig. S1A], neither did the treatment increase pyknosis, a sign of apoptosis, after 48 h [one-way ANOVA; F (4,10) = 0.38, p = 0.82; Figure 3A]. On the other hand, the addition of LDL decreased cell proliferation, as assessed 2 h after the incorporation of BrdU [one-way ANOVA; F (4,10) = 29.94; p < 0.0001; Figure 3B]. In order to estimate cell divisions, we used the fluorescein compound CFSE, which is internalized by the cells at the time of seeding and segregates roughly equally into the daughter cells, thereby being diluted with every cell division. We observed that cells treated with LDL at 100 μg/mL showed fewer cell divisions during the tracing period [t-test; t (4) = 6.14, p = 0.036; Figure 3C].
Figure 3.
Effects of LDL treatment and siRNA-mediated LDLr knock-down on survival and proliferation of adult hippocampal precursor cells in vitro. Precursor cells isolated from C57BL/6J mice were propagated as adherent monolayers and treated with: (A–C) human plasma LDL (0–200 μg/mL); (D–G) transfected with a pool of siRNAs targeting the LDLR gene or a pool of non-targeting siRNAs as a control; (H–I) or 27-HC (0–10 μM). (D) LDLR transcript expression after siRNA transfection was evaluated by qRT-PCR. (A, E, H) 48 h after each treatment, the number of pyknotic nuclei assessed by morphological analysis of Hoechst stainings. To asses cell proliferation, cultures were analyzed 48 h after addition of LDL (B–C), transfection with siRNAs (F–G) or 27-HC treatment (I). (B, F; I) Precursor cells were incubated for 2 h with BrdU. Proportion of BrdU-immunoreactive, actively dividing cells decreased after exposure to LDL (B) LDLR knock-down (F) but not 27-HC treatment (I). (C, G) Fluorescence of CFSE-loaded cells increased after addition of LDL (100 μg/mL; C) or transfection with LDLR siRNA (G) indicating fewer cell divisions compared to control cells. Data are presented as means ± SEM. N = 3 per group. *p < 0.05, t-test (C, D, E, F, G), one-way ANOVA followed by Dunnett's post hoc test (A, B, H, I).
To downregulate the expression of the LDLR mRNA in adherent cultures, cells were transfected with a pool of four siRNAs targeting LDLR or a pool of non-target control siRNAs. A reduction of 73% in LDLR mRNA was confirmed by RT-qPCR [t-test; t (4) = 22.79, p < 0.0001; Figure 3D]. The LDLR siRNA transfection did not affect cell survival after 24 h [t-test; t (4) = 0.30, p = 0.78; Fig. S1D] or pyknosis at 48 h [t-test; t (4) = 1.96, p = 0.12; Figure 3E] but it decreased cell proliferation assessed by BrdU incorporation [t-test; t (4) = 17.27, p < 0.0001; Figure 3F]. The increased CFSE retention after the cell tracing also indicated a lower number of cell divisions [t-test; t (4) = 5.98, p = 0.004; Figure 3G].
In hypercholesterolemia, there is an excessive formation of the cholesterol oxidized metabolite 27-HC, which has been linked to pathogenesis of neurodegenerative diseases [45]. Therefore, we tested whether this compound could impair survival and proliferation of precursor cells. 27-HC decreased cell survival after 24 h [one-way ANOVA: F (3, 8) = 14.16, p = 0.0015; Fig. S1G], and increased pyknosis at 48 h [one-way ANOVA: F (3, 8) = 34.97, p < 0.001; Figure 3H] without affecting cell proliferation [one-way ANOVA: F (3, 8) = 1.11, p = 0.4015; Figure 3I].
Together, our data show that the presence of high concentrations of LDL in the culture medium as well as lowering of LDLr expression reduced cell proliferation of adult hippocampal NPC.
3.5. LDL treatment increased intracellular lipid storage in precursor cells with no effect on LDL trafficking
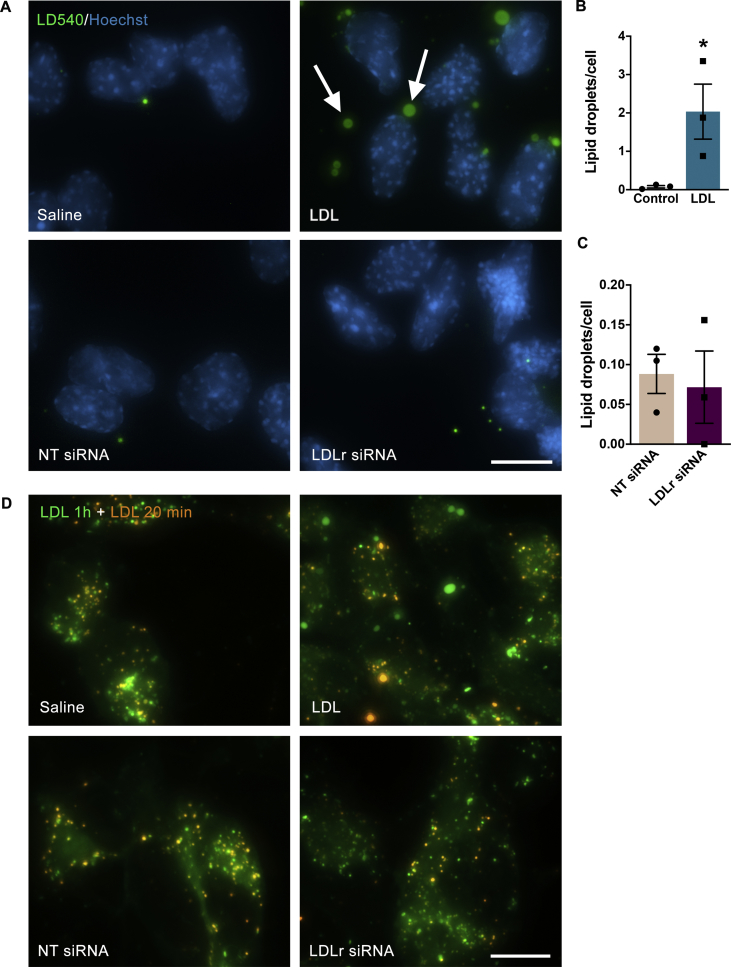
Neurons in the adult brain take up the ApoE-cholesterol complexes via endocytosis through the LRP1 and LDLr receptors and cholesterol-loaded endocytic vesicles are hydrolyzed in lysosomes [46]. To check if the exogenously added LDL cholesterol would accumulate intracellularly, we estimated cholesterol storage by staining lipid droplets with the LD540 dye. Since the loss of LDLr might have altered cellular lipid homeostasis, we also investigated lipid droplets in siRNA transfected cells. We found that the LDL treatment of adult hippocampal NPC in vitro induced an increase in the number of lipid droplets [t-test; t (4) = 9.37, p = 0.0007; Figure 4B]. On the other hand, the knock-down of the LDLr by siRNA did not cause any alteration [t-test; t (4) = 0.44, p = 0.68; Figure 4C].
Figure 4.
Effect of LDL treatment and LDLr siRNA transfection on lipid internalization and storage. (A) Representative images of precursor cells stained with LD540 to visualise lipid droplets (green; white arrows). Cell nuclei were counterstained with Hoechst (blue). (B–C) Quantification of lipid droplets per field in LDL (100 μg/mL) treated (B) and siRNA transfected cells (C). (D) Representative images of internalized fluorescent LDL. LDL-Bodipy conjugate (green) was added 60 min before cell fixation, while the second LDL pulse, LDL-DiI (orange), was added 20 min before cell fixation. NT: non-target control. Scale bars = 20 μm. Data are presented as mean ± SEM. N = 3 per group. *p < 0.05, t-test.
