Abstract
A new class of mitochondrial disease has been identified and characterized as Multiple Mitochondrial Dysfunctions Syndrome (MMDS). Four different forms of the disease have each been attributed to point mutations in proteins involved in iron-sulfur (Fe-S) biosynthesis; in particular, MMDS2 has been associated with the protein BOLA3. To date, this protein has been characterized in vitro concerning its ability to form heterodimeric complexes with two putative Fe-S cluster binding partners: GLRX5 and NFU. However, BOLA3 has yet to be characterized in its own discrete holo form. Herein we describe procedures to isolate and characterize the human holo BOLA3 protein in terms of Fe-S cluster binding and trafficking, and demonstrate that human BOLA3 can form a functional homodimer capable of engaging in Fe-S cluster transfer.
Keywords: BOLA3, iron-sulfur cluster, cluster transfer, reactivity, homodimer
Introduction
Iron-sulfur cofactors are required for a variety of essential functions, such as electron transport, enzymatic catalysis, and DNA replication and repair [5, 6], In recent years, a number of disease conditions have been linked to errors in mitochondrial cofactor biogenesis. Of particular interest has been the cofactor lipoic acid, since defects in its production can be linked not only to enzymes involved in lipoic acid biosynthesis, but also to proteins related to biogenesis of the iron-sulfur (Fe-S) cluster cofactor. In general, mutations in proteins in either pathway give rise to similar downstream deficiencies that result in overall decreased energy metabolism and respiratory failure [7], While many of the specific steps in these biosynthetic pathways and the link between them are somewhat unclear, nevertheless, reports of patient case studies provide a basis for in-depth biochemical investigation.
Beginning in 2011, a new class of mitochondrial disorders was characterized as Multiple Mitochondrial Dysfunctions Syndromes (MMDS). MMDS provided the initial link between Fe-S clusters and lipoic acid because all four types of MMDS so far identified are associated with genes that encode proteins involved in the biosynthesis of Fe-S clusters: namely, IBA57, ISCA2, BOLA3, or NFU1 [8–14], These disorders are typically fatal, as patients are found with a severe impairment of key metabolic pathways and energy production as a consequence of single nucleotide genetic mutations in any one of these Fe-S proteins [9, 10], In particular, the disease phenotypes caused by these mutations have revealed a specific impairment of downstream [4Fe-4S]-cluster-containing proteins [7–9], which results in a deficiency of mitochondrial respiratory complexes and impaired function of lipoic-acid-dependent enzymes, such as pyruvate dehydrogenase (PDH) and protein H of the Glycine Cleavage System (GCS) [7, 10, 15],
Production and incorporation of the Fe-S cluster cofactor is complex, yet highly conserved across evolution and all kingdoms of life [16], Cluster biosynthesis in eukaryotes begins with de novo [2Fe-2S] cluster formation on the IscU/ISCU class of cluster scaffold proteins with subsequent transfer to downstream [2Fe-2S] cluster target proteins via a complex of the heat shock chaperone system and a cluster delivery protein, GLRX5 [17, 18], From GLRX5, the [2Fe-2S] cluster has been suggested to convert to a [4Fe-4S] cluster through the scaffold-like proteins ISCA1 and ISCA2 for delivery to downstream [4Fe-4S] cluster target proteins [19, 20],
To date, the clearest biochemical information available for BOLA3 has been determined from case studies of disease states such as MMDS. Since missense mutations on the cluster binding proteins NFU1 and BOLA3 have resulted in decreased lipoic acid formation in patients [9, 13, 15], they are believed to be the delivery proteins for lipoic acid synthase (LIAS); however, there is no elaborated mechanism for cluster uptake by human LIAS. Although many questions remain regarding the native interaction partner(s) for BOLA3 and its precise cellular role in Fe-S cluster biosynthesis, it is clear that either a deficiency of BOLA3, or a missense mutation, impair both Fe-S cluster biosynthesis and incorporation of a cluster into LIAS [9, 13],
Aside from being a causative agent of MMDS2, very little is known about BOLA3. Humans have three Bol-like proteins [21], with BOLA1 and BOLA3 localized to the mitochondria and BOLA2 present in the cytosol [22, 23], Human BOLA2 and its yeast homo log Bol2 have been studied for their ability to form a heterocomplex with the cytosolic monothiol glutaredoxin 3 [22, 24, 25], which also binds a Fe-S cluster. Additionally, the [2Fe-2S] cluster-bound Bol2-Grx3 complex has been characterized for its role in yeast iron regulation [22], In vitro studies have shown that the bridging [2Fe-2S] cluster of the GLRX3-BOLA2 heterocomplex can be transferred to anamorsin, a protein implicated in the cytosolic iron-sulfur assembly pathway (CIA) [26], an observation that was also supported by in vivo data [27], While the precise role and interacting partners of the mitochondrial BOLA proteins are unclear, initial in vitro studies have examined the ability of BOLA3 to interact with the mitochondrial Fe-S cluster proteins NFU and GLRX5 in the form of heterodimeric complexes [28, 29], The [2Fe-2S] bridged BOLA3-GLRX5 heterocomplex has also been demonstrated to be suitable for cluster trafficking [1, 30]; however, a holo form of the BOLA3 protein has not been previously isolated or characterized. Herein we show that BOLA3 can bind a [2Fe-2S] as a functional homodimer that is capable of exchanging that cluster with a variety of putative partner Fe-S cluster proteins and glutathione. These studies advance our understanding of the physiological role(s) of BOLA3 and the molecular mechanisms of cellular Fe-S cluster biosynthesis and trafficking.
Materials and Methods
Materials
The gene for human BOLA3 in the pET28b(+) vector (UniProt: Q53S33; residues 25–107 [23], with the mitochondrial targeting sequence excluded) was purchased from GenScript. The gene for Homo sapiens glutaredoxin 5 (GLRX5) located in the pET28b(+) vector between the NdeI and HindIII restriction sites and lacking the first 31 amino acids (Δ1–31) that correspond to the mitochondrial targeting sequence [19], was ordered from GenScript. PD-10 desalting columns were purchased from GE Healthcare. Ferric chloride, sodium sulfide, and L-cysteine were obtained from Fisher.
