Abstract
Sepsis remains the primary cause of death in intensive care units and multiple long non-coding RNAs (lncRNAs) have been demonstrated to be dysregulated in samples of patients with sepsis. However, whether lncRNA-HOTAIR is involved in the etiology of sepsis remains unclear. The aim of the present study was to investigate the role of HOTAIR in sepsis and to reveal the associated mechanisms. A bioinformatics analysis and dual-luciferase reporter assay was performed to evaluate the interaction between HOTAIR and miR-211, as well as miR-211 and IL-6R. An animal model of sepsis was established in mice via cecal ligation and puncture. Interferon (IFN)-γ, interleukin (IL)-6, IL-17, tumor necrosis factor (TNF)-α, IL-1β, IL-6 receptor (R), microRNA (miR)-211 and HOTAIR expression was measured using reverse transcription-quantitative PCR. Cellular proliferation and apoptosis of monocytes were assessed using cell counting kit-8 assay and flow cytometry, respectively. miR-211 was revealed to be targeted by HOTAIR and IL-6R. The expression of IFN-γ, IL-6, IL-17, TNF-α, IL-1β, IL-6R and HOTAIR was significantly upregulated in the septic mice, whereas miR-211 expression was downregulated. The overexpression of hox transcript antisense RNA (HOTAIR) and knockdown of miR-211 were associated with an increased expression of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R in monocytes, while the overexpression of miR-211 exhibited the opposite effect. HOTAIR overexpression and miR-211 knockdown significantly inhibited cellular proliferation and promoted monocyte apoptosis, whereas the overexpression of miR-211 exhibited the opposite effects in monocytes. Therefore, HOTAIR may promote the progression of sepsis by indirectly regulating the expression of IL-6R via miR-211.
Keywords: sepsis, hox transcript antisense RNA, microRNA-211, interleukin-6R, apoptosis, cecal ligation and puncture
Introduction
Sepsis is a serious clinical condition caused by multiple agents, including bacterial, viral and fungal infections, which subsequently initiates the inflammatory response, leading to organ failure in the affected host (1). Currently, sepsis remains the primary cause of death in intensive care units (ICU), despite recent advancements in medical technology (2,3). It has been estimated that in 2017, the percentage of admissions to the ICU caused by sepsis is ~25% and is associated with a mortality rate >50% worldwide (2,4). Although the precise pathophysiology of sepsis remains unclear, increasing evidence has indicated that it may move from an early hyper-inflammatory phase characterized by systemic inflammation induced by the excessive release of pro-inflammatory factors, followed by a late immuno-suppressive phase, characterized by the apoptosis of immune cells (including monocytes and lymphocytes) (5,6). Furthermore, apoptosis that occurs in the cells of tissues in the site of primary infection during sepsis can result in microvascular dysfunction, which subsequently leads to organ failure (7,8). Therefore, inhibiting sepsis-induced apoptosis may be a potential therapeutic approach for patients with sepsis.
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are two important ncRNAs, which are characterized by a lack of protein encoding ability (9–12). These ncRNAs have been demonstrated to be involved in multiple biological processes, including apoptosis, proliferation and differentiation (13–15). miRNAs are a type of short RNA molecule consisting of ~20–22 nucleotides that negatively regulate gene expression by binding to the 3′-untranslated regions (3′-UTR) of the mRNA of a target gene (16). Aberrant miRNA expression has been observed in a number of human diseases, including cancer, neurodegenerative disorders and inflammatory-associated disorders (17–19). LncRNAs also participate in the progression of various human diseases, including cancer (20,21), cardiovascular disease (22) and rheumatic diseases (23) by acting as a miRNA sponge (24). LncRNA-hox transcript antisense RNA (HOTAIR) has been previously reported to function as an oncogenic molecule in a number of human malignancies including lung (25), prostate (26), gastric (27) and colorectal cancer (28), etc.. Recently, HOTAIR was observed to be upregulated in mice and cardiomyocytes following lipopolysaccharide (LPS)-induced sepsis, in which silencing HOTAIR protected the cardiac function of septic mice by downregulating tumor necrosis factor-α (TNF-α) via the NF-κB signaling pathway (29).
It has been well documented that sepsis may be mediated by multiple inflammatory cytokines, including TNF-α, interleukin-6 (IL)-6 and IL-1β (30,31). In addition, emerging studies have revealed an association between plasma inflammatory cytokine concentrations and mortality in patients with sepsis (30,32). In particular, the upregulation of IL-6 and its receptor, IL-6R, has been frequently observed in patients with sepsis and the production of IL-6 was demonstrated to be a good prognostic agent in the early phase of sepsis (33,34). These results indicate that IL-6 and its receptor may function as two potential therapeutic targets for patients with sepsis.
In the present study, HOTAIR and IL-6R were revealed to be targeted by miR-211; however, since it remains unknown how the interaction between HOTAIR, IL-6R and miR-211 contribute to the etiology of sepsis, the aim of the present study was to investigate the effects of the HOTAIR/miR-211/IL-6R axis on the pathogenesis of sepsis.
Materials and methods
Establishment of an animal model of sepsis
C57BL/6 mice (age, 8 weeks; mean weight, 23.4±0.92 g; weight range, 22–25 g) were purchased from the Animal Experiment Center of the Institute of Radiation Medicine of the Chinese Academy of Medical Sciences and all of the animal protocols used in the present study were approved by the Institute of Radiation Medicine of the Chinese Academy of Medical Sciences. All the animals were raised for seven days to adapt to the environment prior to experimentation. Animals were raised with sufficient water and feed, at a temperature of 20–24°C, humidity of 50–60% with a 12-h light/dark cycle. A total of 8 male C57BL/6 mice (8 weeks old) were used to induce sepsis via cecal ligation and puncture (CLP). After anesthetizing the mice with 2% pentobarbital sodium (50 mg/kg, intraperitoneally), a small incision was made in the abdomen. The cecum of the mice was then exposed and a sterile 21-gauge needle was used to puncture the cecum twice to extrude fecal matter. Subsequently, the cecum was returned into the abdominal cavity and the incision was closed in two layers. Control mice (n=8) were treated the same as the experimental animals, but without CLP.