To estimate if endocytosis of cholesterol in NPC is perturbed when LDLr is down-regulated or when cells are overloaded with LDL, we investigated the dynamics of LDL intracellular transport by pulse labeling with two different LDL fluorescent conjugates, LDL-Bodipy and LDL-DiI, at 60 and 20 min before fixation (Figure 4D). The qualitative evaluation suggested that after 48 h of LDL treatment, cells were still able to uptake fluorescent LDL, despite intracellular cholesterol overload. Additionally, knock-down of LDLr using siRNA did not prevent the uptake of LDL in adherent hippocampal NPC, because they were still able to internalize the dyes. Finally, the lack of co-localization of the dyes administered at different time points suggested that the intracellular trafficking of LDL cargo through subsequent endocytic compartments was not disrupted.
3.6. Cell differentiation, LDL treatment and LDLr knock-down modulate cholesterol metabolism-related genes
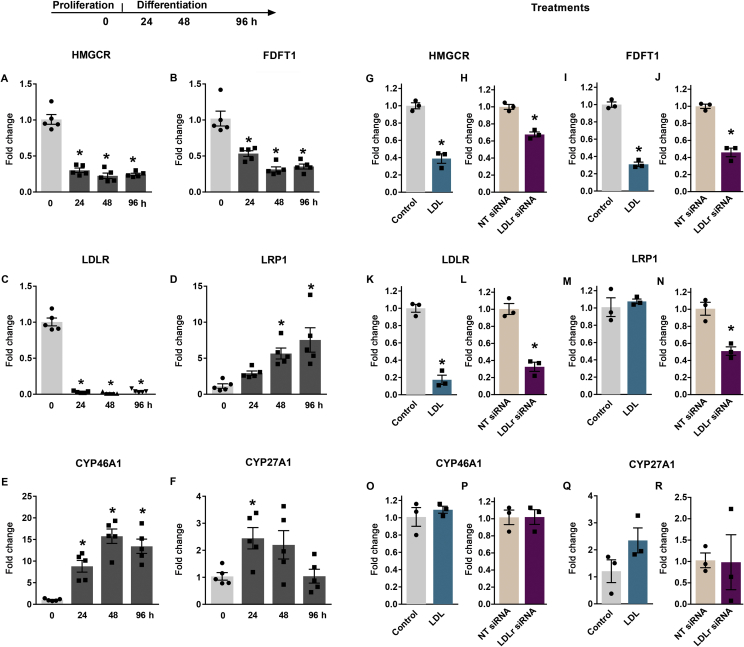
To better understand the roles of LDL cholesterol and LDLr in proliferation and differentiation of hippocampal NPC, we analyzed the dynamics of mRNA expression of genes related to cholesterol metabolism in NPC in vitro in proliferative conditions and at 24, 48 and 96 h after growth factor withdrawal, which induces cell cycle exit and differentiation [34] (Figure 5). Gene expression levels of key enzymes in the cholesterol synthesis pathway, HMG-CoA reductase (HMGCR; Figure 5A) and squalene synthase (FDFT1; Figure 5B), were decreased already after 24 h from the onset of differentiation [one-way ANOVA; HMGCR: F (3,16) = 79.51; p < 0.0001; FDFT1: F (3,16) = 29.06; p < 0.0001, respectively]. The LDLR mRNA expression was dramatically decreased in differentiated cells [one-way ANOVA; F (3,16) = 302.3; p < 0.0001] at all time points analyzed, while the LRP1 mRNA levels were increased [one-way ANOVA; F (3,16) = 8.88; p = 0.0011], with a significant difference from 48 h of differentiation (Figure 5C,D). The mRNA levels of enzymes that catalyze oxidation of cholesterol to oxysterols, CYP46A1 and CYP27A1, were increased after 24 h in differentiation conditions [CYP46A1: F (3,16) = 22.87; p < 0.0001; one-way ANOVA; and CYP27A1: F (3,16) = 4.34; p = 0.02; Figure 5E,F]. While the CYP27A1 mRNA returned to levels similar to the proliferating cells after 96 h of differentiation, CYP46A1 remained upregulated. In conclusion, transition from proliferation to differentiation in adult hippocampal NPC in vitro was associated with remodelling of cholesterol synthesis, uptake and catabolism.
Figure 5.
Expression of cholesterol metabolism-related genes in precursor cells in vitro. (A–F) Time-course evaluation of mRNA levels in proliferating precursor cells (0) and at 24, 48 or 96 h of differentiation. (G–R) Effect of LDL treatment (100 μg/mL) and LDLR siRNA transfection on gene expression in proliferating precursor cells. mRNA levels were measured by qRT-PCR. HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; FDFT1: squalene synthase; LDLR: low-density lipoprotein receptor; LRP1: Low-density lipoprotein receptor-related protein 1; CYP46A1: cholesterol 24-hydroxylase; CYP27A1: sterol 27-hydroxylase; NT: non-target control. Data are presented as means ± SEM. N = 5 per group, time-course assay, N = 3 for treatments. *p < 0.05. One-way ANOVA followed by Dunnett's post hoc (time-course assay) and t-test (treatments).
We next examined the effects of LDL treatment and LDLr knock-down on the expression of genes related to cholesterol metabolism in NPC under proliferative conditions. As shown in Figure 6, 48 h of treatment with LDL at a concentration of 100 μg/mL decreased the expression of mRNAs encoding for HMG-CoA reductase and squalene synthase enzymes, as well as of LDLR and LRP1 (HMGCR: t(4) = 9.38, p = 0.007; FDFT1: t(4) = 17.43, p < 0.0001; LDLR: t(4) = 11.80, p = 0.0003; LRP1; t(4) = 0.60, p = 0.58). Cells transfected with siRNA against LDLR again showed a 70% reduction in the LDLR expression (t (4) = 8.07, p = 0.0013). This downregulation was associated with reduced HMGCR and FDFT1 (HMGCR: t (4) = 7.89, p = 0.0014; FDFT1: t (4) = 9.65, p = 0.0006, respectively). Surprisingly, knock-down of LDLr also resulted in downregulation of the LRP1 receptor gene expression (t (4) = 5.52, p = 0.0053). The mRNA levels of CYP46A1 and CYP27A1 were not affected by either LDL treatment (CYP46A1: t (4) = 0.72, p = 0.51; CYP27A1: t (4) = 1.83, p = 0.14) or LDLR siRNA transfection (CYP46A1: t (4) = 0.03, p = 0.98; CYP27A1: t (4) = 0.06, p = 0.95).