Site Directed Mutagenesis
Site-directed mutagenesis was performed to prepare C59A and H96A single variants and C59A/H96A double variants of BOLA3. Non complementary primers were designed by NEBaseChanger and used for PCR amplification by Phusion polymerase from Fisher Scientific. The protocol involved initial denaturation at 98 °C for 1 min, 30 cycles comprising denaturation at 98 °C for 30 sec, annealing at 62 °C for 1 min, and primer extension for 3 min at 72 °C. Upon completion, KLD enzyme mix (NEB) was added to the mixture and incubated for 2 h at temperature to assist intramolecular cyclization/ligation of amplified DNA and to digest parent plasmid. The reaction mix was then transformed into BL21-DE3 E. coli cells by the heat-shock method, following which the cells were plated on kanamycin plates. Positive clones were identified by isolating plasmid DNA from the colonies and sending them for sequencing.
Protein expression and purification
Purification of human ISCU (Hs ISCU) and Thermatoga maritima NifS (Tm NifS) was performed as previously reported [31–33], The expression vector for human ferredoxin-1 (Hs FDX1) was kindly provided by J. Markley, and the protein was expressed and purified according to literature procedures [34], Purification of human ferredoxin-2 (Hs FDX2) was performed as previously reported [35], The ferredoxins purified as holo proteins and were then subsequently converted to apo forms by treatment with 100 mM EDTA, 5 mM DTT, and 8 M urea in 50 mM HEPES, 100 mM NaCl, pH 7.5. To isolate the apo ferredoxins, the colorless solution was passed through a PD-10 column to remove EDTA, DTT, and urea. Apo protein concentration was determined by use of the Bradford assay, as described at the end of the protein expression and purification section of the experimental.
A construct of human GLRX2 (comprising residues 56–161) with a tobacco etch virus cleavable N-terminal His6 tag in the expression vector pNic-Bsa4 was kindly provided by Drs. Kavanagh, Muller-Knapp and Oppermann, and the protein was expressed and purified as previously reported [36],
The BOLA3 construct in BL21-DE3 E. coli was grown in 10 mL of Luria Bertani (LB) broth media containing kanamycin (50 μM) at 37 °C overnight and then transferred to 3 L of fresh LB media containing kanamycin and incubated at 37 °C until the OD600 reached 0.8. Protein expression was induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) (300 μM), and the cell culture was incubated at 37 °C. Cell pellets were collected by centrifugation at 4330 g for 15 min, suspended in 30 mL of 50 mM HEPES, 100 mM NaCl, pH 7.5, and lysed by use of a dismembranator. The lysate was centrifuged at 28,928 g for 30 min, the supernatant subsequently applied to a Ni-NTA column, and protein eluted with buffer containing 50 mM HEPES, 100 mM NaCl, 0.25 M imidazole, pH 7.5, prior to analysis on a 12% SDS-PAGE gel that was then visualized by Coomassie Blue staining (Figure S1). Dialysis was carried out at 4 °C against 50 mM HEPES, 100 mM NaCl, pH 7.5. Purified BOLA3 was lyophilized and resuspended in 50 mM HEPES, 100 mM NaCl, pH 7.5 to determine the concentration from the absorbance at 274 nm, ε274 = 2548 M−1 cm−1.
In vitro reconstitution of apo proteins
BOLA3 reconstitution was performed by incubating 200 μM protein with 2 μM Tm-NifS and 5 mM DTT, argon purging for 30 min, and making up to 0.6 mM Fe3+ and 0.6 mM L-cysteine. Following incubation for 1 h, the reaction mixture was passed through a PD-10 desalting column, equilibrated with 50 mM HEPES, 100 mM NaCl, pH 7.5, to remove excess reagents. GLRX2 was reconstituted by the same protocol, except that an additional 3 mM glutathione was added along with DTT and Tm-NifS in the initial reconstitution mixture. For ISCU [37], a 200 μM solution of apo protein was incubated with 50 mM DTT and 8 M urea, argon purged for 30 min, and then made up to 1 mM Fe3+ and 1 mM S2-. After 1 h, excess reagents were removed by passing the reconstitution mixture through a PD-10 column. BOLA3-GLRX5 heterodimer was reconstituted as reported previously [30].
Following reconstitution, total protein concentration was determined by use of the Bradford assay. Holo protein concentration was determined by extinction coefficients and confirmed through iron quantitation.
EPR Spectroscopy
Reconstituted holo BOLA3 in degassed 50 mM HEPES, 100 mM NaCl, pH 7.5 was reduced by addition of excess sodium dithionite and rapidly frozen in liquid nitrogen. EPR spectra were acquired using a Brücker EMXPlus EPR spectrometer equipped with a ColdEdge cryostat at 50 K at a microwave frequency of 9.4 GHz. Acquisition parameters were as follows: microwave power: 20 mW, modulation frequency: 100 kHz, modulation amplitude: 10.0 G, and acquisition time constant: 40.96 ms. EPR spectra were also recorded at 20 K and 70 K with the same acquisition parameters.
Detemiination of oligomeric state by analytical ultracentrifugation (AUC)
Apo BOLA3 at 400 μM protein (OD280 = 1.0) was loaded into the ultracentrifugation chambers and sealed, using 50 mM HEPES, 100 mM NaCl, pH 7.5 as a reference. Reconstituted holo BOLA3 at 248 μM (OD420 = 1.2) was loaded in the same manner with the same reference buffer. Samples were centrifuged at 45,000 rpm for 6 hours to reach complete sedimentation. The sedimentation profiles were fit using SEDFIT to the Lamm equation [3, 38].
HPLC of apo and holo BOLA3
Size exclusion chromatography was performed on a Shimadzu HPLC using an Agilent Zorbax GE-250 (4 μm, 4.6 × 250 mm) column in 50 mM HEPES, 100 mM NaCl, pH 7.5 at 0.5 mL/min. Apo and reconstituted holo BOLA3 were injected at equal concentrations (100 μM) and UV absorbance collected between 200–600 nm for 60 min per injection.
[2Fe-2S] (GS)4 synthesis
The cluster used was synthesized as previously reported [39], Briefly, ferric chloride (20 mM) and sodium sulfide (20 mM) were added to 10 mL of 40 mM glutathione solution, pH 8.6. A volume (40 mL) of ethanol was added to the mixture and mixed by vortexing. The precipitate was collected by centrifugation at 13,000 rpm for 10 min, washed twice with ethanol and dried under vacuum.
Cluster exchange with the [2Fe-2S](GS)4 complex measured via UV-Visible spectrophotometry
UV-vis measurements were obtained on a Varian Cary 50 UV-vis spectrophotometer as previously described, by monitoring the change in absorbance between 800 and 200 nm every 2 min over the course of 1 h [40, 41], Fe-S cluster charge-transfer bands exhibit absorbance signals at 330 nm and 415 nm, which can be used to monitor cluster formation, cluster transfer from the [2Fe-2S](GS)4 complex to apo proteins, and cluster extraction from holo proteins by glutathione to form the [2Fe-2S](GS)4 complex [39–41], Since the [2Fe-2S](GS)4 complex and holo proteins differ in extinction coefficient [39, 40, 42, 43], changes in absorbance at those wavelengths can be attributed to cluster transfer.