Monocytes isolation
Following the establishment of the mouse model 48 h following CPL, C57BL/6 mice in the sepsis and control groups were decapitated and the spleen was subsequently removed. After washing with PBS, the spleen was broken by collagenase (cat. no. 17104019; Gibco; Thermo Fisher Scientific, Inc.) for 5 mins at 37°C, filtered through a 74 µm pore size strainer (BD Biosciences) to create a single-cell suspension. Cell concentration was determined using a hemocytometer (Hausser Scientific) and adjusted to 1×108 cells/ml. After which the mouse monocytes were purified using CD11b MicroBeads (cat. no. 130-049-001; Miltenyi Biotec GmbH) according to the manufacturer's protocol.
Cell culture
Monocytes were isolated from the spleens as aforementioned and maintained in DMEM medium (Sigma-Aldrich; Merck KGaA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) with 1% penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5% CO2.
Transfections of miRNA mimics and inhibitors
Negative control (NC or scramble for mimics and inhibitors; 5′-UUCUCCGAACGUGUCACGUTT-3′), miR-211 mimics (5′-UUCCCUUUGUCAUCCUUUGCCU-3′) and miR-211 inhibitors (5′-AGGCAAAGGATGACAAAGGGAA-3′) were synthesized by Shanghai GenePharma Co., Ltd. Monocytes (5×104 cells/well) were seeded in 6-well plates and transfected with NC (50 nM), miR-211 mimics (50 nM) and miR-211 inhibitors (50 nM) using Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer's protocol. Subsequent experiments were performed 48 h following transfection.
Vector construction and transfection
DNA was extracted from the 293T cells using TIANamp Genomic DNA Kit (Tiangen Biotech Co., Ltd.) according to manufacturer's protocol. HOTAIR was amplified using Taq PCR Master Mix Kit (Qiagen, Inc.) with XhoI and BamHI restriction sites. The temperature protocol for the PCR consisted of 94°C for 3 min; followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C 1 min; and final extension at 72°C for 5 min. The primers for amplification were forward, 5′-CCGCTCGAGACATTCTGCCCTGATTTCCGGAACC-3′ and reverse, 5′-CGCGGATCCCCACCACACACACACAACCTACAC-3′. HOTAIR DNAs were inserted into the pcDNA3.0 vector (Invitrogen; Thermo Fisher Scientific, Inc.) according to previous studies (35,36). A total of 1×105 monocytes were seeded into 6-well plates and transfected with the HOTAIR-expression vector or empty vector (control) using Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR) assay
The total RNA isolated from the splenic tissues of septic and control mice, monocytes transfected with HOTAIR, miR-211 mimics, miR-211 inhibitor and corresponding controls were all prepared using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was subsequently synthesized using PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd.) using 50 ng total RNA with the temperature protocol consisting of 95°C for 30 sec and 60°C for 30 mins. The amplification of interferon (IFN)-γ, IL-6, IL-17, TNF-α, IL-1β, IL-6R, miR-211 and HOTAIR was performed using a Bestar® SYBR Green qPCR master mix (DBI Bioscience; Shanghai Xinghan Biotechnology Co., Ltd.) kit using an ABI PRISM 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were: 95°C for 2 min; followed by 30 cycles of 95°C 10 sec, and 60°C 34 sec. The primer sequences used were listed in Table I. The expressions of IFN-γ, IL-6, IL-17, TNF-α, IL-1β, IL-6R and HOTAIR were normalized to the level of GAPDH, whereas miR-211 expression was normalized to the level of U6. The relative expression levels were analyzed using 2−ΔΔCq method (37).
Table I.
Primer sequences for RT-qPCR assay used in this study.
| Gene | Sequence |
|---|---|
| GAPDH-F | 5′-TGTTCGTCATGGGTGTGAAC-3′ |
| GAPDH-R | 5′-ATGGCATGGACTGTGGTCAT-3′ |
| U6-F | 5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R | 5′-AACGCTTCACGAATTTGCGT-3′ |
| IFN-γ-F | 5′-AGCGGATAATGGAACTCTTTTCTTAG-3′ |
| IFN-γ-R | 5′-AAGTTTGAAGTAAAAGAAGACAATTTGG-3′ |
| IL-6-F | 5′-AGTTGCCTTCTTGGGACTGA-3′ |
| IL-6-R | 5′-CAGAATTGCCATTGCACAAC-3′ |
| IL-17-F | 5′-CCGGACTGTGATGGTCAA-3′ |
| IL-17-R | 5′-CTCATTGCGGTGGAGATT-3′ |
| TNF-α-F | 5′-CGGGCAGGTCTACTTTGGAG-3′ |
| TNF-α-R | 5′-CAGGTCACTGTCCCAGCATC-3′ |
| IL-1β-F | 5′-CTTCTTCGACACATGGGATAAC-3′ |
| IL-1β-R | 5′-TTTGGGATCTACACTCTCCAGC-3′ |
| IL-6R-F | 5′-TGAGCTCAGATATCGGGCTGAAC-3′ |
| IL-6R-R | 5′-CGTCGTGGATGACACAGTGATG-3′ |
| miR-211-F | 5′-TTGTGGGCTTCCCTTTGTCATCCT-3′ |
| miR-211-R | 5′-TGCTGTGGGAAGTGACAACTGA-3′ |
| HOTAIR-F | 5′-CAGTGGGGAACTCTGACTCG-3′ |
| HOTAIR-R | 5′-GTGCCTGGTGCTCTCTTACC-3′ |
IFN-γ, interferon-γ; IL, interleukin; TNF-α, tumor-necrosis factor-α; miR, microRNA; HOTAIR, hox transcript antisense RNA.