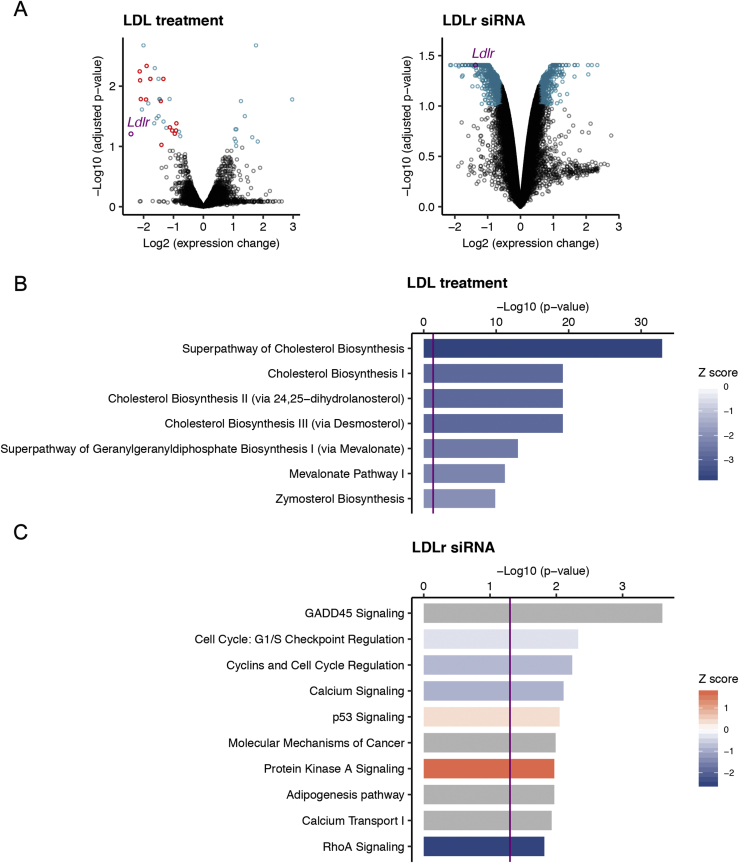
Figure 6.
Gene expression profiling in precursor cells reveals distinct biological effects of LDL and LDLr downregulation. Proliferating adherent precursor cells were treated with 100 μg/mL of LDL and LDLR siRNA, and after treatments gene expression was analyzed by microarrays. (A) Volcano plots showing differentially regulated genes at 10% false discovery rate and an absolute fold change cut-off of 1.5 (blue). Red points indicate genes from the cholesterol biosynthetic pathway changed in LDL treated cells. LDLR is highlighted in purple. (B, C) Selected top ingenuity canonical pathways enriched in LDL treated (B) and LDLR siRNA transfected (C) NPCs. Blue indicates pathways that are predicted to be inhibited in treated cells, while activated pathways are shown in orange. The strength of change (Z score) is indicated by the colour intensity. For pathways shown in grey, insufficient data exist to predict the activity change. Purple vertical line indicates significance of enrichment (p < 0.05). Full lists of differentially expressed genes and enriched pathways are available in the extended data Supplementary Tables 2–4.
3.7. Gene expression analysis revealed cell biological pathways affected by LDL treatment and LDLr downregulation
To get an insight into molecular changes that might underlie altered behavior of NPC upon LDL exposure and LDLr downregulation, we performed gene expression profiling using microarrays. We detected 42 and 1151 differentially expressed transcripts in LDL treated (100 μg/mL) and LDLR siRNA transfected cells, respectively (Figure 6A). We next carried out a functional annotation of differentially expressed genes using Ingenuity Pathway Analysis (IPA). IPA is considering directionality and magnitude of gene expression changes to predict activation or inhibition of each enriched biological pathway. Furthermore, it predicts changes in activity of upstream regulators that are consistent with differences in gene expression [40]. Upstream regulator analysis revealed that the LDL treatment inhibited transcriptional networks dependent on sterol regulatory element-binding proteins (SREBPs) with top enriched canonical pathways related to cholesterol biosynthesis (Figure 6B). Although the knockdown of LDLR induced broad changes in gene expression, the enrichment analysis did not lead to strong conclusions (Supplementary Table 2). In agreement with slower proliferation of LDLR siRNA transfected NPC, several of the top enriched ingenuity canonical pathways were related to cell cycle regulation (Figure 6C and Supplementary Table 3). In both treatments, IPA also identified numerous putative upstream regulators (Table 1 and Supplementary Table 3).
Table 1.
Ingenuity pathway upstream analysis. Selected regulators, for which activation status was predicted from observed gene expression changes in LDL treated (100 μg/mL) and LDLR siRNA transfected NPC. Full lists of identified regulators are available in a table in extended data Supplementary Table 4.
| Upstream regulator | Predicted activation state | Activation Z-score | p-value of overlap |
|---|---|---|---|
| LDL treatment | |||
| SREBF2 | Inhibited | −4.28 | 8.4E-39 |
| SCAP | Inhibited | −4.177 | 2E-38 |
| INSIG1 | Activated | 3.904 | 6.8E-29 |
| SREBF1 | Inhibited | −4.221 | 8.8E-29 |
| INSIG2 | Activated | 2.758 | 1.3E-19 |
| CYP51A1 | Activated | 2.646 | 5.4E-17 |
| INSR | Inhibited | −3.412 | 1.8E-16 |
| SIRT2 | Inhibited | −2.646 | 2E-16 |
| MBTPS1 | Inhibited | −2.433 | 2E-14 |
| PPARGC1B | Inhibited | −2.789 | 2.4E-13 |
| MTORC1 | Inhibited | −2.043 | 1.3E-10 |
| LMNB1 | Activated | 2.433 | 3.3E-10 |
| CYP7A1 | Inhibited | −2.219 | 3.7E-10 |
| HDL-cholesterol | Activated | 2.236 | 0.000000006 |
| ACACB | Activated | 2 | 9E-09 |
| ABCA1 | Activated | 2.019 | 9E-09 |
| Pde4 | Activated | 2 | 1.6E-08 |
| PPARG | Inhibited | −2.489 | 0.00000055 |
| MAPK14 | Activated | 2.207 | 0.0000094 |
| EGF | Inhibited | −2.219 | 0.00013 |
| PTEN | Activated | 2.433 | 0.00022 |
| LDLR siRNA | |||
| NEUROG1 | Activated | 2.53 | 0.0011 |
| FOXO3 | Activated | 2.161 | 0.034 |
| SP1 | Inhibited | −2.568 | 0.00014 |
| ATF4 | Inhibited | −2.997 | 0.00019 |
| TGFB1 | Inhibited | −2.013 | 0.00028 |
| E2F2 | Inhibited | −2 | 0.0067 |
| CREBBP | Inhibited | −2.891 | 0.000053 |
4. Discussion
In this study we have shown that a mutation in the LDLr gene, as found in FH, might have direct effects on the biology of adult neural stem cells and adult neurogenesis, which in turn might be responsible for cognitive impairments observed in cases of hypercholesterolemia [1], [2], [3]. Adult hippocampal neurogenesis is believed to functionally contribute to neural plasticity and cognition throughout life, while its decay might be linked to impaired cognitive and emotional resilience in humans [47], [48], [49].
Our findings show that 3 months old LDLr−/− mice performed poorly in the DG-dependent metric change task, which is in line with previous studies reporting spatial memory deficits in this rodent model of hypercholesterolemia [14], [17], [50]. Additionally, as previously reported, the LDLr−/− mice present hyperlocomotion [51] and, since they cover a greater distance, it is reasonable that they probably find the objects more frequently during the training session, resulting in increased exploration values. Lack of deficit in the CA-related temporal ordering task might be an indication that the previously observed synaptic loss in the CA of LDLr −/− mice at 6 and 14 months does not affect younger animals, or is not compromising its function [13], [14]. In our study, the DG-related behavioral alterations were accompanied by decreased cell proliferation and neurogenesis in this structure. Notably, ablation of adult neurogenesis is well known to impair performance in behavioral tasks that require resolution of similar inputs, such as a subtle metric novelty [52].