For cluster uptake, degassed 20 μM apo BOLA3 in 50 mM HEPES, 100 mM NaCl, pH 7.5 with 5 mM DTT was transferred to an anaerobic cuvette. The [2Fe-2S](GS)4 complex was resuspended in degassed 50 mM HEPES, 100 mM NaCl, pH 7.5 and added to the cuvette via a gas-tight syringe to a desired final concentration. The cluster concentration was varied from 40 – 200 μM, while the protein concentration remained constant. Alternatively, the cluster concentration was held constant at 200 μM, while the protein concentration was varied from 20 to 100 μM.
In the case of GSH extraction from reconstituted BOLA3 to form the [2Fe-2S](GS)4 complex, degassed and reconstituted holo protein in 50 mM HEPES, 100 mM NaCl, pH 7.5 was incubated with GSH from 50 μM to 1 mM in an anaerobic cuvette. Cluster extraction by GSH to form the [2Fe-2S](GS)4 complex was confirmed by use of ESI mass spectrometry on a Brücker Micro-TOF (ESI) spectrometer and data was analyzed by use of Data Analysis software (Brücker) [39–41, 44].
Iron quantitation [37, 45]
Holo protein (50 μM, 200 μL) was acidified by concentrated HC1 (60 μL) and heated to 100 °C for 15 min. The resulting suspension was centrifuged at 14,000 rpm for 2 min and the supernatant (100 μL) was diluted with Tris-HCl (0.5 M, 1.3 mL, pH 8.5). Solutions of sodium ascorbate (0.1 mL, 5%) and bathophenanthroline-disulfonate (0.4 mL, 0.1%) were sequentially added to the neutralized reaction solution with mixing between each addition. The solution was incubated at 25 °C for 1 h and iron was quantitated by measuring the absorbance at 535 nm on a UV-vis spectrophotometer and calculated from a calibration curve made with 0.01 to 0.30 mM FeCl3 standard solutions. Subsequent analysis of the pellet revealed no bound iron.
Cluster transfer experiments monitored by circular dichroism
CD spectra were recorded using a Jasco J815 spectropolarimeter. The time course for cluster transfer from holo iron-sulfur cluster proteins to apo proteins was monitored under anaerobic conditions at 25 °C using UV-vis CD spectroscopy in small volume 1-cm quartz cuvettes. CD scans from 600 to 300 nm were collected to analyze signature peaks from cluster-bound protein at a scan rate of 200 nm/min with a 2 min interval between accumulations. Reactions were carried out in 50 mM HEPES, 100 mM NaCl, pH 7.5, and were prepared by degassing a mixture of apo protein in 5 mM DTT. The reaction was transferred to an anaerobic cuvette via a gas tight syringe and was initiated via the addition of degassed holo protein. The concentrations of [2Fe-2S] cluster for each holo protein were determined via iron quantitation following reconstitution, and are specified for each of the transfer reactions analyzed in the figure legends. Data were processed by use of JASCO Spectramanager II Analysis software and analyzed using the chemical kinetics and equilibria program DynaFit by BioKin [2], The deconvolution function from Spectramanager II analysis software was used for analysis of bands in the spectra that contained overlapping Lorentzian curves having the same full width at half maximum value that accurately distinguishes the peak positions for each band.
A second-order rate constant for cluster transfer was obtained via a method similar to the iron-sulfur cluster transfer method of Johnson and colleagues [43, 46], Proteins were mixed in a 1:1 ratio (20–100 μM) and the peak at 330 nm for ISCU, 445 nm for FDX1 and 440 nm for FDX2 was monitored by CD. The data were converted to percent cluster transfer. The percentage of cluster transferred was determined from a set of control spectra of the holo proteins at a series of known concentrations recorded after reconstitution, and the plot of percent cluster transfer versus time was analyzed using the DynaFit program [2] to determine the second-order rate constants for the various reactions by best-fit simulation to second-order kinetics.
Results and Discussion
BOLA proteins, especially the cytosolic BOLA2, have been examined for their ability to form heterocomplexes for the transfer and delivery of Fe-S clusters to target proteins [21–25], To date, only heterodimeric complexes of BOLA-type proteins with GLRX/Grx proteins have been studied, with the experimental reconstitution of homodimeric human BOLA2 unsuccessful [24], Similar in vitro attempts of reconstitution of the mitochondrial BOLA proteins have not been described in literature; however, we examined the possibility of forming human holo BOLA3 to further characterize its cluster binding properties.
Herein, we describe the successful reconstitution by use of either ferric chloride and sodium sulfide, or iron chloride, L-cysteine, and Tm NifS. Both methods provided similar UV spectra, with charge-transfer bands at 330 and 420 nm, and therefore, only a representative holo spectrum is shown (Figure 1). Moreover, the EPR spectrum obtained for reduced holo BOLA3 (Figure 2) is very similar to that obtained for the [2Fe-2S]+ bridged BOLA3-GLRX5 heterocomplex, both as described herein and in a prior report [1], Temperature dependent EPR measurements are also indicative of a [2Fe-2S]+ cluster [47], as typically [4Fe-4S]+ clusters show an almost exponential decay of signal amplitude with an increase in temperature after reaching a maxima around 10–20 K. [48–50]
Figure 1.
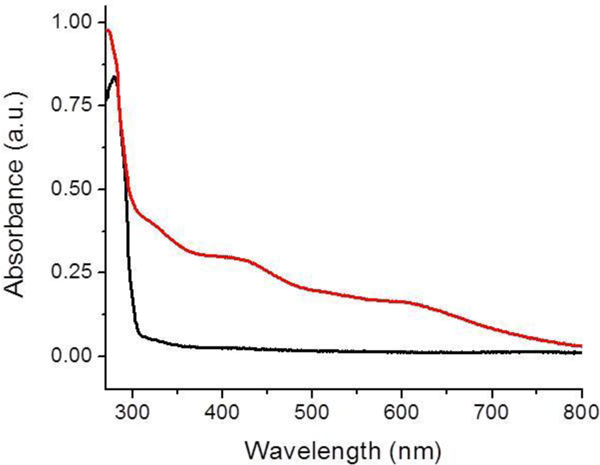
UV spectra of apo BOLA3 (black) and holo BOLA3 reconstituted using ferric chloride, L-cysteine and Tm NifS (red). Charge-transfer bands indicative of a bound Fe-S cluster are present at 330 and 420 nm.
