Abstract
BACKGROUND:
DNA methylation is implicated in coronary heart disease (CHD), but current evidence is based on small, cross-sectional studies. We examined blood DNA methylation in relation to incident CHD across multiple prospective cohorts.
METHODS:
Nine population-based cohorts from the United States and Europe profiled epigenome-wide blood leukocyte DNA methylation using the Illumina Infinium 450k microarray, and prospectively ascertained CHD events including coronary insufficiency/unstable angina, recognized myocardial infarction (MI), coronary revascularization, and coronary death. Cohorts conducted race-specific analyses adjusted for age, sex, smoking, education, body mass index, blood cell type proportions, and technical variables. We conducted fixed-effect meta-analyses across cohorts.
RESULTS:
Among 11,461 individuals (mean age 64 years, 67% women, 35% African-American) free of CHD at baseline, 1,895 developed CHD during a mean follow-up of 11.2 years. Methylation levels at 52 cytosine-phosphate-guanine (CpG) sites were associated with incident CHD or MI (false discovery rate<0.05). These CpGs map to genes with key roles in calcium regulation (ATP2B2, CASR, GUCA1B, HPCAL1), and genes identified in genome- and epigenome-wide studies of serum calcium (CASR), serum calcium-related risk of CHD (CASR), coronary artery calcified plaque (PTPRN2), and kidney function (CDH23, HPCAL1), among others. Mendelian randomization analyses supported a causal effect of DNA methylation on incident CHD; these CpGs map to active regulatory regions proximal to long non-coding RNA transcripts.
CONCLUSION:
Methylation of blood-derived DNA is associated with risk of future CHD across diverse populations, and may serve as an informative tool for gaining further insight on the development of CHD.
Keywords: coronary artery disease, coronary heart disease risk, epigenetics, genomics, gene regulation
INTRODUCTION
Coronary heart disease (CHD) is a major contributor to global morbidity and mortality.1 Despite substantial progress in CHD prevention, improved approaches are needed to further reduce CHD incidence. Methylation of DNA at cytosine-phosphate-guanine (CpG) dinucleotides is a stable yet environmentally responsive epigenetic regulatory mechanism. DNA methylation at a CpG site is dependent on both underlying genetic variation as well as exposures to environmental factors.2 In vitro and animal-based studies provide evidence that DNA methylation changes are involved in the development of CHD,3 and large-scale population-based studies have shown that risk factors for CHD including smoking,4 obesity,5 hypertension,6,7 serum lipids,8,9 and type-2 diabetes10 are linked to persistent differences in leukocyte DNA methylation. Hence DNA methylation, as a molecular bio-archive integrating genetic predisposition and risk factor exposures, may provide further insight on CHD development and identify novel modifiable pathways related to CHD. Prior studies of DNA methylation and CHD in humans11–15 have generally been small in sample size (e.g. n < 300), focused on repetitive elements11,13 or selective genomic regions,15 or have been cross-sectional or case-control in design.11,13–15 Whether blood leukocyte DNA methylation predicts future CHD has not been comprehensively investigated.
We conducted a longitudinal, large-scale, multi-cohort, epigenome-wide investigation of incident CHD events among 11,461 participants in the Cohorts for Heart and Aging Genetic Epidemiology (CHARGE) consortium.16 We first assessed whether leukocyte DNA methylation was associated with risk of CHD. We then combined information on the identified CHD-associated methylation sites with genetic sequence variation to provide an integrated genomic map reflecting CHD risk, and evaluated if there was evidence for causal effects of DNA methylation variation on incident CHD.
METHODS
Study design and population
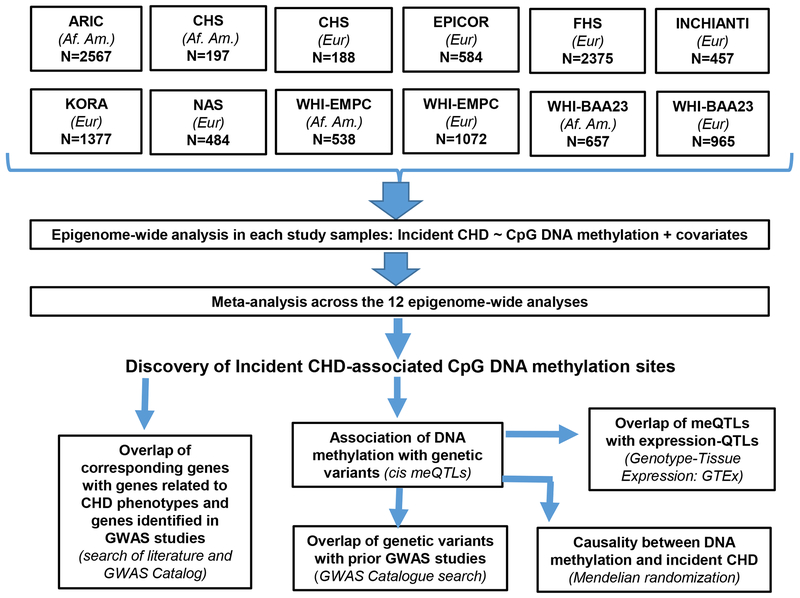
We selected cohorts participating in the CHARGE Consortium in which genome-wide leukocyte DNA methylation was assessed using the Infinium 450k microarray, and CHD events were prospectively ascertained. Nine population-based cohorts comprising a total of 11,461 participants from the United States and Europe were included: the Atherosclerosis Risk in Communities Study (ARIC), Cardiovascular Health Study (CHS), long-tErm follow-up of antithrombotic management Patterns In acute CORonary syndrome patients (EPICOR), the Framingham Heart Study (FHS), the Invecchiare in Chianti study (InCHIANTI), the Kooperative Gesundheitsforschung in der Region Augsburg study (KORA), the Normative Aging Study (NAS), the Women’s Health Initiative “Epigenetic Mechanisms of Particulate Matter-Mediated CVD” (WHI-EMPC) ancillary study, and the “Integrative genomics and risk of CHD and related phenotypes in the Women’s Health Initiative” (WHI-BAA23) Ancillary study (Detailed information in Methods S1). Each cohort study obtained informed consent from participants and protocol approval from its respective institutional review board and ethics committee. DNA methylation data was only collected for African-Americans of the ARIC cohort, from the Jackson, MS and Forsyth County, NC study sites of the cohort. Cohorts comprising participants of both African-American and European Ancestry were analyzed in a race-specific manner. Accordingly, we performed an epigenome-wide analysis within each of the 12 study samples, and then meta-analyzed the resulting summary statistics from the 12 analyses. We also examined the association between DNA methylation and cis-genetic variants (±500 KB) in a subset of the cohorts and conducted Mendelian Randomization to evaluate potential causal relations between DNA methylation and incident CHD (Figure 1). .
Figure 1.
Overall analytic workflow. Af. Am. denotes individuals of African-American ancestry; Eur denotes individuals of European ancestry. meQTLs: methylation quantitative trait loci.
Data and Materials
The DNA methylation datasets from ARIC and CHS data can be requested from the corresponding author. EPICOR data are available upon request from HuGeF a project agreement; requests should be sent to info@hugef-torino.org. The FHS DNA methylation datasets are available from the dbGAP repository: phs000724. The genotype datasets are available from the dbGAP repository: phs000007. The InCHIANTI data are available on request from the corresponding author. The KORA data can be requested at KORA Project Application Self-Service Tool (PASST) from the Helmholtz Zentrum München German Research Center for Environmental Health. The NAS DNA methylation datasets are available at the dbGAP repository: phs000853. The WHI-BAA23 DNA methylation dataset is available at dbGAP repository phs001335. WHI-EMPC data are available on request from the WHI website or the corresponding author.
