Abstract
When available, vaccines are an effective means of disease prevention. Unfortunately, efficacious vaccines have not yet been developed for several major infectious diseases, including HIV and malaria. Vaccine sieve analysis studies whether and how the efficacy of a vaccine varies with the genetics of the pathogen of interest, which can guide subsequent vaccine development and deployment. In sieve analyses, the effect of the vaccine on the cumulative incidence corresponding to each of several possible genotypes is often assessed within a competing risks framework. In the context of clinical trials, the estimators employed in these analyses generally do not account for covariates, even though the latter may be predictive of the study endpoint or censoring. Motivated by two recent preventive vaccine efficacy trials for HIV and malaria, we develop new methodology for vaccine sieve analysis. Our approach offers improved validity and efficiency relative to existing approaches by allowing covariate adjustment through ensemble machine learning. We derive results that indicate how to perform statistical inference using our estimators. Our analysis of the HIV and malaria trials shows markedly increased precision -- up to doubled efficiency in both trials -- under more plausible assumptions compared with standard methodology. Our findings provide greater evidence for vaccine sieve effects in both trials.
Keywords: competing risks, machine learning, vaccine, targeted minimum loss-based estimation, dependent censoring, HIV, malaria
1. Introduction
The development of safe and effective vaccines is one of the greatest public health achievements of the last century. Nevertheless, several infectious pathogens responsible for substantial disease burden still lack an effective preventive vaccine. Vaccine development is often more challenging for genetically heterogeneous pathogens. Indeed, vaccines are typically constructed using only one or several pathogen sequences, and while they may stimulate immune responses that are protective to these few sequences, they may not stimulate immune responses that are more broadly protective. Consequently, vaccines may only be efficacious at preventing infection and disease due to pathogens with sequences similar to the vaccine construct. In an efficacy trial of a preventive vaccine, it is therefore important to study whether and how vaccine efficacy varies by pathogen sequence.
This field of study has been called sieve analysis, nomenclature that derives from imagining a vaccine-induced immune response as a kitchen sieve (Gilbert et al., 1998, 2001). The response presents a barrier to infection or disease, but may have “holes” that allow certain pathogen strains to break through the protective barrier. A sieve effect at a pathogen genetic locus may be defined as differential vaccine efficacy against pathogens with amino acids matched versus mismatched to the vaccine at these residues. Identification of sieve effects can guide selection of antigen sequences in future, potentially multivalent vaccines, which may offer broader protection against genetically diverse pathogens. Sieve analysis can also guide vaccine deployment. If pathogen surveillance data are available, researchers may be able to identify geographic regions where a vaccine will provide a large impact. From a policy perspective, this information is especially important for diseases for which competing partially-effective prevention strategies exist.
Two major diseases caused by highly genetically diverse pathogens for which highly efficacious preventive vaccines have yet to be developed – and therefore diseases for which vaccine sieve analysis is particularly important – are HIV and malaria. For each of these diseases, only a single vaccine has shown efficacy in a Phase III clinical trial. In the case of HIV, this is the ALVAC/AIDSVAX vaccine, whose antigen consists of three HIV-1 envelope protein sequences. The vaccine’s efficacy was evaluated in the RV144 trial, a randomized, placebo-controlled trial in Thailand (Rerks-Ngarm et al., 2009). The vaccine reduced the average instantaneous risk of HIV infection by an estimated 31% over three and a half years follow-up. Efficacy waned over time and was deemed insufficient to gain licensure. For malaria, the most advanced vaccine candidate from a regulatory perspective is the RTS,S/AS01 vaccine. The vaccine’s antigen consists of a portion of the circumsporozoite protein (CS protein) found on the surface of the plasmodium falciparum parasite during the sporozoite phase of its life cycle. The RTS,S/AS01 vaccine reduced the average instantaneous risk of clinical malaria over a follow-up of 12 months by an estimated 63% in 5–17 month old children (RTS,S Clinical Trials Partnership, 2011; Agnandji et al., 2012). As with the RV144 trial, efficacy waned and the vaccine has not been licensed by the United States Food and Drug Administration. However, due to the high morbidity and mortality associated with malaria, the vaccine may still be useful for prevention in areas of high malaria transmission (WHO, 2016).
Statistically, the setting of sieve analysis is an example of a longitudinal data structure with competing risks. After entering the study, trial participants are at risk of experiencing several “competing” endpoints, defined by the genotype of the pathogen causing the study endpoint, with each genotype representing a distinct type of endpoint. To assess the effect of a treatment on the risk of a specific type of endpoint, instantaneous or cumulative parameters may be studied. The choice of parameter depends on the scientific context (Pintilie, 2007), and both types have been used in sieve analysis (Gilbert, 2000). Instantaneous parameters are often based on the cause-specific hazard function (Prentice et al., 1978; Benichou and Gail, 1990; Gaynor et al., 1993; Lunn and McNeil, 1995), defined as the instantaneous rate at which an endpoint of a given type occurs amongst those who have not yet experienced an endpoint. The most common cumulative parameter is cumulative incidence, defined as the probability that an event occurs by a given time and is of a particular type. Cumulative incidence has also been referred to as the subdistribution function (Fine and Gray, 1999), cause-specific failure probability (Gaynor et al., 1993), and absolute cause-specific risk (Benichou and Gail, 1990). While both instantaneous and cumulative parameters are relevant for assessing vaccine sieve effects, given the waning vaccine efficacy observed in the RV144 and RTS,S/AS01 trials, cumulative parameters may have greater public health relevance.
Cumulative vaccine sieve effects are most often estimated with the Aalen-Johansen (AJ) estimator (Aalen, 1978; Aalen and Johansen, 1978; Gilbert, 2000), a generalization of the Kaplan-Meier estimator to settings with competing risks. This estimator requires few assumptions to achieve desirable statistical properties. It is consistent provided participant dropout is non-informative, and is nonparametric efficient if the data do not include any prognostic baseline covariate. However, in many preventive vaccine trials, informative censoring is present and prognostic covariates are available – opportunities for improving on the AJ estimator are therefore common. In the present work, we propose covariate-adjusted estimators that retain a desirable interpretation while offering potential improvements in both bias – by accounting for certain forms of dependent censoring – and in variance – by fully accounting for measured covariates.
In practice, covariate adjustment is often performed using methods based on parametric and semiparametric models. However, when assessing treatment efficacy in randomized trials, statisticians are often reluctant to use these methods due to the risk of model misspecffication. The overwhelming concern is that the potential benefits afforded by covariate adjustment may be offset by bias induced by the use of a misspecified model (cf. Gail et al., 1984). An alternative approach for covariate adjustment is to use flexible regression techniques, such as stacked regression (Wolpert, 1992; Breiman, 1996). Stacking evaluates many regression estimators using cross-validation and selects the best weighted combination of the various candidate estimators. Such estimators have also been referred to as super learners, because they perform essentially as well as the (unknown) best-performing combination of candidate estimators (van der Laan et al., 2004, 2007). The candidate estimators considered may include both parametric regressions, as well as more flexible, machine learning estimators, the latter potentially mitigating concerns of model misspecffication.
While flexible regression techniques are valuable for accurately adjusting for covariates, statistical inference based on the resulting estimate is theoretically challenging. In this paper, we construct flexible targeted minimum loss-based estimators of cumulative incidence for which inference can be facilitated. Relative to the existing approach for estimation of vaccine sieve effects, our method requires weaker assumptions on the censoring mechanism and exhibits markedly improved precision in both trials. Our analysis of the RTS,S/AS01 trial data additionally addresses the challenge of assessing vaccine sieve effects when multiple parasite genotypes may be observed in trial participants. Due to existing privacy policies, we are not able to provide the data used for our analyses; however, the survtmle R package available via CRAN can be used to implement our methods, and we provide an online code repository (https://github.com/benkeser/sieveml) that includes analysis of simulated data sets with identical structure to the real RV144 and RTS,S data.