Figure 2.
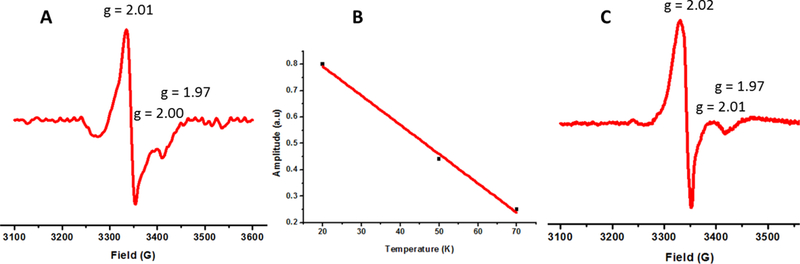
(A) EPR spectrum of holo BOLA3 after dithionite reduction and rapid freezing. (B) Temperature dependence of the EPR signal amplitude for holo BOLA3. (C) EPR spectrum of the BOLA3-GLRX5 holo heterodimer after dithionite reduction and rapid freezing. It is similar to a previously reported spectrum [1] and to the spectrum obtained for holo BOLA3 shown in (A).
Holo BOLA3 did not demonstrate a CD spectrum in the 300–600 nm range, even over a ten-fold concentration range from 40 to 150 μM (data not shown), suggesting that the cluster chromophore is CD silent in this range. BOLA proteins are known to form a [2Fe-2S] cluster-bridged heterodimeric complex with monothiol glutaredoxins and utilize coordination from His (common for all known heterocomplexes) and a cysteine or an undefined N/O ligand [1, 21, 23–25], While [2Fe-2S] cluster-bridged heterodimeric complexes have a well-defined CD signature [23, 26, 51], the low chirality in the local cluster environment of holo BOLA3 results in the absence of any CD signature in the 300–600 nm range, where characteristic ligand-to-metal charge transfer transitions appear in many iron-sulfur cluster proteins. We have previously discussed how the circular dichroic effect reflects local symmetry around the cluster chromophore and low levels of chirality can result in the absence of such a CD signature in some [2Fe-2S] containing proteins, including yeast Grx3 and human GLRX5 when reconstituted by non-GSH thiol sources, and the CD silence of the symmetric glutathione-complexed cluster [52, 53] and other synthetic models of [2Fe-2S] clusters. Nevertheless, reconstitution of human BOLA3 resulted in a cluster reconstitution yield of almost 70%, as determined by iron quantitation, which is in the range typically obtained for chemical reconstitution of Fe-S cluster proteins [40, 41, 43],
Since a holo form of human BOLA3 has not yet been characterized in the absence of a partner Fe-S cluster protein, we next sought to examine the oligomeric state for cluster binding by use of analytical ultracentrifugation (AUC), which has been used for characterization of other Fe-S cluster proteins [41, 54], This would address the question of whether cluster is bound by monomeric BOLA3 or bridged between multimers. AUC of apo BOLA3 demonstrated a mix of oligomeric states (Figure 3), with a monomer as the major species, although dimeric and tetrameric forms were also observed. The holo protein was unstable under the longer and more demanding experimental conditions of the AUC experiment (6 h), resulting in a sample with primarily precipitated protein. However, the holo form was relatively stable for the duration of size exclusion HPLC, which was employed to analyze the oligomeric states present in reconstituted holo BOLA3. Two wavelengths, 218 nm for total protein content and 420 nm for cluster content, were monitored for reconstituted holo protein and compared with that of apo. Since the extinction coefficient at 420 nm is much lower than that for protein backbone absorbance at 218 nm, the traces at these two wavelengths were scaled (between 0 and 1) for analysis (Figure S2). The 218 nm absorbance for the reconstituted BOLA3 shows two distinct peaks, one for the dimer (holo) and one for monomer, which most likely reflects residual apo protein because it correlates with the one major peak observed for apo protein. A 218 nm absorbance peak was observed for apo (red trace), while the 218 nm and 420 nm absorbance traces for holo (blue trace) match at around 5 min confirming a higher order dimeric/tetrameric state. AUC experiments demonstrate a fraction of apo BOLA3 to exist as a tetrameric species (Figure 3), and so a tetrameric solution component of holo BOLA3 is also likely present as a dimer of dimers, as observed for [2Fe-2S]-bridged holo GLRX5 [55],
Figure 3.
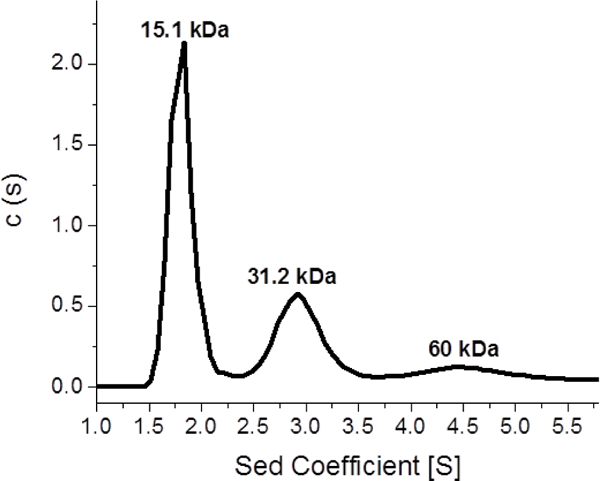
AUC for the determination of the oligomeric states of apo human BOLA3, monitored at 280 nm. The first peak of the black trace at 15.1 kDa accounts for 54% of the sample, the second peak at 31.2 kDa accounts for 30%, and the third peak at 60 kDa corresponds to 14% of the sample. The AUC results were obtained by fitting the sedimentation data to the Lamm equation [3, 4] using a continuous distribution model.
Aside from the UV-vis spectrum, additional evidence for a [2Fe-2S]-bridged homodimeric complex stems from the characteristic EPR signature described earlier, and the presence of the generally-accepted Cys/His ligand pair implicated in formation of cluster-bridged heterodimer species. Formation of monomeric holo BOLA3 is improbable because of the absence of other reasonable candidate ligands, aside from the sole cysteine (Cys59) and histidine (His96) residues implicated in Fe-S cluster binding in heterodimeric complexes [21–25], While a second histidine residue is located near the C-terminus of BOLA3, opposite the cluster binding site [23], it is structurally prevented from participating in binding, and is not conserved across other BOLA3 homologs. All known BOLA-type proteins have an invariant histidine residue that coordinates a bridging [2Fe-2S] cluster of BOLA-Grx heterocomplexes [1, 26, 51, 56], and a recent study with [2Fe-2S]-bridged BOLA3-GLRX5 established Cys59 (the only Cys in BOLA3) as the other ligand [1], The EPR spectrum obtained for holo BOLA3 is similar to that for the holo BOLA3-GLRX5 heterocomplex (Figure 2), supporting a similarity in coordination environments.
