Abstract
Lanthipeptides represent a large class of cyclic natural products defined by the presence of lanthionine (Lan) and methyllanthionine (MeLan) crosslinks. With the advances in DNA sequencing technologies and genome mining tools, new biosynthetic enzymes capable of installing unusual structural features are continuously being discovered. In this study, we investigated an O-methyltransferase that is a member of the most prominent auxiliary enzyme family associated with class I lanthipeptide biosynthetic gene clusters. Despite the prevalence of these enzymes, their function has not been established. Herein, we demonstrate that the O-methyltransferase OlvSA encoded in the olv gene cluster from Streptomyces olivaceus NRRL B-3009 catalyzes the rearrangement of a highly conserved aspartate residue to a β-amino acid, isoaspartate, in the lanthipeptide OlvA(BCSA). We elucidated the NMR solution structure of the GluC-digested peptide, OlvA(BCSA)GluC, which revealed a unique ring topology comprised of four interlocking rings and positions the isoaspartate residue in a solvent exposed loop that is stabilized by a MeLan ring. Gas chromatography-mass spectrometry analysis further indicated that OlvA(BCSA) contains two DL-MeLan rings and two Lan rings with an unusual LL-stereochemistry. Lastly, in vitro reconstitution of OlvSA activity showed that it is a leader peptide-independent and S-adenosyl methionine-dependent O-methyltransferase that mediates the conversion of a highly conserved aspartate residue in a cyclic substrate into a succinimide, which is hydrolyzed to generate an Asp and isoAsp containing peptide. This overall transformation converts an α-amino acid into a β-amino acid in a ribosomally synthesized peptide, via an electrophilic intermediate that may be the intended product.
Graphical Abstract

INTRODUCTION
The structural diversity of natural products continues to expand as new biosynthetic machineries are discovered as a result of the unceasing advancements and innovations in genome mining technologies. Access to genomic information allows the identification of uncharacterized enzymes that catalyze unique and interesting reactions resulting in the formation of new natural products, including ribosomally synthesized and post-translationally modified peptides (RiPPs).1–7 In this study, we characterized a novel O-methyltransferase that has been identified through genome analyses to be the most abundant auxiliary enzyme in class I lanthipeptide gene clusters.8,9
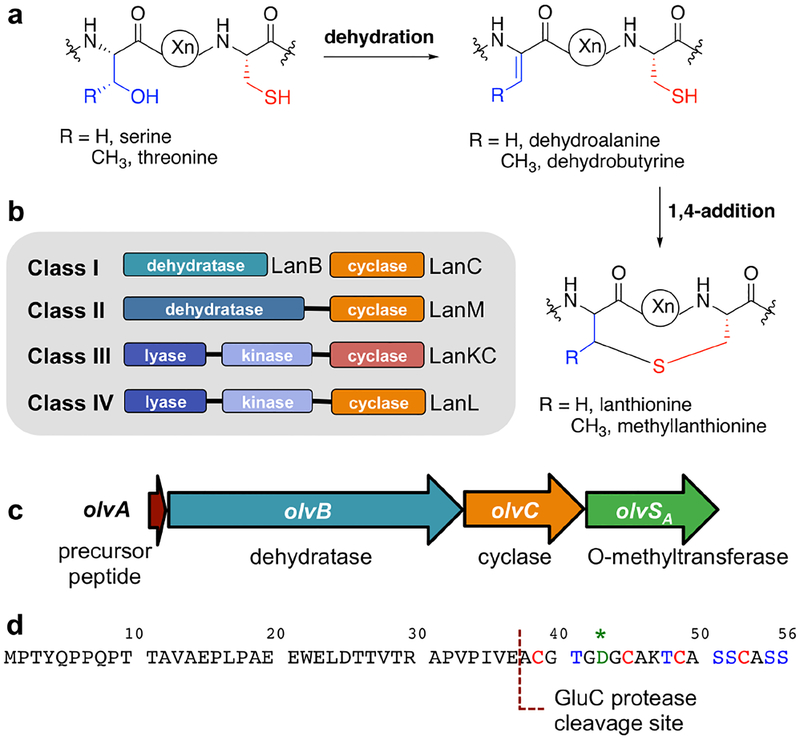
Lanthipeptides are the largest family of RiPPs based on current genome sequences and are defined by the presence of lanthionine (Lan) and/or methyllanthionine (MeLan) crosslinks (Figure 1a).10 They are best known for their antibacterial activities, although other bioactivities have also been reported, including antifungal, antiviral, antinociceptive, antiallodynic, and morphogenetic activities.11–17 Lanthipeptides are initially produced as precursor peptides (LanA) that consist of an N-terminal leader peptide and a C-terminal core peptide. The leader peptide is typically required for enzyme recognition and is eventually removed during maturation, whereas the post-translational modifications are installed in the core peptide.18 The formation of Lan and MeLan crosslinks is initiated by the dehydration of specific Ser and Thr residues to generate dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively (Figure 1a). These dehydro amino acid residues are subsequently attacked by the thiol moiety of Cys via a Michael-type addition mechanism, forming Lan and MeLan from Dha and Dhb, respectively (Figure 1a). Lanthipeptides can be categorized into four classes based on the biosynthetic machineries involved in the formation of Lan and MeLan rings (Figure 1b).10,18,19 For class I lanthipeptides, the core biosynthetic enzymes include a LanB dehydratase and a LanC cyclase. LanB has a glutamylation domain that catalyzes a transesterification reaction between glutamyl-tRNAGlu and the hydroxyl group of Ser or Thr.20,21 The elimination domain of LanB catalyzes the subsequent β-elimination of glutamate to produce the corresponding dehydro amino acid residue. A Cys thiol group then attacks the newly formed double bond catalyzed by the LanC cyclase.22–24 Conversely, class II lanthipeptide synthetases are bifunctional enzymes (LanMs) that catalyze both the dehydration and cyclization steps. Unlike LanB that uses glutamylation for substrate activation, LanM activates the Ser/Thr side chains through phosphorylation with ATP as the phosphoryl group donor.25–27 For classes III and IV lanthipeptides, the lanthipeptide synthetases are referred to as LanKC and LanL, respectively. Both enzymes consist of three domains, an N-terminal lyase, a central kinase, and a C-terminal cyclase.28 The type of cyclase domain differentiates LanKC from LanL. Specifically, LanKC lacks the Zn2+ coordination sites that are present in the cyclase domain of LanL. This metal coordination site is also present in LanC cyclase and the LanM cyclase domain of classes I and II lanthipeptides, respectively. Similar to class II lanthipeptides, the dehydration step for classes III and IV lanthipeptides is accomplished through the ATP-dependent phosphorylation of the hydroxyl group of Ser and Thr, and the subsequent β-elimination of phosphate to yield the dehydro amino acid residues.28,29
Figure 1.
(a) General mechanism of lanthipeptide biosynthesis. (b) Four classes of lanthipeptides based on their biosynthetic machineries. (c) olv gene cluster from Streptomyces olivaceus NRRL B-3009. (d) OlvA precursor peptide amino acid sequence with a GluC protease cleavage site used in this study highlighted. The highly conserved Asp residue is indicated with a green asterisk.
Apart from the abovementioned (methyl)lanthionine biosynthetic enzymes, tailoring enzymes can install additional distinct structural features in the core peptide, including D-amino acids, halogenated residues, and aminovinyl-cysteine residues.30 With the advent of genome mining, uncharacterized enzymes and intriguing lanthipeptide biosynthetic gene clusters are continuously being uncovered.8,9,31 In 2010, Marsh et al. first reported that many actinobacteria-associated class I lanthipeptide clusters contain a gene that encodes an uncharacterized O-methyltransferase.8 A larger scale genome analysis performed in 2015 supported the earlier findings.9 For the latter study, it was further shown that the precursor peptides associated with these O-methyltransferases contain a highly conserved Asp residue in the core peptide region, although the correlation of these Asp residues to the O-methyltransferase activity was undetermined. More recently, our genome mining efforts revealed that these O-methyltransferases are the third most abundant enzymes associated with class I lanthipeptide biosynthetic clusters, next to the core biosynthetic enzymes LanB and LanC (unpublished data). Despite the prevalence of the O-methyltransferases, their function had not been established prior to the current work.
In this study, we investigated the activity of the O-methyltransferase present in the olv biosynthetic gene cluster from Streptomyces olivaceus NRRL B-3009 (NCBI accession WP_031034767.1). We propose the use of “LanS” as the generic name for methyltransferases in lanthipeptide gene clusters, and “LanSA” to refer to the family of O-methyltransferases that are first characterized in this work. A different methyltransferase that methylates the C-terminal carboxylate of a RiPP precursor was recently reported and was termed LanSB.32 In addition to the O-methyltransferase gene (olvSA), the olv gene cluster is comprised of genes encoding the LanA precursor peptide (OlvA), LanB dehydratase (OlvB), and LanC cyclase (OlvC) (Figure 1c). We investigated the olv gene cluster using heterologous expression in Escherichia coli, and show that OlvB, OlvC, and OlvSA convert OlvA into a polycyclic product that bears a β-peptide linkage and displays a unique ring topology and stereochemistry. We compared the NMR solution structures of the product that was generated with and without OlvSA, and clearly demonstrate that OlvSA activity resulted in the incorporation of the β-amino acid isoaspartate (isoAsp). Furthermore, we reconstituted the activity of OlvSA in vitro and delineated its mechanism, which proceeds via a rearrangement reaction of the highly conserved Asp residue to introduce the aforementioned isoAsp residue via an imide intermediate.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Materials.