Measurement of DNA methylation
For all cohorts, DNA was extracted from whole blood samples and bisulfite-converted using a Zymo EZ DNA methylation kit. The Illumina Infinium Human Methylation450K BeadChip (Illumina Inc, San Diego, CA, USA) was used to measure DNA methylation. Quality control, filtering, and normalization of the methylation data were independently conducted for each cohort according to established criteria4,6 and other diagnostics unique to the cohort (details in Supplemental Methods). For each CpG, methylation = M/(M+U+ε), where M and U are the average fluorescence intensity from the probe (i.e., the oligonucleotide that hybridizes to the target CpG) corresponding to the methylated (M) and unmethylated (U) target CpG, respectively, and ε=100 to protect against division by zero. Therefore, the methylation at each CpG is contained in the interval 0-1, with 0 indicating no methylation and 1 indicating 100% methylation at the target CpG across DNA from cells in the sample.
Definition of coronary heart disease (CHD) and myocardial infarction (MI) events
Our primary outcome of interest was incident CHD, defined as any of the following: recognized nonfatal or fatal MI (hospitalization with diagnostic electrocardiographic (ECG) changes and/or biomarkers of MI), coronary insufficiency/unstable angina, coronary revascularization, or coronary death. We also conducted a secondary meta-analyses restricted to incident MI-only (recognized nonfatal or fatal MI), in order to evaluate whether analysis with this more homogenous outcome measure supported robustness of the results.
STATISTICAL ANALYSES
Individual study epigenome-wide analyses
Baseline was defined as the time of blood sampling for DNA methylation assays, and all cohorts excluded individuals with prevalent CHD at baseline. Seven cohorts conducted time-to-event analyses using Cox proportional hazard models, and three of these cohorts adapted Firth’s penalized Cox regression17 due to a low number of CHD events. Two prospective cohorts, EPICOR and WHI-BAA23, employed a nested case-control design with incident CHD events and performed logistic regression analyses, which—under specific assumptions—provide risk estimates that are unbiased in relation to the estimates derived from Cox regression.18,19 All analyses were race-specific, and adjusted for age, sex, body mass index (kg/m2), smoking status (current, former, never), education (as years of education or categorical levels of school degrees completed), differential cell counts,20 family structure (if present), and batch-related technical variables (Supplemental Methods).
Meta-analysis
We performed an inverse variance-weighted fixed-effects meta-analysis using the metafor package in R. The fixed-effects method is standard practice in genome-wide studies21,22 and has been consistently used in prior large-scare epigenome-wide studies in CHARGE and other consortia.4,6 It is well-documented that using the Random-effects approach leads to substantially diminished power,23 which has major implications for such genome-wide meta-analysis studies that are more exploratory than standard epidemiological studies and are aimed at uncovering new loci previously not possible. However, we also include, as sensitivity analyses, results with random-effects models (described further in the results section). We accounted for multiple-testing by controlling the false discovery rate (FDR) at 5%. Of the CpGs that exceeded this a priori multiple-testing threshold, we excluded CpGs where the CpG probe sequence harbored a single nucleotide polymorphism [SNP] assayed in the 1000 Genomes Project with a minor allele frequency >0.01 (given that the frequency of underlying genetic variation differs between race/ancestry groups, we excluded CpGs with the potential for underlying SNPs specific to that cohorts race/ancestry), and CpGs that had high inter-study heterogeneity assessed using Cochran’s Q test (Q <0.05).
Identification of associated genetic variants and Mendelian Randomization analyses
We investigated whether genetic variants within ±500 kb (cis) of the incident CHD- and MI-associated CpGs contributed to variation in methylation levels, i.e., were methylation-quantitative trait loci (meQTLs). The discovery analysis was conducted on 3,868 individuals from the FHS, followed by replication in 1,731 individuals from KORA. Genotyping was conducted with the Affymetrix 500K and MIPs 50K platforms in FHS, and the Affymetrix Axiom array in KORA, and imputation was performed using the 1000 Genomes reference panel in both cohorts. meQTLs detected at P <1x10−4 in the discovery stage, followed by P<bonferroni threshold (i.e. P<0.05/number of significant discovery stage meQTLs) at the replication stage were selected for conducting MR using a two-sample instrumental variable approach (implemented with MRbase24) to infer causal relations between DNA methylation and incident CHD. Single strongest cis-meQTLs were utilized: 1) to minimize potential of horizontal pleiotropy, and 2) due to lack of sufficient meQTLs from independent loci in low linkage disequilibrium. Genotype associations for CHD and MI were obtained from the CARDIoGRAMplusC4D 2015 GWAS (n= 60,801 cases and n= 123,504 controls).25 We used the meQTLs for differential methylation at CpGs that were significant for a causal relationship between DNA methylation and incident CHD in Mendelian randomization analyses (p < 0.05) for further inquiry on putative effects on gene expression, by overlapping meQTLs with expression-QTLs from the Genotype-Tissue Expression (GTEx) resource (The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 11/25/2018 and 02/10/2019).
RESULTS
Participant characteristics
Among the 11,461 participants, mean age at baseline was 64 years, 67% were female, and 35% were of African-American ancestry (Table 1). During a mean follow-up of 11.2 years, a total of 1,895 CHD events and 1,183 MI events occurred.
Table 1.
Demographic characteristics of participants across the nine cohorts included in the study
| Total N | No. CHD events | No. MI- only events | Mean age (SD) | Female (%) | Smoking status %former;%current | Mean BMI (SD) | Mean years of education or % >HS | |
|---|---|---|---|---|---|---|---|---|
| ARIC-Af. Am. | 2567 | 389 | 233 | 56.5 (5.8) | 64% | 30%;25% | 30.1 (6.3) | 13.3 (5.2) |
| CHS-Af. Am. | 197 | 57 | 26 | 72.9 (5.4) | 66% | 33%;50% | 28.8 (5.0) | 11.4 (3.7) |
| CHS-White | 188 | 54 | 26 | 76.1(5.1) | 55% | 41%;13% | 27.1 (4.9) | 12.5 (3.2) |
| EPICOR*† | 584 | 292 | 292 | 52.9 (7.4) | 36% | 33%;31% | 26.6 (3.8) | 11.3 (5.0) |
| FHS | 2375 | 116 | 70 | 65.8 (8.8) | 57% | 55%;8% | 28.1 (5.3) | 14.3 (2.6) |
| InChianti† | 457 | 50 | 50 | 62.1 (15.8) | 53% | 24%;19% | 27.1 (3.9) | 12% |
| KORA† | 1377 | 32 | 32 | 54.2 (8.8) | 51% | 37%;17% | 27.7 (4.5) | 11.4 (2.3) |
| NAS | 484 | 102 | 50 | 72.0 (7.1) | 0% | 65%;4% | 28.0 (4.1) | 15.2 (2.9) |
| WHI (EMPC‡)-Af. Am. | 538 | 38 | -- | 62.7 (7.0) | 100% | 40%;10% | 31.5 (6.0) | 62% |
| WHI (EMPC‡)-Eur, | 1072 | 88 | 19 | 64.7 (7.1) | 100% | 41%;8% | 28.8 (5.9) | 62% |
| WHI (BAA23*‡)-Af. Am. | 657 | 254 | 112 | 62.9 (6.7) | 100% | 51%; 2% | 31.8 (6.6) | 28% |
| WHI (BAA23*‡)-Eur. | 965 | 423 | 273 | 68.3 (6.3) | 100% | 45%;0% | 28.8 (5.7) | 30% |
Abbreviations: Af. Am.: African American ancestry; BMI, body-mass index; CHD, coronary heart disease; Eur: European ancestry; HS: high school; MI, myocardial infarction; SD, standard deviation.