Western blot assay
The proteins were isolated from the spleens of septic and control mice, monocytes transfected with HOTAIR, miR-211 mimics οr miR-211 inhibitor and corresponding control using RIPA Lysis Buffer System (Santa Cruz Biotechnology, Inc.) supplemented with 1.5 mM PMSF (Sigma-Aldrich; Merck KGaA). The lysates were then subjected to centrifugation at 12,000 × g for 15 min at 4°C, after which the supernatants were collected. The protein concentration was determined using a bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.). A total of 30 µg proteins per lane were isolated using 10% SDS-PAGE and transferred onto nitrocellulose membranes (EMD Millipore; Merck KGaA) and incubated with 5% skimmed milk at room temperature for 2 h. The membranes were then incubated with primary rabbit antibodies against IFN-γ (1:1,000; cat. no. ab77246), IL-6 (1:2,000; cat. no. ab6672), IL-17 (1:500; cat. no. ab136668), TNF-α (1:500; cat. no. ab6671), IL-1β (1:1,000; cat. no. ab200478), and IL-6R (1:200; cat. no ab128008; all Abcam) at 4°C for 8 h. The membranes were incubated with horseradish peroxidase-conjugated donkey anti-rabbit secondary antibodies (1:2,000; cat. no. ab7083; Abcam) at room temperature for 2 h. Finally, the signals were detected using enhanced chemiluminescent (ECL) kit (Pierce; Thermo Fisher Scientific, Inc.). The grayscale values of the membranes were counted using an ImageJ software (ver. 1.51d; National institutes of Health).
Bioinformatics analysis
TargetScan (http://targetscan.org/) was applied to analyze the possible binding sites between HOTAIR and miR-211 and between miR-211 and IL-6; TargetScan (http://targetscan.org/) (38), StarBase v2.0 (http://starbase.sysu.edu.cn/) (39) and miRDB (http://mirdb.org/miRDB/) (40) databases were utilized to analyze the possible binding site of IL-6 as the downstream target of miR-211.
Dual-luciferase reporter assay
The interaction between miR-211 and HOTAIR, as well as miR-211 and IL-6R were verified with a dual-luciferase reporter assay. Wild-type (WT) HOTAIR, mutant type (Mut) HOTAIR, WT IL-6R and Mut IL-6R were purchased from Hanbio Co., Ltd. (Hanbio Biotechnology Co., Ltd.). Briefly, total DNA was extracted from 293T cells using TIANamp Genomic DNA Kit (Tiangen Biotech Co., Ltd.), and the 3′-untranslated regions (3′-UTR) of the wild type (WT) HOTAIR containing the miR-211 binding sites were amplified using Taq PCR Master Mix Kit (Qiagen GmbH; cat. no. 201443). The temperature protocol for the PCR consisted of 94°C for 3 min; followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C 1 min; and final extension at 72°C for 5 min. The mutant (Mut) 3′-UTR of HOTAIR was generated by changing the sequence from ‘AAAGGGAA’ to ‘UUUCCCUU’. The DNA products were sub-cloned into the luciferase vector, psi-CHECK2 (Promega Corporation) to form a recombinant reporter plasmid. The WT IL-6R (WT-IL-6R) and Mut IL-6R (MUT-IL-6R) were constructed in the same manner as WT-HOTAIR and Mut-HOTAIR. For the miR-211 and HOTAIR dual-luciferase reporter assay, 293T cells were seeded into 24-well plates at a density of 1×104 cells/well. After culturing overnight at 37°C, 293T cells were co-transfected with WT-HOTAIR or Mut-HOTAIR combined with miR-211 mimics or its negative control using a Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The collected 293T cells were seeded into 24-well plates at a concentration of 2×105 cells/well, and cultured at 37°C overnight. Based on manufacturer's instructions, the luciferase activities were measured with a Dual Luciferase Assay System (Promega Corporation) to verify that the interaction between miR-211 and IL-6R was the same as miR-211 and HOTAIR. All luciferase activities were normalized to Renilla luciferase activity.
Cell proliferation analysis
Cell Counting Kit-8 (CCK-8; Sigma-Aldrich; Merck KGaA) was used to evaluate the effects of HOTAIR and miR-211 on monocyte proliferation. The transfected monocytes (1×103 cell/well) were seeded into 96-well plates and transfected with HOTAIR, miR-211 mimics or miR-211 inhibitors for 48 h at 37°C in a humidified incubator with 5% CO2. The optical density (OD) was then measured at 450 nm using a microtiter plate reader (SpectraMax; Molecular Devices, LCC).
Cell apoptosis analysis
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining and flow cytometry were performed to determine the effects of HOTAIR and miR-211 on monocyte apoptosis. After culturing in DMEM for 48 h at 37°C, monocytes transfected with either HOTAIR, miR-211 mimic or miR-211 inhibitor were harvested by centrifugation (1,000 × g for 5 min) and washed twice with PBS. The monocytes were fixed in 70% ethanol for 2 h at room temperature and then incubated with annexin V-FITC and PI (Keygentec) for 10 min in the dark. Finally, the apoptotic cells were evaluated using flow cytometry (BD Biosciences), and analyzed using BD CellQuest software (Version 3.3; BD Biosciences).
Statistical analysis
SPSS software (version 22.0; IBM, Corp.) was used for all statistical analyses. Each experiment was repeated at least three times and the data were expressed as the mean ± standard deviation (SD). A Student's t-test was used for the statistical analyses between two groups and the statistical differences between more than two groups were analyzed using a one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
HOTAIR is directly targeted by miR-211
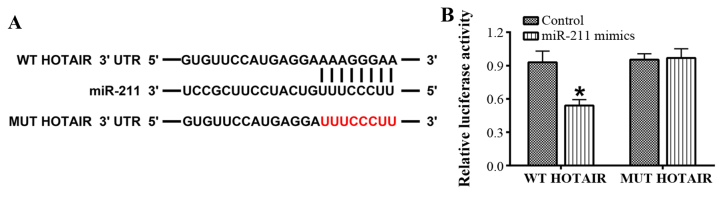
The interaction between miR-211 and HOTAIR was evaluated using online bioinformatics analysis and a dual-luciferase reporter assay in 293T cells. The bioinformatics analysis revealed that there were putative binding sites for miR-211 in HOTAIR (Fig. 1). Further analysis confirmed that luciferase activity was driven by WT-HOTAIR as it was significantly attenuated by the miR-211 mimics. However, no significant difference was observed with MUT-HOTAIR (P<0.05; Fig. 1).