E. coli DH5α was used for cloning and plasmid propagation, while Escherichia coli BL21 Star (DE3) was used for protein/peptide expression. The expression vectors pCDFDuet-1, pETDuet-1, and pET28b were obtained from Novagen. Oligonucleotides were purchased either from Integrated DNA Technologies (IDT) or Twist Bioscience. Restriction endonucleases, DNA polymerases, and Gibson Assembly Master Mix were acquired from New England Biolabs (NEB). GluC protease was purchased from Worthington Biochemical Corporation. Media components were obtained from Difco Laboratories. The SAH nucleosidase Pfs from E. coli was expressed and purified as reported previously.33
General Methods.
Polymerase chain reactions were performed on a C1000 thermal cycler (Bio-Rad), while DNA sequencing was carried out by ACGT, Inc. For the reversed-phase high performance liquid chromatography (RP-HPLC) purifications, water with 0.1% trifluoroacetic acid (TFA) was used as solvent A, while acetonitrile with 0.1% TFA was used as solvent B. The detector was set at 220 nm. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) data were obtained using a Bruker Daltonics UltrafleXtreme MALDI-TOF/TOF MS (Bruker) and super 2,5-dihydroxybenzoic acid (Sigma) in 80% acetonitrile as matrix. Samples were desalted using ZipTip C18 (Millipore) pipet tips. LC-ESI-QTOF (ESI, electrospray ionization; Q, quadrupole) MS and tandem MS (MS/MS) were carried out on a Synapt ESI-QTOF-MS system (Waters).
Construction of Co-expression Plasmids.
The synthetic genes for OlvA, OlvB, OlvC, and OlvSA were codon-optimized for E. coli expression (Table S1). They were amplified using the primers listed in Table S2 to introduce 5′ overhangs and allow subsequent Gibson Assembly. For pCDFDuet-1-olvA-olvB, olvA was first cloned in MCS1 of pCDFDuet-1 that was digested with EcoRI and HindIII, producing pCDFDuet-1-olvA. This procedure introduced an N-terminal His6-tag in OlvA. The olvB gene was then cloned in MCS2 after digestion of pCDFDuet-1-olvA with NdeI and XhoI. For pETDuet-1-olvC-olvSA, olvC was first cloned in MCS1 of pETDuet-1 that was digested with NcoI and HindIII, yielding pETDuet-1-olvC. The olvSA gene was then cloned in MCS2 of the NdeI- and XhoI-digested pETDuet-1-olvC plasmid.
Expression and Purification of OlvA(BCSA) and its Variants.
E. coli BL21 Star (DE3) cells were transformed with the plasmids listed in Table S3 and plated on LB plates with the appropriate antibiotic(s). The nucleotides in the tRNAGlu acceptor stem that have been previously shown to be essential for LanB dehydratase recognition34 are the same in Thermobispora bispora and S. olivaceus (Figure S1a). Hence, the plasmid pTrc33-T.bispora-GluRS-tRNAGlu that was designed in an earlier study35 was used for co-expression experiments that included OlvB dehydratase. OlvB was indeed shown to be less active without the pTrc33-T.bispora-GluRS-tRNAGlu plasmid (Figure S1b,c). For each transformant, 1 L terrific broth was inoculated with an overnight starter culture (1% v/v) and grown at 37 °C with shaking at 200 rpm to an optical density (OD600) of 0.5–0.6. Appropriate antibiotics were added for selective pressure as specified in Table S3. The cultures were cooled on ice for 30 min, and protein expression was induced with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). The cells were further incubated at 18 °C for 20 h at 200 rpm before harvesting (5,000×g, 10 min, 4 °C). The cell pellet was resuspended in buffer 1 (6 M guanidine hydrochloride, 20 mM NaH2PO4, pH 7.5, 500 mM NaCl, 0.5 mM imidazole), lysed by sonication, and centrifuged at 50,000×g for 30 min. The supernatant was loaded onto 3 mL pre-equilibrated His60 Ni Superflow resin cartridges (Takara Bio USA, Inc.) and allowed to flow by gravity. The resin was washed with 20 column volumes (CV) buffer 1, 10 CV each of buffer 2 (4 M guanidine hydrochloride, 20 mM NaH2PO4, pH 7.5, 300 mM NaCl, 30 mM imidazole) and buffer 3 (20 mM NaH2PO4, pH 7.5, 300 mM NaCl, 30 mM imidazole), and eluted with 5 CV of buffer 4 (20 mM NaH2PO4, pH 7.5, 300 mM NaCl, 500 mM imidazole). The eluted peptides were detected by MALDI-TOF MS. The peptides that were used for in vitro assays (His6-OlvA precursor peptide, His6-OlvA co-expressed with OlvB, and His6-OlvA co-expressed with OlvB and OlvC) were further purified by RP-HPLC using a Hypersil Gold C8 column (5 μm particle size, 175 Å pore size, 4.6 mm×250 mm). Solvent B was programmed to increase from 2% to 70% in 17.5 min and ramped up to 100% for 2 min at a flow rate of 1 mL/min. Peptides eluted at 14–15 min, and were lyophilized and stored at −20 °C.
Leader Cleavage and Purification of OlvA(BCSA)GluC and OlvA(BC)GluC.
The His6-OlvA(BCSA) sample (i.e. His6-OlvA co-expressed with OlvB, OlvC, and OlvSA) that was obtained from Ni affinity chromatography purification was incubated for 18 h at 37 °C to allow complete imide hydrolysis (see Results and Discussion). The sample was then purified by semi-preparatory RP-HPLC using a Phenomenex Luna C18(2) column (10 μm particle size, 100 Å pore size, 10 mm×250 mm). The His6-OlvA(BC) sample, which refers to the OlvB- and OlvC- modified peptide, was purified in the same manner. For the semi-preparatory RP-HPLC purification, solvent B was set at 2% for 10 min, ramped up to 70% B in 20 min, and increased to 100% B for 2 min at a flow rate of 7 mL/min. The peptides eluted at 23 min, were lyophilized, and subsequently digested with GluC protease at 100:1 peptide:GluC w/w ratio in 50 mM Tris-HCl buffer (pH 7.5). GluC-digested peptides are labelled with “GluC” in superscript from here onwards. The cleavage cocktail was incubated at 37 °C for 18 h, and ran through an analytical RP-HPLC Macherey-Nagel NUCLEODUR 100–5 C18 ec column (5 μm particle size, 100 Å pore size, 4.6 mm×250 mm). Solvent B was ramped up from 2% to 20% in the span of 20 min. OlvA(BCSA)GluC eluted at 14.5 min, while OlvA(BC)GluC eluted at 14.2 min. The purified peptides were collected, lyophilized, and stored at −20 °C.
Dithiothreitol (DTT) Assay.
To check for the presence of dehydro amino acid residues, the peptide samples were adjusted to pH 7.5 and treated with DTT to a final concentration of 250 mM. The samples were incubated at 37 °C for 18 h and monitored by MALDI-TOF MS.
N-ethylmaleimide (NEM) Assay.
To determine the cyclization state of the peptides, free thiols were reacted with NEM. Prior to addition of NEM, the samples were treated with 3 mM tris(2-carboxyethyl)phosphine in 50 mM HEPES buffer (pH 7) for 20 min in order to ensure the complete reduction of free cysteines. Freshly prepared NEM solution (50 mM in ethanol) was then added to a final concentration of 3 mM. The reaction was run for 30 min at room temperature. Samples were then desalted, and checked by MALDI-TOF MS. Unmodified OlvA, which has four free thiols, was used as positive control.
LC-ESI-QTOF MS and MS/MS Analyses.
Samples were loaded onto a Macherey-Nagel EC 125/2 NUCLEODUR 100–5 C18 ec column (5 μm particle size, 100 Å pore size, 2 mm×125 mm) and run through a solvent gradient of 3% to 97% solvent B (acetonitrile with 0.1% formic acid) for 10 min at 0.2 mL/min. Water with 0.1% formic acid was used as solvent A. The fractionated sample was analyzed by ESI-QTOF-MS (ESI positive mode) using a Waters Synapt mass spectrometer. Instrument calibration was done prior to sample injection. The capillary voltage was set to 3.0–3.5 kV, while the ionization source and desolvation gas were heated to 120 °C and 300 °C, respectively. The cone and desolvation gases were set to 80 L/h and 600 L/h, respectively. The transfer collision energy used for both MS and MS/MS analyses was 4 V. For MS analysis, the trap collision energy was set to 6 V. For MS/MS analysis, a 15 to 30 V trap collision energy ramp was applied on multiply charged parent ions to achieve fragmentation. [Glu1]-Fibrinopeptide B (Sigma) was directly infused for lock mass recalibration. Spectra were processed using MaxEnt3 software and analyzed using MassLynx V4.1 (Waters).
NMR Spectroscopy.
To obtain sufficient material for NMR data acquisition, His6-tagged OlvA(BC) and OlvA(BCSA) were each expressed in 8 L of TB. The samples were purified and digested with GluC protease as described in the earlier section, yielding ~1.5 mg of the C-terminal 19-mer of OlvA(BC)GluC or OlvA(BCSA)GluC. Each peptide was dissolved in 300 μL of 30 mM sodium phosphate buffer (pH 5.8) in 9:1 H2O:D2O, and loaded in a 5-mm D2O-matched Shigemi tube. DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) was added for referencing, with a standard peak for its methyl protons set to 0.0 ppm. One dimensional 1H NMR, and two-dimensional homonuclear 1H–1H-TOCSY (total correlation spectroscopy) and -NOESY (nuclear Overhauser effect spectroscopy) spectra were acquired on a Varian INOVA 750 MHz spectrometer equipped with a 5 mm triple resonance (1H–13C–15N) triaxial gradient probe and pulse-shaping capabilities, and with the use of VNMRJ 2.1B software and BioPack suite of pulse sequences. Temperature screening was performed to identify the best condition, wherein maximal signal dispersion could be achieved. The optimum temperature was determined to be 15 °C. The water signal was suppressed either by presaturation during the relaxation delay or through water gradient tailored excitation. After data acquisition, the peptides were lyophilized, re-dissolved in 100% D2O, and the same set of experiments were acquired. The experimental details are summarized in Table S4. The data were processed using NMRPipe, and analyzed in NMRView.36,37 Chemical shifts were assigned following previously described procedures.38,39 The chemical shift assignments are shown in Tables S5 and S6.