EPICOR and WHI-BAA23 had a case-control design nested within their respective prospective cohorts; all cases and controls were adjudicated during follow-up
EPICOR, InChianti, and KORA only contributed MI events to the meta-analyses; thus, all CHD events are MI by definition, and the numbers of events in columns 2 and 3 are the same
EMPC refers to the WHI ancillary study “Epigenetic Mechanisms of Particulate Matter-Mediated CVD”; BAA23 refers to another sample of WHI in the substudy “Integrative genomics and risk of CHD and related phenotypes in the Women’s Health Initiative”
The dashed line indicates that WHI (EMPC)-Af. Am. was not included in the MI meta-analyses because the number of MI-only cases was too low (n <10)
Association of DNA methylation with risk of Coronary heart disease (CHD)
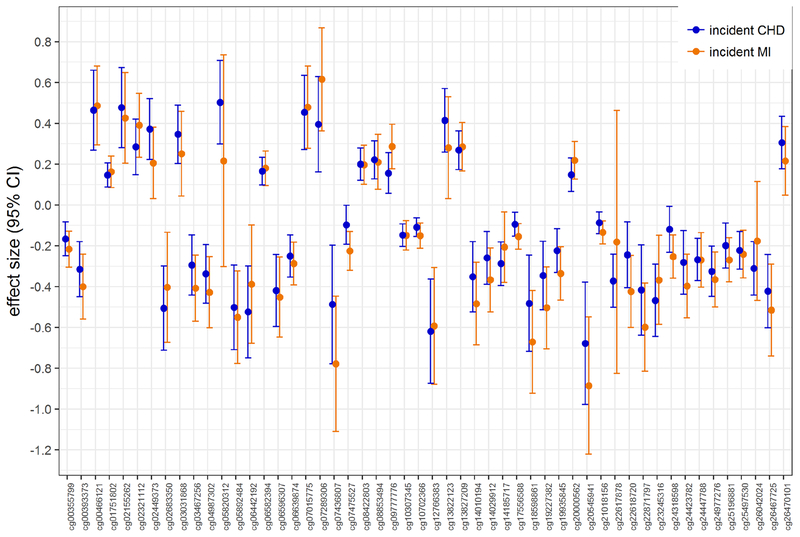
Among 442,192 CpGs analyzed, methylation levels at 30 CpGs were associated (FDR <0.05) with incident CHD (Table 2; individual forest plots for each CpG in Supplemental Figure 1), after excluding CpGs with underlying SNPs that could interfere with probe binding (n=7) and CpGs that demonstrated substantial heterogeneity (Q < 0.05) in the meta-analyses (n=8). Methylation levels at 29 CpGs were associated with our secondary outcome of incident MI at a FDR <0.05 (Table 3; individual forest plots for each CpG in Supplemental Figure 2), after similarly excluding CpGs with underlying SNPs (n=4) and high heterogeneity (n=5). Additional genomic information on these CpGs are provided in Supplemental Tables 1 and 2. Among these 30 and 29 CpGs identified in the incident CHD and incident MI-only meta-analyses, respectively, seven CpGs met the FDR<0.05 threshold in both analyses, resulting in 52 unique CpGs identified across the two meta-analyses. We found that the direction, magnitude, and precision of estimated effects for these 52 CpGs were highly concordant when comparing results from the two different meta-analyses (Figure 2). Manhattan plots indicated that significant associations were distributed across the genome (Supplemental Figure 3). Neither meta-analysis was strongly influenced by inflation from technical or batch effects, and both had a uniform distribution of p-values and symmetry in the coefficient direction of effect (Supplemental Figure 3). As results obtained from the secondary, incident MI-only meta-analysis did not materially differ from the primary, CHD meta-analysis, we henceforth combined the results from the two meta-analyses and simply refer to all 52 CpGs as CHD-associated CpGs.
Table 2.
DNA methylation at 30 CpG sites associated with the risk of coronary heart disease (CHD).* CpGs with false discovery rate <0.05 in are shown (Bonferroni-significant sites in bold).
| CpG name | beta coefficient† | Nominal p-value | Hazard Ratio (95% CI) | Gene‡ |
|---|---|---|---|---|
| cg22617878 | −0.3719 | 1.99E-08 | 0.69 (0.61,0.79) | ATP2B2 |
| cg13827209 | 0.2680 | 3.76E-08 | 1.31 (1.19,1.44) | TGFBR1 |
| cg14185717 | −0.2878 | 1.38E-07 | 0.75 (0.67,0.83) | BNC2 |
| cg10307345 | −0.1480 | 1.86E-07 | 0.86 (0.82,0.91) | PTPN5 |
| cg13822123 | 0.4138 | 2.03E-07 | 1.51 (1.29,1.77) | PSME4 |
| cg23245316 | −0.4674 | 2.17E-07 | 0.63 (0.53,0.75) | TSSC1 |
| cg24977276 | −0.3256 | 2.54E-07 | 0.72 (0.64,0.82) | GTF2I |
| cg24447788 | −0.2679 | 4.33E-07 | 0.76 (0.69,0.85) | (PTBP1**) |
| cg08422803 | 0.1994 | 7.52E-07 | 1.22 (1.13,1.32) | ITGB2 |
| cg01751802 | 0.1473 | 9.35E-07 | 1.16 (1.09,1.23) | KANK2 |
| cg02449373 | 0.3715 | 9.98E-07 | 1.45 (1.25,1.68) | FUT1 |
| cg02683350 | −0.5062 | 1.55E-06 | 0.60 (0.49,0.74) | ADAMTS2 |
| cg05820312 | 0.5031 | 1.60E-06 | 1.65 (1.35,2.03) | TRAPPC9 |
| cg06639874 | −0.2506 | 1.83E-06 | 0.78 (0.7,0.86) | MLPH |
| cg06582394 | 0.1657 | 1.90E-06 | 1.18 (1.1,1.26) | CASR |
| cg02155262 | 0.4770 | 1.97E-06 | 1.61 (1.32,1.96) | AGA |
| cg12766383 | −0.6194 | 1.98E-06 | 0.54 (0.42,0.69) | UBR4 |
| cg05892484 | −0.5020 | 2.01E-06 | 0.61 (0.49,0.74) | MAD1L1 |
| cg03031868 | 0.3461 | 2.29E-06 | 1.41 (1.22,1.63) | ESD |
| cg25497530 | −0.2225 | 2.62E-06 | 0.80 (0.73,0.88) | PTPRN2 |
| cg06596307 | −0.4198 | 2.99E-06 | 0.66 (0.55,0.78) | IGF1R |
| cg10702366 | −0.1093 | 3.09E-06 | 0.90 (0.86,0.94) | FGGY |
| cg26470101 | 0.3052 | 3.09E-06 | 1.36 (1.19,1.54) | (DLX2**) |
| cg26042024 | −0.3109 | 3.13E-06 | 0.73 (0.64,0.84) | ZFAT |
| cg00466121 | 0.4646 | 3.16E-06 | 1.59 (1.31,1.93) | ZNHIT6 |
| cg04987302 | −0.3378 | 3.71E-06 | 0.71 (0.62,0.82) | (OTX2-AS1**) |
| cg08853494 | 0.2210 | 4.03E-06 | 1.25 (1.14,1.37) | RCHY1;THAP6 |
| cg26467725 | −0.4225 | 4.22E-06 | 0.66 (0.55,0.78) | SLCO3A1 |
| cg06442192 | −0.5241 | 4.89E-06 | 0.59 (0.47,0.74) | ZNF541 |
| cg00393373 | −0.3156 | 4.91E-06 | 0.73 (0.64,0.84) | ZNF518B |
The CpGs reported as significant do not include X,Y chromosome probes, cross-reactive probes, single nucleotide polymorphism (SNP)-associated probes, or probes that were significant for heterogeneity in the meta-analysis (i.e., QEp <0.05)
effect estimates represent log hazard ratio per 5% increase in DNA methylation
Gene information is based on Illumina annotation (February 2009 - GRCh37/hg19) assembly). For CpG sites annotated to inter-genic regions, information on nearest annotated gene (shown with **) is from R Bioconductor package FDb.InfiniumMethylation.hg19
Table 3.