2. Notation and assumptions
The RV144 and RTS,S/AS01 trials had similar designs: trial participants were enrolled, baseline covariates W were measured, and participants were randomly assigned either to vaccine (Z = 1) or placebo (Z = 0) – see Table 1 for details. After randomization, RV144 participants were asked to attend biannual clinic visits to be tested for HIV, and the endpoint of interest was time to HIV diagnosis. In the RTS,S/AS01 trial, the primary endpoint was the first episode of clinical malaria. We use T to denote the time from randomization until the study endpoint is observed. For RV144, a natural unit for time is number of clinic visits. For RTS,S the most granular unit of time available is days, although for computational simplicity we use months as the unit for the RTS,S analysis.
Table 1:
Summary of two vaccine efficacy trials. The sample size is shown indicates the number of trial participants with complete covariate and genetic sequencing information. AA = amino acid; CS = circumsporozoite.
| Trial | Covariates | Endpoint | Pathogen locus |
|---|---|---|---|
| RV144 (n=15,955) | year of trial enrollment, age, gender, behavioral risk score | first diagnosis of infection with HIV-1 | V2 envelope, A A 169 |
| RTS,S/AS01 (n=6,890) | study site, age, distance from the nearest outpatient/inpatient facility, height-for-age z-score, weight-for-age z-score, weight-for-height z-score, head circumference, hemoglobin, rainy/dry season at enrollment | first episode of clinical malaria with parasite density>5000/mm3 | CS protein |
Regardless, our methodology applies with an arbitrarily fine time-scale. We use τ to refer to the final time-point of the analysis, which was three years post-vaccination for RV144 and one year post-vaccination for RTS,S. We use Y to denote the time until right-censoring. If a participant completed follow-up without experiencing any event, we set Y = τ; if a participant dropped out during follow-up, then Y ∈ {1, 2, …, τ − 1}. For participants who experienced the study endpoint, the genotype of the infecting pathogen was obtained. Genotypes were categorized as matched to the vaccine at the given pathogen protein locus (J = 1) or mismatched in that region (J = 2).
In the absence of right-censoring, the full data unit is available for each trial participant. The cumulative incidence of type j ∈ {1, 2} endpoints at time t0 ∈ {1, 2, …, τ} under vaccine assignment z ∈ {0, 1} is defined as and denoted Fj,z(t0). The cumulative vaccine efficacy against genotype j is defined as the multiplicative reduction in cumulative incidence comparing vaccine to control,
In vaccine sieve analysis, we are interested in testing the null hypothesis that V E1(t0) = V E2(t0) against various alternatives. We define the vaccine sieve effect as the ratio (match vs. mismatched) of ratios (control vs. vaccine) of cumulative incidence,
To estimate the sieve effect, we estimate the cumulative incidence of each endpoint type in each treatment arm. For simplicity, we consider the statistical problem of estimating cumulative incidence of matched endpoints (i.e., of type j = 1) under assignment to the vaccine (i.e., z = 1). Because the labeling of events and treatments is arbitrary, our developments immediately apply to estimating the cumulative incidence of every other combination of endpoint and treatment arm. Our choice of t0 is also arbitrary and our results directly apply to pointwise estimation of cumulative incidence, vaccine efficacy, and vaccine sieve effects over time.
Cumulative incidence cannot be estimated directly from the observed data because of right-censoring. In practice, we observe , where , , and P0 denotes the true distribution of O. Our goal is to develop an estimator of F1,1 (t0) based on n independent realizations O1, … , On from P0. It will be useful to express the observed data unit in terms of discrete counting processes, i.e., O = (W, Z, {N(t), M(t), C(t) : t = 1. 2, …, τ}). where , and .
3. Identification and estimation of cumulative incidence
Under conditions described below, cumulative incidence can be identified by the longitudinal G-computation formula (Robins, 1999; Bang and Robins, 2005), a functional of the observed data distribution P0. Using this approach, we may identify cumulative incidence using iteratively-defined conditional means of the discrete counting process, hereafter referred to as iterated means. We define H(t) := (N(t), M(t), W) to denote a participant’s observed history through time t. We define the iterated means as
and so on. In words, μt0 (h(t − 1)) is the true conditional probability of a matched endpoint at t0 given vaccine, no censoring up to t0 − 1, and history h(t − 1). Thereafter, the iterated mean at t is defined recursively as the conditional mean of μt+1 (H(t)) given vaccine, no censoring up to t − 1, and history h(t − 1). For example, μt0−2(h(t0 − 3)) with n(t0 − 3) = 0, m(t0 − 3) = 0 and w = w0 is the probability a vaccinated participant with baseline covariate vector value w0 experienced a J = 1 endpoint at times t0 − 2, t0 − 1 or t0 given no endpoint prior to t0 − 2. While each iterated mean is implicitly dependent on the final time-point t0, we suppress this to simplify notation. We note that μ1 is only a function of the value of W, since no participant has yet experienced an endpoint.
We can relate the iterated means above to cumulative incidence under two key conditions. The first condition is that the observed data unit O is a random coarsening of the full data unit X (see, e.g., van der Laan and Robins, 2003). This is satisfied, for example, if (T, J) is independent of C given W and Z; in words, there are no unmeasured confounders of the relationship between the censoring mechanism and outcome process. The second condition essentially states that the conditional probability of remaining uncensored throughout the study given W = w is strictly positive for each value of w that arises in the population. This is often referred to as the positivity condition (see, e.g., Petersen et al., 2010). In Appendix A, we show that under these two conditions, for each t = 1, … , t0, μt(h(t − 1)) with h(t − 1) = (0, 0, w) equals the conditional probability of a J = 1 endpoint between t and t0 for a vaccinated participant with covariates W = w and no endpoint observed prior to t. Thus, because no participant has experienced an event at time zero, μ1 is equal to the conditional probability of a J = 1 endpoint between 1 and t0 for a vaccinated participant with W = w, that is, the covariate conditional cumulative incidence of matched endpoints in vaccinated participants. The marginal cumulative incidence of matched endpoints in the vaccine arm can be obtained by integrating μ1 over the distribution of covariates. Defining G(w) := prP (W ≤ w) the cumulative distribution function of covariates at w and , we have that under coarsening at random and positivity assumptions.
A heuristic justification of the identification result is that μt describes what happens in the stratum of vaccinated participants under observation at t with covariate value W = w. The positivity assumption ensures that this stratum indeed exists for each choice of w, while coarsening at random ensures that at-risk participants with W = w are on average similar to previously censored participants with W = w. Thus, at any given time, participants at risk can be used to infer, on average, what would have happened to other participants in the same stratum had they remained under study.
The identification result implies that an estimator of F1,1 (t0) may be obtained by estimating μ := {μt : t = 1, …, t0} and subsequently marginalizing the estimator of μ1 using an estimator of the distribution of baseline covariates. When parametric regression models are used to estimate μ, this estimator is commonly referred to as a G-computation estimator, and it is an example of a plug-in estimator. For concreteness, we outline the practical construction of the G-computation estimator based on the use of the empirical distribution of covariates to estimate G and a modified version of logistic regression to estimate μ. We modify standard logistic regression in two ways. First, we account for the fact that for any observation with H(t − 1) = (1, 0, w), indicating a matched endpoint prior to t, N(t) = 1 necessarily, and thus our regression estimator μn,t of μt should equal 1 for all such observations. Similarly, if H(t − 1) = (0, 1, w), indicating a mismatched endpoint prior to t, then N(t) = 0 and so μn,t should equal zero for all such observations. Our second modification of logistic regression is that the outcome of our regressions may take values between zero and one. Below, we refer to such regressions as quasi-logistic. For simplicity of exposition, we assume momentarily that W is univariate. The algorithm includes the following steps:
Set μn,t0 (Hi(t0 − 1)) = 1 for all observations with Ni(t0 − 1) = 1 and set μn,t0 (Hi(t0 − 1)) = 0 for all observations with Mi(t0 − 1) = 1. Define a logistic regression model, logit {μt0(h(t0 − 1))} = αt0 + β,t0w, for the remaining observations. Fit the logistic regression model by regressing outcome Ni(t0 − 1) on Wi using all Oi with Zi = 1, Ci(t − 1) = 0, Ni(t − 1) = 0, and Mi(t − 1) = 0. Let αn,t0 and βn,t0 denote the estimated parameters of the model. Set μn,t0 (Hi(t0 − 1)) = expit(αn,t0 + βn,t0 Wi) for all observations with Mi(t0 − 1) = 0 and Ni(t0 − 1) = 0.