Structure Calculations.
A combination of automatically and manually assigned NOEs was used to calculate the structures of OlvA(BCSA)GluC and OlvA(BC)GluC using CYANA 2.1.40 Structural calculations involved 7 cycles with 10,000 steps per cycle. Through simulated annealing, 100 conformers were calculated per cycle. The 20 lowest energy conformers from the final cycle were used for further analysis, which included determination of the root-mean-square deviation (rmsd) and generation of Ramachandran plots. The coordinates were deposited in the Protein Data Bank (accession numbers 6PQF for OlvA(BCSA)GluC and 6PQG for OlvA(BC)GluC), while the chemical shift assignments were deposited in the Biological Magnetic Resonance Data Bank (accession numbers 30630 for OlvA(BCSA)GluC and 30631 for OlvA(BC)GluC). PyMOL41 was used to generate figures of the three-dimensional (3D) structures and hydrophobic surface maps.
Gas Chromatography.
Purified OlvA(BCSA)GluC (~0.5 mg) was dissolved in 3 mL of 6 M DCl in D2O and transferred to a sealed pressure tube. The sample was then heated to 110 °C for 24 h and allowed to cool before the removal of solvent by rotary evaporation. In a separate flask, acetyl chloride (1.5 mL) was added dropwise to 5 mL methanol in an ice-water bath. This solution (3 mL) was then added to the hydrolyzed sample residue and brought to 110 °C for 1 h with reflux. The solution was allowed to cool to room temperature prior to the removal of solvent by rotary evaporation. The resulting residue was then suspended in 3 mL of dichloromethane and cooled under nitrogen in an ice-water bath. Pentafluoropropionic anhydride (1 mL) was slowly added to the reaction vessel and the mixture was heated to 110 °C for 30 min with reflux. The solution was allowed to cool before removing the solvent under a gentle stream of nitrogen. The sample residue was then dissolved in 0.1 mL of methanol, transferred to a clean vial, and stored at −20 °C prior to analysis.
Gas chromatography-MS (GC-MS) analysis was conducted on an Agilent HP 6890N instrument equipped with a CP-Chirasil-L-Val (Agilent) column (25 m × 0.25 mm × 0.12 μm). Samples in methanol (1–5 μL) were applied to the column via splitless injection (230 °C injector inlet temperature) with a helium flow at a rate of 1.7 to 2.0 mL/min. Runs were held at 80 °C for 5 min, then raised to 200 °C at a rate of 4 °C/min, and then held at 200 °C for an additional 3 min. Selected-ion monitoring (SIM) mode was used to detect characteristic mass fragments for derivatized Lan, MeLan, and aspartic acid (365, 379, and 206 m/z, respectively). Confirmation of stereochemistry was achieved through the co-injection of isomerically pure synthetic standards. Derivatized DL-MeLan, LL-MeLan, DL-Lan, and LL-Lan residues were prepared according to previously reported procedures.42,43 Derivatized D- and L-aspartic acid standards were prepared from isomerically pure aspartic acids (Aldrich) according to the procedure described in the previous paragraph.
Construction of pET28b-olvSA-His6.
The olvSA gene, codon-optimized for E. coli expression (Table S1), was amplified using the primers JZA_98_28bOMF (ATTTTGTTTAACTTTAAGAAGGAGATATACATGTCTCAAACCCCGACTGCC) and JZA_97_28bOMR (ATCTCAGTGGTGGTGGTGGTGGTGCGAGCCACCGTAGGTCAGCGT GATACGGGTTTC). The pET28b vector was amplified using the primers JZA_96_28VF (GCACCACCACCACCACCACTGAGATC) and JZA_95_28VR (GTATATCTCCTTCTTAA AGTTAAACAAAATTATTTCTAGAGGGGAATTG). The PCR products were gel-purified and olvSA was cloned into pET28b using Gibson Assembly in a manner that introduced a C-terminal His6-tag.
Expression and Purification of OlvSA-His6.
E. coli BL21 Star (DE3) cells were transformed with pET28b-olvSA-His6 and plated on LB agar with 50 mg/L kanamycin. A single colony was picked and grown in a 15 mL overnight starter culture at 37 °C. A liter of LB was then inoculated with 1% v/v of the overnight starter culture and grown at 37 °C with shaking at 200 rpm to an optical density (OD600) of 0.5–0.6. The culture was cooled on ice for 30 min, 0.1 mM IPTG was added (final concentration), and the culture was incubated at 18 °C for 20 h at 200 rpm. The cells were harvested (5,000×g, 10 min, 4 °C) and the pellet was resuspended in buffer A (20 mM Tris, pH 7.8, 0.5 M NaCl). The sample was lysed using a high-pressure homogenizer (Avestin, Inc.) and clarified by centrifugation at 50,000×g for 30 min. The supernatant was loaded onto a pre-equilibrated 5 mL HisTrap HP column, washed with 5 CV of buffer A, and the column was attached to an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare). The column was washed using a linear gradient of buffer B (20 mM Tris, pH 7.8, 0.5 M NaCl, 500 mM imidazole) in buffer A at a flow rate of 2.5 mL/min. Buffer B was initially set at 6% for 5 CV, increased to 10% for 3 CV, and ramped up to 25% for 0.5 CV. Sample elution was monitored at 280 nm and fractions were analyzed by SDS-PAGE (Bio-Rad). The fractions containing OlvSA were combined, concentrated using an Amicon Ultra-15 Centrifugal Filter Unit (10 kDA MWCO, Millipore), and further purified by size-exclusion chromatography using a Superdex 75 Increase 10/300 GL (GE Healthcare) column. Buffer A was used as run buffer set at a flow rate of 0.5 mL/min. The purified peptide was concentrated using an Amicon Ultra-15 Centrifugal Filter Unit (10 kDA MWCO, Millipore), glycerol was added to 10%, and the sample was aliquoted, flash-frozen, and stored at −80 °C until use.
OlvSA In Vitro Assays.
The in vitro assays consisted of 50 μM substrate, 10 μM OlvSA, 1 mM S-adenosyl-L-methionine (SAM) in 50 mM HEPES buffer (pH 7) with a final volume of 50 μL. The reactions were set up either at room temperature or 37 °C and monitored at different time points by MALDI-TOF MS. In order to investigate whether the imide formation and hydrolysis steps are enzyme-catalyzed, the in vitro assay was repeated using the conditions described above, and 3 M guanidine hydrochloride (pH 7.5, 20 mM phosphate buffer) was added after 1 h reaction time, when the methyl ester had accumulated with no concurrent imide production. The sample was incubated at room temperature for 4 h and checked by MALDI-TOF MS to monitor imide formation in the absence of OlvSA. The sample was further incubated at 37 °C overnight and checked by MALDI-TOF MS to monitor imide hydrolysis.
Hydrazine Test.
The OlvSA in vitro assay was performed as described above, and upon complete conversion of the methyl ester to the imide species (i.e. 24 h reaction time at room temperature), hydrazine was added to a final concentration of 2 M. The sample was incubated at room temperature for 30 min and checked by MALDI-TOF MS.
18O-Labeling Assay.
The OlvSA in vitro assays, comprised of 50 μM substrate, 10 μM OlvSA, 1 mM SAM in 50 mM HEPES buffer (pH 7), were performed in buffer made with H218O in a final volume of 50 μL. The substrates that were tested included the unmodified His6-OlvA, dehydrated His6-OlvA(B), and cyclized His6-OlvA(BC). The reactions were set up at 37 °C for 24 h. GluC was then added at 20:1 w/w peptide:GluC ratio, and the sample was incubated at 37 °C for 18 h. The digests were analyzed by LC-ESI-QTOF MS.
RESULTS AND DISCUSSION
In Vivo OlvSA Activity.
We initially attempted detection of the product of the olv gene cluster produced by the native organism, but despite trying various growth conditions, we were unable to observe the anticipated product(s). Similar observations were made for organisms encoding related clusters (Frankia sp. EUN1f, Micromonospora aurantiaca, Nocardiopsis alba ATCC BAA-2165, Streptomyces cattleya NRRL 8057, Streptomyces clavuligerus ATCC 27064, and Streptomyces coelicolor A3(2) (Figure S1d,e). Therefore, to investigate the function of the O-methyltransferase OlvSA, His6-OlvA was co-expressed in E. coli with OlvB dehydratase and OlvC cyclase in the presence and absence of OlvSA. Furthermore, based on previous studies regarding substrate recognition of LanB dehydratases,34,35 a tRNAGlu from an actinobacterium (T. bispora) was co-expressed with OlvB, which improved the OlvB-catalyzed dehydration of His6-OlvA (Figure S1b,c). OlvA consists of 56 amino acids and has a predicted core peptide segment with two Thr, four Ser, and four Cys residues (Figure 1d). The co-expression products were purified by nickel affinity chromatography and analyzed by MALDI-TOF MS. In the absence of OlvSA, the observed mass of the OlvB- and OlvC-modified peptide [termed His6-OlvA(BC)] corresponded to a four-fold dehydrated peptide (Figure 2a). On the other hand, when OlvSA was present, the peptide products included a four-fold dehydrated peptide as the major product and a five-fold dehydrated peptide as a minor product (Figure 2b). Upon sample incubation at 37 °C for 18 h, the five-fold dehydrated peptide disappeared, resulting in a single product with a mass corresponding to four dehydrations (Figure 2c). The OlvB-, OlvC- and OlvSA-modified peptide was named OlvA(BCSA). The identical masses of the predominant peptide products from the co-expression experiments with and without OlvSA and the absence of any methylated products called into question whether OlvSA was active or not.