DNA methylation at 30CpG sites associated with the risk of myocardial infarction (MI).* CpGs with false discovery rate <0.05 in are shown (Bonferroni-significant sites in bold).
| CpG name | beta coefficient† | Nominal p-value | Hazard Ratio (95% CI) | Gene‡ |
|---|---|---|---|---|
| cg22871797 | −0.599 | 5.29E-08 | 0.55(0.44,0.68) | CYFIP1 |
| cg24977276 | −0.366 | 9.97E-08 | 0.69(0.61,0.79) | GTF2I |
| cg18598861 | −0.671 | 1.61E-07 | 0.51 (0.4,0.66) | IRF9 |
| cg09777776 | 0.287 | 2.25E-07 | 1.33 (1.19,1.48) | ZNF254 |
| cg20545941 | −0.885 | 2.47E-07 | 0.41 (0.29,0.58) | MPPED1 |
| cg19935845 | −0.336 | 4.65E-07 | 0.71 (0.63,0.81) | TNXB |
| cg24423782 | −0.398 | 5.37E-07 | 0.67 (0.58,0.78) | MIR182 |
| cg00393373 | −0.401 | 7.68E-07 | 0.67 (0.57,0.79) | ZNF518B |
| cg00466121 | 0.487 | 7.79E-07 | 1.63 (1.34,1.97) | ZNHIT6 |
| cg19227382 | −0.504 | 8.12E-07 | 0.60 (0.49,0.74) | CDH23 |
| cg03467256 | −0.408 | 8.33E-07 | 0.67 (0.57,0.78) | HPCAL1 |
| cg25196881 | −0.269 | 1.05E-06 | 0.76 (0.69,0.85) | (THBS1**) |
| cg02321112 | 0.390 | 1.08E-06 | 1.48 (1.26,1.73) | (MNX1-AS1**) |
| cg00355799 | −0.216 | 1.40E-06 | 0.81 (0.74,0.88) | (LOC339529**) |
| cg17556588 | −0.154 | 1.45E-06 | 0.86(0.8,0.91) | PRRG4 |
| cg04987302 | −0.428 | 1.50E-06 | 0.65 (0.55,0.78) | (OTX2-AS1**) |
| cg07289306 | 0.616 | 1.71E-06 | 1.85 (1.44,2.38) | (MIR138-1**) |
| cg05892484 | −0.551 | 1.84E-06 | 0.58 (0.46,0.72) | MAD1L1 |
| cg10702366 | −0.150 | 2.11E-06 | 0.86 (0.81,0.92) | FGGY |
| cg22618720 | −0.424 | 2.37E-06 | 0.65 (0.55,0.78) | (MIR5095**) |
| cg14010194 | −0.484 | 2.71E-06 | 0.62 (0.5,0.75) | GUCA1B |
| cg13827209 | 0.285 | 2.71E-06 | 1.33 (1.18,1.5) | TGFBR1 |
| cg24318598 | −0.254 | 2.79E-06 | 0.78 (0.7,0.86) | ANO1 |
| cg07015775 | 0.479 | 3.13E-06 | 1.61 (1.32,1.97) | ZNHIT6 |
| cg21018156 | −0.135 | 3.17E-06 | 0.87 (0.83,0.92) | (LINC01312**) |
| cg07475527 | −0.225 | 3.77E-06 | 0.80 (0.73,0.88) | (RCAN3**) |
| cg20000562 | 0.218 | 3.93E-06 | 1.24 (1.13,1.36) | SFTA3 |
| cg07436807 | −0.779 | 4.10E-06 | 0.46 (0.33,0.64) | STAMBPL1; ACTA2 |
| cg14029912 | −0.367 | 4.29E-06 | 0.69 (0.59,0.81) | (BHLHE40**) |
The CpGs reported as significant do not include X,Y chromosome probes, cross-reactive probes, single nucleotide polymorphism (SNP)-associated probes, or probes that were significant for heterogeneity in the meta-analysis (i.e., QEp <0.05)
effect estimates represent log hazard ratio per 5% increase in DNA methylation
Gene information is based on Illumina annotation (February 2009 - GRCh37/hg19) assembly). For CpG sites annotated to inter-genic regions, information on nearest annotated gene (shown with **) is from R Bioconductor package FDb.InfiniumMethylation.hg19
Figure 2.
Plot of effect sizes (i.e., log hazard ratios) and 95% confidence intervals (CIs) for the 52 coronary heart disease (CHD)-associated CpG DNA methylation sites, comparing results from the incident CHD meta-analysis (blue) vs the incident MI-only meta-analysis (orange).
Sensitivity-analyses and race-specific meta-analyses
We observed highly consistent results when comparing associations for the 52 CHD-associated CpGs from all cohorts (n=11,461) to results from the subset meta-analysis of the seven cohorts that performed Cox regression (n=9,255) (Supplemental Figure 4). Similarly, we performed four additional meta-analyses, each time excluding one of the four largest cohorts (FHS, NAS, ARIC, KORA), and found similar results across these meta-analyses for a majority of the 52 CHD-associated CpGs (Supplemental Figure 5).However associations appear to be notably driven by results from ARIC and FHS for 20 of the CpGs. In race-specific meta-analyses, the effect size and direction of effects for the majority of the CHD-associated CpGs were similar when comparing those of European versus African-American ancestry (Supplemental Figure 6). However, 11 of the 52 CpGs showed race-specific differences in the association of DNA methylation with incident CHD (p-value <0.05 for difference in t-statistic; Supplemental Table 3). Finally, we also conducted a random-effects sensitivity meta-analysis on the 30 CpGs that were associated with incident CHD (i.e. those reported in Table 2). When comparing results from models run under a fixed-effects meta-analysis vs. a random-effects meta-analysis,the majority of these 30 CpGs had either exactly the same or very similar effect sizes (CI), and many had the same p-value as well. Exceptions to this include: cg22617878, cg02155262, cg06596307, and cg08853494 (Supplemental Table 4).
Associations of DNA methylation with genetic variants
For 10 of the 52 CHD-associated CpGs, we were able to detect and replicate multiple meQTLs for each CpG, comprising 1,634 unique SNPs total (full list available as online supplementary Excel File). Across these 1,634 meQTLs, we observed overlap with SNPs identified in prior GWAS studies on diabetic kidney disease, age-related macular degeneration, prostate cancer, neutrophil count, multiple sclerosis, follicular lymphoma, diffuse large B cell lymphoma, multiple sclerosis, and kidney stones (Supplemental Table 5).
Identifying causal associations between DNA methylation and incident CHD using Mendelian Randomization
For each of the 10 CpGs with replicated meQTLs, we proceeded to select the cis-meQTL(+/−500kb) with the lowest p-value, to utilize as an instrumental variable to model the causal exposure of differential methylation at the 10 CpGs on development of incident CHD (Table 4). For two of the 10 CpGs, Mendelian Randomization (MR) analyses supported a causal effect of DNA methylation on incident CHD: cg26470101 (β [95% CI] for 1% increase in DNA methylation = 0.042 [0.002, 0.08]; P = 0.037) and cg07289306 (β [95% CI] for 1% increase in DNA methylation = −0.148 [−0.288, −0.009]; P = 0.04) on CHD (Table 4). Both CpGs map to regulatory active intergenic regions within CpG islands, and cg07289306 is located proximal to two long non-coding RNA transcripts26 (Figure 3).
Table 4.