To further explore the cluster coordination environment, both C59A and H96A variants of BOLA3 were prepared. Each derivative was readily reconstituted by standard reconstitution protocols and display EPR features similar to native (Figure S3). Clusters were also readily transferred to apo acceptors such as FDX2 with rate constants comparable to native (2200 M−1 cm−1 for native BOLA3 vs 1820 M−1 cm−1 for C59A, Figures S4 and S5). Prior studies with [2Fe-2S] BOLA3-GLRX5 heterodimer have reported successful reconstitution with similar mutants, demonstrating that neighboring alternate residues can participate in cluster binding [1], For example, BOLA3-GLRX5 demonstrates Cys59 and His96 as cluster ligands that were alternately taken up by an unknown O/N ligand in C59A and GSH coordination in H96A [1], Furthermore, reported studies with holo BOLA3 have demonstrated very rapid formation of the holo BOLA3-GLRX5 heterocomplex following addition of apo GLRX5 [30], and so it is unlikely that the BOLA3-derived cluster-binding residues would be different in the homo- and heterodimeric forms. The double variant C59A/H96A could not be reconstituted under similar reconstitution conditions, confirming that the Cys59 and His96 are indeed important residues for cluster coordination.
To eliminate the possible involvement of DTT as an ancillary ligand in holo native BOLA3 we have also performed a sulfide-mediated reconstitution of BOLA3 in the absence of any DTT, with the apo protein pre-reduced with TCEP. Such reconstitution also results in a holo BOLA3 that can transfer its cluster to a [2Fe-2S] cluster acceptor such as FDX1 (Figure S6), similar to reconstitutions conducted with DTT. This rules out any role of DTT in cluster coordination. This combined body of evidence leads us to propose that the cluster-coordinating residues in holo homodimer native BOLA3 are most likely the Cys59 and His96 pairs from each protein subunit in a symmetric fashion. Following substitution, the C59A and H96A derivatives result in alternate cluster coordination from a nearby serine, S56, or alternate residue while the double variant C59A/H96A cannot bind cluster.
We have previously demonstrated that a glutathione-complexed [2Fe-2S] cluster, [2Fe-2S](GS)4, can efficiently and selectively reconstitute a variety of Fe-S cluster proteins, and this complex can be formed via GSH extraction from holo proteins [40, 41, 57], in particular from the scaffold protein ISCU, and serve as a substrate for the ABC transporter Atmlp for cluster export from the mitochondria [58, 59], Reconstitution of BOLA3 by this complex was attempted, but no significant uptake of cluster was observed (data not shown). However, glutathione was found to extract the cluster from chemically reconstituted BOLA3 (Figure 4), with the absorbance at 420 nm exhibiting a decay, relative to the control experiment performed in the absence of glutathione, in line with our reported results for other Fe-S cluster proteins [60].
Figure 4.
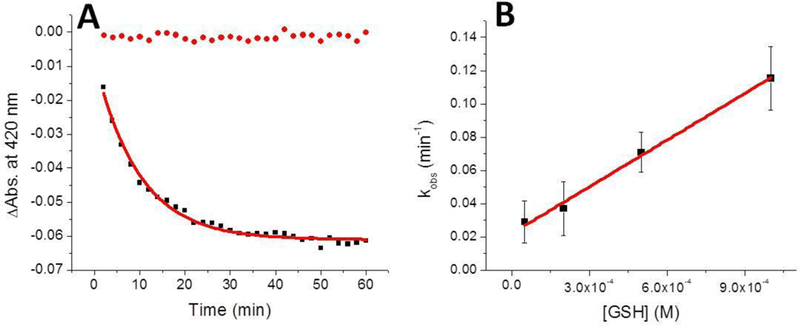
Extraction of the [2Fe-2S] cluster, by use of excess glutathione, from reconstituted holo human BOLA3 (15 μM in cluster) to form the [2Fe-2S](GS)4 complex, monitored by UV-Vis at 420 nm. (A) Stability of the holo protein was examined in the absence of glutathione as a control (red), while the experimental time-dependence for cluster extraction in the presence of 1 mM GSH is shown in black. For the latter fit, the equation includes both exponential and linear terms, where the linear term accommodates trace background cluster degradation, to yield a kobs for each concentration of GSH examined. (B) The concentration of GSH was plotted against the k0bs and fit linearly to obtain a second-order rate constant of 93 ± 5 M−1 min−1.
The BOLA3 holo homodimer is also functional in cluster transfer reactions, with the ability to deliver a [2Fe-2S] cluster to a variety of [2Fe-2S] cluster target proteins. Because monothiol glutaredoxins such as GLRX3 and GLRX5 are known to form a cluster-bridged heterodimeric species with BOLA3, these were not included in our investigation to identify probable acceptor proteins that are able to take up cluster from a BOLA3 homodimer. Direct and quantitative cluster transfer was observed for the human ferredoxins, ISCU, and GLRX2 (a dithiol glutaredoxin) with relatively large second-order rate constants (Table 1). Cluster transfer from holo BOLA3 was monitored under anaerobic conditions using CD, and spectra were recorded every 2 min over the course of 80 min for the cluster transfer reaction with the ferredoxin proteins to reach completion (Figure 5). Holo BOLA3 was able to deliver a [2Fe-2S] cluster to apo ferredoxin 1 with a second-order rate constant of 4,800 ± 1,800 M−1min−1, which is approximately two-fold faster than transfer to apo FDX2 with a second-order rate constant of 2,200 ± 430 M−1 min−1 (Figure 5; Table 1), and indicates a slight preference for delivery to ferredoxin 1 over ferredoxin 2. However, both of these rate constants are in line with those that we have previously observed for [2Fe-2S] cluster transfer to the apo human ferredoxin proteins from other Fe-S cluster proteins, such as NFU1 and ISCU [41, 61].
Table 1.
Second-order rate constants for cluster uptake and delivery involving BOLA3.
| Second-Order Rate Constant (M−1min−1) | |
|---|---|
| human BOLA3 to human FDX1 | 4,800 ± 1,800 |
| human BOLA3 to human FDX2 | 2,200 ± 430 |
| human BOLA3 to human GLRX2 | 4,150 ± 600 |
| human BOLA3 to human ISCU | 11,100 ± 330 |
| human BOLA3 to GSH | 93 ± 5 |
| human GLRX2 to human BOLA3 | no transfer |
| human ISCU to human BOLA3 | no transfer |
Figure 5.