Figure 2.
MALDI-TOF mass spectra showing the [M+H]+ of (a) His6-OlvA co-expressed with OlvB and OlvC [His6-OlvA(BC)], (b) full-length OlvA(BCSA) after nickel affinity purification, and (c) after incubation at 37 °C for 18 h. Samples were reacted with N-ethylmaleimide to alkylate incompletely cyclized peptides and retain only the completely modified peptides in the 7100–7200 m/z observation range. (d) RP-HPLC profile of OlvA(BC)GluC, OlvA(BCSA)GluC, and co-injection of OlvA(BC)GluC and OlvA(BCSA)GluC. (e) ESI tandem mass spectrum of OlvA(BCSA)GluC.
The exact location of the leader peptide cleavage site could not be identified based on sequence analysis. Moreover, a gene encoding a putative leader peptide protease has yet to be identified, as such a gene is not found near the biosynthetic gene cluster. Fortuitously, a GluC endoprotease-cleavage site is located at the −2 position from the most N-terminal Cys residue (Figure 1d). The leader peptides of His6-OlvA(BC) and His6-OlvA(BCSA) were therefore cleaved using GluC protease, releasing a C-terminal peptide that contained 19 amino acid residues and contained all the post-translational modifications. Surprisingly, despite having identical masses, purification by RP-HPLC showed that the major products of the OlvA(BC)GluC and OlvA(BCSA)GluC peptides eluted at different retention times (Figure 2d). Co-injection of the two products further confirmed that they were not identical (Figure 2d).
Structural Studies on OlvA(BC)GluC and OlvA(BCSA)GluC.
In order to establish the difference between OlvA(BC)GluC and OlvA(BCSA)GluC that would explain the observations from the RP-HPLC analysis, we pursued structural analysis of both peptides. The cyclization states of OlvA(BC) and OlvA(BCSA) were first determined by treating the peptides with N-ethylmaleimide, a thiol-alkylating reagent. For both peptides, no alkylated products were observed for the four- and five-fold dehydrated products, signifying that all four Cys residues were involved in the formation of Lan/MeLan (Figures S2a and S3a). This observation was in agreement with the complementary results of a dithiothreitol (DTT) assay, wherein no DTT adducts were detected, confirming that all dehydro amino acid residues reacted with the four Cys residues (Figures S2b and S3b). OlvA(BCSA)GluC was then analyzed by tandem MS, which revealed that the peptide was recalcitrant to fragmentation. The only ions observed were b18 and a18 ions, which indicated that the C-terminal Ser was not dehydrated (Figure 2e). The results suggested that the four Lan/MeLan rings were interlocking, which would hamper MS/MS fragmentation. Since no further detailed structural information could be derived using MS, NMR studies were performed.
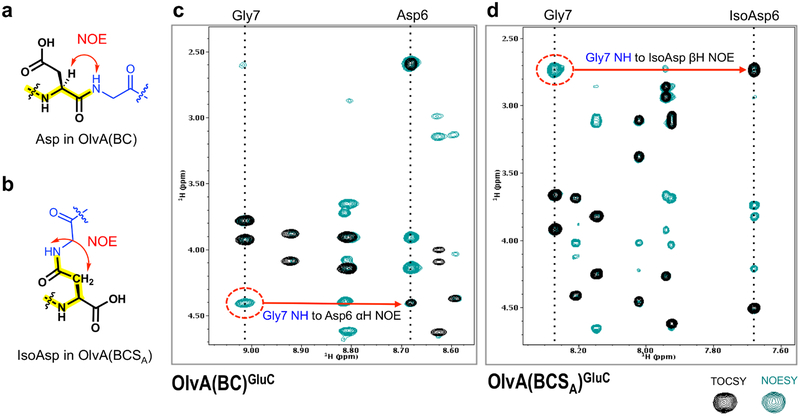
TOCSY and NOESY spectra were acquired and used to sequentially assign the chemical shifts of the hydrogens in OlvA(BC)GluC and OlvA(BCSA)GluC. The chemical shift assignments are tabulated in Tables S5 and S6. The spin systems in the TOCSY spectra are presented in Figures S4 and S5. The hydrogens in OlvA(BC)GluC and OlvA(BCSA)GluC have similar chemical shifts (within ± 0.2 ppm), except for the amide hydrogens of residues 6 and 7. Specifically, the amide hydrogens of Asp6 and Gly7 in OlvA(BC)GluC at 8.7 and 9.0 ppm, respectively, were shifted to 7.7 and 8.3 ppm, respectively, in OlvA(BCSA)GluC. The most pronounced chemical shift deviation was observed for the amide hydrogen of Asp6, which appeared to be the site of OlvSA modification as further supported by the NOESY data. This Asp residue corresponds to the previously identified highly conserved Asp residue that is present in precursor peptides associated with gene clusters encoding a LanSA enzyme (Figure S1e).9
A strong NOE between the amide hydrogen of Gly7 and the alpha hydrogen of Asp6 is observed in OlvA(BC)GluC, and indicates the presence of a native peptide bond (Figure 3a,c). OlvA(BCSA)GluC, on the other hand, does not exhibit the aforementioned NOE, and instead displays a strong NOE between the amide hydrogen of Gly7 and the beta hydrogens of residue 6 (Figure 3d). This unusual NOE is indicative of the presence of a β-peptide linkage between Gly7 and Asp6, wherein the peptide backbone bond is formed between the side chain carboxylate group of Asp6 and the amino group of Gly7 (Figure 3b). This transformation of Asp to isoAsp introduces an additional methylene group in the peptide backbone and an unusual carboxyl side chain, thereby explaining the observed difference in the behavior of OlvA(BCSA)GluC and Olv(ABC) GluC in the RP-HPLC analysis.
Figure 3.
(a) Characteristic NOE between the amide hydrogen of a C-terminal Gly and the α hydrogen of an N-terminal Asp, as was found in OlvA(BC)GluC. (b) Characteristic NOE between the amide hydrogen of a C-terminal Gly and the β hydrogens of an N-terminal isoAsp, as was found in OlvA(BCSA)GluC. (c) Overlay of the TOCSY and NOESY spectra highlighting the described characteristic NOEs of OlvA(BC)GluC and (d) OlvA(BCSA)GluC. TOCSY crosspeaks are shown in black, while NOESY crosspeaks are in green.
The NMR data were also used to determine the ring topology of OlvA(BCSA)GluC. Former Ser/Thr residues involved in the formation of Lan/MeLan rings displayed more shielded βHs relative to those present in unmodified Ser/Thr. This change is due to the replacement of the hydroxyl oxygen with a less electronegative sulfur atom in the thioether crosslink. The βH to βH NOE correlations, along with characteristic chemical shift changes, revealed that OlvA(BCSA)GluC contains two Lan and two MeLan residues (Figure 4a). The isoAsp residue is located within the N-terminal MeLan ring that is formed between Cys8 and Thr4, and is flanked by two glycine residues (Figure 4a,c and Figure S6). The second MeLan involves Cys16 and Thr11, producing a cyclic structure that is comprised of six amino acid residues (Figure S7). The C-terminal Lan ring is formed between Cys18 and Ser12, and is interlocking with the previously described C-terminal MeLan ring (Figure S8). Lastly, the second Lan links the N-terminal Cys2 to a central Ser14 residue (Figure S9). The described ring topology of OlvA(BCSA)GluC (Figure 4a) has not been observed in other lanthipeptides that have been reported thus far.
Figure 4.
(a) Schematic diagram of the OlvA(BCSA)GluC containing two DL-MeLan and two LL-Lan. The isoaspartate residue (isoD) is shown in green. Cys residues that now constitute the (Me)Lan rings are colored red. Ser and Thr residues that are now part of the Lan and MeLan rings are colored blue and are labelled as A (alanine) and Abu (2-aminobutyrate), respectively. (b) Superimposition of the 3D structures of OlvA(BCSA)GluC in gray and OlvA(BC)GluC in red. The isoaspartate (isoAsp) in OlvA(BCSA)GluC, and Asp in OlvA(BC)GluC are shown in green. The N- and C- termini are specified. (c) Superimposition of the 20 lowest energy conformers of OlvA(BCSA)GluC and (d) OlvA(BC)GluC as calculated by CYANA 2.1. (e) NMR solution structure and (f) hydrophobic surface map of OlvA(BCSA)GluC.
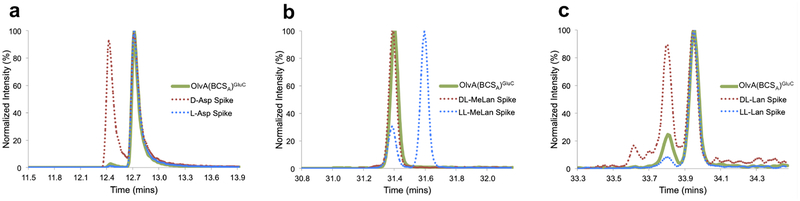
In order to more fully characterize the structure of OlvA(BCSA)GluC, the stereochemistry of the isoAsp, Lan, and MeLan residues in the peptide were determined. Following the hydrolysis of OlvA(BCSA)GluC and derivatization of the resulting amino acids (isoAsp gives Asp upon complete peptide hydrolysis), GC-MS analysis was performed as described previously.42,44 The stereochemistry of the isoAsp residue was determined by comparison to derivatized D- and L-aspartic acid standards; only L-aspartic acid could be detected (Figure 5a).
Figure 5.