The higher incidence of early cognitive deficits in FH patients, when compared to patients with sporadic hypercholesterolemia [5], [6], draws attention to inherited factors, such as mutations in the LDLr gene. In the LDLr−/- mice, high plasma cholesterol levels and the absence of LDLr might both have a role in impairing the neurogenic process. High levels of cholesterol in the blood of the LDLr−/− mice are associated with increased BBB permeability [50]. It has been demonstrated that apoB100, the apolipoprotein constituent of LDL, found in the brain of hypercholesterolemic animals, was associated with structural and functional damages to neuronal cells [9], [11], [53]. As the neurogenic niche is a largely vascularized area [54], this region is more likely to be vulnerable to the entrance of LDL and other molecules from hypercholesterolemic plasma [55]. Recent studies that used dietary strategies, resembling the sporadic form of the disease, enforce the hypothesis of hypercholesterolemia disturbing neurogenesis and hippocampus-dependent behaviors in adult mice [4], [56]. However, the present study demonstrates for the first time an impairment in adult neurogenesis in the LDLr−/− model of FH.
We showed here that plasma cholesterol exposure and LDLr downregulation can individually affect proliferation of NPC in vitro. Primary NPC challenged with high concentration of plasma LDL decreased proliferation and reduced differentiation towards the neuronal lineage (Fig. S2). Moreover, we observed a prominent effect on astrocytic morphology after LDL treatment. This observation might be linked to previously shown astrocytic alterations in the LDLr−/− mice [15], [50]. Additionally, it was recently reported that astrocytes are set with a very distinct transcriptome to manage lipid particles in CNS, and such LDL overload might be disrupting this machinery [57]. We found that the exogenous LDL is taken up by the proliferating cells and stored intracellularly as lipid droplets. It was shown before that primary neurons treated with LDL exhibited increased cholesterol accumulation in endolysosomes and concomitantly decreased levels of synaptophysin [53]. A recent study suggested that accumulation of lipids in cells from the neurogenic niche of the lateral ventricles, as found in brains from Alzheimer's disease patients, could contribute to reduced NPC proliferation [58]. In our study, it is conceivable that the increase in intracellular cholesterol content contributed to the effect on cell proliferation and led to the observed down regulation of the expression of sterol metabolism related genes (Table 1 and Supplementary Table 1). Specifically, the down-regulated genes contain the sterol regulatory element (SRE) promoter region, activated by the SRE-binding proteins (SREBPs), which are suppressed by high intracellular sterol content [59]. In contrast, the expression of LRP1 gene, which doesn't have the SRE promoter region [60], was not altered. In agreement, IPA upstream analysis of differentially expressed genes suggested inhibition of both SREBF1 and SREBF2. Inhibition of SREBPs and downregulation of HMGCR may result in decreased supply of mevalonate pathway end products, like isoprenoids, which are added to proteins involved in signaling to modify their localization and function and consequently cell differentiation and proliferation [61], [62]. In this context, it is intriguing that knock-down of LDLr also resulted in downregulation of HMGCR and FDFT1, albeit to a smaller extent. Finally, the increase in lipid droplet content per se could reshape intracellular signaling. These organelles play active roles in maturation, sequestration and turnover of many proteins, and even in transcriptional regulation [63].
De novo lipogenesis and fatty acid oxidation have a regulatory role in cell proliferation [23], [64] and maintenance of quiescence of adult NPC [65]. Several studies have demonstrated the important role of cholesterol metabolism in the development of the CNS and dependence of mature neurons on astrocytic cholesterol supply [12], [66], [67], [68]. Although there is a scarcity of information about cholesterol metabolism in adult neurogenic niches, regarding the LDLr family, interesting studies demonstrated the expression of LRP2 [67], [68] and LRP1 [69], [70] by the precursor cells from the subventricular zone of the lateral ventricles. In addition, the LRP6 protein was found to be expressed in the hippocampal adult NPC and manipulation of its expression affected the proliferative capacity of cells in vitro [70].
Herein we showed that the gene expression of cholesterol biosynthesis enzymes and LDLR are higher in proliferating than in the differentiating cells, resembling the high cholesterol requirement during CNS development, when both synthesis and uptake are highly stimulated [71]. Accordingly, the downregulation of the LDLR expression reduced NPC proliferation. The LDLr knock-down reduced mRNA levels of LRP1 as well. These two proteins are the main receptors for ApoE, which in turn is required for maintenance of the DG adult progenitor pool [72]. Fluorescent LDL pulse-chase suggested that the internalization and intracellular dynamics dependent of the LDL receptors are intact, so it is unlikely that the cells are experiencing a restriction in extracellular cholesterol supply or disruption of endocytosis. It is important to note that none of the treatments completely eliminated LDLr. Even after siRNA transfection, cells maintained 30% of LDLR mRNA. LDLR downregulation, rather than knock-out, is concurrent with the phenotype expressed by most of the FH patients (heterozygous form of the disease, loss in receptor activity up to 50%) [73].
Remarkably, the LDL treatment also strongly inhibited the LDLR expression. One could suggest that lack of the LDLr downstream signaling due to its down regulation is a convergent mechanism between the effect of the two treatments. Supporting LDLr importance in proliferative cells, cancer studies have shown that the LDLr inhibition potentiates tumor regression induced by chemotherapy [74], [75]. LDLR knock-down resulted in extensive changes in gene expression, distinct from the gene expression profiles after LDL treatment, indicative of some specificity of molecular regulation in both paradigms. IPA exploratory analysis implied modulation of several pathways and upstream regulators related to cell proliferation, including GADD45 signaling, important in adult hippocampal NPC proliferation and growth [76]. It is noteworthy that LDL treatment led to very few changes at the mRNA level, limited to the cholesterol synthesis pathway. This suggests that metabolic signaling triggered by intracellular accumulation of cholesterol induced post-transcriptional modifications in activity of regulatory proteins. In agreement, IPA upstream analysis pointed to changes in activation status of proteins and complexes known as hubs in orchestration of cellular metabolism and fate decisions, such as PTEN, MTORC1 or PPARG. Alternatively, the robust effect of LDL treatment over sterol related pathways might have mitigated the statistical highlight of other regulatory systems, involved in the cell cycle control, for example. In this regard, it was previously reported that in vitro exposure to LDL induces chromosome instability and mis-segregation resulting in aneuploidy [77], [78], and that it can affect cell-cycle progression via STAT5B mediated induction of cell-cycle arrest [79].
Although we did not find important alterations in glycolysis or OXPHOS after LDLR siRNA and LDL treatments (Fig. S3), a redox imbalance in the first hours of LDL treatment [19] might be conditioning the mitochondria to deal with produced oxidants (observed as a shift in the maximal respiratory capacity; Fig. S3C). From a number of studies, mitochondria bioenergetics emerges as an important regulator of NPC quiescence, proliferation, cell cycle exit, and neuronal differentiation [80], [81], [82].
5. Conclusions
In summary, the present study suggests that impairment in hippocampal neurogenesis might contribute to the behavioral phenotype in the LDLr−/− mice. Although we do not exclude niche-derived or systemic factors in the cognitive deterioration or negative regulation of adult neurogenesis, our in vitro data showed that high LDL levels and the downregulation of LDLr might directly contribute to a disruption of adult hippocampal NPC proliferation. The present findings also draw attention to the critical role of cholesterol metabolism in these processes and add to the accumulating evidence that complex metabolic networks orchestrate adult neural stem cell behavior and, accordingly, perturbation of the metabolic balance may have profound consequences for the cognitive capacity of the organism, in extreme cases leading to neuropathology.