For t = t0 − 1, …, 1, recursively construct an estimate μn,t of μt as follows. First, set μn,t(Hi(t − 1)) = 1 for all observations with Ni(t − 1) = 1 and set μn,t0 (Hi(t − 1)) = 0 for all observations with Mi(t − 1) = 1. Define a quasi-logistic regression model, logit{μt(h(t − 1))} = αt + βtw, for the remaining observations. Fit the quasi-logistic regression model by regressing outcome μn,t+1(Hi(t)) on Wi using all Oi with Zi = 1, Ci(t − 1) = 0, Ni(t − 1) = 0, and Mi(t − 1) = 0. Let αn,t and βn,t denote the estimated parameters of the model. Set μn,t(Hi(t − 1)) = expit(αn,t + βn,t Wi) for all observations with Mi(t − 1) = 0 and Ni(t − 1) = 0.
Construct estimate by averaging μn,1 over observed values of W.
If the fitted logistic regression approximates μt well for each t, then relative to the AJ estimator, we expect this plug-in estimator to have improved bias when censoring is dependent, and reduced variability when covariates are predictive of the outcome process. Intuitively, there are two sources of variation in the time-to-endpoint of trial participants: variation due to measured versus unmeasured characteristics, the latter often referred to as noise. Covariate-averaged estimators of the type exhibited above often mitigate the former source of variation and can therefore have improved precision. Among others, Zhang et al. (2008) and Moore and van der Laan (2009) discuss efficiency improvements in randomized trials.
These arguments point to the potential for possibly important gains from using a covariate-averaged estimator of marginal cumulative incidence. However, to realize these gains, the fitted regression estimators must adequately approximate the true iterated mean at each time-point. Moore and van der Laan (2009) showed that, in studies with no right-censoring, adjusting for covariates, even via misspecified regression models, can yield unbiased treatment effect estimates that are more precise than unadjusted estimates. However, the same is not true in the presence of right-censoring. This motivates the use of flexible regression techniques to ensure that estimates of the iterated means are as close to the truth as possible. As discussed in the introduction, stacking or super learning (Wolpert, 1992; Breiman, 1996; van der Laan et al., 2007) based on a library of candidate learners may be useful towards this end. Numerous studies illustrate the advantages of this approach in practice (van der Laan et al., 2007; Rose, 2013; Pirracchio et al., 2015). In order to approximate the truth as well as possible, it appears strategic in the context of super learning to consider a large set of candidate regression estimators including both parametric and machine learning-based estimators. However, when flexible regression techniques are used, performing valid statistical inference is a more challenging task. Whereas plug-in estimators based on a parametric MLE typically have an asymptotic normal distribution, the same is not true when adaptive procedures, like the super learner, are used. This complicates greatly the construction of valid confidence intervals and hypothesis tests. The targeted minimum loss-based estimation (TMLE) framework provides a means of addressing this problem.
4. Targeted minimum loss-based estimation
To understand why plug-in estimators based on adaptive regression estimators behave poorly, we require the form of the efficient influence function (EIF), a key element in efficiency theory (Bickel et al., 1997). We define ζ(w) := prP0(Z = 1 ∣ W = w) as the conditional probability of receiving vaccine given covariates. For t = 1, 2,…, t0 − 1, we define πt(w) := prP0{C(t) = 0 ∣ Z = 1, N(t − 1) = 0, M(t − 1) = 0, C(t − 1) = 0, W = w} as the conditional probability of remaining uncensored at time t given vaccine, no endpoint nor censoring through t − 1, and baseline covariates, and we write π := {πt : t}. The EIF of cumulative incidence at P0 relative to a nonparametric model evaluated on a typical observation Oi is
| (1) |
where, for t = 1, 2,…, t0, we define Dt(μ, ζ, π) as the mapping
and DW(μ, G) as the mapping o ↦ μ1(w) − ∫ μ1(u)dG(u). This is also the EIF in a model with arbitrary constraints imposed on the conditional treatment or censoring distribution. If an estimator of is such that , it follows that has a normal distribution in large samples, with variance at least as small as any other regular, asymptotically linear estimator of . Valid inference based on such an estimator may be facilitated by knowledge of this asymptotic distribution.
The construction of a plug-in estimator of only requires estimators of μ and G. However, estimators of the treatment probability ζ and censoring distribution π are also generally needed to construct efficient estimators of . Under conditions stated in Appendix B, the estimation error of any plug-in estimator of can be expressed as
| (2) |
where ζn and πn are estimators of ζ and π, respectively. Sufficient regularity conditions include that ζn and πn tend in probability to ζ and π, respectively, at a sufficiently fast rate. In the case of a randomized trial, the treatment assignment probability ζ is known exactly; however, the conditional censoring distribution π is unknown and must be estimated from the data. To ensure that our estimate of the censoring distribution is as close to the truth as possible, we resort to super learning. Because in our proposed implementation μn and πn are super learner-based estimators, the second summand on the right-hand side of (2), , generally does not converge to zero faster than n−1/2. This renders inefficient and prevents it from having a tractable distributional limit theory. Practically, this implies that will exhibit large finite-sample bias, making it difficult, if not impossible, to perform valid inference based on .
In the previous section, we argued that flexible estimation techniques may be useful for obtaining an accurate estimate of the iterated means. However, equation (2) makes clear that there are repercussions vis-à-vis statistical inference for adopting such an approach. The challenge is thus to construct regression estimators and πn that (i) are flexible enough to provide a good approximation to μ and π, respectively, and (ii) have minimal finite-sample bias relative to , in the sense that . If such estimators can be constructed, then the plug-in estimator based on is consistent, asymptotically normal and efficient.
The TMLE framework provides a way to update an initial estimator μn into a revised estimator that satisfies a set of user-specified equations. Remarkably, the idea of TMLE was first suggested in Stein (1956) as a means of constructing nonparametric efficient estimators, though, to our knowledge, examples of these estimators did not appear in the literature until Scharfstein et al. (1999). The concept was developed in generality and formally studied in van der Laan and Rubin (2006), which led to the development of the field of targeted learning (van der Laan and Rose, 2011, 2018). In the longitudinal data setting, TMLE estimators of average treatment effects based on parametric regressions were described in Robins (1999) and implemented in Bang and Robins (2005). TMLE estimators using super learning-based estimators were described and evaluated in van der Laan and Gruber (2012).
In the present problem, we use TMLE to map initial estimates (μn, ζn, πn) into an estimate of μ that satisfies
| (3) |
Here, we describe how such a procedure can be implemented in practice – theoretical details follow from van der Laan and Gruber (2012) and are omitted. The targeted plug-in estimator is implemented similarly as the G-computation estimator described in Section 3, though it includes an additional univariate model-fitting step after each regression.
If the conditional treatment probability ζ0 is known, set ζn = ζ0; otherwise, construct estimate ζn of ζ0. Construct estimate πn of the censoring probabilities π0.
- Set for i = 1,…, n. For t = t0, t0 − 1,…, 1, perform recursively:
- construct and evaluate an initial estimate μn,t of μt. First, set μn,t(Hi(t − 1)) = 1 for all observations with Ni(t − 1) = 1. Similarly, set μn,t(Hi(t − 1)) = 0 for all observations with Mi(t − 1) = 1. For the remaining observations fit a super learner regression of the outcome on covariates Wi using Oi with Zi = 1, Ci(t − 1) = 0, Ni(t − 1) = 0, and Mi(t − 1) = 0. Note that at t = t0 this outcome is simply Ni(t0), while for other time points it is a vector of predictions from the quasi-logistic regression generated in step 2b) at time t + 1. Set μn,t(Hi(t − 1)) equal to the super learner-predicted value for all Oi with Ni(t − 1) = 0 and Mi(t − 1) = 0. The estimate μn,t has been evaluated on all observations and we have a vector of n observations containing predicted values of μt.
-
fit a quasi-logistic regression with outcome , offset term logit{μn,t(Hi(t − 1))} and covariate using only observations with Zi = 1, Ci(t − 1) = 0, Ni(t − 1) = 0, and Mi(t − 1) = 0. Denote by ϵn,t the estimated regression coefficient in this model. For observations with Ni(t − 1) = Mi(t − 1) = 0, definefor all other observations set . Thus, we again obtain a vector of n predicted values for μt, which will serve as outcome values in the regression at t − 1.