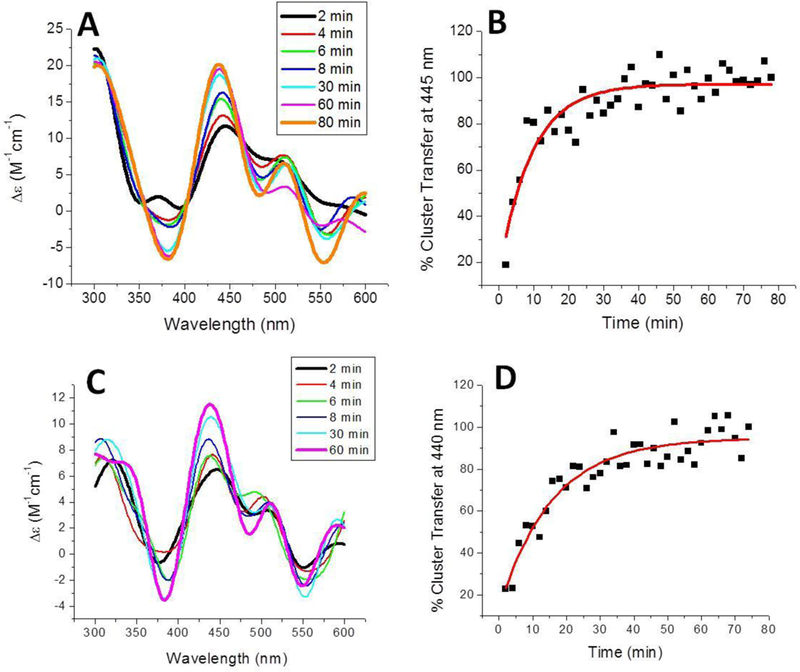
Time course of [2Fe-2S] cluster transfer from holo human BOLA3 to apo human ferredoxins. (A) Cluster transfer to apo ferredoxin 1 in 50 mM HEPES, 100 mM NaCl, pH 7.5. Spectra were recorded every 2 min after the addition of holo BOLA3, which was added to yield a final protein concentration ratio of 1:1, and converted to percent cluster transfer (B) to calculate the second-order rate constant of 4800 ± 1800 M1 min’1 using the DynaFit simulation software [2], based on the concentration of the [2Fe-2S] cluster. (C) Cluster transfer to apo ferredoxin 2 was monitored in the same way. (D) CD signal was again converted to percentage of cluster transferred to calculate the second-order rate constant of 2200 ± 430 M−1 min−1 using the DynaFit simulation software [2], based on the concentration of the [2Fe-2S] cluster.
Successful transfer reactions were also observed when reconstituted holo BOLA3 was added to either apo glutaredoxin 2 (GLRX2) or apo ISCU. Cluster transfer to apo GLRX2 was monitored over the course of an hour at 360 nm and exhibited a second-order rate constant of 4,150 ± 600 M−1 min−1 (Figure 6C and Table 1), which is again similar to previous rate constants determined from [2Fe-2S] cluster transfer from human NFU1 or S. pombe Isal [41, 61], CD signal changes were also observed for this transfer reaction in the 500–600 nm region but are weaker than the data obtained at 360 nm that was used for analysis.
Figure 6.
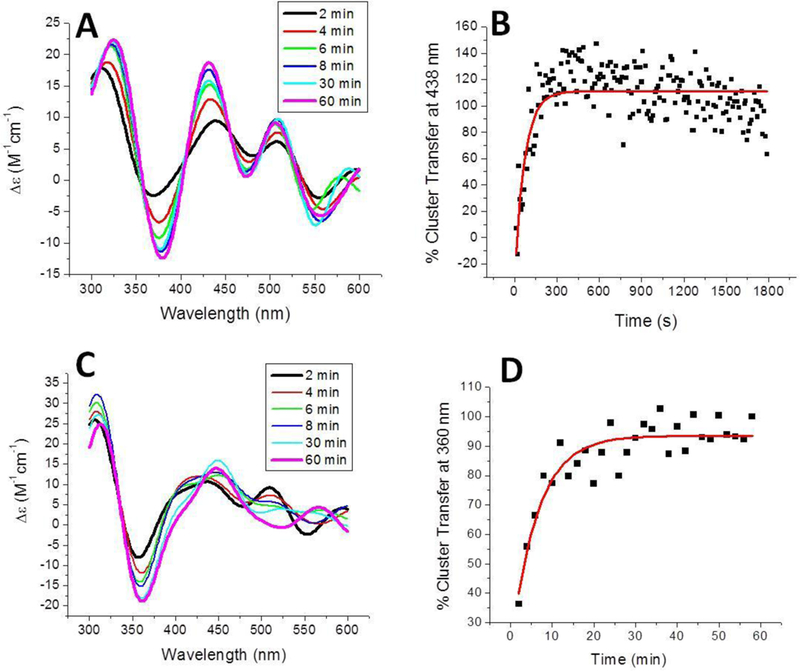
Time course of [2Fe-2S] cluster transfer from holo human BOLA3 to additional mitochondrial target proteins. (A) Cluster transfer to apo human ISCU in 50 mM HEPES, 100 mM NaCl, pH 7.5. Spectra were recorded every 2 min after the addition of holo BOLA3, which was added to yield a final protein concentration ratio of 1:1, and converted to percent cluster transfer (B) to calculate the second-order rate constant of 11,100 ± 330 M−1 min−1 using the DynaFit simulation software [2], based on the concentration of the [2Fe-2S] cluster. (C) Cluster transfer to apo human glutaredoxin 2 was monitored in the same way. (D) CD signal was again converted to percentage of cluster transferred to calculate the second-order rate constant of 4,150 ± 600 M−1 min−1 using the DynaFit simulation software [2], based on the concentration of the [2Fe-2S] cluster.