Author contributions
Conceptualization, D.F.E., A.F.B., P.S.B., G.K., A.N.G., and T.L.W.; Methodology, D.F.E., A.F.B., P.S.B., G.K., A.N.G., and T.L.W.; In vivo experiments, D.F.E., J.O., and P.S.B.; In vitro experiments, D.F.E., A.N.G., and A.P.; Analyses, D.F.E., A.F.B., P.S.B., G.K., and A.N.G.; Writing, D.F.E., A.F.B., P.S.B., G.K., and A.N.G.
Acknowledgments
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - 424809/2018-4, PVE/CNPq 401065/2014-6), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Apoio à Pesquisa do Estado de Santa Catarina (FAPESC), INCT-NIM (Instituto Nacional de Ciência e Tecnologia – NeuroImunoModulação - 485489/2014-1), and L'Oreal-UNESCO-ABC: Women in Science. JO, AFB and PB are supported by research fellowships from CAPES and CNPq. DFE received CAPES/PDSE scholarship.
The authors would like to thank Dr. Yannis Kalaidzidis for scientific advice in experimental design of LDL pulse and chase assay and Dr. Muhammad Ichwan for preparation of adherent precursor cells. Part of Images were acquired and processed using equipment of the Imaging Platform of the Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE) with the help of Silke White.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.09.002.
Contributor Information
Daiane F. Engel, Email: daiane.engel01@gmail.com.
Andreza F. de Bem, Email: debemandreza@gmail.com.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Statistical analysis for all parameters measured using a one-way ANOVA or t-test in the study.
Differentially expressed genes in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC. Genes were considered as significantly different between the treatment and control at 10% FDR and absolute fold change of 1.5 cut-offs. Transcripts corresponding to pseudogenes or unknown genes were removed. logFC, log2(expression in control cells/expression in treated cells).
IPA canonical pathways enriched in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC. –log(p-value), –log10 of p-value from Fisher exact test; Ratio, (# differentially expressed genes in the pathway)/(total # of genes in the pathway in the reference set); Z-score, prediction of activation (Z-score > 0) or inhibition (Z-score < 0) of a given pathway.
IPA upstream analysis of differentially expressed genes in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC.
Fig. S1.
Effects of LDL treatment and siRNA-mediated LDLr knock-down on survival of adult hippocampal precursor cells in vitro. Precursor cells isolated from C57BL/6J mice were propagated as adherent monolayers and treated with: (A–C) human plasma LDL (0-200 μg/mL); (D–F) transfected with a pool of siRNAs targeting the LDLR gene or a pool of non-targeting siRNAs as a control; (G–I) or 27-HC (0–10 μM). (A, D, G) 24 h after each treatment, cell viability was measured by the resazurin assay. Hoechst staining allowed morphological analysis of apoptosis, as integrated density and average nucleus size after exposure to LDL (B–C) LDLR knock-down (E–F) and 27-HC treatment (H–I). Data are presented as means ± SEM. N = 3 per group. *p < 0.05, t- test (D–F), one-way ANOVA followed by Dunnett’s post hoc test (A–C, G–I).
Fig. S2.
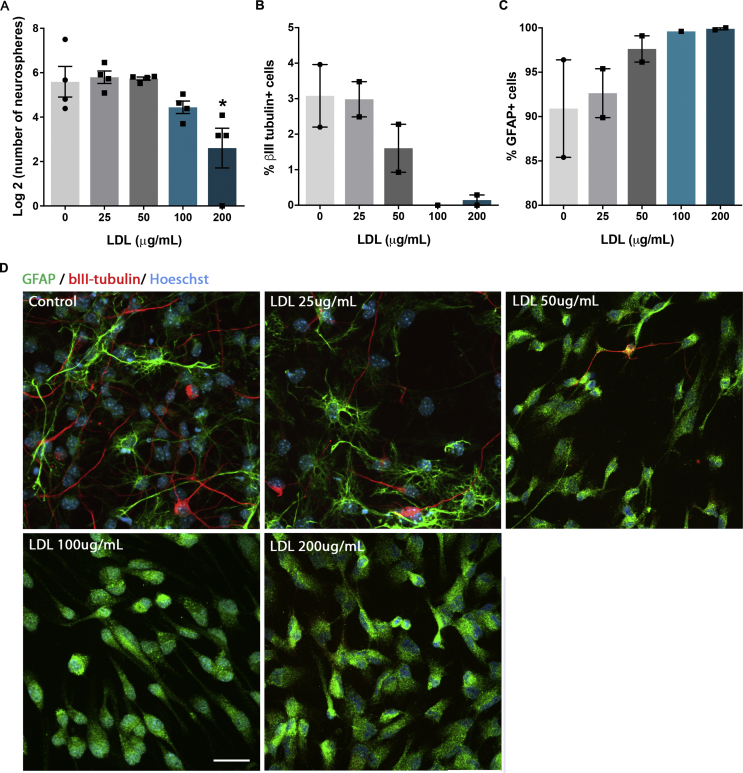
Effect of LDL treatment on proliferation and differentiation potential of precursor cells in vitro. (A) Cell proliferation was estimated by quantifying the number of primary neurospheres after 12 days exposure to LDL (0–200 μg/mL). (B-D) Following, neurospheres were collected and placed on PDL-laminin coated slides and allowed to differentiate for 7 days in absence of growth factors and no further LDL addition. Data in (A) were log2 transformed to visualize concentration-dependent response to LDL. Decrease of 1 indicates drop in neurosphere numbers by half. (D) Immunostaining against βIII- tubulin (red) and GFAP (green) identified neurons and astrocytes, respectively. LDL treatment decreased neuronal differentiation (B) and concomitantly increased proportion of astrocytes (C). *p < 0.05. Neurosphere numbers were analyzed with one-way ANOVA followed by Dunnett’s post hoc. The ratios of bIII-tubulin- and GFAP-positive cells were analyzed using generalized linear model with logit link function. Likelihood ratio test against the model omitting treatment showed significant effect of LDL on differentiation potential of NPCs [LRT, Chisq(4) = 195.23; p < 2.2e-16]. Post-hoc comparisons were performed using Dunnett’s test. The significance of the difference between control and 100 mg treatment group could not be estimated due to lack of variation in the 100 mg group (there were no neurons). N = 4 preparations (A); N = 84 neurospheres in two experimental batches (B). Scale bar = 20 μm. Data are presented as means from experiments ± SEM.
Fig. S3.
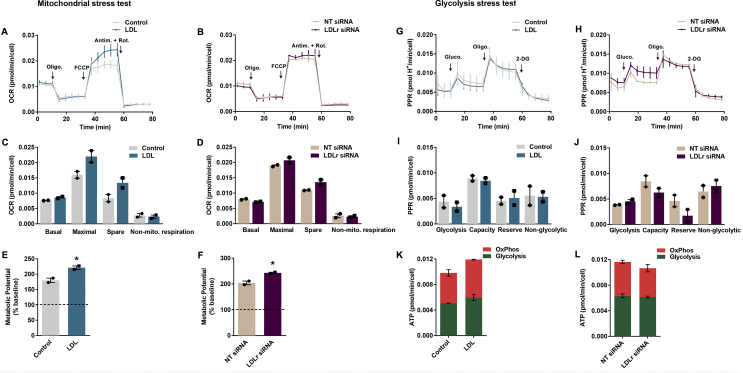
Effect of LDL treatment and LDLr siRNA transfection on mitochondrial bioenergetics. In the real-time analysis of OCR (A and B), ATP synthase inhibitor oligomycin, mitochondrial uncoupler FCCP and mitochondrial complex I and III inhibitors rotenone and antimycin A were injected sequentially at the indicated time points into each well after baseline rate measurement. A representative experiment of two independent is shown. (C, D) The resulting basal, maximal, spare and non-mitochondrial respiration OCR’s are similar between LDL and LDLr siRNA treatments and their controls. (E, F) Both treatments increased metabolic potential. (G, H) In the real-time analysis of ECAR (depicted in the graphs as proton pump rate, PPR), glucose, oligomycin and 2-deoxy glucose (2-DG; a glycolytic inhibitor) were injected sequentially at the indicated time points into each well after baseline rate measurement. A representative experiment of two independent is shown. (I, J) The resultant glycolysis, capacity, reserve and non-glxcolytic ECAR’s are similar between LDL and LDLr siRNA treatments and their controls. (K, L) No differences were seen in ATP production in from glycolysis and OxPhos, for LDL (K) or LDLr siRNA treatments (L). Data are presented as means ± SEM. N = 2 per group (experiment consisted of 8 technical replicates), *p < 0.05, t-test.