Construct estimate by averaging over observed values of W.
The targeted estimator has desirable large-sample properties. If each of ζn, μn and πn are consistent for ζ, μ and π, respectively, at a sufficiently fast rate, is a nonparametric efficient estimator of , and tends to a mean-zero normal distribution with variance consistently estimated by . Additionally, is doubly-robust in the sense that it is consistent if at least one of μn and (πn, ζn) is consistent (van der Laan and Gruber, 2012). If μn is inconsistent but the treatment and censoring mechanisms are known exactly, is no longer efficient but tends to a normal distribution with variance still consistently estimated by . However, if μn is inconsistent, ζ is known and π is consistently estimated using the observed data, the asymptotic variance is more complicated. Nevertheless, in the special case that πn is an efficient estimator within a parametric model, then is a conservative estimate of the asymptotic variance (see, e.g., Theorem 2.3 of van der Laan and Robins, 2003). A similar result holds for the case in which πn or ζn is inconsistent but μn is consistently estimated.
Use of the Wald interval ( , ), where zβ is the β-quantile of the standard normal distribution, is motivated by these large-sample results. This interval will typically have asymptotic coverage no smaller than 1 − α under a range of scenarios, and asymptotically exact coverage either if each of μn, ζn and πn are consistent, or (π, ζ) is exactly known. Given a fixed , a test of the null hypothesis versus the two-sided complement alternative with asymptotic size no larger than α can be constructed by rejecting the null whenever . In Section 2 of the web supplement, we illustrate how the delta method can be used to obtain estimates of the asymptotic variance of V Ej(t0) and V S E(t0).
5. Simulation study
We evaluated the performance of the super learner-based TMLE estimators via simulation studies. The baseline covariate vector W had independent components W1 ~ Uniform(−2, 2) and W2 ~ Bernoulli(0.5). Vaccine was randomly assigned with equal probability, that is, Z ~ Bernoulli(0.5) independently of W. Given W = w and Z = z, the time T of interest was simulated as T ~ Geometric{λβ(w, z)}, where λβ(w, z) = expit(−2 + βw1 − 2βw1w2 + z), and the time C to censoring was simulated as C ~ Geometric{ηγ(w, z)}, where ηγ(w, z) = expit(−3 + γw1 − 2γw1w2 + z). The type of endpoint was randomly assigned with equal probability to types J = 1 and J = 2. The observed failure time was taken to be the minimum of T and C, with ties labeled as events.
We compared the performance of our proposed TMLE estimator using super learning to estimate the iterated means and censoring distribution to: (1) the AJ estimator, which does not adjust for covariates, and (2) a G-computation estimator that adjusts for covariates via main terms logistic regression. The super learner library included four algorithms: a main terms logistic regression, a logistic regression with forward stepwise variable selection including all two-way interactions, an intercept-only logistic regression, and multivariate adaptive regression splines (Friedman, 1991) – this represents a combination of traditional regression techniques and machine learning algorithms.
We evaluated the performance of estimators of at two sample sizes, n ∈ {500, 5000}, in five different scenarios that varied how covariates related to endpoints and censoring (Table 3). In the first scenario, we set β = 0 and γ = 0, and covariates were related neither to endpoints nor censoring (W ↛ T, W ↛ C). In this scenario, all three estimators considered are consistent and asymptotically efficient, and indeed we found that the three estimators performed equally well at both sample sizes.
Table 3:
Left: theoretical properties of the Aalen-Johansen (AJ), main terms logistic regression-based G-computation (GCOMP), and super learner-based targeted minimum loss-based estimator (TMLE). Right: Monte Carlo evaluation of finite-sample performance of estimators of cumulative incidence in terms of bias, coefficient of variation, mean squared-error (MSE) and coverage probability of nominal 95% confidence intervals. Results are based on 1,000 samples from the data generating mechanism for two sample sizes and four relationships between covariates (W), time-to-endpoint (T) and time-to-censoring (C). The fifth scenario includes an unmeasured confounder U.
| Large-sample properties |
n = 500 |
n = 5000 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AJ | GCOMP | TMLE | AJ | GCOMP | TMLE | AJ | GCOMP | TMLE | |||
| W ↛ T, W ↛ C | Consistent | ✓ | ✓ | ✓ | Relative Bias (%) | −0.12 | −0.12 | −0.95 | 0.14 | 0.14 | 0.03 |
| Coef. Var. (×10) | 0.86 | 0.86 | 0.88 | 0.26 | 0.26 | 0.26 | |||||
| = 0.42 | Efficient | ✓ | ✓ | ✓ | MSE (×100) | 0.74 | 0.74 | 0.78 | 0.07 | 0.07 | 0.07 |
| Coverage (%) | 92.9 | 92.9 | 92.4 | 95.3 | 95.0 | 95.1 | |||||
| W ↛ T, W → C | Consistent | ✓ | ✓ | ✓ | Relative Bias (%) | 0.02 | 0.26 | −2.83 | 0.05 | 0.07 | −0.73 |
| Coef. Var. (×10) | 0.91 | 0.90 | 1.08 | 0.28 | 0.28 | 0.33 | |||||
| = 0.42 | Efficient | ✓ | ✓ | ✓ | MSE (×100) | 0.83 | 0.82 | 1.24 | 0.08 | 0.08 | 0.11 |
| Coverage (%) | 95.8 | 95.8 | 93.0 | 95.8 | 95.6 | 98.0 | |||||
| W → T, W ↛ C | Consistent | ✓ | ✓ | ✓ | Relative Bias (%) | −0.63 | −0.62 | −1.29 | 0.08 | 0.08 | −0.00 |
| Coef. Var. (×10) | 0.95 | 0.96 | 0.92 | 0.31 | 0.31 | 0.30 | |||||
| = 0.33 | Efficient | ✕ | ✕ | ✓ | MSE (×100) | 0.91 | 0.92 | 0.86 | 0.10 | 0.10 | 0.09 |
| Coverage (%) | 95.1 | 95.1 | 94.8 | 95.3 | 95.3 | 95.3 | |||||
| W → T, W → C | Consistent | ✕ | ✕ | ✓ | Relative Bias (%) | −7.96 | −7.65 | −1.69 | −7.64 | −7.60 | 0.52 |
| Coef. Var. (×10) | 0.99 | 0.99 | 0.99 | 0.33 | 0.33 | 0.32 | |||||
| = 0.33 | Efficient | ✕ | ✕ | ✕ | MSE (×100) | 1.74 | 1.67 | 1.01 | 0.80 | 0.79 | 0.10 |
| Coverage (%) | 84.8 | 85.1 | 95.8 | 24.8 | 25.2 | 94.0 | |||||
| (W, U) → T, (W, U) → C | Consistent | ✕ | ✕ | ✕ | Relative Bias (%) | −6.50 | −6.50 | −6.81 | −6.87 | −6.87 | −6.96 |
| Coef. Var. (×10) | 0.09 | 0.09 | 0.09 | 0.29 | 0.29 | 0.30 | |||||
| = 0.35 | Efficient | ✕ | ✕ | ✕ | MSE (×100) | 0.14 | 0.15 | 0.15 | 0.07 | 0.07 | 0.07 |
| Coverage (%) | 88.1 | 88.4 | 87.9 | 31.5 | 31.5 | 31.6 | |||||
In the second scenario, we set β = 0 and γ = 2, and covariates were related to censoring but not to endpoints (W ↛ T, W → C). Again, all estimators are consistent and asymptotically efficient, and we found that the three estimators performed well. The TMLE estimator had slightly more bias than the other two estimators, which may be due to practical violations of the positivity assumption, a known concern when implementing efficient estimators in small sample sizes (Petersen et al., 2010). In particular, for this data-generating mechanism, there were observations with < 0.0001 probability of remaining uncensored until the last time point. These observations receive large weight in both the TMLE estimation procedure and in the variance estimates for the TMLE, which may contribute to the slight deterioration in performance.