Figure 1.
HOTAIR is directly targeted by miR-211. (A) The putative binding sites of miR-211 in HOTAIR were predicted using online bioinformatics analysis. (B) The interaction between miR-211 and HOTAIR was verified using a dual-luciferase reporter assay. *P<0.05 vs. control group. Each experiment was repeated three times. HOTAIR, hox transcript antisense RNA; miR/miRNA, microRNA; WT, wild type; UTR, untranslated region; MUT, mutant.
The 3′-UTR of IL-6R is targeted by miR-211
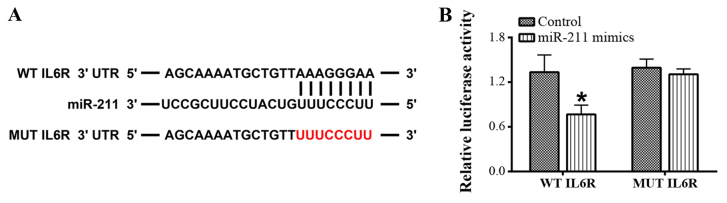
The interaction between miR-211 and IL-6 using an online bioinformatics analysis tool and dual-luciferase reporter assay. The bioinformatics analysis indicated that there were putative binding sites in the 3′-UTR of IL-6 for miR-211 (Fig. 2). Subsequently, a dual-luciferase reporter assay was performed to verify the interaction between miR-211 and IL-6 and the results revealed that the luciferase activity driven by WT-IL-6R was significantly reduced by the miR-211 mimics; however, there was no significant difference in luciferase activity with MUT-IL-6R following treatment with the miR-211 mimics (P<0.05; Fig. 2).
Figure 2.
The 3′-UTR of the IL-6R is targeted by miR-211. (A) The putative binding sites of miR-211 in IL-6R were predicted using online bioinformatics analysis. (B) The interaction between miR-211 and IL-6R was verified using a dual-luciferase reporter assay. *P<0.05 vs. control group. Each experiment was repeated three times. UTR, untranslated region; IL, interleukin; miR/miRNA, microRNA; MUT, mutant; WT, wild type.
miR-211 and HOTAIR expression is significantly upregulated in the spleens of mice with CLP-induced sepsis
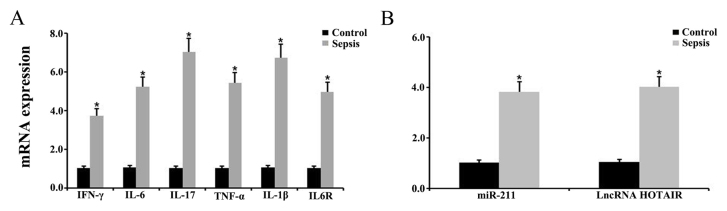
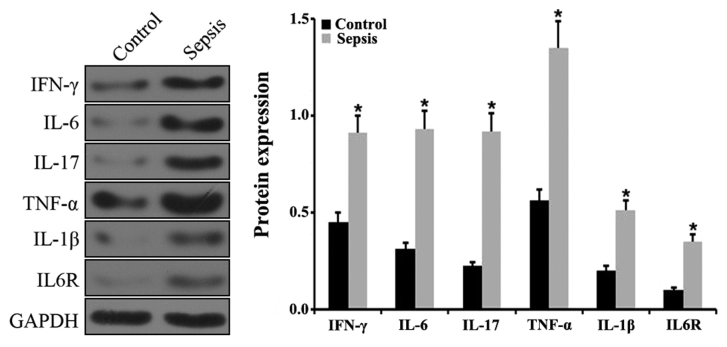
To explore whether miR-211 and HOTAIR were involved in the pathogenesis of sepsis, a CLP-induced mouse model of sepsis was established. The animal model of sepsis was initially verified by detecting the levels of various inflammatory factors in the spleens via RT-qPCR. The results indicated that the levels of IFN-γ, IL-6, IL-17, TNF-α and IL-1β expression were significantly upregulated in septic mice compared with control mice (Fig. 3A). In addition, there was a significant upregulation in IL-6R expression in the septic mice compared with control mice (P<0.05; Fig. 3A). The level of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R expression was further examined via western blotting. The results demonstrated a significant increase in IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R in the spleens of septic mice compared with control mice (P<0.05; Fig. 4). The expression of miR-211 and HOTAIR in the septic mice was subsequently examined using RT-qPCR. The results indicated that the relative levels of miR-211 and HOTIAR were significantly increased in the splenic tissues from the septic mice compared with control mice (P<0.05; Fig. 3B).
Figure 3.
Expression level of various inflammatory factors, miR-211, and HOTAIR in the spleen. Relative expression level of (A) IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R and (B) miR-211 and HOTAIR in the spleens of control and septic mice, as determined via reverse transcription-quantitative PCR. *P<0.05 vs. control group. Each experiment was repeated three times. miR, microRNA; HOTAIR, hox transcript antisense RNA; IL, interleukin; IFN, interferon; TNF, tumor necrosis factor.
Figure 4.