References
- 1.Solomon A., Kivipelto M., Wolozin B., Zhou J., Whitmer R.A. Midlife serum cholesterol and increased risk of Alzheimer's and vascular dementia three decades later. Dementia and Geriatric Cognitive Disorders. 2009;28(1):75–80. doi: 10.1159/000231980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Vliet P. Cholesterol and late-life cognitive decline. Journal of Alzheimer's Disease JAD. 2012;30(Suppl 2):S147–S162. doi: 10.3233/JAD-2011-111028. [DOI] [PubMed] [Google Scholar]
- 3.Kivipelto M., Helkala E.L., Laakso M.P., Hänninen T., Hallikainen M., Alhainen K. Midlife vascular risk factors and Alzheimer's disease in later life: longitudinal, population based study. BMJ (Clinical Research Ed) 2001;322(7300):1447–1451. doi: 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim I.Y., Hwang I.K., Choi J.W., Yoo K.-Y., Kim Y.N., Yi S.S. Effects of high cholesterol diet on newly generated cells in the dentate gyrus of C57BL/6N and C3H/HeN mice. The Journal of Veterinary Medical Science. 2009;71(6):753–758. doi: 10.1292/jvms.71.753. [DOI] [PubMed] [Google Scholar]
- 5.Zambón D., Quintana M., Mata P., Alonso R., Benavent J., Cruz-Sánchez F. Higher incidence of mild cognitive impairment in familial hypercholesterolemia. The American Journal of Medicine. 2010;123(3):267–274. doi: 10.1016/j.amjmed.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariza M., Cuenca N., Mauri M., Jurado M.A., Garolera M. Neuropsychological performance of young familial hypercholesterolemia patients. European Journal of Internal Medicine. 2016;34:e29–e31. doi: 10.1016/j.ejim.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Hobbs H.H., Brown M.S., Goldstein J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Human Mutation. 1992;1(6):445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 8.Dietschy J.M. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. The Journal of Lipid Research. 2004;45(8):1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Wagener J.F., Morgan D.H., Hui L., Ghribi O., Geiger J.D. Endolysosome mechanisms associated with Alzheimer's disease-like pathology in rabbits ingesting cholesterol-enriched diet. Journal of Alzheimer's Disease: JAD. 2010;22(4):1289–1303. doi: 10.3233/JAD-2010-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acharya N.K., Levin E.C., Clifford P.M., Han M., Tourtellotte R., Chamberlain D. Diabetes and hypercholesterolemia increase blood-brain barrier permeability and brain amyloid deposition: beneficial effects of the LpPLA2 inhibitor darapladib. Journal of Alzheimer's Disease: JAD. 2013;35(1):179–198. doi: 10.3233/JAD-122254. [DOI] [PubMed] [Google Scholar]
- 11.Löffler T., Flunkert S., Havas D., Sántha M., Hutter-Paier B., Steyrer E. Impact of ApoB-100 expression on cognition and brain pathology in wild-type and hAPPsl mice. Neurobiology of Aging. 2013;34(10):2379–2388. doi: 10.1016/j.neurobiolaging.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Pfrieger F.W., Ungerer N. Cholesterol metabolism in neurons and astrocytes. Progress in Lipid Research. 2011;50(4):357–371. doi: 10.1016/j.plipres.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Mulder M., Koopmans G., Wassink G., Al Mansouri G., Simard M.-L., Havekes L.M. LDL receptor deficiency results in decreased cell proliferation and presynaptic bouton density in the murine hippocampus. Neuroscience Research. 2007;59(3):251–256. doi: 10.1016/j.neures.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Mulder M., Jansen P.J., Janssen B.J.A., van de Berg W.D.J., van der Boom H., Havekes L.M. Low-density lipoprotein receptor-knockout mice display impaired spatial memory associated with a decreased synaptic density in the hippocampus. Neurobiology of Disease. 2004;16(1):212–219. doi: 10.1016/j.nbd.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Thirumangalakudi L., Prakasam A., Zhang R., Bimonte-Nelson H., Sambamurti K., Kindy M.S. High cholesterol-induced neuroinflammation and amyloid precursor protein processing correlate with loss of working memory in mice. Journal of Neurochemistry. 2008;106(1):475–485. doi: 10.1111/j.1471-4159.2008.05415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Oliveira J., Hort M.A., Moreira E.L.G., Glaser V., Ribeiro-do-Valle R.M., Prediger R.D. Positive correlation between elevated plasma cholesterol levels and cognitive impairments in LDL receptor knockout mice: relevance of cortico-cerebral mitochondrial dysfunction and oxidative stress. Neuroscience. 2011;197:99–106. doi: 10.1016/j.neuroscience.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Moreira E.L.G., de Oliveira J., Nunes J.C., Santos D.B., Nunes F.C., Vieira D.S.C. Age-related cognitive decline in hypercholesterolemic LDL receptor knockout mice (LDLr–/–): evidence of antioxidant imbalance and increased acetylcholinesterase activity in the prefrontal cortex. Journal of Alzheimer's Disease: JAD. 2012;32(2):495–511. doi: 10.3233/JAD-2012-120541. [DOI] [PubMed] [Google Scholar]
- 18.Moreira E.L.G., Aguiar A.S., de Carvalho C.R., Santos D.B., de Oliveira J., de Bem A.F. Effects of lifestyle modifications on cognitive impairments in a mouse model of hypercholesterolemia. Neuroscience Letters. 2013;541:193–198. doi: 10.1016/j.neulet.2013.02.043. [DOI] [PubMed] [Google Scholar]
- 19.Engel D.F., de Oliveira J., Lopes J.B., Santos D.B., Moreira E.L.G., Farina M. Is there an association between hypercholesterolemia and depression? Behavioral evidence from the LDLr−/− mouse experimental model. Behavioural Brain Research. 2016;311:31–38. doi: 10.1016/j.bbr.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 20.Sahay A., Hen R. Adult hippocampal neurogenesis in depression. Nature Neuroscience. 2007;10(9):1110–1115. doi: 10.1038/nn1969. [DOI] [PubMed] [Google Scholar]
- 21.Kempermann G., Kronenberg G. Depressed new neurons--adult hippocampal neurogenesis and a cellular plasticity hypothesis of major depression. Biological Psychiatry. 2003;54(5):499–503. doi: 10.1016/s0006-3223(03)00319-6. [DOI] [PubMed] [Google Scholar]
- 22.Kempermann G., Song H., Gage F.H. Neurogenesis in the adult Hippocampus. Cold Spring Harbor Perspectives in Biology. 2015;7(9):a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knobloch M., Pilz G.-A., Ghesquière B., Kovacs W.J., Wegleiter T., Moore D.L. A fatty acid oxidation-dependent metabolic shift regulates adult neural stem cell activity. Cell Reports. 2017;20(9):2144–2155. doi: 10.1016/j.celrep.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishibashi S., Brown M.S., Goldstein J.L., Gerard R.D., Hammer R.E., Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. The Journal of Clinical Investigation. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper-Kuhn C.M., Georg Kuhn H. Is it all DNA repair? Developmental Brain Research. 2002;134(1–2):13–21. doi: 10.1016/s0165-3806(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 26.Goodrich-Hunsaker N.J., Hunsaker M.R., Kesner R.P. Dissociating the role of the parietal cortex and dorsal hippocampus for spatial information processing. Behavioral Neuroscience. 2005;119(5):1307–1315. doi: 10.1037/0735-7044.119.5.1307. [DOI] [PubMed] [Google Scholar]