In the third simulation scenario, we set β = 2 and γ = 0, and covariates were related to endpoints but not to censoring (W → T, W ↛ C). In this scenario, all estimators are consistent; however, only the TMLE estimator, which correctly adjusts for the covariate-endpoint relationship, is asymptotically efficient. We found low bias for all estimators in this scenario, but slightly lower variability in the TMLE estimates. Though the improvements in variability are modest, they come near to achieving the theoretical efficiency gains possible for this data-generating process. The theoretical efficiency gains can be computed by contrasting the efficiency bound for covariate-adjusted estimators and for covariate-unadjusted estimators. This can be achieved by computing the variance of the efficient influence function of μ based on a data structure that does or does not include the available baseline covariate vector. We numerically evaluated these efficiency bounds – these are lower bounds for asymptotic variances – and found 0.54 and 0.48 for unadjusted and adjusted estimators, respectively. Thus, it is only theoretically possible to improve the efficiency of the unadjusted estimator in this simulation study by about 11%. The TMLE estimators nearly achieve this efficiency improvement at the sample sizes we considered.
In the fourth scenario, we set β = 2 and γ = 2, and covariates were related both to endpoints and censoring (W → T, W → C). In this case, the G-computation and AJ estimators are inconsistent, while the TMLE estimator remains consistent and asymptotically efficient. We found that the confidence intervals based on the G-computation and AJ estimators suffered from poor performance, while the TMLE-based intervals achieved approximately nominal coverage.
Finally, in the fifth scenario, we studied the performance of estimators when the coarsening at random assumption is violated. In this scenario, we additionally simulated an unobserved confounder U of T and C. Given W1 = w1, we drew U from a Normal(w1, 1) distribution. Given W = w, U = u and Z = z, and time to endpoint was simulated as , where , time to censoring was simulated as , where . In this situation, all estimators are inconsistent and had equally poor performance.
6. RV144 HIV Vaccine Efficacy Trial Sieve Analysis
We studied the efficacy of the ALVAC/AIDSVAX vaccine to prevent HIV infection by viruses matched and mismatched to the vaccine at amino acid position 169 of the V2 loop of the envelope protein. We estimated the cumulative incidence, vaccine efficacy and sieve effects against matched and mismatched infections every six months from the time of vaccination until three years post vaccination. We adjusted for baseline patient characteristics (year of enrollment, age, gender, behavioral risk score) by using the super learner to estimate of the conditional censoring distribution and the iterated means. The super learner library included both parametric techniques and machine learning algorithms – details are provided in Appendix C.
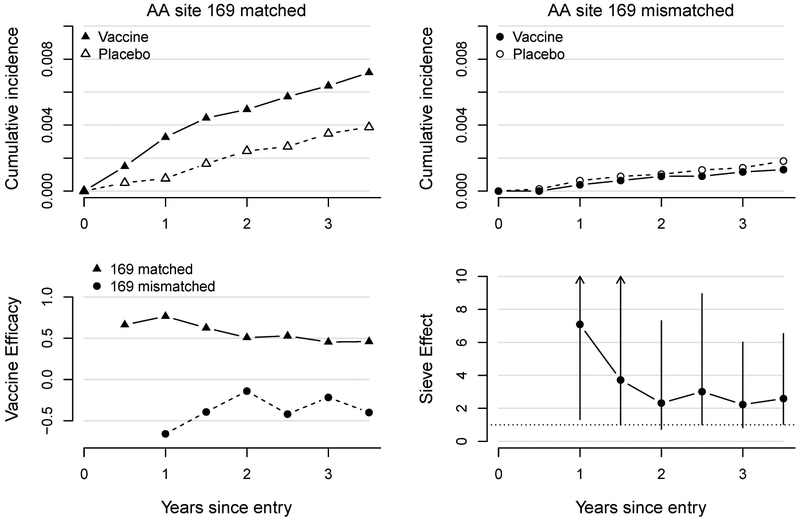
There was a clear separation between the estimated vaccine and control cumulative incidence curves for matched infections, but not for mismatched infections (top row, Figure 1), which led to positive estimates of vaccine efficacy against matched infections and negative efficacy against mismatched infections (bottom left). The efficacy against matched infections appeared to wane over time but was greater than zero at all time-points. The sieve effect comparing these efficacies is thus strongest at early time-points, when the efficacy against matched infections is highest (bottom right). At one and a half years post-vaccination, the vaccine sieve effect was estimated to be 3.0 (95% CI: 1.0, 8.9; p-value=0.05), indicating higher vaccine efficacy against matched rather than mismatched infections. Over time, the sieve effect waned along with the efficacy against matched infections. At three years post vaccination, the vaccine sieve effect was estimated to be 2.3 (95% CI: 0.9, 6.0; p-value=0.12).
Figure 1:
Results from the TMLE analysis of the RV144 trial. Top: Cumulative incidence in vaccine and control groups of HIV-1 infections matched (left) and mismatched (right) to the vaccine at AA 169. Bottom left: Vaccine efficacy against matched (triangle) and mismatched (circle) infections. Bottom right: Vaccine sieve effect with 95% confidence intervals.
We also analyzed the data using the AJ estimator for comparison. We found the point estimates to be nearly identical to those of the TMLE, which might indicate a lack of dependent censoring based on the measured covariates. The confidence intervals about the TMLE-estimated sieve effects were considerably narrower than those for the AJ estimator, as illustrated in Figure 2. By year 3 the efficiency gain is large, with a 33% narrower confidence interval, equating to a 1 − 0.672 = 45% increase in efficiency. These results are consistent with our theory and simulation results, which predict efficiency gains when adjusting for prognostic covariates. The greater precision strengthens the evidence for vaccine sieve effects at site 169 that wane over time; we interpret the scientific impact of these results in the discussion.
Figure 2:
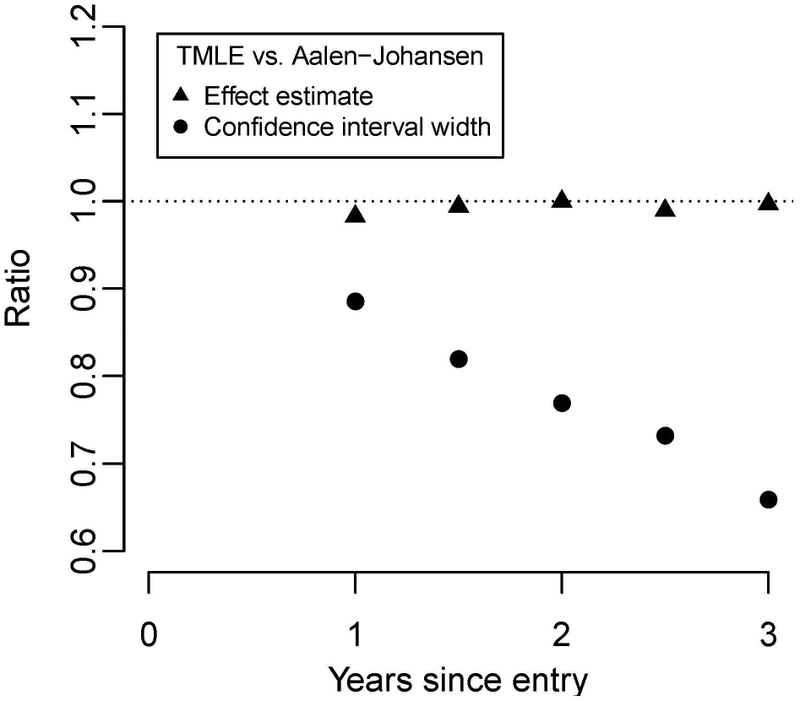
Relative sieve effect estimate (triangle) and confidence interval width (circle) from RV144 trial comparing TMLE to AJ. Values less than one indicate lower point estimate/variance for TMLE.
7. RTS,S/AS01 Malaria Vaccine Efficacy Trial Sieve Analysis
The majority of malaria endpoints in the RTS,S/AS01 trial had multiple parasite genotypes, which occurred because most children were infected by malaria parasites multiple times before presenting with clinical disease. Each strain in a multiple-genotype infection is likely caused by a separate transmission event from a different mosquito. Only clinical cases were registered as study endpoints and all of the associated parasite genomes were subsequently sequenced. The fact that many cases had both matched and mismatched genotypes along the CS protein led to an interesting question: how should vaccine efficacy and vaccine sieve effect parameters be defined?