Because recent reports suggest a possible influence for DTT in promoting cluster transfer [62], we addressed this possibility by performing reactions over a ten-fold range of DTT concentrations (0.5 to 5 mM) for cluster transfer from holo BOLA3 to apo FDX2 and confirmed that the transfer is independent of DTT concentration in the concentration range tested (Table S1), and consistent with the absence of any DTT effect from the work that we and others have reported [26, 63],
In the case of cluster transfer to ISCU, a significantly more rapid change was observed, such that the signature ISCU peak was monitored over 10 s intervals (Figures 6A and 6B) to yield a second-order rate constant of 11,100 ± 330 M−1min−1 (Table 1). Specifically, the cluster transfer reaction to apo ISCU is the most rapid reconstitution mechanism for ISCU that we have observed to date [40, 61], Furthermore, aside from the [2Fe-2S](GS)4 complex, holo homodimeric BOLA3 is the first component of the human mitochondrial system that we have found to be capable of providing a cluster to ISCU. This complicates determination of the precise role of BOLA3 in Fe-S cluster trafficking and suggests that it may be playing a multifunctional and varied role in cluster biogenesis pathways.
Interestingly, transfer of a [2Fe-2S] cluster from the various mitochondrial donors that were examined proved unsuccessful in promoting the formation of the holo BOLA3 homodimer. Neither holo human ISCU nor holo human GLRX2 demonstrated a change in CD spectra when added to apo BOLA3 over the typical time frame examined (data not shown), suggesting that these clusters were not transferred from the holo donor to form the CD silent holo BOLA3 homodimer. BOLA proteins have a demonstrated precedence to form heterodimeric complexes, particularly with monothiol glutaredoxins [21, 23–25], However, in this paper we have demonstrated that human BOLA3 can be reconstituted to form a functional holo homodimer that can transfer [2Fe2S] clusters to select target proteins.
An equilibrium concentration of holo dimer could have some functional utility in light of our prior reports [64, 65] that have revealed substantial redundancy in the functional roles of many of the proteins involved in Fe-S cluster biosynthesis and trafficking. The BOLA3 homodimer could possibly an ancillary role in providing alternative cluster trafficking pathways, either in disease states where preferred pathways become dysfunctional, or under conditions of cellular stress. Redundancy in functional roles provides a ready explanation for why so many deletions of “essential” Fe-S biosynthesis/trafficking proteins are not lethal.
The possible physiological role(s) of this holo form of BOLA3 and the overall implications for the role of BOLA3 in the molecular mechanisms of Fe-S cluster biosynthesis and trafficking require further investigation. While holo BOLA3 may not be an intrinsically physiologically active species it could be of potential relevance under disease conditions where MMDS-causing mutations could inactivate heterodimer formation while retaining activity for the homodimer. Studies to evaluate these ideas are currently underway
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health [AI072443].
Abbreviations
- GLRX5
(Glutaredoxin 5)
- (CD)
Circular Dichroism
- (DTT)
Dithiothreitol
- (FDX)
Ferredoxin
- (GSH)
Glutathione
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Nasta V, Giachetti A, Ciofi-Baffoni S and Banci L (2017) Biochim Biophys Acta 1861:2119–2131 [DOI] [PubMed] [Google Scholar]
- 2.Kuzmic P (1996) Anal Biochem 237:260–273 [DOI] [PubMed] [Google Scholar]
- 3.Lamm O (1929) Ark Mat Astr Fys: 1–4 [Google Scholar]
- 4.Schuck P (2000) Biophys J 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuss JO, Tsai C-L, Ishida JP and Tainer JA (2015) Biochim Biophys Acta 1853:1253–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DC, Dean DR, Smith AD and Johnson MK (2005) Annu Rev Biochem 74:247–281 [DOI] [PubMed] [Google Scholar]
- 7.Tort F, Ferrer-Cortes X and Ribes A (2016) J Inherit Metab Dis 39:781–793 [DOI] [PubMed] [Google Scholar]
- 8.Ajit Bolar N, Vanlander AV, Wilbrecht C, Van der Aa N, Smet J, De Paepe B, Vandeweyer G, Kooy F, Eyskens F, De Latter E, Delanghe G, Govaert P, Leroy JG, Loeys B, Lill R, Van Laer L and Van Coster R (2013) Hum Mol Genet 22:2590–2602 [DOI] [PubMed] [Google Scholar]
- 9.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA and Robinson BH (2011) Am J Hum Genet 89:486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stehling O, Wilbrecht C and Lill R (2014) Biochimie 100:61–77 [DOI] [PubMed] [Google Scholar]
- 11.Debray F-G, Stumpfig C, Vanlander AV, Dideberg V, Josse C, Caberg J-H, Boemer F, Bours V, Stevens R, Seneca S, Smet J, Lill R and van Coster R (2015) J Inherit Metab Dis 38:1147–1153 [DOI] [PubMed] [Google Scholar]
- 12.Al-Hassnan ZN, Al-Dosary M, Alfadhel M, Faqeih EA, Alsagob M, Kenana R, Almass R, Al-Harazi OS, Al-Hindi H, Malibari OI, Almutari FB, Tulbah S, Alhadeq F, Al-Sheddi T, Alamro R, AlAsmari A, Almuntashri M, Alshaalan H, Al-Mohanna FA, Colak D and Kaya N (2015) J Med Genet 52:186–194 [DOI] [PubMed] [Google Scholar]
- 13.Baker PR, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, Freckmann ML, Shih LY, Wasserstein M, Rasmussen MB, Lund AM, Procopis P, Cameron JM, Robinson BH, Brown GK, Brown RM, Compton AG, Dieckmann CL, Collard R, Coughlin CR, Spector E, Wempe MF and Van Hove JLK (2013) Brain 137:366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lossos A, Stümpfig C, Stevanin G, Gaussen M, Zimmerman B-E, Mundwiller E, Asulin M, Chamma L, Sheffer R, Misk A, Dotan S, Gomori JM, Ponger P, Brice A, Lerer I, Meiner V and Lill R (2015) Neurology 84 [DOI] [PubMed] [Google Scholar]
- 15.Navarro-Sastre A, Tort F, Stehling O, Uzarska Marta A., Arranz José A., del Toro M, Labayru MT, Landa J, Font A, Garcia-Villoria J, Merinero B, Ugarte M, Gutierrez-Solana Luis G., Campistol J, Garcia-Cazorla A, Vaquerizo J, Riudor E, Briones P, Elpeleg O, Ribes A and Lill R (2011) Am J Hum Genet 89:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lill R Muehlenhoff U(2008) Ann Rev Biochem 77:669–700 [DOI] [PubMed] [Google Scholar]