- 27.Goodrich-Hunsaker N.J., Hunsaker M.R., Kesner R.P. The interactions and dissociations of the dorsal hippocampus subregions: how the dentate gyrus, CA3, and CA1 process spatial information. Behavioral Neuroscience. 2008;122(1):16–26. doi: 10.1037/0735-7044.122.1.16. [DOI] [PubMed] [Google Scholar]
- 28.Hunsaker M.R., Wenzel H.J., Willemsen R., Berman R.F. Progressive spatial processing deficits in a mouse model of the fragile X premutation. Behavioral Neuroscience. 2009;123(6):1315–1324. doi: 10.1037/a0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunsaker M.R., Goodrich-Hunsaker N.J., Willemsen R., Berman R.F. Temporal ordering deficits in female CGG KI mice heterozygous for the fragile X premutation. Behavioural Brain Research. 2010;213(2):263–268. doi: 10.1016/j.bbr.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholzen T., Gerdes J. The Ki-67 protein: from the known and the unknown. Journal of Cellular Physiology. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Christie B.R., Cameron H.A. Neurogenesis in the adult hippocampus. Hippocampus. 2006;16(3):199–207. doi: 10.1002/hipo.20151. [DOI] [PubMed] [Google Scholar]
- 32.Franklin K.B.J., Paxinos G. 4th ed. Academic Press, an imprint of Elsevier; Amsterdam: 2013. Paxinos and Franklin's the mouse brain in stereotaxic coordinates. [Google Scholar]
- 33.Kempermann G., Gast D., Kronenberg G., Yamaguchi M., Gage F.H. Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development (Cambridge, England) 2003;130(2):391–399. doi: 10.1242/dev.00203. [DOI] [PubMed] [Google Scholar]
- 34.Babu H., Cheung G., Kettenmann H., Palmer T.D., Kempermann G. Enriched monolayer precursor cell cultures from micro-dissected adult mouse dentate gyrus yield functional granule cell-like neurons. PLoS One. 2007;2(4):e388. doi: 10.1371/journal.pone.0000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babu H., Claasen J.-H., Kannan S., Rünker A.E., Palmer T., Kempermann G. A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Frontiers in Neuroscience. 2011;5 doi: 10.3389/fnins.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathews E.S., Mawdsley D.J., Walker M., Hines J.H., Pozzoli M., Appel B. Mutation of 3-hydroxy-3-methylglutaryl CoA synthase I reveals requirements for isoprenoid and cholesterol synthesis in oligodendrocyte migration arrest, Axon wrapping, and myelin gene expression. Journal of Neuroscience. 2014;34(9):3402–3412. doi: 10.1523/JNEUROSCI.4587-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bem A.F., Farina M., de Lima Portella R., Nogueira C.W., Dinis T.C.P., Laranjinha J.A.N. Diphenyl diselenide, a simple glutathione peroxidase mimetic, inhibits human LDL oxidation in vitro. Atherosclerosis. 2008;201(1):92–100. doi: 10.1016/j.atherosclerosis.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 38.Spandl J., White D.J., Peychl J., Thiele C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic (Copenhagen, Denmark) 2009;10(11):1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krämer A., Green J., Pollard J., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (Oxford, England) 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert P.E., Kesner R.P., Lee I. Dissociating hippocampal subregions: double dissociation between dentate gyrus and CA1. Hippocampus. 2001;11(6):626–636. doi: 10.1002/hipo.1077. [DOI] [PubMed] [Google Scholar]
- 42.Hoge J., Kesner R.P. Role of CA3 and CA1 subregions of the dorsal hippocampus on temporal processing of objects. Neurobiology of Learning and Memory. 2007;88(2):225–231. doi: 10.1016/j.nlm.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoang L.T., Kesner R.P. Dorsal hippocampus, CA3, and CA1 lesions disrupt temporal sequence completion. Behavioral Neuroscience. 2008;122(1):9–15. doi: 10.1037/0735-7044.122.1.9. [DOI] [PubMed] [Google Scholar]
- 44.Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. Journal of Immunology (Baltimore, Md.: 1950) 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- 45.Marwarha G., Ghribi O. Does the oxysterol 27-hydroxycholesterol underlie Alzheimer's disease-Parkinson’s disease overlap? Experimental Gerontology. 2015;68:13–18. doi: 10.1016/j.exger.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrov A.M., Kasimov M.R., Zefirov A.L. Brain cholesterol metabolism and its defects: linkage to neurodegenerative diseases and synaptic dysfunction. Acta Naturae. 2016;8(1):58–73. [PMC free article] [PubMed] [Google Scholar]
- 47.Boldrini M., Fulmore C.A., Tartt A.N., Simeon L.R., Pavlova I., Poposka V. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22(4):589–599. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kempermann G., Gage F.H., Aigner L., Song H., Curtis M.A., Thuret S. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018;23(1):25–30. doi: 10.1016/j.stem.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno-Jiménez E.P., Flor-García M., Terreros-Roncal J., Rábano A., Cafini F., Pallas-Bazarra N. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer's disease. Nature Medicine. 2019;25(4):554–560. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 50.de Oliveira J., Moreira E.L.G., dos Santos D.B., Piermartiri T.C., Dutra R.C., Pinton S. Increased susceptibility to amyloid-β-induced neurotoxicity in mice lacking the low-density lipoprotein receptor. Journal of Alzheimer's Disease: JAD. 2014;41(1):43–60. doi: 10.3233/JAD-132228. [DOI] [PubMed] [Google Scholar]
- 51.Elder G.A., Ragnauth A., Dorr N., Franciosi S., Schmeidler J., Haroutunian V. Increased locomotor activity in mice lacking the low-density lipoprotein receptor. Behavioural Brain Research. 2008;191(2):256–265. doi: 10.1016/j.bbr.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolls E.T., Kesner R.P. A computational theory of hippocampal function, and empirical tests of the theory. Progress in Neurobiology. 2006;79(1):1–48. doi: 10.1016/j.pneurobio.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Hui L., Chen X., Geiger J.D. Endolysosome involvement in LDL cholesterol-induced Alzheimer's disease-like pathology in primary cultured neurons. Life Sciences. 2012;91(23–24):1159–1168. doi: 10.1016/j.lfs.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer T.D., Willhoite A.R., Gage F.H. Vascular niche for adult hippocampal neurogenesis. The Journal of Comparative Neurology. 2000;425(4):479–494. doi: 10.1002/1096-9861(20001002)425:4<479::aid-cne2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Stokes K.Y., Cooper D., Tailor A., Granger D.N. Hypercholesterolemia promotes inflammation and microvascular dysfunction: role of nitric oxide and superoxide. Free Radical Biology & Medicine. 2002;33(8):1026–1036. doi: 10.1016/s0891-5849(02)01015-8. [DOI] [PubMed] [Google Scholar]