Mendelian Randomization analysis for assessing causality between DNA methylation and incident coronary heart disease (CHD), using identified methylation quantitative trait loci (meQTLs) as genetic proxies
| Exposure = DNA methylation | Genetic proxy as instrumental variable = SNP associated with DNA methylation (meQTL) | Outcome = CHD (CC4D 2015) | Outcome = MI (CC4D 2015) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | CpG genomic position | Gene or nearest gene* | Associated SNP (meQTL)† | SNP genomic position | SNP Fx Allele | Causal estimates from Mendelian Ranomization | Causal estimates from Mendelian Ranomization | ||||
| β | se | p-value | β | se | p-value | ||||||
| cg01751802 | 19:11309639 | KANK2 | rs3745682 | 19:11313256 | G | 0.03 | 0.91 | 0.978 | −0.45 | 1.02 | 0.660 |
| cg06582394 | 3:121902622 | CASR | rs9883099 | 3:121902945 | A | 0.94 | 0.78 | 0.228 | 0.37 | 0.87 | 0.667 |
| cg06639874 | 2:238417703 | MLPH | rs10187185 | 2:238416524 | C | −0.40 | 1.27 | 0.751 | 0.29 | 1.37 | 0.832 |
| cg07289306 | 3:44039357 | (MIR138-1‡) | rs28731098 | 3:44019816 | A | −14.84 | 7.13 | 0.037 | −17.58 | 7.96 | 0.027 |
| cg08422803 | 21:46341067 | ITGB2 | rs4050931 | 21:46346135 | T | 1.24 | 1.13 | 0.273 | 1.70 | 1.24 | 0.170 |
| cg10307345 | 11:18771567 | PTPN5 | rs12575661 | 11:18771648 | C | −0.41 | 0.63 | 0.518 | −0.38 | 0.70 | 0.588 |
| cg13827209 | 9:101912842 | TGFBR1 | rs1013186 | 9:101884337 | C | −0.26 | 2.73 | 0.925 | −3.64 | 3.01 | 0.226 |
| cg14010194 | 6:42152817 | GUCA1B | rs2395805 | 6:42163839 | T | 2.15 | 1.96 | 0.275 | −1.08 | 2.16 | 0.618 |
| cg25497530 | 7:158059944 | PTPRN2 | rs6953878 | 7:157943641 | G | 1.88 | 2.12 | 0.374 | 1.70 | 2.34 | 0.467 |
| cg26470101 | 2:173099597 | (DLX2‡) | rs2054832 | 2:173080117 | C | 4.24 | 2.04 | 0.037 | 4.13 | 2.16 | 0.056 |
Abbreviations: CHD: MeQTL: methylation quantitative trait loci; MI: myocardial infarction; SNP: single nucleotide polymorphism; CHR: chromosome
Gene information are based on Illumina annotation (February 2009 - GRCh37/hg19) assembly). For CpG sites annotated to inter-genic regions, information on nearest annotated gene (shown with ‡) is from R Bioconductor package FDb.InfiniumMethylation.hg19
For each CpG, we observed multiple meQTLs, but only selected the meQTL with the lowest p-value / highest R2-explained for reporting, and for conducting the Mendelian Randomization analyses
Mendelian Randomization analyses produce causal estimates for 1 unit increase in DNA methylation, which corresponds to 100% increase in DNA methylation. For a more appropriate interpretation, these estimates, shown here, are reported and discussed in the manuscript after converting them to correspond to a 1% increase in DNA methylation.
Figure 3.
Adapted UCSC genome browser image - for genomic location of CpGs cg07289306 and cg26470101. The red zoom-in triangles are our addition to the UCSC image (http://genome.ucsc.edu), and represent a magnified region corresponding to the red marked region on each chromosome. The yellow highlights are also our addition, and highlight the exact genomic location of each CpG
Expression-QTLs overlapping with methylation-QTLs of CpGs showing causal associations between DNA methylation and incident CHD
For methylation at CpGs cg07289306 and cg26470101, which had evidence for causal effects on CHD development in MR analyses, as described above, we took all corresponding meQTLs (n=26 and 261, respectively) to identify whether these meQTLs overlapped with expression-QTLs (eQTLs), using the GTEx catalogue. For cg07289306, we found that 26 of 28 meQTLs for this CpG overlap with eQTLs for a long non-coding RNA downstream of cg07289306: lncRNA RP4-555D20.2. Similarly, we found that for the 261 meQTLs detected and replicated for cg26470101, 84 overlapped with an eQTL for the Integrin Subunit Alpha 6 (ITGA6) gene.
DISCUSSION
We conducted a large-scale analysis of DNA methylation in relation to incident CHD and MI among 11,461 adults across multiple cohort studies. Methylation at 52 CpGs across the genome were associated with future risk of CHD and MI. A 5% increase in methylation of identified CpGs was related to differences in CHD risk of a clinically relevant magnitude, ranging from a 46% decrease in the risk of CHD (cg12766383) to a 65% increase in risk (cg05820312), independent of age, sex, and other known CHD risk factors. In exploratory analyses to highlight candidates for functional experimentation, Mendelian randomization analyses revealed that methylation at two loci had a causal effect on incident CHD, potentially via non-coding RNA regulation and tissue structural elements.
Biological relevance and clinical implications
Several of the 52 CHD-associated CpGs in our study map to genes with roles in calcium regulation, as well as genes that have been identified in association with calcium levels and kidney function in prior GWAS and DNA methylation studies. CpG cg2261787 maps to the ATPase plasma membrane calcium transporter 2 (ATP2B2) gene from the plasma membrane calcium transporter family with critical roles in intracellular calcium homeostasis. Similarly, CpG cg06582394 maps to the calcium sensing receptor (CASR) gene, which has a key role in calcium homeostasis. SNPs in CASR have been consistently associated with serum calcium in populations from several different ancestries.27–29 Furthermore, in a recent Mendelian randomization analysis of 184,305 individuals, Larsson et al.28 reported that a genetic variant at the CASR locus showed strong associations with coronary artery disease and MI. Similarly, CpGs cg14010194 and cg03467256 map to guanylate cyclase activator 1B (GUCA1B) and hippocalcin-like 1 (HPCAL1), respectively, both with roles in calcium-dependent regulation.30,31
We also identified CHD-associated CpG loci linked to renal function. CpGs cg19227382, cg03467256, and cg25497530 map to genes cadherin-related 23 (CDH23), HPCAL1, and protein tyrosine phosphatase receptor type N2 (PTPRN2), respectively. Both CDH23 and HPCAL1 were identified in a GWAS of kidney function in approximately 64,000 participants of European decent.32 An epigenome-wide study of 400 individuals showed differential blood DNA methylation at the PTPRN2 locus in chronic kidney disease cases relative to controls.33 However, genetic variants in PTPRN2 were also associated with coronary artery calcified atherosclerotic plaque in a meta-analysis GWAS among African-Americans with type-2 diabetes.34
Observational studies and calcium supplementation randomized clinical trials provide evidence of associations between serum calcium levels and increased risk of CHD and MI.35,36 Our results provide the first evidence that epigenetic regulation of calcium homeostasis may be involved in calcium-related CHD risk, an underdeveloped area of therapeutics. Similarly, kidney function is a well-recognized risk factor for CVD, with a recent AHA report highlighting that individuals with an estimated glomerular filtration rate (eGFR) of 15 to 30 mL/min per 1.73 m2 have the highest adjusted relative risk of CVD mortality.37,38 Our results suggest that epigenetic regulation may be involved in pathways linking kidney function to CHD risk. We do note that since our analyses were adjusted for major risk factors such as smoking and BMI, we may not have identified BMI- and smoking-specific methylation signatures related to incident HD. Our goal was to identify methylation signatures related to CHD beyond major known risk factors such as smoking and BMI, and we direct readers to major epigenome-wide meta-analyses studies on BMI39 and smoking4 that have previously been published.
