A novel approach to generate a dominant-negative bZIP mutant with high specificity is developed and successfully applied to characterize Arabidopsis bZIP53 and its dimerization partners.
Keywords: Arabidopsis thaliana, bZIP transcription factor, diurnal metabolic regulation, dimerization, DNA binding, dominant negative mutant, functional redundancy, low energy signaling, nuclear localization, stress responses
Abstract
bZIP transcription factors regulate diverse processes in eukaryotic cells. Arabidopsis bZIP members of the C and S1 groups form heterodimers and synergistically control metabolic reprogramming during stress responses. However, their functional characterization is complicated due to an overlapping heterodimerization network and high redundancy. In this study, we develop a simple but powerful approach for generating dominant negative mutants of bZIP factors with high specificity. By applying in vitro DNA-binding, reporter gene and protoplast two-hybrid assays, and plant mutant analysis, we show that phosphorylation-mimicking substitution of conserved serines in the DNA-binding domain of bZIP monomeric subunits suffices for the disruption of the interaction of both bZIP homo- and heterodimers with cognate DNA. This results in the transcriptional inactivation of target genes. The dominant-negative effect is achieved by the unaltered function of the intrinsic nuclear localization signal and dimerization properties of the mutated bZIP protein. Our findings not only reveal an additional regulatory mechanism of bZIP10 intracellular localization, but also provide evidence of the involvement of bZIP53 in the diurnal adjustments of amino acid metabolism. Our data demonstrate the advantages and the suitability of this new approach for the artificial inactivation of bZIP transcription factors in plants, and it may also be of use for other organisms.
Introduction
As sessile organisms, plants continually adjust their growth and development to ever-changing environmental conditions. They have developed sensory systems and signal transduction pathways to convert environmental signals into metabolic reactions and growth responses. Metabolic adjustments allow plants to withstand transient energy deprivation during both the day–night rhythm and diverse stresses (Baena-González and Sheen, 2008; Stitt and Zeeman, 2012; Smeekens and Hellmann, 2014). Acute or lasting energy deprivation induces the so-called low-energy stress, which correlates with reduced photosynthesis, hydrolysis of polysaccharides, degradation of proteins, and arrest of growth, which consequently limit yield. The SNF1-related protein kinase 1 (SnRK1) kinases have been proposed to function as the central integrators that link low energy status to growth (Baena-González et al., 2007). They are structurally similar to sucrose non-fermenting 1 (SNF1) kinase in yeasts and AMP-kinase (AMPK) in mammals. Activation of SnRK1 by energy depletion antagonizes trehalose-6-phosphate (T6P) and Target Of Rapamycin (TOR)-dependent processes and leads to the inhibition of anabolism and promotion of catabolism (Smeekens and Hellmann, 2014; Baena-González and Hanson, 2017; Shi et al., 2018). In Arabidopsis, two genes, KIN10 and KIN11, encode the catalytic α-subunit of the SnRK1 protein complex. By comparison of gene expression profiles in protoplasts with or without KIN10 expression, over 1000 putative SnRK1-dependent genes have been identified. A subset of bZIP transcription factors has been shown to partially mediate primary KIN10 signaling (Baena-González et al., 2007).
The bZIP family is present in all eukaryotes. In plants, they are involved in diverse biological processes such as abiotic stress signaling, pathogen defense, photomorphogenesis, seed maturation, and senescence (Jakoby et al., 2002). In Arabidopsis, there are 78 members that are subdivided into 10 to 13 groups based on structural homology (Jakoby et al., 2002; Dröge-Laser et al., 2018). Only S1-group members (which are small in size) have been shown to be able to activate the DARK-INDUCED 6 (DIN6) gene in transiently transformed protoplasts, this being one of the genes most strongly induced by energy depletion and KIN10 expression. The activation of DIN6, which codes for the glutamine-dependent ASPARAGINE SYNTHETASE 1 (ASN1), has been shown to be mediated by a G-box element, a typical binding site of bZIP transcription factors (Baena-González et al., 2007). The S1 group consists of five members, namely bZIP1, 2, 11, 44, and 53. They are known to induce the expression of key metabolic genes such as ASN1, genes involved in the catabolism of branched-chain amino acids, and PROLINE DEHYDROGENASE (ProDH), which catalyses the first step in proline degradation. They also induce the expression of the endosperm-specific genes of the seed-storage albumins 2S1 and 2S2 (Alonso et al., 2009; Dröge-Laser and Weiste, 2018; Pedrotti et al., 2018). Through direct interaction with G-box or ACTCAT cis-elements in the promoters of these genes, bZIP1 and 53 have been shown to mediate starvation responses in Arabidopsis plants (Weltmeier et al., 2006; Dietrich et al., 2011).