GC-MS traces of hydrolyzed and derivatized (a) isoAsp, (b) MeLan, and (c) Lan residues from OlvA(BCSA)GluC. Traces of the co-injection of the hydrolyzed and derivatized OlvA(BCSA)GluC with synthetic standards are indicated in broken lines. A small amount of epimerization occurs for Lan as previously reported.45,46
Next, synthetic DL- and LL-(Me)Lan diastereomers were individually spiked into the derivatized sample to confirm the stereochemistry of (Me)Lan residues found in OlvA(BCSA)GluC. As shown in Figure 5b, both MeLan residues present in OlvA(BCSA)GluC were found to be of the DL-diastereomeric configuration. Interestingly, analysis of the Lan composition revealed mostly the presence of the LL-diastereomer (Figure 5c). This finding was somewhat surprising, as LL-Lan has not been widely reported in lanthipeptides. Furthermore, the OlvA core peptide lacks any sequence resembling the Dhb-Dhb-Xxx-Xxx-Cys motif (Xxx = any residue except Dha or Dhb) that was previously shown to impart LL-(Me)Lan stereochemistry in the class II lanthipeptides haloduracin and cytolysin,44 geobacillin II,47 and carnolysin.48 OlvA(BCSA)GluC is the first class I lanthipeptide reported to contain LL-lanthionine.
Taking into account the ring topology and stereochemistry, we then elucidated the NMR solution structures of OlvA(BCSA)GluC and compared it with those of OlvA(BC)GluC (Figure 4b–e). Three-dimensional structural calculations were pursued because the wide dispersion of the proton chemical shifts and the presence of a number of medium- and long-range NOEs indicated that both peptides were well structured under the employed solvent conditions (30 mM sodium phosphate buffer, pH 5.8). NOE restraints were generated and imported into CYANA 2.1, together with the chemical shifts and definitions of the non-canonical stereocenters and thioether bridges. For OlvA(BCSA)GluC, a total of 104 NOE restraints were used for the final calculation, 61 of which were short-range, 21 medium-range, and 22 long-range NOEs (Table 1). Similarly, 103 NOE crosspeaks were used for OlvA(BC)GluC; 48 of which were short-range, 31 medium-range, and 24 long-range NOEs. The 20 lowest energy conformers for each peptide were superimposed and are presented in Figures 4c and 4d for OlvA(BCSA)GluC and OlvA(BC)GluC, respectively. The structural statistics are summarized in Table 1. The low values for the root-mean-square deviation (rmsd), 0.95 Å for OlvA(BCSA)GluC and 0.85 Å for OlvA(BC)GluC, confirmed the highly structured nature of these peptides. The Ramachandran plots for OlvA(BCSA)GluC and OlvA(BC)GluC are presented in Figures S10a and S10b, respectively. There were no φ and ψ backbone angles found in the disallowed regions for both peptides (Table 1).
Table 1.
Structure calculation statistics for OlvA(BCSA)GluC and OlvA(BC)GluC.
| OlvA(BCSA)GluC | OlvA(BC)GluC | |
|---|---|---|
| Total NOE peak assignments | 104 | 103 |
| short-range, |i-j|≤1 | 61 | 48 |
| medium-range, 1<|i-j|<5 | 21 | 31 |
| long-range, |i-j|≥5 | 22 | 24 |
| RMSD for full peptide | ||
| backbone atoms (Å) | 0.95 | 0.85 |
| heavy atoms (Å) | 1.21 | 1.11 |
| Ramachandran Plot | ||
| Φ/Ψ in most favored regions | 76.2% | 74.0% |
| Φ/Ψ in additionally allowed regions | 23.8% | 26.0% |
| Φ/Ψ in generously allowed regions | 0.0% | 0.0% |
| Φ/Ψ in disallowed regions | 0.0% | 0.0% |
The calculated structures of OlvA(BCSA)GluC and OlvA(BC)GluC were superimposed (Figure 4b) and resulted in an rmsd of 0.914 Å over 181 atoms. As expected, the main difference between their 3D structures is found in the loop region that contains the Asp/isoAsp residue. The Asp/isoAsp residue is situated in a readily accessible and solvent exposed loop region that is formed by the N-terminal MeLan ring involving Cys8 and Thr4. The abovementioned loop is further stabilized by the Lan linkage between Cys2 and Ser14. The C-terminal end of the peptides displays interlocking Lan and MeLan rings (Figure 4a,e). A surface map of OlvA(BCSA)GluC was generated using PyMOL (Figure 4f) and this C-terminal section exhibits a relatively deep pocket. Based on the red gradient color scheme used for the hydrophobic surface map (Figure 4f) wherein hydrophilic patches are shown in white and increasing red color signifies increasing hydrophobic character, OlvA(BCSA)GluC appears to be mainly hydrophilic. Moreover, the free carboxylate group of isoAsp and the side chain of Lys10 stand out as white hydrophilic patches clustered in one region of the molecule’s surface. The observed surface properties explain the high water solubility of OlvA(BCSA)GluC.
In vitro Reconstitution of OlvSA Activity.
Upon establishing the structure of OlvA(BCSA)GluC, we investigated the mechanism by which the Asp residue was rearranged into isoAsp by reconstituting the activity of OlvSA in vitro. OlvSA was expressed with a C-terminal His6-tag in E. coli BL21 (DE3), and purified by a combination of nickel affinity chromatography and size-exclusion chromatography (Figure S11). The purified four-fold cyclized His6-OlvA(BC) was used as substrate. First, we examined whether OlvSA requires SAM for activity by incubating His6-OlvA(BC) with OlvSA in the presence and absence of SAM. The formation of the five-fold dehydrated product was only observed in the presence of SAM, indicating that SAM is indeed a cosubstrate of OlvSA (Figure S12a,b).
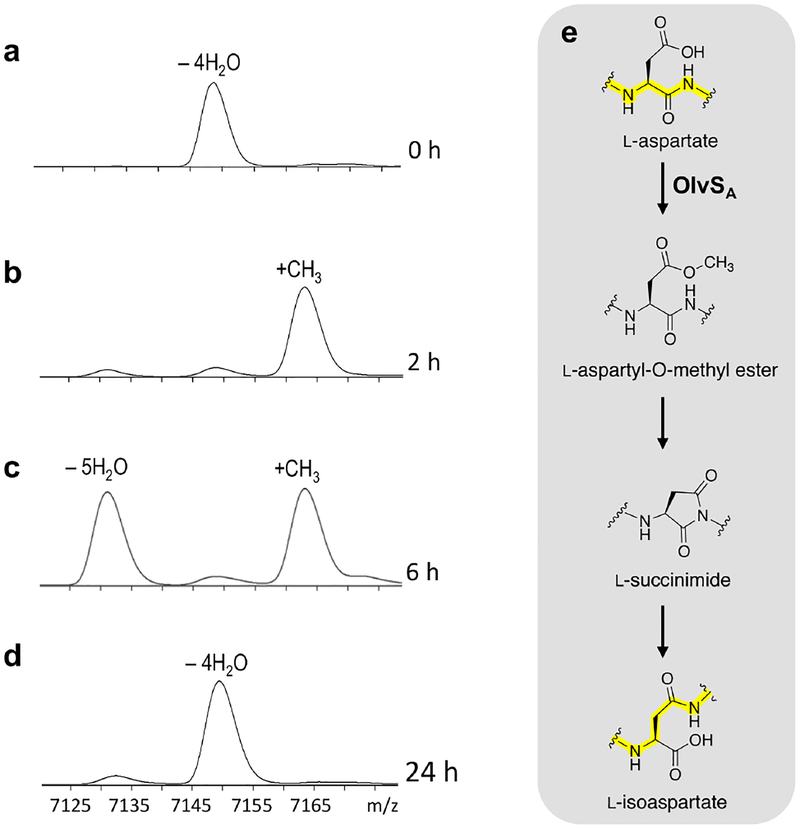
To probe the possible reaction mechanism, a time course assay was set up and the reaction was monitored by MALDI-TOF MS. Methylation (+14 m/z mass shift) was observed in 15 min at room temperature (Figure S12c), and proceeded to completion in 2 h (Figure 6a,b). A five-fold dehydrated product, consistent with the formation of a succinimide, was observed in 4 h (Figure S12c). The amount of putative imide continually increased and reached a 1:1 methylated peptide:imide ratio after 6 h of incubation (Figure 6c). When left at room temperature, the methylated product was completely converted to the imide-containing species after 24 h of incubation, with the concomitant appearance of a four-fold dehydrated product (Figure S12c). On the other hand, when the sample was placed at 37 °C, the imide-containing peptide was completely converted to the four-fold dehydrated product after the 24 h observation time (Figure 6d). Based on RP-HPLC analysis, the four-fold dehydrated final product consisted of 70–80% isoAsp-containing peptide (OlvA(BCSA)) and 20–30% Asp-containing peptide (OlvA(BC)) (Figure S13). The proposed order of events to explain the observed temporal changes in mass is summarized in Figure 6e.
Figure 6.
MALDI-TOF mass spectra of His6-OlvA(BC) reacted with OlvSA in (a) 0 h (b) 2 h, (c) 6 h, and (d) 24 h. Data shown correspond to [M+H]+. The sample was initially set up at room temperature and then incubated at 37 °C after the 6 h observation time. (e) Reaction scheme for the OlvSA-mediated rearrangement of L-aspartate to L-isoaspartate, consistent with the corresponding MS data shown on the left.