Other gene loci identified include the insulin growth factor 1 receptor (IGF1R), transforming growth factor beta receptor 1 (TGFBR1), and integrin subunit beta 2 (ITGB2). The roles of IGF1R and the TGF-beta signaling in cardiac remodeling and function are well recognized,40,41 and recently TGFBR1 gene expression levels in blood samples from acute MI patients strongly predicted left-ventricular dysfunction.42 Furthemore, ITGB2 encodes a leukocyte cell-surface adhesion molecule that directly facilitates leukocyte transendothelial migration, a key step in formation of atherosclerosis.43
DNA methylation at CpGs cg26470101 and cg07289306 showed evidence of a causal effect on CHD. Both of these CpGs are located within CpG islands in intergenic regions, with cg07289306 located proximal to two long non-coding RNAs (lncRNAs). Furthermore, meQTLs for cg07289306 overlap with the expression-QTLs (eQTLs) for a long non-coding RNA downstream of cg07289306: lncRNA RP4-555D20.2. This suggests that methylation at cg07289306 may be part of regulatory pathways involving lncRNAs. Increasing evidence indicates that lncRNAs are key components of transcriptional regulatory pathways that govern cardiac development and cardiovascular pathophysiology.44,45 Similarly, meQTls we identified for CpG cg26470101 overlapped with eQTLs for Integrin Subunit Alpha 6 (ITGA6) transcript expression. In a study of left ventricular myocardium tissue in mice, Lodder et al.46 assessed collagen levels combined with genome-wide genotyping and cardiac expression analyses, and found that eQTLs for ITGA6 transcripts overlapped with QTLs related to cardiac collagen deposition. They report their findings to suggest that ITGA6 is an important part of the molecular network modulating collagen deposition in the heart. In another study of murine cardiac tissue,47 ITGA6 was one of six identified (immune response) genes with decreased expression profiles in cardiac tissue macrophages from older mice compared to that of young mice . Our findings in the context of the findings from these other studies may suggest that DNA methylation plays a role in premature cardiovascular aging and risk of CHD via non-coding RNA as well as tissue cellular structural elements.
Findings in the context of prior evidence
Our epigenome-wide study identifies numerous loci and related genes and pathways that have not been identified in incident CHD genome-wide association studies alone. Furthermore, our findings did not overlap with those of previous epigenome-wide studies of CHD, as prior studies were small and predominantly cross-sectional. Cross-sectional studies may identify CpGs and associated genes and pathways that are altered as a result of disease state, rather than the prospective design employed in our study which may be identifying loci involved in pathways preceding manifest disease. Prior studies were also often composed of select populations geographically and ethnically distinct from the populations in our meta-analysis. For example, Sharma et al. identified differentially methylated regions (DMRs) near or within genes C1QL4, CCDC47, and TGFBR3 in a study of 36 men (18 CAD, 18 controls) from India.15 Nakatochi et al.14 compared 192 MI cases with 192 controls in an epigenome-wide whole-blood analysis on elderly Japanese individuals, and reported DNA methylation at two CpGs, located in the ZFHX3 and SMARCA4 genes, to be associated with MI. In the prospective Italian EPICOR cohort, Guarrera et al.12 compared 292 MI cases with 292 matched controls ascertained prospectively during follow-up, and reported that a differentially-methylated region (DMR), within the Zinc Finger And BTB Domain Containing 2 (ZBTB12) gene body was associated with MI.
Study limitations
We used well-established statistical procedures to remove the effect of cell-type heterogeneity as a source of confounding,20,48 however residual confounding is still possible. Further, while we used a stringent threshold to exclude any results for which there was between-study heterogeneity, some degree of heterogeneity is likely and may affect the results observed. However, some element of the heterogeneity likely reflects racial and environment specific sources of methylation differences. Additionally, our Mendelian randomization analyses provide evidence supporting a causal role of methylation at two CpGs but this does not prove causality, and thus, follow-up experimental work is needed. Currently, leukocyte-specific and trans-tissue meQTL datasets are limited with relatively small sample sizes, thus limiting our ability to use multiple independent meQTL loci for multi-SNP instrumental variables in Mendelian randomization analyses Another limitation is the relatively large contribution from cohorts based in primarily Western countries in Europe and the United States due to the current limited availability of DNA methylation and incident CHD data in more ethnically diverse cohorts.
Study strengths
Our study is by far the largest of its kind, with nearly 12,000 participants. We also made use of incident cases that were stringently adjudicated over a long-term follow-up. Furthermore, we used Mendelian randomization to build evidence regarding causal effects of DNA methylation on incident CHD.
CONCLUSION
We present novel and robust findings on associations of leukocyte DNA methylation with risk of CHD, with effect sizes that are of a clinically relevant magnitude. In addition, our findings highlight known as well as under-recognized pathways to CHD, including calcium regulation, kidney function, and gene regulation mechanisms involving non-coding RNAs. Overall, the findings provide a deeper understanding of the molecular landscape of incident CHD and may present novel avenues for targeting disease pathways and development of therapeutic interventions.
Supplementary Material
CLINICAL PERSPECTIVES.
What’s New?
Among 11,461 individuals across nine population-based cohorts from the United States and Europe, differences in blood leukocyte DNA methylation at 52 CpG loci were robustly associated with incident CHD.
Several of the differentially-methylated loci map to genes related to calcium regulation and kidney function.
Exploratory analyses with Mendelian randomization supported a causal effect of DNA methylation on incident CHD at loci in active regulatory regions, with links to non-coding RNAs and genes involved in cellular and tissue structural components.
What are the clinical implications?
Leukocyte genome regulatory mechanisms, via DNA methylation, are robustly linked to risk of developing CHD.
Our findings provide the first evidence that genomic regulation via epigenetic modifications in kidney function- and calcium homeostasis-related pathways may be involved in the development of CHD.
Our findings of epigenetic loci related to non-coding RNAs highlight pathways that have not emerged in genome-wide studies of CHD, and may represent novel therapeutic targets which are thus far unexplored.
ACKNOWLEDGEMENTS
GA, AAB, MMM, CWC, RJ, and DL were involved in designing the study and preparing the analysis plan. AAB, DL, EAW, TLA, MF, AP, GM, NS, LF, LH were involved in the acquisition of DNA methylation data. GA and RJ were involved in preparing the analytic programming code. GA, CWC, TH, RG, ES, JAB, GF, JB, BHC were involved in conducting the individual cohort epigenome-wide analyses. CWC, MMM, RJ, TH, GA were involved in additional analyses of gene variants and Mendelian randomization in FHS and KORA. GA, AAB, MMM, CWC, RJ were involved in drafting of the manuscript.
All authors listed contributed to interpretation of the data and results, enhancement of the analyses, multiple revisions of the manuscript, and final approval of the submitted manuscript.
The authors would like to thank the KORA participants for their participation in the KORA cohort. The authors wish to thank all who participated in, or collaborated with, EPIC; the Italian AVIS blood donor organization, AIRE-ONLUS di Ragusa, and the Sicilian Government.
ROLE OF THE FUNDING SOURCE
ARIC has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HSN268201700005I). The authors thank the staff and participants of the ARIC study for their important contributions. Funding was also supported by 5RC2HL102419 and R01NS087541. CHS was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, U01HL130114, UL1TR001881, with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS), R01AG023629, UL1TR000124, DK063491.
EPICOR is supported by the Compagnia di San Paolo for the EPIC-Italy and EPICOR projects, the Human Genetics Foundation-Turin (HuGeF) / Italian Institute for Genomic Medicine (IIGM), and by Ministero dell’Istruzione, dell’Università e della Ricerca – MIUR project “Dipartimenti di Eccellenza 2018 – 2022” to Department of Medical Sciences, University of Turin. FHS is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD. InCHIANTI was supported as a targeted project of the Italian Ministry of Health (grant ICS110.1/RF97.71) and the US National Institute on Aging (grants 263MD 9164 and 263 MD 821336).
KORA is supported by the Helmholtz Zentrum München, which is funded by the German Federal Ministry of Education and Research (BMBF) and the State of Bavaria. This work was supported in part by the BMBF within the framework of the e:Med research (e:AtheroSysMed, grant 01ZX1313A-2014) and the Munich Center of Health Sciences, Ludwig Maximilians-Universität, as part of LMUinnovativ. NAS is supported by grants from the National Institute of Environmental Health Sciences (NIEHS) (R01ES021733, R01ES025225, R01ES027747, R01ES015172, R01ES014663, R21ES020010, P30ES009089), Environmental Protection Agency (EPA) grant RD832416, as well as by the Cooperative Studies Program/ERIC, US Department of Veterans Affairs, and is a research component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). Additional support was provided by the US Department of Agriculture, Agricultural Research Service (contract 53-K06-510). The views expressed in this paper are those of the authors and do not necessarily represent the views of the US Department of Veterans Affairs. WHI-EMPC was funded by the National Institute of Environmental Health Sciences (R01-ES020836; Whitsel, Baccarelli, Hou). LH was funded by AHA grant “14SFRN20790000”. WHI-BAA23 was supported by NHLBI’s Broad Agency Announcement contract HHSN268201300006C. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible.