One approach is to define each endpoint as matched to the vaccine if any of the parasites strains are matched to the vaccine. However, a vaccine sieve effect defined in this way is a mixture of the effect of (i) the vaccine on the number of parasite strains present, and (ii) genetic sieve effects, and the latter are of primary interest in our analysis. To estimate the genetic component of the sieve effect, it may be most relevant to define a parameter describing the vaccine’s effect on a randomly sampled founder parasite of a clinical malaria case. Let Ai := (A1,i,…, AKi,i) denote the Ki different genotypes found in participant i, with Ai = ∅ if participant i is not a case. Let Ri denote a random index drawn from a discrete uniform distribution on {1, 2,…, Ki}, with Ri = 0 if Ai = ∅. Let Ji,Ri = 1 if ARi,i is matched to the vaccine and Ji,Ri = 2 otherwise. Let denote the cumulative incidence of clinical malaria associated with genotype r in treatment arm z. The average cumulative vaccine efficacy against parasites of type j is defined using the geometric mean over random draws of R as
| (4) |
The geometric mean is used so that the averaging process occurs on a scale with greater symmetry.
The multiple outputation approach of Follmann et al. (2003) can be used to estimate such parameters. In this procedure, a dataset is constructed by randomly sampling a single founder genotype from each clinical malaria endpoint, and a statistical method designed for a single genotype per endpoint is applied. For each resample r, we apply our TMLE procedure to obtain a point estimate Fn,jr,z(t0) of Fjr,z(t0) for j ∈ {1, 2} and z ∈ {0, 1}. The resampling process is repeated a large number B times. For this analysis, based on the guidelines of Follmann et al. (2003), the estimated number of outputations required for stable inference was B = 3, 677. The overall estimate is calculated as the average of point estimates across the resampled data sets. Specifically, the overall estimate of the true average cumulative vaccine efficacy provided in (4) is computed as
for j = 1, 2, while the corresponding estimate of sieve effect is given by
The asymptotic variance of and can easily be approximated via the delta method by appropriately combining the estimated influence functions for each outputation sample (details in Section 2 of the web supplement). These influence function-based variance estimators offer an improvement over the nonparametric variance estimators suggested in Follmann et al. (2003), which are not guaranteed to be positive.
Similarly as in the RV144 example, we estimated genotype-specific vaccine efficacy for each genotype and tested for differential vaccine efficacy. We used the super learner with a large library to generate initial estimates of the conditional censoring distribution and the iterated conditional means. Further details are included in Table C1.
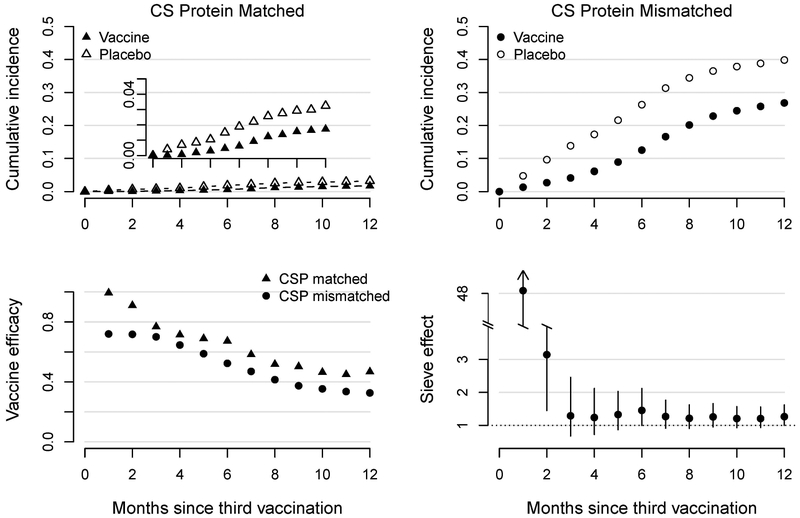
The results are displayed in Figure 3. In the top row, we illustrate the estimated cumulative incidence of disease with malaria parasites matched (left) and mismatched (right) to the RTS,S/AS01 vaccine along the CS protein (left). The cumulative incidence of matched malaria is much lower than mismatched malaria at each time-point for each treatment group. However, for both genotypes there is a clear separation between the vaccine and control curves, indicating vaccine efficacy against both types of endpoints. The vaccine efficacy wanes over time (bottom left) but remains significant through 12 months of follow-up. Vaccine efficacy is significantly higher against matched versus mismatched malaria at each time-point. For example, at 12 months the multiplicative effect of the vaccine is estimated to be 1.3 (95% CI: 1.0, 1.6; p=0.05) times stronger for preventing clinical disease caused by CS protein-matched vs. mismatched parasites.
Figure 3:
Results from the TMLE analysis of the RTS,S/AS01 trial. Top: Multiple outputation TMLE estimates in vaccine and control groups of the incidence of clinical malaria cases matched (left) and mismatched (right) to the vaccine along the CS protein. Bottom left: Vaccine efficacy against matched (triangle) and mismatched (circle) infections. Bottom right: Vaccine sieve effect with 95% confidence intervals.
Once more, our TMLE-based results yielded similar point estimates as the AJ estimator but narrower confidence intervals for the TMLE-estimated sieve effects, achieving a doubled efficiency (1 − 0.72 = 0.51) at the 10-month time-point (Figure 4).
Figure 4:
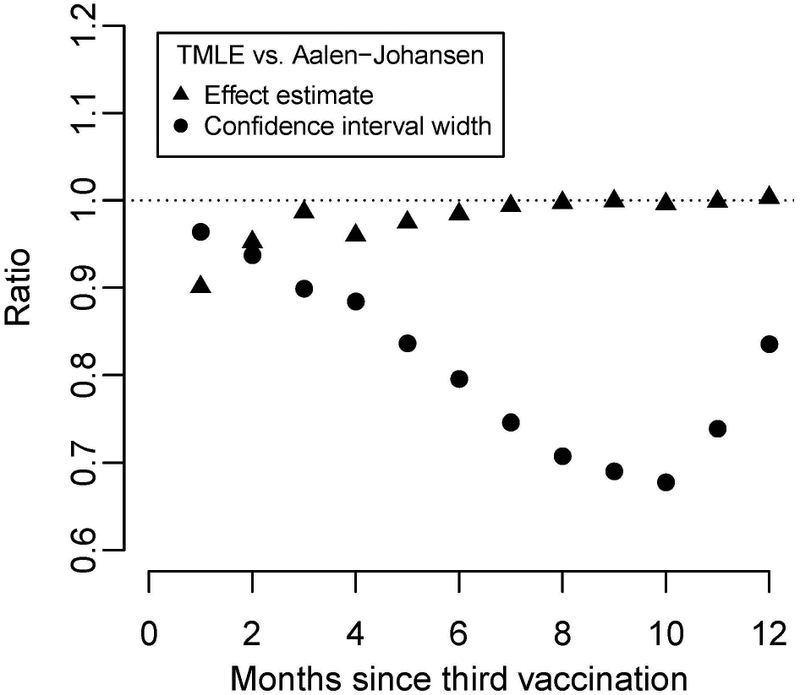
Relative sieve effect estimate (triangle) and confidence interval width (circle) from RTS,S/AS01 trial comparing TMLE to AJ. Values less than one indicate lower point estimate/variance for TMLE.
8. Discussion
For both studies we have analyzed, the identified sieve effects contribute to vaccine science in several ways. First, they motivate basic science studies of mechanisms of protection associated with antibody epitopes at site 169 for the HIV vaccine and in CSP for the malaria vaccine. Second, they support building refined versions of the vaccine that include additional antigen sequences. Third, they have generated hypotheses about sieve effects that will be tested in follow-up efficacy trials of refined vaccine regimens now being planned for both HIV and malaria.
Accurate and precise estimates of vaccine efficacy are important from a regulatory and public health perspective. In preventive vaccine trials, we can never be certain that censoring is completely random and therefore should be concerned that estimators based on this assumption may be systematically biased for efficacy in the enrolled population. Bias due to non-random participant dropout can be mitigated to some extent through the use of measured covariates that predict both dropout and events. As demonstrated in the simulation study, our proposed estimators account for such covariates and thereby reduce bias. However, the simulation also demonstrated that unmeasured confounding can have an impact on estimation, which makes developing relevant sensitivity analyses (e.g., Rotnitzky et al., 2001; Díaz and van der Laan, 2013) an important next step.