Protein expression of various inflammatory factors in the spleen. The relative levels of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R protein expression was determined in the spleens of control and septic animals using western blot analysis. *P<0.05 vs. control group. Each experiment was repeated three times. IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
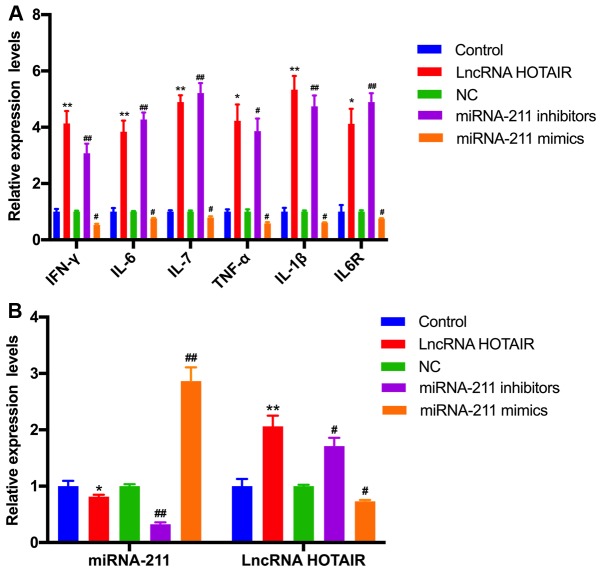
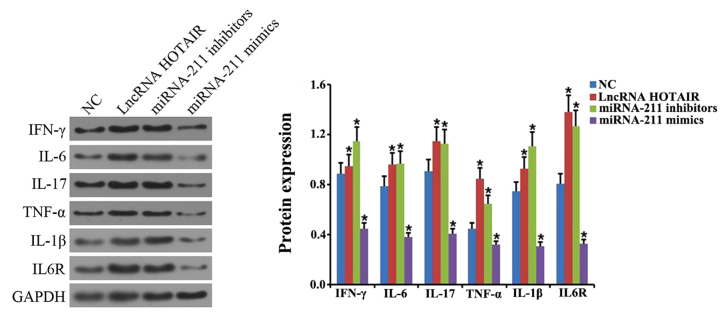
Overexpression of HOTIAR and knockdown of miR-211 promotes the inflammatory response in monocytes
To further explore the biological function of miR-211 and HOTAIR in sepsis, RT-qPCR was performed to determine the levels of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R in the monocytes transfected with HOTAIR, the miR-211 mimics and miR-211 inhibitors. The relative level of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R mRNA and protein expression was demonstrated to be significantly upregulated in the HOTAIR and miR-211 inhibitor-treated groups compared with that in the negative control group (P<0.05; Figs. 5A and 6). However, there was a significant downregulation in the level of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R expression in monocytes treated with the miR-211 mimics compared with the negative control (P<0.05; Figs. 5A and 6). In addition, miR-211 expression was significantly decreased in the HOTAIR overexpressed group, and HOTAIR expression was significantly increased in the miR-211-silenced group. These results indicated that there was a negative association between the expression of miR-211 and HOTAIR in monocytes (P<0.05; Fig. 5B).
Figure 5.
Effects of HOTAIR and miR-211 overexpression or knockdown on the mRNA expression of various inflammatory factors, miR-211 and HOTAIR. The relative level of (A) IFN-γ, IL-6, IL-17, TNF-α, IL-1β, and IL-6R and (B) miR-211, and HOTAIR expression in monocytes transfected with HOTAIR, miR-211 mimics, and miR-211 inhibitor was examined using reverse transcription-quantitative PCR. *P<0.05, **P<0.01 vs. control group; #P<0.05, ##P<0.01 vs. NC group. Each experiment was repeated three times. HOTAIR, hox transcript antisense RNA; miR, microRNA; IL, interleukin; TNF, tumor necrosis factor; IFN, interferon; lncRNA, long non-coding RNA; NC, negative control for miRNA mimic and inhibitor; Control, empty vector that do not express HOTAIR.
Figure 6.
Effects of HOTAIR and miR-211 overexpression or miR-211 knockdown on the protein expression of various inflammatory factors. Western blot analysis was performed to determine the level of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R protein expression in cells transfected with HOTAIR, miR-211 mimics and miR-211 inhibitor. *P<0.05 vs. NC group. Each experiment was repeated three times. HOTAIR, hox transcript antisense RNA; miR, microRNA; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor; lncRNA, long non-coding RNA; NC, negative control.
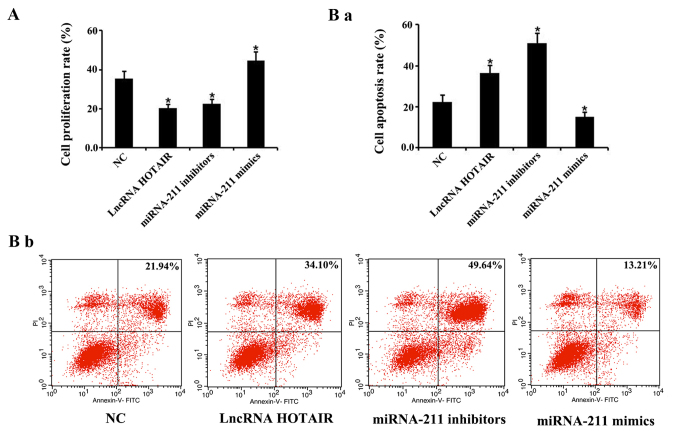
Overexpression of HOTAIR and knockdown of miR-211 inhibits proliferation and promotes apoptosis in monocytes
A CCK-8 assay and flow cytometry were performed to investigate the effects of HOTAIR and miR-211 on cellular proliferation and apoptosis, respectively, in monocytes. The results revealed that the rate of cellular monocyte proliferation transfected with HOTAIR and miR-211 inhibitors was significantly reduced, whereas the cells transfected the miR-211 mimics exhibited a significant increase in proliferation compared with the negative control groups, respectively (P<0.05; Fig. 7A). Furthermore, monocytes transfected with the HOTAIR and miR-211 inhibitors demonstrated a significant increase in apoptosis, whereas those transfected with the miR-211 mimics showed a significant decrease in apoptosis, compared with the negative control group (P<0.05, Fig. 7Ba and Bb).
Figure 7.
Effects of HOTAIR and miR-211 overexpression or miR-211 knockdown on cellular proliferation and apoptosis. (A) A cell counting kit-8 assay was performed to evaluate the level of cellular proliferation of cells treated with HOTAIR, miR-211 mimics, and miR-211 inhibitor. Data represents the mean ± SD. (B) Monocytes were transfected with HOTAIR, miR-211 mimics or miR-211 inhibitor and the rate of apoptosis was determined using flow cytometry, as presented (Ba) statistically and (Bb) as plots. Data represents the mean ± SD. *P<0.05 vs. NC group. Each experiment was repeated three times. HOTAIR, hox transcript antisense RNA; miR, microRNA; lncRNA, long non-coding RNA; NC, negative control.