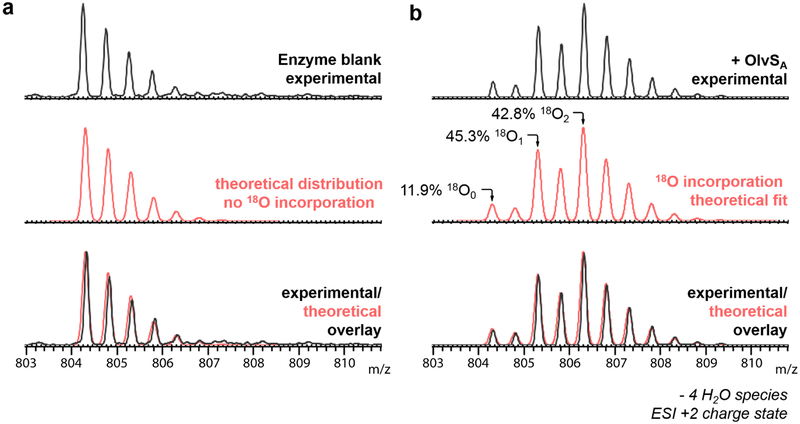
To verify formation of the imide species, hydrazine trapping and 18O-labelling experiments were performed.49 For the hydrazine test, the in vitro assay was allowed to proceed until it reached complete conversion of the methylated product to the imide-containing peptide. To the sample was then added 2 M hydrazine. After 30 min, a +32 m/z mass shift was observed, consistent with the nucleophilic addition of hydrazine to the imide (Figure S12d). For the 18O-labelling experiments, in vitro assays of OlvA(BC) with OlvSA were performed in H218O, and the products were digested with GluC protease and analyzed by LC-ESI-QTOF MS. A mass shift of +2 m/z was observed in the product, consistent with the order of events in Figure 6e including imide hydrolysis. In the absence of OlvSA, no 18O incorporation was observed (Figure 7a). Furthermore, a subset of the products contained two 18O atoms, suggesting that multiple cycles of methylation, imide formation, and hydrolysis occurred over the course of the assay (Figure 7a,b). A fit of the data using calculated spectra of peptide containing one, two, and three 18O labels indicates the distribution is about 12% peptide without label, 45% peptide with one 18O label and 43% of peptide with two 18O labels. Collectively, the NMR structure of the final product with an isoAsp, the trapping of the five-fold dehydrated peptide with hydrazine, and the observation of 18O incorporation upon hydrolysis of the five-fold dehydrated peptide is consistent with succinimide formation.
Figure 7.
ESI-QTOF mass spectra (+2 charge state) of OlvA(BC)GluC from the 18O-labeling experiments; (a) OlvA(BC)GluC substrate incubated in buffer made in H218O, and (b) 50 μM OlvA(BC)GluC reacted with 10 μM OlvSA and 1 mM SAM in 50 mM Hepes made in H218O, pH 7. The spectrum in panel b is from a mixture of Asp and isoAsp containing peptides as explained in the text.
The in vitro assay in H218O was also performed using the unmodified His6-OlvA and the dehydrated His6-OlvA(B) peptide (i.e. His6-OlvA co-expressed with OlvB only) as substrates. In both cases, no methylation and no 18O incorporation was observed (Figure S14), demonstrating that OlvSA requires the cyclized version of the substrate. We were surprised that the product ratio of isoAsp to Asp-containing products was ~4:1 after both 24 h of in vitro reaction or 24 h of coexpression. Instead, we had expected the process to go nearly completely to the isoAsp product. We first checked the possibility of feedback inhibition by the product S-adenosyl homocysteine (SAH) in the in vitro process. Indeed, the reaction was faster in the presence of the SAH nucleosidase Pfs from E. coli (Figure S15),33 but such inhibition does not explain the ratio after co-expression in E. coli. Instead, we wondered whether the isoAsp-containing OlvA(BCSA) was perhaps also a substrate for OlvSA, which could explain the product ratios. Indeed, complete conversion of full length OlvA(BCSA) (consisting of a ~70:30 mixture of isoAsp and Asp-containing peptides) as well as purified isoAsp-containing OlvA(BCSA)Glu to the succinimide was observed (Figure S16). Thus, OlvSA methylates both Asp and isoAsp in the context of the tricyclic scaffold. We note that this observation predicts that upon very long incubation of OlvA(BC) with OlvSA in H218O three 18O labels will be incorporated in the peptide (two in the carboxylate of Asp/isoAsp and one in the amide backbone) provided the enzyme remains active during the assay. Attempts to further investigate the substrate scope by mutations to OlvA were hampered by incomplete or abolished OlvBC activity with the mutants.
The observation that both Asp and isoAsp containing peptides are substrates for LahSA could set up a futile cycle of methylation and hydrolysis ultimately resulting in a ratio of the two peptides that is governed by the regioselectivity of hydrolysis, which favors isoAsp.50 The observed 70–80% isoAsp content after 24 h incubations in vitro (Figure S13) or after 24 h of co-expression in E. coli (Figure 2d) is consistent with this scenario. Of course, this situation would waste resources in the form of SAM. We anticipate that in the producing organism a subsequent step might be selective for the isoAsp-containing peptide, such as leader peptide removal or transport. Alternatively, regioselective imide hydrolysis by a currently unknown enzyme could be a mechanism to prevent wasteful utilization of SAM provided one of the aforementioned subsequent steps were faster than methylation of the isoAsp-containing peptide. We also recognize that the observation that both Asp and isoAsp containing tetracyclic peptides are substrates for LahSA could be an indication that the succinimide might be the actual intended product.
We next investigated the role of the leader peptide in OlvSA activity by using OlvA(BC)GluC as substrate. OlvSA was still able to modify the resulting peptide, albeit at a lower efficiency (Figure S17). Leader peptide-independent modifications catalyzed by other lanthipeptide tailoring enzymes have been previously reported.10,18 Altogether, the results demonstrate that OlvSA does not need the leader peptide as recognition motif, but requires a cyclized core peptide.
Lastly, we looked into the enzyme-dependence of the imide formation and hydrolysis steps. The in vitro assay was run for 1 h to achieve methylation of the His6-OlvA(BC) substrate while ensuring that the imide species was not yet formed. The sample was then treated with 3 M guanidine hydrochloride at pH 7.5 in order to denature the enzyme.51 Despite successfully denaturing the enzyme as demonstrated by a control experiment, the formation of the imide was still observed, followed by its hydrolysis (Figure S18). The results suggest that OlvSA catalyzes the methylation of Asp to the corresponding methyl ester, which spontaneously forms a succinimide followed by non-enzymatic hydrolysis as previously observed in other systems.52–54
To test the potential function of OlvA(BCSA), antimicrobial screening assays were performed with OlvA(BCSA)GluC, OlvA(BC)GluC, and the imide-containing species, however, no activity has been observed thus far under the conditions tested with a range of gram-positive and gram-negative bacteria. Recent examples have been reported of lanthipeptides that have bioactivities other than antimicrobial (e.g. antiviral, antifungal, antinociceptive, antiallodynic, and morphogenetic activities).11–17 Evaluation of such a broader range of bioactivities for OlvA(BCSA) is currently underway.
β-Amino Acids in Ribosomally Synthesized Natural Products.
The incorporation of β-amino acids in bacterial natural products, which contributes to structural diversification and unique bioactivities, has typically been observed in nonribosomal peptides.5,55 To the best of our knowledge, currently only one system has been reported that incorporates β-amino acids in ribosomal natural products, wherein α-keto β-amides are installed in certain proteusins by a radical SAM splicase enzyme, PlpX, of the plp cluster from Pleurocapsa sp. PCC 7319.5 PlpX acts on an “Xxx-Tyr-Gly” motif in the precursor peptide, and a β-linkage is installed through excision of tyramine from Tyr and rejoining of the remaining segments of the peptide. In this study, we describe another family of enzymes (LanSA) that mediate the incorporation of β-amino acids in ribosomal peptides via a different mechanism involving a rearrangement reaction of L-Asp to L-isoAsp that proceeds through the methylation of Asp and the subsequent imide formation and hydrolysis. In mammalian systems, an alternative mechanism for conversion of Asp to isoAsp has been described. O-linked β-N-acetylglucosamine transferase can mediate this transformation via the glycosylation of Asp, followed by imide formation, and hydrolysis.54 While isoAsp formation has typically been discussed in terms of the result of unwanted damage to Asp/Asn residues, hypothetical roles of isoAsp residues have been proposed.56
LanSA vs. Protein L-Isoaspartate (D-Aspartate) O-Methyltransferases.
The genome mining study by Zhang et al. indicated that the O-methyltransferases that are ubiquitous in class I lanthipeptide gene clusters in actinobacteria, now referred to as LanSA enzymes, have low homology with protein L-isoaspartate (D-aspartate) O-methyltransferases (PIMTs), also known as L-isoaspartyl/D-aspartyl protein carboxyl methyltransferases (PCMT).9 PIMTs are repair enzymes that convert L-isoAsp and D-Asp in proteins to L-Asp. L-isoAsp and D-Asp are formed in proteins through L-Asn deamination or L-Asp hydrolysis via a spontaneous process that is accelerated by various stresses (e.g. extreme pH values, temperature, oxidants).57–62 The accumulation of L-isoAsp and D-Asp has been associated with aging, neurodegenerative diseases, and cancer, as a consequence of protein misfolding that compromises structural integrity and protein function.63–67 PIMTs restore or protect biological systems from these detrimental effects by converting L-isoAsp and D-Asp residues to L-Asp using a similar sequence of reactions of methylation, imide formation, and hydrolysis. OlvSA differs from PIMTs because OlvSA recognizes L-Asp and isoAsp as substrate, specifically in a cyclic peptide, while PIMTs typically accepts L-isoAsp and D-Asp,58,68 although in some cases Asp methylation has been reported.69 PIMTs go through multiple rounds of catalysis to fully repair isoAsp-containing proteins because the imide hydrolysis step generates both Asp and isoAsp.70 The data in Figure 2d and Figure 7 suggest that this sub-optimal process, because of incomplete regioselectivity of imide hydrolysis, is also the case for the process of converting Asp to isoAsp catalyzed by OlvSA. An amino acid sequence alignment of OlvSA with PIMT enzymes of E. coli, Homo sapiens (human), Mus musculus (mouse), Arabidopsis thaliana (mouse-ear cress plant), and Triticum aestivum (wheat) reveals that OlvSA is approximately twice as long as the PIMT enzymes (Figure S19). OlvSA consists of an N-terminal region that is homologous to PIMTs, and a unique C-terminal segment. These in silico data, together with our experimental results, highlight that LanSA represent a new family of O-methyltransferases related to, yet distinct from PIMTs.