DISCLOSURES
Bruce M. Psaty serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the steering committee of the Yale Open Data Access Project funded by Johnson & Johson. Brian H. Chen was an employee of the US National Institutes of Health during this study, but is currently employed by Life Epigenetics, Inc,, which had no influence on the analysis, interpretation, or reporting of this publication.
NON-STANDARD ABBREVIATIONS AND ACRONYMS
- DMR
Differentially Methylated Region
- e-QTL
expression-quantitative trait loci
- FDR
false discovery rate
- GTEx
Genotype-Tissue Expression
- GWAS
genome-wide association study
- LncRNA
long non-coding RNA
- RNA
ribonucleic acid
- SNP
single-nucleotide polymorphism
References
- 1.Butler D Un targets top killers. Nature. 2011;477:260–261. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. [DOI] [PubMed] [Google Scholar]
- 3.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: Basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O’Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SL, Melzer D, Baccarelli AA, van Meurs JB, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Tregouet DA, Deloukas P, Samani NJ. DNA methylation and body-mass index: A genome-wide analysis. Lancet. 2014;383:1990–1998. [DOI] [PubMed] [Google Scholar]
- 6.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, Colicino E, Waite LL, Joehanes R, Guan W, Brody JA, Elks C, Marioni R, Jhun MA, Agha G, Bressler J, Ward-Caviness CK, Chen BH, Huan T, Bakulski K, Salfati EL, Investigators W-E, Fiorito G, Disease CeoCH, Wahl S, Schramm K, Sha J, Hernandez DG, Just AC, Smith JA, Sotoodehnia N, Pilling LC, Pankow JS, Tsao PS, Liu C, Zhao W, Guarrera S, Michopoulos VJ, Smith AK, Peters MJ, Melzer D, Vokonas P, Fornage M, Prokisch H, Bis JC, Chu AY, Herder C, Grallert H, Yao C, Shah S, McRae AF, Lin H, Horvath S, Fallin D, Hofman A, Wareham NJ, Wiggins KL, Feinberg AP, Starr JM, Visscher PM, Murabito JM, Kardia SL, Absher DM, Binder EB, Singleton AB, Bandinelli S, Peters A, Waldenberger M, Matullo G, Schwartz JD, Demerath EW, Uitterlinden AG, van Meurs JB, Franco OH, Chen YI, Levy D, Turner ST, Deary IJ, Ressler KJ, Dupuis J, Ferrucci L, Ong KK, Assimes TL, Boerwinkle E, Koenig W, Arnett DK, Baccarelli AA, Benjamin EJ, Dehghan A. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard MA, Huan T, Ligthart S, Gondalia R, Jhun MA, Brody JA, Irvin MR, Marioni R, Shen J, Tsai PC, Montasser ME, Jia Y, Syme C, Salfati EL, Boerwinkle E, Guan W, Mosley TH Jr., Bressler J, Morrison AC, Liu C, Mendelson MM, Uitterlinden AG, van Meurs JB, Consortium B, Franco OH, Zhang G, Li Y, Stewart JD, Bis JC, Psaty BM, Chen YI, Kardia SLR, Zhao W, Turner ST, Absher D, Aslibekyan S, Starr JM, McRae AF, Hou L, Just AC, Schwartz JD, Vokonas PS, Menni C, Spector TD, Shuldiner A, Damcott CM, Rotter JI, Palmas W, Liu Y, Paus T, Horvath S, O’Connell JR, Guo X, Pausova Z, Assimes TL, Sotoodehnia N, Smith JA, Arnett DK, Deary IJ, Baccarelli AA, Bell JT, Whitsel E, Dehghan A, Levy D, Fornage M. DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet. 2017;101:888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun KVE, Dhana K, de Vries PS, Voortman T, van Meurs JBJ, Uitterlinden AG, consortium B, Hofman A, Hu FB, Franco OH, Dehghan A. Epigenome-wide association study (ewas) on lipids: The rotterdam study. Clin Epigenetics. 2017;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, Thibeault KS, Patel N, Day K, Jones LW, Liang L, Chen BH, Yao C, Tiwari HK, Ordovas JM, Levy D, Absher D, Arnett DK. Epigenome-wide association study of fasting blood lipids in the genetics of lipid-lowering drugs and diet network study. Circulation. 2014;130:565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H, Wolffenbuttel BHR, van Vliet-Ostaptchouk JV. DNA methylation markers associated with type 2 diabetes, fasting glucose and hba1c levels: A systematic review and replication in a case-control sample of the lifelines study. Diabetologia. 2018;61:354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, Sparrow D, Vokonas P, Schwartz J. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarrera S, Fiorito G, Onland-Moret NC, Russo A, Agnoli C, Allione A, Di Gaetano C, Mattiello A, Ricceri F, Chiodini P, Polidoro S, Frasca G, Verschuren MW, Boer JM, Iacoviello L, van der Schouw YT, Tumino R, Vineis P, Krogh V, Panico S, Sacerdote C, Matullo G. Gene-specific DNA methylation profiles and line-1 hypomethylation are associated with myocardial infarction risk. Clil Epigenetics. 2015;7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PloS one. 2010;5:e9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatochi M, Ichihara S, Yamamoto K, Naruse K, Yokota S, Asano H, Matsubara T, Yokota M. Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin Epigenetics. 2017;9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma P, Garg G, Kumar A, Mohammad F, Kumar SR, Tanwar VS, Sati S, Sharma A, Karthikeyan G, Brahmachari V, Sengupta S. Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene. 2014;541:31–40. [DOI] [PubMed] [Google Scholar]
- 16.Psaty BM, Sitlani C. The cohorts for heart and aging research in genomic epidemiology (charge) consortium as a model of collaborative science. Epidemiology. 2013;24:346–348. [DOI] [PubMed] [Google Scholar]
- 17.Heinze G, Schemper M. A solution to the problem of monotone likelihood in cox regression. Biometrics. 2001;57:114–119. [DOI] [PubMed] [Google Scholar]
- 18.Lubin JH, Gail MH. Biased selection of controls for case-control analyses of cohort studies. Biometrics. 1984;40:63–75 [PubMed] [Google Scholar]
- 19.Robins JM, Gail MH, Lubin JH. More on “biased selection of controls for case-control analyses of cohort studies”. Biometrics. 1986;42:293–299. [PubMed] [Google Scholar]
- 20.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D, Turner R. Power analysis for random-effects meta-analysis. Research synthesis methods. 2017;8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The mr-base platform supports systematic causal inference across the human phenome. eLife. 2018;7e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJ, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O’Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M, Consortium CAD. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapur K, Johnson T, Beckmann ND, Sehmi J, Tanaka T, Kutalik Z, Styrkarsdottir U, Zhang W, Marek D, Gudbjartsson DF, Milaneschi Y, Holm H, Diiorio A, Waterworth D, Li Y, Singleton AB, Bjornsdottir US, Sigurdsson G, Hernandez DG, Desilva R, Elliott P, Eyjolfsson GI, Guralnik JM, Scott J, Thorsteinsdottir U, Bandinelli S, Chambers J, Stefansson K, Waeber G, Ferrucci L, Kooner JS, Mooser V, Vollenweider P, Beckmann JS, Bochud M, Bergmann S. Genome-wide meta-analysis for serum calcium identifies significantly associated snps near the calcium-sensing receptor (casr) gene. PLoS Genet. 