In addition to controlling bias due to informative dropout, the proposed TMLE estimators can gain precision by accounting for predictive covariates. Such gains in precision are particularly important in vaccine sieve analysis, since efficacy trials are designed to ensure adequate power to assess overall vaccine efficacy but not genotype-specific vaccine efficacy. The utilization of covariate information thus provides a means of increasing the power to detect sieve effects. For both the HIV and malaria motivating studies, use of the covariate-adjusted TMLE appears to provide sieve effect estimates that are more precise than the standard unadjusted method. On the other hand, our simulation study demonstrates that efficiency gains are not always substantial. The theoretical improvement in variance that is achievable through covariate adjustment is a function both of the covariate-conditional event process and the censoring process. Thus, when evaluating the potential for gains in a given problem, both these processes need to be taken into account.
A common alternative for identification of cumulative incidence is via cause-specific hazard functions. We also derived a TMLE estimator for this approach (details in Section 1 of the web supplement). We found that the two approaches performed similarly in simulations, and in data analyses. The iterated mean approach may be more appealing in that incorporation of time-varying covariates to the TMLE procedure is straightforward (van der Laan and Gruber, 2012), whereas this is quite challenging with the hazards-based approach (Stitelman et al., 2012).
Our simulation studies highlighted that there may be room to improve the TMLE estimators in finite samples when there are practical positivity violations. Collaborative targeted minimum loss-based estimation (C-TMLE) builds upon the TMLE framework and incorporates data-adaptive selection of estimates of the censoring distribution in order to produce iterated means estimates with reduced bias. C-TMLE appears to be a potential avenue for improving performance of the proposed estimators in the context of practical positivity violations (Sekhon et al., 2011).
Supplementary Material
Table 2:
Key notation used in theoretical development. Full data parameters refer to summaries of the population when no right-censoring is present. Observed data parameters refer to summaries of the observed population, which includes participants with right-censored outcome data. In the text, a subscript n denotes an estimate of the relevant quantity.
| Quantity | Expression | Description |
|---|---|---|
| Full data parameters | ||
| Fj,z(t0) | cumulative incidence at time t0 of type j endpoints in arm z | |
| V Ej(t0) | 1 − Fj,1(t0)/Fj,0(t0) | vaccine efficacy against type j endpoints at time t0 |
| V S E(t0) | (1 − V E1(t0))/(1 − V E2(t0)) | vaccine sieve effect at time t0 |
| Observed data parameters | ||
| μt(h(t − 1)) |
EP0{μt + 1(H(t)) ∣ Z = 1, C(t − 1) = 0, H(t − 1) = h(t − 1)} |
iterated mean at time t given history h(t − 1) |
| G(w) | prP0{W ≤ w} | distribution function of baseline covariates |
| ∫ μ1 (w)dG(w) | mean of μ1; under assumptions, equals F1,1 (t0) | |
| ζ(w) | prP0{Z = 1 ∣ W = w} | probability of receiving vaccine given covariate value w |
| πt(w) | prP0 {C(t) = 0 ∣ Z = 1, N(t − 1) = 0, M(t − 1) = 0, C(t − 1) = 0, W = w} |
probability of remaining under study at time t given at risk at t − 1 and covariate value w |
Acknowledgements
The authors thank the participants, investigators, and sponsors of the RV144 and RTS,S/ AS01 trials. For RV144 these include Morgane Rolland and the U.S. Military HIV Research Program (MHRP); U.S. Army Medical Research and Materiel Command; NIAID; U.S. and Thai Components, Armed Forces Research Institute of Medical Science Ministry of Public Health, Thailand; Mahidol University; SanofiPasteur; and Global Solutions for Infectious Diseases. For RTS,S/AS01 these include Christian Ockenhouse for the Path Malaria Vaccine Initiative; Dyann Wirth for the Harvard School of Public Health; GlaxoSmithKline; and the Broad Institute Genomics Platform. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The views expressed are those of the authors and should not be construed to represent the positions of the US Army or the Department of Defense. David Benkeser was partially supported by Bill and Melinda Gates Foundation grant OPP1147962. Marco Carone was partially supported by NIAID grant 3UM1AI068635-09 and by the Career Development Fund of the Department of Biostatistics at the University of Washington. Peter Gilbert was partially supported by contract # 792087 from the Henry Jackson Foundation for the MHRP and by NIH NIAID grant R37AI054165.
Appendix A. Relating iterated means to incidence
Under the assumptions stated above, the iterated mean μt can be expressed as an indicator plus the probability of failing due to a matched endpoint between t and t0 given vaccine receipt and no endpoint prior to time t. We derive this result for the vaccine arm (Z = 1) and for matched endpoints (J = 1), but note that the labeling of treatment arms and endpoint type is arbitrary so that these results can also be applied to any combination of Z ∈ {0, 1} and J ∈ {1, 2}.
Defining
as the conditional cause-specific hazard functions for matched and mismatched endpoints, respectively, we have that μt(n(t − 1), m(t − 1), w) equals
for t = 1, 2, …, t0. Since under the identification assumptions, we have that
| (5) |
The case t = 1 shows that μ1 is indeed equivalent to the covariate-conditional cumulative incidence.
Appendix B. Conditions for asymptotic efficiency
Equation (2) will hold for estimators μn, Gn, ζn and πn that satisfy the following conditions:
D*(μn, Gn, ζn, πn) falls into a P0-Donsker class with probability tending to one;
EP0 [{D*(μn, Gn, ζn, πn)(O) − D*(μ, G, ζ, π)(O)}2] converges to zero in probability;
-
the second-order term
is asymptotically negligible – that is, Rn = oP(n−1/2). In the above equation, for the sake of brevity, we let πn,0 and π denote ζn and ζ, respectively, and set empty products equal to one.
Condition 3 will generally be satisfied if for t = 1,…, t0 both
Table C1:
Candidate regressions included in super learner analysis of the RV144 data. Tuning parameters for random forest and regression trees were selected from a grid of eight possible combinations using 10-fold cross-validation.
| Algorithm | Tuning parameters |
|---|---|
| GLM | intercept only |
| GLM | main terms |
| Random forest | 10-fold CV |
| Regression tree | 10-fold CV |
Table C2:
Candidate regressions included in super learner analysis of the RTS,S/AS01 data for estimation of censoring distribution. GLM = logistic regression; GAM = generalized additive model; GBM = gradient boosted machine. A variable selection was included in some candidate regression algorithms (third column). In some cases, variables were selected a-priori; in others, variables were selected based on their correlation with the outcome.
| Algorithm | Tuning parameters | Covariates |
|---|---|---|
| GLM | intercept only | none |
| GLM | main terms | all |
| GLM | main terms | five highest correlations |
| GLM | main terms | study site, treatment, time |
| GLM | all two-way interactions | study site, treatment, time |
| GAM | main terms, df=3 | five highest correlations |
| GAM | main terms | five highest correlations |
| GAM | main terms | study site, treatment, time |
| GAM | all two-way interactions | study site, treatment, time |
| Random forest | mtry=5,ntree=1000 | All |
| GBM | int.depth=2, ntree=1000 | All |
Appendix C. Details of RV144 and RTS,S analyses
The super learner library for the RV144 analysis included four algorithms to estimate both the iterated means and the censoring distribution (Table C1). The RTS,S analysis used considerably more algorithms and slightly different libraries to estimate the iterated means and censoring distribution (Tables C2 and C3).