- 56.Vinuesa A., Pomilio C., Menafra M., Bonaventura M.M., Garay L., Mercogliano M.F. Juvenile exposure to a high fat diet promotes behavioral and limbic alterations in the absence of obesity. Psychoneuroendocrinology. 2016;72:22–33. doi: 10.1016/j.psyneuen.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Ioannou M.S., Jackson J., Sheu S.-H., Chang C.-L., Weigel A.V., Liu H. Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell. 2019;177(6):1522–1535. doi: 10.1016/j.cell.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton L.K., Dufresne M., Joppé S.E., Petryszyn S., Aumont A., Calon F. Aberrant lipid metabolism in the forebrain niche suppresses adult neural stem cell proliferation in an animal model of Alzheimer's disease. Cell Stem Cell. 2015;17(4):397–411. doi: 10.1016/j.stem.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Progress in Lipid Research. 2001;40(6):439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- 60.Kütt H., Herz J., Stanley K.K. Structure of the low-density lipoprotein receptor-related protein (LRP) promoter. Biochimica et Biophysica Acta. 1989;1009(3):229–236. doi: 10.1016/0167-4781(89)90107-3. [DOI] [PubMed] [Google Scholar]
- 61.Cartocci V., Segatto M., Di Tunno I., Leone S., Pfrieger F.W., Pallottini V. Modulation of the isoprenoid/cholesterol biosynthetic pathway during neuronal differentiation in vitro. Journal of Cellular Biochemistry. 2016;117(9):2036–2044. doi: 10.1002/jcb.25500. [DOI] [PubMed] [Google Scholar]
- 62.Prendergast G.C., Oliff A. Farnesyltransferase inhibitors: antineoplastic properties, mechanisms of action, and clinical prospects. Seminars in Cancer Biology. 2000;10(6):443–452. doi: 10.1006/scbi.2000.0335. [DOI] [PubMed] [Google Scholar]
- 63.Welte M.A., Gould A.P. Lipid droplet functions beyond energy storage. Biochimica et Biophysica Acta Molecular and Cell Biology of Lipids. 2017;1862(10 Pt B):1260–1272. doi: 10.1016/j.bbalip.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knobloch M., Braun S.M.G., Zurkirchen L., von Schoultz C., Zamboni N., Araúzo-Bravo M.J. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2012;493(7431):226–230. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17(3):329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 66.Funfschilling U., Jockusch W.J., Sivakumar N., Mobius W., Corthals K., Li S. Critical time window of neuronal cholesterol synthesis during neurite outgrowth. Journal of Neuroscience. 2012;32(22):7632–7645. doi: 10.1523/JNEUROSCI.1352-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gajera C.R., Emich H., Lioubinski O., Christ A., Beckervordersandforth-Bonk R., Yoshikawa K. LRP2 in ependymal cells regulates BMP signaling in the adult neurogenic niche. Journal of Cell Science. 2010;123(Pt 11):1922–1930. doi: 10.1242/jcs.065912. [DOI] [PubMed] [Google Scholar]
- 68.Zywitza V., Misios A., Bunatyan L., Willnow T.E., Rajewsky N. Single-cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Reports. 2018;25(9):2457–2469. doi: 10.1016/j.celrep.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Safina D., Schlitt F., Romeo R., Pflanzner T., Pietrzik C.U., Narayanaswami V. Low-density lipoprotein receptor-related protein 1 is a novel modulator of radial glia stem cell proliferation, survival, and differentiation. Glia. 2016;64(8):1363–1380. doi: 10.1002/glia.23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kannan S., Nicola Z., Overall R.W., Ichwan M., Ramírez-Rodríguez G.N., Grzyb A. Systems genetics analysis of a recombinant inbred mouse cell culture panel reveals wnt pathway member Lrp6 as a regulator of adult hippocampal precursor cell proliferation: lrp6 regulates hippocampal neurogenesis. Stem Cells. 2016;34(3):674–684. doi: 10.1002/stem.2313. [DOI] [PubMed] [Google Scholar]
- 71.Farese R.V., Jr., Herz J. Cholesterol metabolism and embryogenesis. Trends in Genetics. 1998;14(3):115–120. doi: 10.1016/s0168-9525(97)01377-2. [DOI] [PubMed] [Google Scholar]
- 72.Yang C.-P., Gilley J.A., Zhang G., Kernie S.G. ApoE is required for maintenance of the dentate gyrus neural progenitor pool. Development. 2011;138(20):4351–4362. doi: 10.1242/dev.065540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rader D.J., Cohen J., Hobbs H.H. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. The Journal of Clinical Investigation. 2003;111(12):1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furuya Y., Sekine Y., Kato H., Miyazawa Y., Koike H., Suzuki K. Low-density lipoprotein receptors play an important role in the inhibition of prostate cancer cell proliferation by statins. Prostate International. 2016;4(2):56–60. doi: 10.1016/j.prnil.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guillaumond F., Bidaut G., Ouaissi M., Servais S., Gouirand V., Olivares O. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(8):2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma D.K., Jang M.-H., Guo J.U., Kitabatake Y., Chang M.-L., Pow-Anpongkul N. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science (New York, N.Y.) 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Granic A., Potter H. Mitotic spindle defects and chromosome mis-segregation induced by LDL/Cholesterol—implications for Niemann-pick C1, Alzheimer's disease, and atherosclerosis. PLoS One. 2013;8(4):e60718. doi: 10.1371/journal.pone.0060718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brizzi M.F., Dentelli P., Pavan M., Rosso A., Gambino R., Grazia De Cesaris M. Diabetic LDL inhibits cell-cycle progression via STAT5B and p21(waf) The Journal of Clinical Investigation. 2002;109(1):111–119. doi: 10.1172/JCI13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gonçalves J.T., Schafer S.T., Gage F.H. Adult neurogenesis in the Hippocampus: from stem cells to behavior. Cell. 2016;167(4):897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 80.Khacho M., Harris R., Slack R.S. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nature Reviews Neuroscience. 2019;20(1):34–48. doi: 10.1038/s41583-018-0091-3. [DOI] [PubMed] [Google Scholar]
- 81.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. ELife. 2016;5 doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beckervordersandforth R., Ebert B., Schäffner I., Moss J., Fiebig C., Shin J. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93(6):1518. doi: 10.1016/j.neuron.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis for all parameters measured using a one-way ANOVA or t-test in the study.
Differentially expressed genes in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC. Genes were considered as significantly different between the treatment and control at 10% FDR and absolute fold change of 1.5 cut-offs. Transcripts corresponding to pseudogenes or unknown genes were removed. logFC, log2(expression in control cells/expression in treated cells).
IPA canonical pathways enriched in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC. –log(p-value), –log10 of p-value from Fisher exact test; Ratio, (# differentially expressed genes in the pathway)/(total # of genes in the pathway in the reference set); Z-score, prediction of activation (Z-score > 0) or inhibition (Z-score < 0) of a given pathway.
IPA upstream analysis of differentially expressed genes in LDL treated (100 μg/ml) and LDLR siRNA transfected NPC.