2010;6:e1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson SC, Burgess S, Michaelsson K. Association of genetic variants related to serum calcium levels with coronary artery disease and myocardial infarction. JAMA. 2017;318:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Seaghdha CM, Wu H, Yang Q, Kapur K, Guessous I, Zuber AM, Kottgen A, Stoudmann C, Teumer A, Kutalik Z, Mangino M, Dehghan A, Zhang W, Eiriksdottir G, Li G, Tanaka T, Portas L, Lopez LM, Hayward C, Lohman K, Matsuda K, Padmanabhan S, Firsov D, Sorice R, Ulivi S, Brockhaus AC, Kleber ME, Mahajan A, Ernst FD, Gudnason V, Launer LJ, Mace A, Boerwinckle E, Arking DE, Tanikawa C, Nakamura Y, Brown MJ, Gaspoz JM, Theler JM, Siscovick DS, Psaty BM, Bergmann S, Vollenweider P, Vitart V, Wright AF, Zemunik T, Boban M, Kolcic I, Navarro P, Brown EM, Estrada K, Ding J, Harris TB, Bandinelli S, Hernandez D, Singleton AB, Girotto G, Ruggiero D, d’Adamo AP, Robino A, Meitinger T, Meisinger C, Davies G, Starr JM, Chambers JC, Boehm BO, Winkelmann BR, Huang J, Murgia F, Wild SH, Campbell H, Morris AP, Franco OH, Hofman A, Uitterlinden AG, Rivadeneira F, Volker U, Hannemann A, Biffar R, Hoffmann W, Shin SY, Lescuyer P, Henry H, Schurmann C, Consortium S, Consortium G, Munroe PB, Gasparini P, Pirastu N, Ciullo M, Gieger C, Marz W, Lind L, Spector TD, Smith AV, Rudan I, Wilson JF, Polasek O, Deary IJ, Pirastu M, Ferrucci L, Liu Y, Kestenbaum B, Kooner JS, Witteman JC, Nauck M, Kao WH, Wallaschofski H, Bonny O, Fox CS, Bochud M. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLoS Genet. 2013;9:e1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korkmaz S, Radovits T, Barnucz E, Hirschberg K, Neugebauer P, Loganathan S, Veres G, Pali S, Seidel B, Zollner S, Karck M, Szabo G. Pharmacological activation of soluble guanylate cyclase protects the heart against ischemic injury. Circulation. 2009;120:677–686. [DOI] [PubMed] [Google Scholar]
- 31.Lanfear DE, Yang JJ, Mishra S, Sabbah HN. Genome-wide approach to identify novel candidate genes for beta blocker response in heart failure using an experimental model. Discov Med. 2011;11:359–366. [PMC free article] [PubMed] [Google Scholar]
- 32.Gorski M, Tin A, Garnaas M, McMahon GM, Chu AY, Tayo BO, Pattaro C, Teumer A, Chasman DI, Chalmers J, Hamet P, Tremblay J, Woodward M, Aspelund T, Eiriksdottir G, Gudnason V, Harris TB, Launer LJ, Smith AV, Mitchell BD, O’Connell JR, Shuldiner AR, Coresh J, Li M, Freudenberger P, Hofer E, Schmidt H, Schmidt R, Holliday EG, Mitchell P, Wang JJ, de Boer IH, Li G, Siscovick DS, Kutalik Z, Corre T, Vollenweider P, Waeber G, Gupta J, Kanetsky PA, Hwang SJ, Olden M, Yang Q, de Andrade M, Atkinson EJ, Kardia SL, Turner ST, Stafford JM, Ding J, Liu Y, Barlassina C, Cusi D, Salvi E, Staessen JA, Ridker PM, Grallert H, Meisinger C, Muller-Nurasyid M, Kramer BK, Kramer H, Rosas SE, Nolte IM, Penninx BW, Snieder H, Fabiola Del Greco M, Franke A, Nothlings U, Lieb W, Bakker SJ, Gansevoort RT, van der Harst P, Dehghan A, Franco OH, Hofman A, Rivadeneira F, Sedaghat S, Uitterlinden AG, Coassin S, Haun M, Kollerits B, Kronenberg F, Paulweber B, Aumann N, Endlich K, Pietzner M, Volker U, Rettig R, Chouraki V, Helmer C, Lambert JC, Metzger M, Stengel B, Lehtimaki T, Lyytikainen LP, Raitakari O, Johnson A, Parsa A, Bochud M, Heid IM, Goessling W, Kottgen A, Kao WH, Fox CS, Boger CA. Genome-wide association study of kidney function decline in individuals of european descent. Kidney Int. 2015;87:1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth LJ, McKay GJ, Maxwell AP, McKnight AJ. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Divers J, Palmer ND, Langefeld CD, Brown WM, Lu L, Hicks PJ, Smith SC, Xu J, Terry JG, Register TC, Wagenknecht LE, Parks JS, Ma L, Chan GC, Buxbaum SG, Correa A, Musani S, Wilson JG, Taylor HA, Bowden DW, Carr JJ, Freedman BI. Genome-wide association study of coronary artery calcified atherosclerotic plaque in african americans with type 2 diabetes. BMC Genet. 2017;18:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolland MJ, Grey A, Avenell A, Gamble GD, Reid IR. Calcium supplements with or without vitamin d and risk of cardiovascular events: Reanalysis of the women’s health initiative limited access dataset and meta-analysis. BMJ. 2011;342:d2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohrmann S, Garmo H, Malmstrom H, Hammar N, Jungner I, Walldius G, Van Hemelrijck M. Association between serum calcium concentration and risk of incident and fatal cardiovascular disease in the prospective amoris study. Atherosclerosis. 2016;251:85–93. [DOI] [PubMed] [Google Scholar]
- 37.Correction to: Heart disease and stroke statistics-2017 update: A report from the american heart association. Circulation. 2017;136:e196. [DOI] [PubMed] [Google Scholar]
- 38.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P, American Heart Association Statistics C, Stroke Statistics S. Heart disease and stroke statistics-2017 update: A report from the american heart association. Circulation. 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman AK, Aslibekyan S, Demerath EW, Guan W, Zhi D, Yao C, Huan T, Willinger C, Chen B, Courchesne P, Multhaup M, Irvin MR, Cohain A, Schadt EE, Grove ML, Bressler J, North K, Sundstrom J, Gustafsson S, Shah S, McRae AF, Harris SE, Gibson J, Redmond P, Corley J, Murphy L, Starr JM, Kleinbrink E, Lipovich L, Visscher PM, Wray NR, Krauss RM, Fallin D, Feinberg A, Absher DM, Fornage M, Pankow JS, Lind L, Fox C, Ingelsson E, Arnett DK, Boerwinkle E, Liang L, Levy D, Deary IJ. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: A mendelian randomization approach. PLoS Med. 2017;14:e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (tgf)-beta signaling in cardiac remodeling. J Mol Cell Cardiol. 2011;51:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laustsen PG, Russell SJ, Cui L, Entingh-Pearsall A, Holzenberger M, Liao R, Kahn CR. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol Cell Biol. 2007;27:1649–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devaux Y, Bousquenaud M, Rodius S, Marie PY, Maskali F, Zhang L, Azuaje F, Wagner DR. Transforming growth factor beta receptor 1 is a new candidate prognostic biomarker after acute myocardial infarction. BMC Med Genomics. 2011;4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, Segre AV, Ardlie KG, Khera AV, Kaushik VK, Natarajan P, Consortium CAD, Kathiresan S. Genetic analysis in uk biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizki G, Boyer LA. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ Res. 2015;117:192–206. [DOI] [PubMed] [Google Scholar]
- 45.Thum T, Condorelli G. Long noncoding rnas and micrornas in cardiovascular pathophysiology. Circ Res. 2015;116:751–762. [DOI] [PubMed] [Google Scholar]
- 46.Lodder EM, Scicluna BP, Beekman L, Arends D, Moerland PD, Tanck MW, Adriaens ME, Bezzina CR. Integrative genomic approach identifies multiple genes involved in cardiac collagen deposition. Circ Cardiovasc Genet. 2014;7:790–798. [DOI] [PubMed] [Google Scholar]
- 47.Pinto AR, Godwin JW, Chandran A, Hersey L, Ilinykh A, Debuque R, Wang L, Rosenthal NA. Age-related changes in tissue macrophages precede cardiac functional impairment. Aging. 2014;6:399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.