CONCLUSIONS
In this study, we describe an enzyme-mediated rearrangement reaction of L-Asp to L-isoAsp that introduces a β-amino acid in the peptide backbone. We first performed co-expression experiments of the S. olivaceus NRRL B-3009 olvABCSA cluster, and demonstrated that although the major peptide products, OlvA(BCSA)GluC and OlvA(BC)GluC, produced with and without OlvSA O-methyltransferase, respectively, have the same molecular masses, the two peptides are not identical. The NMR solution structures of OlvA(BCSA)GluC and OlvA(BC)GluC were elucidated and the structures revealed that the main difference between the two peptides lies in the loop region that contains the Asp/isoAsp residue. OlvA(BCSA)GluC consists of two MeLan and two Lan containing rings positioned in a unique interlocking topology, and exhibits a hydrophilic surface. GC-MS analysis showed that both MeLan rings have the canonical DL-stereochemistry, while the Lan rings has the uncommon LL-stereochemistry. In addition to the structural characterization of OlvA(BCSA)GluC, we reconstituted the activity of OlvSA in vitro. OlvSA was found to be a leader-independent, SAM-dependent O-methyltransferase that requires the cyclic version of the precursor peptide as substrate. OlvSA methylates the side chain carboxylate group of a highly conserved Asp residue, followed by the formation of an imide intermediate. The imide then hydrolyzes non-enzymatically to 70–80% L-isoAsp as confirmed by the RP-HPLC, GC-MS, and NMR data. 18O-labelling experiments further indicated that multiple methylation cycles occur via the succinimide intermediate and that OlvSA methylates both the Asp and isoAsp containing cyclic peptides. This observation could indicate that the electrophilic succinimide might be the intended product. OlvA(BCSA)GluC, OlvA(BC)GluC, and the imide-containing peptide did not exhibit any antimicrobial activity under the conditions tested in this study. Hence, future work will be directed towards the determination of the bioactivity of these lanthipeptide that may first require the identification of the leader peptide protease cleavage site.
In summary, this work presents the characterization of OlvA(BCSA), a lanthipeptide with a unique ring topology and stereochemistry, and describes, for the first time, the function and mechanism of a LanSA enzyme. The established foundational knowledge on LanSA activity will guide future investigations on other intriguing gene clusters that contain LanSA enzymes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Zhengan Zhang for providing the pTrc33_T.bispora_GluRS_tRNA plasmid, Dr. Lingyang Zhu (NMR laboratory, UIUC) for her assistance in the acquisition of NMR data, Dr. Alex V. Ulanov (Carver Biotechnology Center at UIUC) for help with the GC-MS experiments, and Dr. Leah Martin-Visscher and Dr. Pascal Mercier for their advice on CYANA. This work was funded by the National Institutes of Health (R37 GM058822 to W.A.V.; F32 GM117765 to I.R.B.). J.Z.A. was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). A.T. was a Snyder Scholar, while C.F. was a Chemical Biology Scholar of the University of Illinois Urbana-Champaign.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Gene sequences, NMR experiment parameters, chemical shift assignments, supporting figures, sequence alignments
REFERENCES
- (1).Luo S; Dong SH Recent advances in the discovery and biosynthetic study of eukaryotic RiPP natural products Molecules 2019, 24, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).O’Neill EC; Schorn M; Larson CB; Millan-Aguinaga N Targeted antibiotic discovery through biosynthesis-associated resistance determinants: target directed genome mining Crit. Rev. Microbiol 2019, 20191. [DOI] [PubMed] [Google Scholar]
- (3).Poorinmohammad N; Bagheban-Shemirani R; Hamedi J Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria Antonie van Leeuwenhoek 2019, 112, 1477. [DOI] [PubMed] [Google Scholar]
- (4).Shang Z; Winter JM; Kauffman CA; Yang I; Fenical W Salinipeptins: integrated genomic and chemical approaches reveal unusual D-amino acid-containing ribosomally synthesized and post-translationally modified peptides (RiPPs) from a Great Salt Lake Streptomyces sp. ACS Chem. Biol 2019, 14, 415. [DOI] [PubMed] [Google Scholar]
- (5).Morinaka BI; Lakis E; Verest M; Helf MJ; Scalvenzi T; Vagstad AL; Sims J; Sunagawa S; Gugger M; Piel J Natural noncanonical protein splicing yields products with diverse beta-amino acid residues Science 2018, 359, 779. [DOI] [PubMed] [Google Scholar]
- (6).Acedo JZ; Chiorean S; Vederas JC; van Belkum MJ The expanding structural variety among bacteriocins from Gram-positive bacteria FEMS Microbiol. Rev 2018, 42, 805. [DOI] [PubMed] [Google Scholar]
- (7).Hetrick KJ; van der Donk WA Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era Curr. Opin. Chem. Biol 2017, 38, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Marsh AJ; O’Sullivan O; Ross RP; Cotter PD; Hill C In silico analysis highlights the frequency and diversity of type 1 lantibiotic gene clusters in genome sequenced bacteria BMC Genomics 2010, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhang Q; Doroghazi JR; Zhao X; Walker MC; van der Donk WA Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria Appl. Environ. Microbiol 2015, 81, 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; Cotter PD; Craik DJ; Dawson M; Dittmann E; Donadio S; Dorrestein PC; Entian KD; Fischbach MA; Garavelli JS; Göransson U; Gruber CW; Haft DH; Hemscheidt TK; Hertweck C; Hill C; Horswill AR; Jaspars M; Kelly WL; Klinman JP; Kuipers OP; Link AJ; Liu W; Marahiel MA; Mitchell DA; Moll GN; Moore BS; Müller R; Nair SK; Nes IF; Norris GE; Olivera BM; Onaka H; Patchett ML; Piel J; Reaney MJ; Rebuffat S; Ross RP; Sahl HG; Schmidt EW; Selsted ME; Severinov K; Shen B; Sivonen K; Smith L; Stein T; Süssmuth RD; Tagg JR; Tang GL; Truman AW; Vederas JC; Walsh CT; Walton JD; Wenzel SC; Willey JM; van der Donk WA Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature Nat. Prod. Rep 2013, 30, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mohr KI; Volz C; Jansen R; Wray V; Hoffmann J; Bernecker S; Wink J; Gerth K; Stadler M; Müller R Pinensins: the first antifungal lantibiotics Angew. Chem. Int. Ed 2015, 54, 11254. [DOI] [PubMed] [Google Scholar]
- (12).Kodani S; Hudson ME; Durrant MC; Buttner MJ; Nodwell JR; Willey JM The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor Proc. Natl. Acad. Sci. U.S.A 2004, 101, 11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kodani S; Lodato MA; Durrant MC; Picart F; Willey JM SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the Streptomycetes Mol. Microbiol 2005, 58, 1368. [DOI] [PubMed] [Google Scholar]
- (14).Férir G; Petrova MI; Andrei G; Huskens D; Hoorelbeke B; Snoeck R; Vanderleyden J; Balzarini J; Bartoschek S; Brönstrup M; Süssmuth RD; Schols D The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications PLoS One 2013, 8, e64010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Iorio M; Sasso O; Maffioli SI; Bertorelli R; Monciardini P; Sosio M; Bonezzi F; Summa M; Brunati C; Bordoni R; Corti G; Tarozzo G; Piomelli D; Reggiani A; Donadio S A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity ACS Chem. Biol 2014, 9, 398. [DOI] [PubMed] [Google Scholar]
- (16).Meindl K; Schmiederer T; Schneider K; Reicke A; Butz D; Keller S; Guhring H; Vertesy L; Wink J; Hoffmann H; Bronstrup M; Sheldrick GM; Süssmuth RD Labyrinthopeptins: a new class of carbacyclic lantibiotics Angew. Chem. Int. Ed 2010, 49, 1151. [DOI] [PubMed] [Google Scholar]
- (17).Smith TE; Pond CD; Pierce E; Harmer ZP; Kwan J; Zachariah MM; Harper MK; Wyche TP; Matainaho TK; Bugni TS; Barrows LR; Ireland CM; Schmidt EW Accessing chemical diversity from the uncultivated symbionts of small marine animals Nat. Chem. Biol 2018, 14, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Repka LM; Chekan JR; Nair SK; van der Donk WA Mechanistic understanding of lanthipeptide biosynthetic enzymes Chem. Rev 2017, 117, 5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).van der Donk WA; Nair SK Structure and mechanism of lanthipeptide biosynthetic enzymes Curr. Opin. Struct. Biol 2014, 29, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Garg N; Salazar-Ocampo LM; van der Donk WA In vitro activity of the nisin dehydratase NisB Proc. Natl. Acad. Sci. U. S. A 2013, 110, 7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ortega MA; Hao Y; Zhang Q; Walker MC; van der Donk WA; Nair SK Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB Nature 2015, 517, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lubelski J; Rink R; Khusainov R; Moll GN; Kuipers OP Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin Cell. Mol. Life Sci 2008, 65, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yang X; van der Donk WA Michael-type cyclizations in lantibiotic biosynthesis are reversible ACS Chem. Biol 2015, 10, 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li B; Yu JP; Brunzelle JS; Moll GN; van der Donk WA; Nair SK Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis Science 2006, 311, 1464. [DOI] [PubMed] [Google Scholar]