- 17.Maio N and Rouault TA (2015) Biochim Biophys Acta 1853:1493–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stehling O, Wilbrecht C and Lill R (2014) Biochimie 100:61–77 [DOI] [PubMed] [Google Scholar]
- 19.Banci L, Brancaccio D, Ciofi-Baffoni S, Del Conte R, Gadepalli R, Mikolajczyk M, Neri S, Piccioli M and Winkelmann J (2014) Proc Natl Acad Sci USA 111:6203–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brancaccio D, Gallo A, Mikolajczyk M, Zovo K, Palumaa P, Novellino E, Piccioli M, Ciofi-Baffoni S and Banci L (2014) J Am Chem Soc 136:16240–16250 [DOI] [PubMed] [Google Scholar]
- 21.Li H and Outten CE (2012) Biochemistry 51:4377–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM and Kaplan J (2008) J Biol Chem 283:10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzarska MA, Nasta V, Weiler BD, Spantgar F, Ciofi-Baffoni S, Saviello MR, Gonnelli L, Muehlenhoff U, Banci L and Lill R (2016) eLife 5:e15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banci L, Camponeschi F, Ciofi-Baffoni S and Muzzioli R (2015) J Am Chem Soc 137:16133–16143 [DOI] [PubMed] [Google Scholar]
- 25.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK and Outten CE (2009) Biochemistry 48:9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banci L, Camponeschi F, Ciofi-Baffoni S and Muzzioli R (2015) J Am Chem Soc 137:16133–16143 [DOI] [PubMed] [Google Scholar]
- 27.Frey AG, Palenchar DJ, Wildemann JD and Philpott CC (2016) J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzarska MA, Nasta V, Weiler BD, Spantgar F, Ciofi-Baffoni S, Saviello MR, Gonnelli L, Miihlenhoff U, Banci L and Lill R (2016) eLife 5:e16673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melber A, Na U, Vashisht A, Weiler BD and Lill R (2016) 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen S, Rao B, Wachnowsky C and Cowan JA (2018) Metallomics 10:1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster M, Mansy S, Hwang J, Penner-Hahn J, Surerus K and Cowan J (2000) J Am Chem Soc 122:6805–6806 [Google Scholar]
- 32.Mansy SS, Xiong Y, Hemann C, Hille R, Sundaralingam M and Cowan JA (2002) Biochemistry 41:1195–1201 [DOI] [PubMed] [Google Scholar]
- 33.Nuth M, Yoon T and Cowan JA (2002) J Am Chem Soc 124:8774–8775 [DOI] [PubMed] [Google Scholar]
- 34.Xia B, Cheng H, Bandarian V, Reed GH and Markley JL (1996) Biochemistry 35:9488–9495 [DOI] [PubMed] [Google Scholar]
- 35.Qi W, Li J and Cowan JA (2013) Dalton Trans 42:3088–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi W and Cowan JA (2011) Chem Commun 47:4989–4991 [DOI] [PubMed] [Google Scholar]
- 37.Wu S-p, Wu G, Surerus KK and Cowan JA (2002) Biochemistry 41:8876–8885 [DOI] [PubMed] [Google Scholar]
- 38.Schuck P (2000) Biophys J 78:1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF and Cowan JA (2012) J Am Chem Soc 134:10745–10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fidai I, Wachnowsky C and Cowan JA (2016) J Biol Inorg Chem 21:887–901 [DOI] [PubMed] [Google Scholar]
- 41.Wachnowsky C, Fidai I and Cowan JA (2016) J Biol Inorg Chem 21:825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandyopadhyay S, Naik SG, O’Carroll IP, Huynh BH, Dean DR, Johnson MK and Dos Santos PC (2008) J Biol Chem 283:14092–14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao H, Subramanian S, Couturier J, Naik SG, Kim S-K, Leustek T, Knaff DB, Wu H-C, Vignols F, Huynh BH, Rouhier N and Johnson MK (2013) Biochemistry 52:6633–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF and Cowan JA (2013) Chem Commun 49:6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moulis J-M and Meyer J (1982) Biochemistry 21:4762–4771 [DOI] [PubMed] [Google Scholar]
- 46.Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N and Johnson MK (2013) Dalton transactions (Cambridge, England : 2003) 42:3107–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cammack R and MacMillan F (2010) In: Hanson G and Berliner L (eds) Metals in biology: Applications of high-resolution epr to metalloenzymes. Springer New York, New York, NY, pp. 11–44 [Google Scholar]
- 48.Jung YS, Bonagura CA, Tilley GJ, Gao-Sheridan HS, Armstrong FA, Stout CD and Burgess BK (2000) J Biol Chem 275:36974–36983 [DOI] [PubMed] [Google Scholar]
- 49.Sweeney WV and Rabinowitz JC (1980) Annu Rev Biochem 49:139–161 [DOI] [PubMed] [Google Scholar]
- 50.Hsueh K-L, Yu L-K, Chen Y-H, Cheng Y-H, Hsieh Y-C, Ke S-c, Hung K-W, Chen C-J and Huang T-h (2013) J Bactenol 195:4726–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK and Outten CE (2009) Biochemistry 48:9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen S, Bonfio C, Mansy SS and Cowan JA (2018) J Biol Inorg Chem 23:241–252 [DOI] [PubMed] [Google Scholar]
- 53.Sen S and Cowan JA (2017) JBIC Journal of Biological Inorganic Chemistry 22:1075–1087 [DOI] [PubMed] [Google Scholar]
- 54.Wachnowsky C, Wesley NA, Fidai I and Cowan JA (2017) J Mol Biol 429:790–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansson C, Roos Annette K., Montano Sergio J., Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U and Kavanagh Kathryn L. (2011) Biochem J 433:303–311 [DOI] [PubMed] [Google Scholar]
- 56.Roret T, Tsan P, Couturier J, Zhang B, Johnson MK, Rouhier N and Didierjean C (2014) J Biol Chem 289:24588–24598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF and Cowan JA (2012) J Am Chem Soc 134:10745–10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J and Cowan JA (2015) Chem Commun 51:2253–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qi W, Li J and Cowan JA (2014) Chem Commun 50:3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fidai I, Wachnowsky C and Cowan JA (2016) J Biol Inorg Chem 21:887–901 [DOI] [PubMed] [Google Scholar]
- 61.Fidai I, Wachnowsky C and Cowan JA (2016) Metallomics 8:1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vranish JN, Russell WK, Yu LE, Cox RM, Russell DH and Barondeau DP (2015) J Am Chem Soc 137:390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banci L, Ciofi-Baffoni S, Gajda K, Muzzioli R, Peruzzini R and Winkelmann J (2015) Nat Chem Biol 11:772–778 [DOI] [PubMed] [Google Scholar]
- 64.Fidai I, Wachnowsky C and Cowan JA (2016) Metallomics 8:1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wachnowsky C, Fidai I and Cowan JA (2016) FEBS Lett 590:4531–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