bZIP proteins contain two characteristic domains, namely a region highly enriched in basic amino acids, which is responsible for both nuclear localization and DNA-binding, and a so-called zipper domain that participates in dimerization (Alber, 1992; Lupas, 1996). Protein dimerization studies have revealed that S1-group members preferably form selective heterodimers with the C-group rather than to homodimerize. The C-group member bZIP63 has been shown to be directly phosphorylated by SnRK1 both in vitro and in vivo. The phosphorylation on one Ser residue in the N-terminus and two residues in the C-terminus is induced by energy deprivation during extended night conditions, thereby causing significant enhancement of bZIP63 heterodimerization with bZIP1 and 11, and leading to increased bZIP63-dependent activation of ProDH1 and ASN1 (Mair et al., 2015). However, a certain level of promiscuity in the dimerization and DNA binding behavior makes the clarification of specific roles and of action mechanisms of individual bZIPs especially challenging.
The basic region involved in DNA binding is the most conserved domain between different bZIP factors. By crystallographic analysis of animal bZIP proteins, the positions of amino acids that are either involved in specific recognition of DNA bases or in direct contact with the DNA backbone have been identified (Fujii et al., 2000; Miller et al., 2003). Interestingly, a high level of Cys, Ser, or Tyr conservation has been found at position 19 in the DNA-binding domain (DBD) of animal bZIP factors, which is in direct contact with the phosphodiester bond. The reversible modification of residues at this position can influence the binding of the bZIPs to their cognate promoter elements, thus enabling the functional modification of the transcription factors through either their redox state or phosphorylation (Amoutzias et al., 2006).
In contrast to animals and fungi, the plant bZIP DBD has a remarkable level of Ser conservation, which in Arabidopsis is 92% at position 19. In addition, another Ser, which is also in contact with the DNA backbone, occupies position 15 in almost 80% of plant bZIPs (Kirchler et al., 2010). Using in silico modeling, we have previously shown that a phosphorylation of bZIP63 at Ser15 and Ser19, or the introduction of a negative charge by substitution of these residues through Asp, leads to the elimination of the interaction with cognate DNA (Kirchler et al., 2010). Indeed, the mutated bZIP63Ser15,19Asp as well as the H-group member HY5Ser15Asp and G-group GBF1Ser15,19Asp are unable to bind to corresponding cis-elements in DNA-binding studies in vitro (Kirchler et al., 2010; Smykowski et al., 2015). Thus far, the disruption of DNA recognition by substitution of Ser19 and/or Ser15 in several bZIPs from different groups has been demonstrated only in vitro. We do not know, however, how such substitutions influence the recognition of the cognate DNA and thus the activity of the bZIP factor inside the cell. Since the substituted amino acids are located in the basic region, the dimerization properties of mutated proteins might remain unaffected. Provided that the introduction of negatively charged amino acids at positions 15 and 19 of the DBD can indeed prevent in vivo DNA binding, this would open a great possibility for the generation of specific dominant negative mutants and thus help in understanding signal cascades operating in the bZIP dimerization networks even at high levels of functional redundancy.
Using bZIP53, 10, and 25 as representatives for the bZIP heterodimerization network, we show here that substitution of conserved Ser residues with Asp in the DBD of bZIP factors completely inhibits their transactivation capacity. This inhibition is obviously caused by the disruption of DNA binding through the introduction of a negative charge at the site of contact with the DNA backbone. The dimerization preferences of bZIP proteins are not diminished by such substitutions, and the nuclear localization signal (NLS) is maintained as functional. The ectopic expression of the inactive bZIP53Ser15,19Asp in the wild-type background leads to the inhibition of the intrinsic bZIP53 activity. Thus, mimicking Ser phosphorylation through Asp substitution in the DBD seems to be sufficient to inactivate any bZIP factor as homo- and heterodimers.
Materials and methods
Plant material and growth conditions
All Arabidopsis thaliana lines were in the Col-0 background. Plants were grown at 22 °C and 50% humidity under a 12-h light and 12-h dark photoperiod. We selected bZIP53, 10, and 25 for study as representatives for the bZIP heterodimerization network.
Constructs containing bZIP coding sequences (wild-type and mutant forms) were used to generate transgenic Arabidopsis lines by infiltration with Agrobacterium tumefaciens strain GV3101 pMP90. To select transformed plants, seedlings were tested under a microscope for green fluorescent protein (GFP) signal.