Discussion
HOTAIR is transcribed by the antisense strand of the HOXC gene located on chromosome 12 and is an important lncRNA that was first identified by Rinn et al (41) in 2007, using a microarray assay. In addition, evidence has indicated that HOTAIR regulates chromatin dynamics and induces gene silencing by interacting with histone methylase and histone demethylase (42). Additionally, HOTAIR has been reported to be involved in the etiology of multiple types of human cancer, including hepatocellular, breast and lung cancer (43–45). HOTAIR has also been demonstrated to regulate the expression of miRNAs by acting as a competitive endogenous RNA (ceRNA) (42). For example, HOTAIR was revealed to possess the binding sites for miR-130a, which were demonstrated to be critical for the modulation of miR-130a by HOTAIR (46). In the present study, HOTAIR was identified to function as a ceRNA of miR-211, and the expression of HOTAIR and miR-211 were negatively associated in monocytes.
As a major public health issue, sepsis is frequently accompanied by microbial infection, systemic inflammation and cellular dysfunction, which can ultimately result in tissue damage, organ failure and even death (47). There are currently three main hypotheses used to explain the pathogenesis of sepsis: i) Pro-inflammatory response; ii) impaired compensatory anti-inflammatory responses; and iii) immune-paralysis, all of which involve the excessive release of inflammatory mediators responsible for the initiation and development of systemic inflammation (1,48). TNF-α was considered to be a central regulator of the immune response and involved in the pathophysiological alterations associated with sepsis (49,50). In addition, TNF-α was demonstrated to promote the release of inflammatory mediators, including IL-6, IL-8, IL-17 and IL-1β, which initiate the host inflammatory response (51,52). IL-6 is primarily released by activated monocytes and has been demonstrated to be negatively associated with the prognosis of patients with sepsis (53). In the present study, since miR-211 was observed to bind to the 3′-UTR of IL-6R, it was hypothesized whether HOTAIR was involved in the pathogenesis of sepsis by indirectly regulating the expression of IL-6R through miR-211.
Currently, various animal models including zebrafish (54), rat (55) and mice (56) have been established to investigate the etiology of sepsis, including toxin treatments such as LPS, zymosan or endotoxins, viable pathogens (including bacteria), as well as altering the endogenous protective barrier in animals (including the induction of colonic permeability leading to bacterial translocation). In addition, CLP is the most frequently applied model in rodents (57,58), which can be used to create sepsis-inducing animal models (59–61). Research has also demonstrated that CLP-induced murine sepsis does not cause lung injury (62), therefore CLP was used to induce murine sepsis in the present study.
Recently, the abnormal expression of lncRNAs has been found in a number of animal models of sepsis, including mice (29) and rat (63), indicating that lncRNAs may be involved in the pathogenesis of sepsis (64,65). In the present study, an animal model of sepsis was established using CLP in mice, which was verified by detecting the increase in mRNA and protein levels of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R. In addition, HOTAIR expression was significantly upregulated, whereas the expression of miR-211 was substantially downregulated in the spleens of septic mice. Furthermore, both HOTAIR overexpression and miR-211 knockdown upregulated the expression of IFN-γ, IL-6, IL-17, TNF-α, IL-1β and IL-6R in monocytes. Treatment with the miR-211 mimics exhibited the opposite effect in monocytes.
Immune suppression caused by corresponding apoptosis (in monocytes and lymphocytes), has been reported to be associated with the pathogenesis of sepsis (5). Therefore, the abrogation of immune cell apoptosis is considered to be a potential therapeutic measure for patients with sepsis. In the present study, HOTAIR overexpression and miR-211 knockdown were revealed to inhibit cellular proliferation and promote apoptosis in monocytes, whereas miR-211 overexpression was demonstrated to induce the opposite effect in monocytes.
In conclusion, the results of the current study indicated that HOTAIR promoted the progression of sepsis indirectly by regulating IL-6R expression via miR-211. Therefore, HOTAIR may be a potential therapeutic target for patients with sepsis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.
Authors' contributions
JC, FC, XG and LZ conceived, designed and coordinated the study and prepared the draft of the manuscript. SW, LZ and YH performed literature research, collected the data, participated in the design of the study and performed the statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All of the animal protocols in the present study were approved by Institute of Radiation Medicine of the Chinese Academy of Medical Sciences.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sagy M, Al-Qaqaa Y, Kim P. Definitions and pathophysiology of sepsis. Curr Probl Pediatr Adolesc Health Care. 2013;43:260–263. doi: 10.1016/j.cppeds.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Mohamed AKS, Mehta AA, James P. Predictors of mortality of severe sepsis among adult patients in the medical Intensive Care Unit. Lung India. 2017;34:330–335. doi: 10.4103/lungindia.lungindia_54_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Vught LA, Wiewel MA, Hoogendijk AJ, Frencken JF, Scicluna BP, Klein Klouwenberg PMC, Zwinderman AH, Lutter R, Horn J, Schultz MJ, et al. The host response in sepsis patients developing intensive care unit-acquired secondary Infections. Am J Respir Crit Care Med. 2017;196:458–470. doi: 10.1164/rccm.201606-1225OC. [DOI] [PubMed] [Google Scholar]