- (25).Xie LL; Miller LM; Chatterjee C; Averin O; Kelleher NL; van der Donk WA Lacticin 481: In vitro reconstitution of lantibiotic synthetase activity Science 2004, 303, 679. [DOI] [PubMed] [Google Scholar]
- (26).Chatterjee C; Miller LM; Leung YL; Xie L; Yi M; Kelleher NL; van der Donk WA Lacticin 481 synthetase phosphorylates its substrate during lantibiotic production J. Am. Chem. Soc 2005, 127, 15332. [DOI] [PubMed] [Google Scholar]
- (27).Dong SH; Tang W; Lukk T; Yu Y; Nair SK; van der Donk WA The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold eLife 2015, 4, e07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Goto Y; Li B; Claesen J; Shi Y; Bibb MJ; van der Donk WA Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights PLoS Biol 2010, 8, e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Müller WM; Schmiederer T; Ensle P; Süssmuth RD In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification Angew. Chem., Int. Ed 2010, 49, 2436. [DOI] [PubMed] [Google Scholar]
- (30).Funk MA; van der Donk WA Ribosomal natural products, tailored to fit Acc. Chem. Res 2017, 50, 1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Singh M; Sareen D Novel LanT associated lantibiotic clusters identified by genome database mining PLoS One 2014, 9, e91352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Huo L; Zhao X; Acedo JZ; Estrada P; Nair SK; van der Donk WA Characterization of a dehydratase and methyltransferase in the biosynthesis of a ribosomally-synthesized and post-translationally modified peptide in Lachnospiraceae ChemBioChem 2019, 10.1002/cbic.201900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Choi-Rhee E; Cronan JE A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase Chem. Biol 2005, 12, 589. [DOI] [PubMed] [Google Scholar]
- (34).Ortega MA; Hao Y; Walker MC; Donadio S; Sosio M; Nair SK; van der Donk WA Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis Cell Chem. Biol 2016, 23, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hudson GA; Zhang Z; Tietz JI; Mitchell DA; van der Donk WA In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin J. Am. Chem. Soc 2015, 137, 16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A NMRPipe: a multidimensional spectral processing system based on UNIX pipes J. Biomol. NMR 1995, 6, 277. [DOI] [PubMed] [Google Scholar]
- (37).Johnson BA Using NMRView to visualize and analyze the NMR spectra of macromolecules Methods Mol. Biol 2004, 278, 313. [DOI] [PubMed] [Google Scholar]
- (38).Wider G; Macura S; Kumar A; Ernst R; Wüthrich K Homonuclear two-dimensional 1H NMR of proteins. Experimental procedures J. Magn. Reson 1984, 56(2), 207. [Google Scholar]
- (39).Wüthrich K NMR of proteins and nucleic acids 1986, The George Fisher Baker non-resident lectureship in chemistry at Cornell University (USA). [Google Scholar]
- (40).Guntert P; Mumenthaler C; Wuthrich K Torsion angle dynamics for NMR structure calculation with the new program DYANA J. Mol. Biol 1997, 273, 283. [DOI] [PubMed] [Google Scholar]
- (41).DeLano W The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- (42).Liu W; Chan ASH; Liu H; Cochrane SA; Vederas JC Solid supported chemical syntheses of both components of the lantibiotic lacticin 3147 J. Am. Chem. Soc 2011, 133, 14216. [DOI] [PubMed] [Google Scholar]
- (43).Tang W; van der Donk WA Structural characterization of four prochlorosins: a novel class of lantipeptides produced by planktonic marine cyanobacteria Biochemistry 2012, 51, 4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tang W; Jiménez-Osés G; Houk KN; van der Donk WA Substrate control in stereoselective lanthionine biosynthesis Nat. Chem 2015, 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kido Y; Hamakado T; Yoshida T; Anno M; Motoki Y; Wakamiya T; Shiba T Isolation and characterization of ancovenin, a new inhibitor of angiotensin I converting enzyme, produced by actinomycetes J. Antibiot 1983, 36, 1295. [DOI] [PubMed] [Google Scholar]
- (46).Ross AC; Liu H; Pattabiraman VR; Vederas JC Synthesis of the lantibiotic lactocin S using peptide cyclizations on solid phase J. Am. Chem. Soc 2010, 132, 462. [DOI] [PubMed] [Google Scholar]
- (47).Garg N; Goto Y; Chen T; van der Donk WA Characterization of the stereochemical configuration of lanthionines formed by the lanthipeptide synthetase GeoM Biopolymers 2016, 106, 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Lohans CT; Li JL; Vederas JC Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with D-amino acids J. Am. Chem. Soc 2014, 136, 13150. [DOI] [PubMed] [Google Scholar]
- (49).Grossenbacher H; Marki W; Coulot M; Muller D; Richter WJ Characterization of succinimide-type dehydration products of recombinant hirudin variant 1 by electrospray tandem mass spectrometry Rapid Commun. Mass Spectrom 1993, 7, 1082. [DOI] [PubMed] [Google Scholar]
- (50).Guttler BH; Cynis H; Seifert F; Ludwig HH; Porzel A; Schilling S A quantitative analysis of spontaneous isoaspartate formation from N-terminal asparaginyl and aspartyl residues Amino Acids 2013, 44, 1205. [DOI] [PubMed] [Google Scholar]
- (51).Liu M; Cheetham J; Cauchon N; Ostovic J; Ni W; Ren D; Zhou ZS Protein isoaspartate methyltransferase-mediated 18O-labeling of isoaspartic acid for mass spectrometry analysis Anal Chem 2012, 84, 1056. [DOI] [PubMed] [Google Scholar]
- (52).Geiger T; Clarke S Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation J. Biol. Chem 1987, 262, 785. [PubMed] [Google Scholar]
- (53).McFadden PN; Clarke S Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins Proc. Natl. Acad. Sci. U. S. A 1987, 84, 2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Janetzko J; Walker S Aspartate glycosylation triggers isomerization to isoaspartate J. Am. Chem. Soc 2017, 139, 3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kudo F; Miyanaga A; Eguchi T Biosynthesis of natural products containing beta-amino acids Nat. Prod. Rep 2014, 31, 1056. [DOI] [PubMed] [Google Scholar]
- (56).Corti A; Curnis F Isoaspartate-dependent molecular switches for integrin-ligand recognition J. Cell Sci 2011, 124, 515. [DOI] [PubMed] [Google Scholar]
- (57).Reissner KJ; Aswad DW Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell. Mol. Life Sci 2003, 60, 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Skinner MM; Puvathingal JM; Walter RL; Friedman AM Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair Structure 2000, 8, 1189. [DOI] [PubMed] [Google Scholar]
- (59).MacLaren DC; Clarke S Expression and purification of a human recombinant methyltransferase that repairs damaged proteins Protein Expr. Purif 1995, 6, 99. [DOI] [PubMed] [Google Scholar]
- (60).Brennan TV; Clarke S Spontaneous degradation of polypeptides at aspartyl and asparaginyl residues: effects of the solvent dielectric Protein Sci 1993, 2, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Brennan TV; Clarke S Effect of adjacent histidine and cysteine residues on the spontaneous degradation of asparaginyl- and aspartyl-containing peptides Int. J. Pept. Protein Res 1995, 45, 547. [DOI] [PubMed] [Google Scholar]
- (62).Hicks WM; Kotlajich MV; Visick JE Recovery from long-term stationary phase and stress survival in Escherichia coli require the L-isoaspartyl protein carboxyl methyltransferase at alkaline pH Microbiology 2005, 151, 2151. [DOI] [PubMed] [Google Scholar]
- (63).Chondrogianni N; Petropoulos I; Grimm S; Georgila K; Catalgol B; Friguet B; Grune T; Gonos ES Protein damage, repair and proteolysis Mol. Aspects Med 2014, 35, 1. [DOI] [PubMed] [Google Scholar]
- (64).Saito H; Yamashita M; Ogasawara M; Yamada N; Niisato M; Tomoyasu M; Deguchi H; Tanita T; Ishida K; Sugai T; Yamauchi K Chaperone protein L-isoaspartate (D-aspartyl) O-methyltransferase as a novel predictor of poor prognosis in lung adenocarcinoma Hum. Pathol 2016, 50, 1. [DOI] [PubMed] [Google Scholar]
- (65).Warmack RA; Mansilla E; Goya RG; Clarke SG Racemized and isomerized proteins in aging rat teeth and eye lens Rejuvenation Res 2016, 19, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ouazia D; Levros LC Jr.; Rassart E; Desrosiers RR The protein l-isoaspartyl (D-aspartyl) methyltransferase protects against dopamine-induced apoptosis in neuroblastoma SH-SY5Y cells Neuroscience 2015, 295, 139. [DOI] [PubMed] [Google Scholar]
- (67).Shimizu T; Watanabe A; Ogawara M; Mori H; Shirasawa T Isoaspartate formation and neurodegeneration in Alzheimer’s disease Arch. Biochem. Biophys 2000, 381, 225. [DOI] [PubMed] [Google Scholar]
- (68).Lowenson JD; Clarke S Recognition of D-aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis J. Biol. Chem 1992, 267, 5985. [PubMed] [Google Scholar]
- (69).Biterge B; Richter F; Mittler G; Schneider R Methylation of histone H4 at aspartate 24 by protein L-isoaspartate O-methyltransferase (PCMT1) links histone modifications with protein homeostasis Sci. Rep 2014, 4, 6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Lindquist JA; McFadden PN Incorporation of two 18O atoms into a peptide during isoaspartyl repair reveals repeated passage through a succinimide intermediate J Protein Chem 1994, 13, 553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.