Table C3:
Candidate regressions included in super learner analysis of the RTS,S/AS01 data for estimation of iterated means. GLM = logistic regression; GAM = generalized additive model; GBM = gradient boosted machine. A variable selection was included in some candidate regression algorithms (third column). In some cases, variables were selected a-priori; in others, variables were selected based on their correlation with the outcome. Gradient boosted machine tuning parameters selected based on out-of-bag (OOB) error rate.
| Algorithm | Tuning parameters | Covariates |
|---|---|---|
| GLM | main terms | all |
| GLM | intercept only | none |
| GLM | main terms | treatment |
| GLM | main terms | study site, treatment |
| GLM | two-way interactions | study site, treatment |
| Stepwise GLM | two-way interactions | five highest correlations |
| Forward stepwise GLM | main terms | all |
| GAM | main terms, df=3 | five highest correlations |
| GAM | main terms, df=3 | ten highest correlations |
| Random Forest | mtry=5,ntree=1000 | all |
| GBM | OOB error rate | all |
References
- Aalen O (1978). Nonparametric inference for a family of counting processes. The Annals of Statistics, 6(4):701–726. [Google Scholar]
- Aalen OO and Johansen S (1978). An empirical transition matrix for non-homogeneous markov chains based on censored observations. Scandinavian Journal of Statistics, 5(3): 141–150. [Google Scholar]
- Agnandji S, Lell B, Fernandes J, Abossolo B, Methogo B, Kabwende A, Adegnika A, Mordmiiller B, Issifou S, et al. (2012). A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. New England Journal of Medicine, 367(24) :2284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang H and Robins JM (2005). Doubly robust estimation in missing data and causal inference models. Biometrics, 61 (4):962–973. [DOI] [PubMed] [Google Scholar]
- Benichou J and Gail MH (1990). Estimates of absolute cause-specific risk in cohort studies. Biometrics, 46(3) :813–826. [PubMed] [Google Scholar]
- Bickel P, Klaassen C, Ritov Y, and Wellner J (1997). Efficient and Adaptive Estimation for Semiparametric Models. Johns Hopkins University Press; Baltimore. [Google Scholar]
- Breiman L (1996). Stacked regressions. Machine Learning, 24(l):49–64. [Google Scholar]
- Díaz I and van der Laan MJ (2013). Sensitivity analysis for causal inference under unmeasured confounding and measurement error problems. International Journal of Biostatistics, 19(2):149–60. [DOI] [PubMed] [Google Scholar]
- Fine JP and Gray RJ (1999). A proportional hazards model for the subdistribution of a competing risk. Journal of the American Statistical Association, 94(446):496–509. [Google Scholar]
- Follmann D, Proschan M, and Leifer E (2003). Multiple outputation: Inference for complex clustered data. Biometrics, 59:420–429. [DOI] [PubMed] [Google Scholar]
- Friedman J (1991). Multivariate adaptive regression splines (with discussion). Annals of Statistics, 19(1) :79–141. [Google Scholar]
- Gail MH, Wieand S, and Piantadosi S (1984). Biased estimates of treatment effect in randomized experiments with nonlinear regressions and omitted covariates. Biometrika, 71(3):431–444. [Google Scholar]
- Gaynor JJ, Feuer EJ, Tan CC, Wu DH, Little CR, Straus DJ, Clarkson BD, and Brennan MF (1993). On the use of cause-specific failure and conditional failure probabilities: examples from clinical oncology data. Journal of the American Statistical Association, 88(422) :400–409. [Google Scholar]
- Gilbert PB (2000). Comparison of competing risks failure time methods and time-independent methods for assessing strain variations in vaccine protection. Statistics in Medicine, 19(22):3065–3086. [DOI] [PubMed] [Google Scholar]
- Gilbert PB, Self SG, and Ashby MA (1998). Statistical methods for assessing differential vaccine protection against human immunodeficiency virus types. Biometrics, 54(3):799–814. [PubMed] [Google Scholar]
- Gilbert PB, Self SG, Rao M, Naficy A, and Clemens J (2001). Sieve analysis: Methods for assessing from vaccine trial data how vaccine efficacy varies with genotypic and phenotypic pathogen variation. Journal of Clinical Epidemiology, 54(l):68–85. [DOI] [PubMed] [Google Scholar]
- Lunn M and McNeil D (1995). Applying Cox regression to competing risks. Biometrics, 51:524–532. [PubMed] [Google Scholar]
- Moore KL and van der Laan MJ (2009). Covariate adjustment in randomized trials with binary outcomes: Targeted maximum likelihood estimation. Statistics in Medicine, 28(1):39–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen ML, Porter KE, Gruber S, Wang Y, and van der Laan MJ (2010). Diagnosing and responding to violations in the positivity assumption. Statistical Methods in Medical Research, 21 (1) :31–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie M (2007). Analysing and interpreting competing risk data. Statistics in Medicine, 26(6) :1360–1367. [DOI] [PubMed] [Google Scholar]
- Pirracchio R, Petersen ML, Carone M, Rigon MR, Chevret S, and van der Laan MJ (2015). Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. The Lancet Respiratory Medicine, 3(1):42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice RL, Kalbfleisch JD, Peterson AV Jr, Flournoy N, Farewell V, and Breslow N (1978). The analysis of failure times in the presence of competing risks. Biometrics, 34(4):541–554. [PubMed] [Google Scholar]
- Rerks-Ngarm S ? Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, and Adams E (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. New England Journal of Medicine, 361(23):2209–2220. [DOI] [PubMed] [Google Scholar]
- Robins JM (1999). Robust estimation in sequentially ignorable missing data and causal inference models. In Proceedings of the American Statistical Association Section on Bayesian Statistical Science, pages 6–10. [Google Scholar]
- Rose S (2013). Mortality risk score prediction in an elderly population using machine learning. American Journal of Epidemiology, 177(5)443–452. [DOI] [PubMed] [Google Scholar]
- Rotnitzky A, Scharfstein D, Su T-L p and Robins J (2001). Methods for conducting sensitivity analysis of trials with potentially nonignorable competing causes of censoring. Biometrics, 57(1)403–113. [DOI] [PubMed] [Google Scholar]
- RTS S Clinical Trials Partnership (2011). First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. New England Journal of Medicine, 365(20)4863–1875. PMID: 22007715. [DOI] [PubMed] [Google Scholar]
- Scharfstein DO, Rotnitzky A, and Robins JM (1999). Adjusting for nonignorable dropout using semiparametric nonresponse models. Journal of the American Statistical Association, 94(448)4096–1120. [Google Scholar]
- Sekhon J, Gruber S, Porter K, and van der Laan MJ (2011). Propensity-score-based estimators and C-TMLE In van der Laan M and Rose S, editors, Targeted Learning: Causal Inference for Observational and Experimental Data, pages 343–364. Springer; New York. [Google Scholar]
- Stein C (1956). Efficient nonparametric testing and estimation In Proceedings of the Third Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Contributions to the Theory of Statistics, pages 187–195, Berkeley, Calif: University of California Press. [Google Scholar]
- Stitelman OM, De Gruttola V, and van der Laan MJ (2012). A general implementation of tmle for longitudinal data applied to causal inference in survival analysis. The International Journal of Bio statistics, 8(1). [DOI] [PubMed] [Google Scholar]
- van der Laan M and Rubin D (2006). Targeted maximum likelihood learning. The International Journal of Biostatistics, 2(1):1–40. [Google Scholar]
- van der Laan MJ, Dudoit S, and Keles S (2004). Asymptotic optimality of likelihood-based cross-validation. Statistical Applications in Genetics and Molecular Biology, 3(1):1–23. [DOI] [PubMed] [Google Scholar]
- van der Laan MJ and Gruber S (2012). Targeted minimum loss based estimation of causal effects of multiple time point interventions. The International Journal of Biostatistics, 8(1) :1–34. [DOI] [PubMed] [Google Scholar]
- van der Laan MJ, Polley EC, and Hubbard AE (2007). Super learner. Statistical Applications in Genetics and Molecular Biology, 6(1):1–23. [DOI] [PubMed] [Google Scholar]
- van der Laan MJ and Robins JM (2003). Unified Methods for Censored Longitudinal Data and Causality. Springer; New York. [Google Scholar]
- van der Laan MJ and Rose S (2011). Targeted Learning: Causal Inference for Observational and Experimental Data. Springer. [Google Scholar]
- van der Laan MJ and Rose S (2018). Targeted Learning in Data Science. Springer International Publishing. [Google Scholar]
- WHO (2016). Weekly epidemiological record. World Health Organization, 91:33–52. [Google Scholar]
- Wolpert D (1992). Stacked generalization. Neural Networks, 5(2):241–259. [Google Scholar]
- Zhang M, Tsiatis A, and Davidian M (2008). Improving efficiency of inferences in randomized clinical trials using auxiliary covariates. Biometrics, 64(3):707–715. PMCID: PMC2574960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.