- 4.Sánchez B, Ferrer R, Suarez D, Romay E, Piacentini E, Gomà G, Martínez ML, Artigas A, Edusepsis Study Group Declining mortality due to severe sepsis and septic shock in Spanish intensive care units: A two-cohort study in 2005 and 2011. Med Intensiva. 2017;41:28–37. doi: 10.1016/j.medin.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA. The sepsis seesaw: Tilting toward immunosuppression. Nat Med. 2009;15:496–497. doi: 10.1038/nm0509-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W, Zhong W, Deng Y, Chen C, Wang Q, Zhou M, Li X, Sun C, Zeng H. Evaluation of a combination ‘lymphocyte apoptosis model’ to predict survival of sepsis patients in an intensive care unit. BMC Anesthesiol. 2018;18:89. doi: 10.1186/s12871-018-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill SE, Rohan M, Mehta S. Role of pulmonary microvascular endothelial cell apoptosis in murine sepsis-induced lung injury in vivo. Respir Res. 2015;16:109. doi: 10.1186/s12931-015-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pool R, Gomez H, Kellum JA. Mechanisms of organ dysfunction in sepsis. Crit Care Clin. 2018;34:63–80. doi: 10.1016/j.ccc.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halimulati M, Duman B, Nijiati J, Aizezi A. Long noncoding RNA TCONS_00024652 regulates vascular endothelial cell proliferation and angiogenesis via microRNA 21. Exp Ther Med. 2018;16:3309–3316. doi: 10.3892/etm.2018.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li TT, He RQ, Ma J, Li ZY, Hu XH, Chen G. Long non-coding RNAs in small cell lung cancer: A potential opening to combat the disease (Review) Oncol Rep. 2018;40:1831–1842. doi: 10.3892/or.2018.6635. [DOI] [PubMed] [Google Scholar]
- 11.Ling Z, Liu D, Zhang G, Liang Q, Xiang P, Xu Y, Han C, Tao T. miR-361-5p modulates metabolism and autophagy via the Sp1-mediated regulation of PKM2 in prostate cancer. Oncol Rep. 2017;38:1621–1628. doi: 10.3892/or.2017.5852. [DOI] [PubMed] [Google Scholar]
- 12.Ni X, Liao Y, Li L, Zhang X, Wu Z. Therapeutic role of long non-coding RNA TCONS_00019174 in depressive disorders is dependent on Wnt/β-catenin signaling pathway. J Integr Neurosci. 2018;17:125–132. doi: 10.3233/JIN-170052. [DOI] [PubMed] [Google Scholar]
- 13.Zhang TN, Li D, Xia J, Wu QJ, Wen R, Yang N, Liu CF. Non-coding RNA: A potential biomarker and therapeutic target for sepsis. Oncotarget. 2017;8:91765–91778. doi: 10.18632/oncotarget.21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamantopoulos MA, Tsiakanikas P, Scorilas A. Non-coding RNAs: The riddle of the transcriptome and their perspectives in cancer. Ann Transl Med. 2018;6:241. doi: 10.21037/atm.2018.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons A, Udawela M, Dean B. Non-coding RNA as novel players in the pathophysiology of schizophrenia. Noncoding RNA. 2018;4(pii):E11. doi: 10.3390/ncrna4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 18.Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. MicroRNAs in cancer treatment and prognosis. Am J Cancer Res. 2012;2:414–433. [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 20.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, Zhou T, Yu X, Xue Z, Shen N. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol. 2017;13:657–669. doi: 10.1038/nrrheum.2017.162. [DOI] [PubMed] [Google Scholar]
- 24.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling Z, Wang X, Tao T, Zhang L, Guan H, You Z, Lu K, Zhang G, Chen S, Wu J, et al. Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J Exp Clin Cancer Res. 2017;36:159. doi: 10.1186/s13046-017-0629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X, Liu Z, Ning X, Huang L, Jiang B. The long noncoding RNA HOTAIR promotes colorectal cancer progression by sponging miR-197. Oncol Res. 2018;26:473–481. doi: 10.3727/096504017X15105708598531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, Liu J, Li W, Liu G, Li Z. LncRNA-HOTAIR promotes TNF-α production in cardiomyocytes of LPS-induced sepsis mice by activating NF-κB pathway. Biochem Biophys Res Commun. 2016;471:240–246. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- 30.Bosmann M, Ward PA. The inflammatory response in sepsis. Trends Immunol. 2013;34:129–136. doi: 10.1016/j.it.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walborn A, Hoppensteadt D, Syed D, Mosier M, Fareed J. Biomarker profile of sepsis-associated coagulopathy using biochip assay for inflammatory cytokines. Clin Appl Thromb Hemost. 2018;24:625–632. doi: 10.1177/1076029617709084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulte W, Bernhagen J, Bucala R. Cytokines in sepsis: Potent immunoregulators and potential therapeutic targets-an updated view. Mediators Inflam. 2013;2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao WX, Yu DJ, Zhang WY, Wang XJ. Clinical significance of IL-6 in the diagnosis of sepsis and discriminating sepsis induced by gram-negative bacteria. Pediatr Infect Dis J. 2018;37:801–805. doi: 10.1097/INF.0000000000001904. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, He J. Interleukin-6 is a key factor for immunoglobulin-like transcript-4-mediated immune injury in sepsis. J Intensive Care. 2018;6:22. doi: 10.1186/s40560-018-0294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, Kung HF. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46:200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 36.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Li JH, Wu J, Zhou KR, Zhou H, Yang JH, Qu LH. StarScan: A web server for scanning small RNA targets from degradome sequencing data. Nucleic Acids Res. 2015;43:W480–W486. doi: 10.1093/nar/gkv524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39 (Database Issue) 2011:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43 (Database Issue) 2015:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu ML, Wang XY, Chen WM. TGF-β1 upregulates the expression of lncRNA UCA1 and its downstream HXK2 to promote the growth of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:4846–4854. doi: 10.26355/eurrev_201808_15620. [DOI] [PubMed] [Google Scholar]