Statistical analyses of rosette diameters were performed with R (http://www.r-project.org/) using the lme4 (http://CRAN.R-project.org/package=lme4) and multcomp (Hothorn et al., 2008) packages. The independent transformation lines (three homozygous lines per genotype) were considered as random integration events in the genome of Arabidopsis, and mixed models were used to design a linear model to describe the data. To test the effect of the genotype in general, we performed a ratio likelihood test using ANOVA of a null model against a full model, which included the genotype as a fixed effect. For the mixed model design, we used the maximum likelihood approach. In order to compare among genotypes, we established a general linear hypothesis for performing multiple comparisons of the genotype in our mixed model, and used the glht function in the multcomp package (Hothorn et al., 2008).
Cloning strategy
The C-group bZIP clones were constructed as described by Veerabagu et al. (2014). bZIP53 clones were generated using Gateway™ technology (Invitrogen); the respective primer pairs are listed in Supplementary Table S1 at JXB online. Site-directed mutagenesis was performed in entry clones according to the Quick-Reference Protocol (Stratagene). Oligonucleotides encoding the desired mutation (listed in Supplementary Table S1) were used for the amplification of the plasmid by Phusion® DNA polymerase (New England Biolabs, NEB).
P (ACTCAT)2x ::GUS, PGAL4::GUS, P35S::NAN, pHBTL-ADGW, and pHBTL-BDGW were kindly provided by Wolfgang Dröge-Laser (University of Würzburg, Germany; Ehlert et al., 2006), and the latter two were used in the LR reaction to generate AD- and BD-fusion proteins for the protoplast two-hybrid analyses.
To create GFP and mCherry fusion proteins, the corresponding DNA sequences were recombined via LR reactions into the binary plant expression vectors pUBC-Dest(eGFP) (C-terminal GFP fusion, UBI10 promoter; Grefen et al., 2010), pH7WGF2.0 (C-terminal GFP fusion, 35S promoter; Karimi et al., 2002), or pEXSG-mCherry (C-terminal mCherry fusion, 35S promoter; personal gift of Niko Tintor, MPI Cologne, Germany).
Reporter gene and protoplast two-hybrid assays in Arabidopsis protoplasts
Arabidopsis cell culture protoplasts were isolated and transfected according to Schütze et al. (2009). The amount of each plasmid used was constant through all the experiments. To keep the total DNA quantity at the same level in each sample, unrelated DNA was added when required. Transfected protoplasts were incubated in darkness overnight at 23 °C. The P35S::NAN construct was used as an internal control to normalize the variations of each transfection due to transformation efficiency and cell viability. GUS and NAN enzyme assays were performed as described by Weltmeier et al. (2006). All experiments were repeated at least three times. Data were analysed by paired t-tests.
Quantitative RT-PCR
Total RNA was isolated from plant material using an RNeasy Plant Mini Kit (Roboklon). The cDNA was generated using oligo-dT primer with AMV Reverse Transcriptase (Roboklon). The amplification of cDNA was performed using PerfeCta SYBR Green SuperMix (Quanta Biosciences, distributed by VWR) according to the manufacturer’s instructions. The PCR reactions were performed in a CFX384 Real Time PCR system (Bio-Rad). Expression levels were normalized using EF-1-alpha (AT5G60390) and Ubi10 (AT4G05320) and quantified using the CFX Manager software (Bio-Rad). All primer sequences for qPCR are listed in Supplementary Table S2. The plant material was collected from two independent experiments; three technical replicates were performed.
Amino acid analysis
For the amino acid analysis, leaves of 7-week-old plants were harvested in the morning immediately after the onset of light (under dim light). Leaf rosettes from three plants were pooled together, immediately frozen in liquid nitrogen, ground with mortar and pestle, and lyophilized without thawing. Dry material was extracted with 80% methanol followed by a second extraction with 20% methanol. The combined supernatants were dried in a speedvac. The sediments were re-suspended in lithium-buffer (Pickering Laboratories) and filtered (0.2-µm Minisart RC15 filters) before analysis. Amino acids were analysed by ion-exchange chromatography (cation exchange column 5 μm, 4.6×75 mm; Pickering Laboratories) with post-column ninhydrin derivatization (Hirner et al., 2006).