- 44.Perrot-Applanat M, Kolf-Clauw M, Michel C, Beausoleil C. Alteration of mammary gland development by bisphenol a and evidence of a mode of action mediated through endocrine disruption. Mol Cell Endocrinol. 2018;475:29–53. doi: 10.1016/j.mce.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Han L, Zhang HC, Li L, Li CX, Di X, Qu X. Downregulation of long noncoding RNA HOTAIR and EZH2 induces apoptosis and inhibits proliferation, invasion, and migration of human breast cancer cells. Cancer Biother Radiopharm. 2018;33:241–251. doi: 10.1089/cbr.2017.2432. [DOI] [PubMed] [Google Scholar]
- 46.Huang J, Ke P, Guo L, Wang W, Tan H, Liang Y, Yao S. Lentivirus-mediated RNA interference targeting the long noncoding RNA HOTAIR inhibits proliferation and invasion of endometrial carcinoma cells in vitro and in vivo. Int J Gynecol Cancer. 2014;24:635–642. doi: 10.1097/IGC.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 47.Yuki K, Murakami N. Sepsis pathophysiology and anesthetic consideration. Cardiovasc Hematol Disord Drug Targets. 2015;15:57–69. doi: 10.2174/1871529X15666150108114810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bone RC, Grodzin CJ, Balk RA. Sepsis: A new hypothesis for pathogenesis of the disease process. Chest. 1997;112:235–243. doi: 10.1378/chest.112.1.235. [DOI] [PubMed] [Google Scholar]
- 49.Ge L, Hu Q, Chen J, Shi M, Yang H, Zhu G. Inhibition of TNF-α sepsis of lipopolysaccharide induction using nano cerium oxide system. Mater Sci Eng C Mater Biol Appl. 2017;77:405–410. doi: 10.1016/j.msec.2017.03.207. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Cui X, Ning L, Wei D. The effects of tumor necrosis factor-α (TNF-α) rs1800629 and rs361525 polymorphisms on sepsis risk. Oncotarget. 2017;8:111456–111469. doi: 10.18632/oncotarget.22824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao H, Liu L, Zhao Y, Hara H, Chen P, Xu J, Tang J, Wei L, Li Z, Cooper DKC, et al. Human IL-6, IL-17, IL-1β, and TNF-α differently regulate the expression of pro-inflammatory related genes, tissue factor and swine leukocyte antigen class I in porcine aortic endothelial cells. Xenotransplantation. 2017;24 doi: 10.1111/xen.12291. [DOI] [PubMed] [Google Scholar]
- 52.Feng S, Yu H, Yu Y, Geng Y, Li D, Yang C, Lv Q, Lu L, Liu T, Li G, Yuan L. Levels of inflammatory cytokines IL-1β, IL-6, IL-8, IL-17A, and TNF-α in aqueous humour of patients with diabetic retinopathy. J Diabetes Res. 2018;2018:8546423. doi: 10.1155/2018/8546423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuire TR, Reardon NT, Bogard K, Plumb TJ, Bultsma CJ, Nissen SW, Fuller PD, Olsen KM. IL6 plasma concentrations in patients with sepsis receiving SLED and antibiotics: A predictor for survival. In Vivo. 2014;28:1131–1134. [PubMed] [Google Scholar]
- 54.Philip AM, Wang Y, Mauro A, El-Rass S, Marshall JC, Lee WL, Slutsky AS, dosSantos CC, Wen XY. Development of a zebrafish sepsis model for high-throughput drug discovery. Mol Med. 2017;23:134–148. doi: 10.2119/molmed.2016.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Sun Z, Zhao F, Wang W, Zhang W, Zhu H. Expression of c-FLIP in a rat model of sepsis and its effects on endothelial apoptosis. Mol Med Rep. 2017;16:231–237. doi: 10.3892/mmr.2017.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buras JA, Holzmann B, Sitkovsky M. Animal models of sepsis: Setting the stage. 2005;4:854–865. doi: 10.1038/nrd1854. [DOI] [PubMed] [Google Scholar]
- 57.Rittirsch D, Hoesel LM, Ward PA. The disconnect between animal models of sepsis and human sepsis. J Leukoc Biol. 2007;81:137–143. doi: 10.1189/jlb.0806542. [DOI] [PubMed] [Google Scholar]
- 58.Chang R, Holcomb JB, Johansson PI, Pati S, Schreiber MA, Wade CE. Plasma resuscitation improved survival in a cecal ligation and puncture rat model of sepsis. Shock. 2018;49:53–61. doi: 10.1097/SHK.0000000000000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberbaum M, Spira RM, Lukasiewicz E, Armon Y, Samuels N, Singer SR, Barak V, Izbicki G, Einav S, Hersch M. Effect of Traumeel S on cytokine profile in a cecal ligation and puncture (CLP) sepsis model in rats. J Altern Complement Med. 2011;17:909–913. doi: 10.1089/acm.2011.0205. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Bi J, Liu S, Pang Q, Zhang R, Wang S, Liu C. 5-HT Drives Mortality in Sepsis Induced by Cecal Ligation and Puncture in Mice. Mediators Inflamm. 2017;2017:6374283. doi: 10.1155/2017/6374283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakai M, Suzuki T, Tomita K, Yamashita S, Palikhe S, Hattori K, Yoshimura N, Matsuda N, Hattori Y. Diminished responsiveness to dobutamine as an inotrope in mice with cecal ligation and puncture-induced sepsis: Attribution to phosphodiesterase 4 upregulation. Am J Physiol Heart Circ Physiol. 2017;312:H1224–H1237. doi: 10.1152/ajpheart.00828.2016. [DOI] [PubMed] [Google Scholar]
- 62.Iskander KN, Craciun FL, Stepien DM, Duffy ER, Kim J, Moitra R, Vaickus LJ, Osuchowski MF, Remick DG. Cecal ligation and puncture-induced murine sepsis does not cause lung injury. Crit Care Med. 2013;41:159–170. doi: 10.1097/CCM.0b013e3182676322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang ZJ, Zhang MY, Fan ZW, Sun WL, Tang Y. Influence of lncRNA HOTAIR on acute kidney injury in sepsis rats through regulating miR-34a/Bcl-2 pathway. Eur Rev Med Pharmacol Sci. 2019;23:3512–3519. doi: 10.26355/eurrev_201904_17717. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Wang X, Yan X, Cheng X, He X, Zheng W. LncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFκB. Int Immunopharmacol. 2018;55:69–76. doi: 10.1016/j.intimp.2017.11.038. [DOI] [PubMed] [Google Scholar]
- 65.Huang W, Lan X, Li X, Wang D, Sun Y, Wang Q, Gao H, Yu K. Long non-coding RNA PVT1 promote LPS-induced septic acute kidney injury by regulating TNFα and JNK/NF-κB pathways in HK-2 cells. Int Immunopharmacol. 2017;47:134–140. doi: 10.1016/j.intimp.2017.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.