ELISA-based DNA-binding assay (DPI-ELISA)
For DNA-binding assays, bZIP coding sequences in pET-Dest42 (Invitrogen) were expressed as described previously (Kirchler et al., 2010). BL21(DE3)-RIL E. coli cells were lysed in DNA-binding buffer [4 mM HEPES, pH 7.5, 100 mM KCl, 0.2% (w/v) BSA, 8% (v/v) glycerol, 5 mM DTT] using FastPrep FP120 (Thermo Electron Corporation) and 0.1-mm glass beads (Sigma). The extract was cleared by centrifugation and used in DNA–protein interaction (DPI)-ELISAs. The oligonucleotide sequences used in the assays are provided in Supplementary Table S3. The DNA-binding assay was performed as described previously (Brand et al., 2010; Veerabagu et al., 2014).
Accession numbers
Accession numbers of gene sequences used in this study are given in Supplementary Table S4 and can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases.
Results
Ser15,19Asp mutation in the DBD of bZIP53 abolishes its DNA binding and transactivity
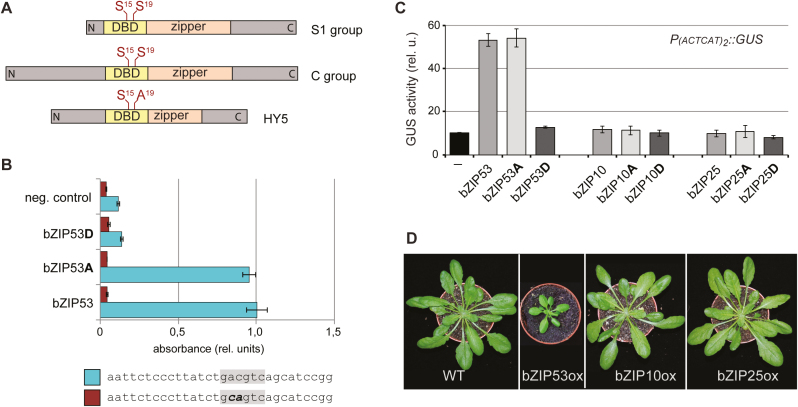
We have previously shown that mutation of Ser19 and/or Ser15 in the DBD to phospho-mimicking Asp completely abolishes the in vitro binding of C-group bZIP63 and H-group HY5 to their cognate DNA motifs (Kirchler et al., 2010). Due to the small size of S1-group factors, the DBD in bZIP53 is situated at the very N-terminus and not in the middle of the protein sequence, as is the case in bZIP63 and HY5 (Fig. 1A). To find out whether this different architecture influences its DNA-binding properties, we performed in vitro DPI-ELISAs (Brand et al., 2010) using the wild-type and mutated versions of bZIP53. Whereas the binding of bZIP53Ser15,19Ala to the C-box did not significantly differ from that of the wild-type bZIP53, binding of bZIP53Ser15,19Asp was reduced to the level of the negative control (Fig. 1B). The specificity of bZIP53-DNA binding was verified by mutation of the core DNA sequence. These results indicated that the imitation of phosphoserine by mutation of Ser to Asp leads to conformational changes that impair the in vitro bZIP53 DNA-binding activity.
Fig. 1.
Effect of the substitutions of Ser15 and 19 within the DNA-binding domain (DBD) of Arabidopsis bZIP factors on their DNA-binding and transcriptional activity. (A) Schematic representation of bZIP factors. The DBDs and leucine zipper (zipper) domains are indicated. S15 and S19 indicate conserved Ser residues within the DBDs. (B) Binding of bZIP53, bZIP53Ser15,19Ala (bZIP53A), bZIP53Ser15,19Asp (bZIP53D), and unspecific protein (neg. control) to the C-box (lower bars) and mutated C-box (upper bars). Data are means (±SD), n=2. (C) Reporter gene assays in Arabidopsis protoplasts. P(ACTCAT)2x::GUS was used as the reporter. A, substitution of Ser15 and Ser19 to Ala; D, substitution of Ser15 and Ser19 to Asp. Data are means (±SD), n=3. (D) Images of representative plants from each transgenic line expressing PUBI10::bZIP53-GFP (bZIP53ox), P35S::bZIP10-GFP (bZIP10ox), and P35S::bZIP25-GFP (bZIP25ox). (This figure is available in colour at JXB online.)
We further questioned whether Ser15,19Asp substitutions (Fig. 1A) might affect the in vivo transactivation capacity of bZIP53. It has previously been shown that bZIP53 directly recognizes an ACTCAT-motif in the promoter of ProDH1 (Weltmeier et al., 2006). Accordingly, a GLUCURONIDASE (GUS) reporter, P(ACTCAT)2x::GUS, was used to test the transcriptional activity of bZIP53-GFP and its phospho-mimicking form carrying the Ser15,19Asp mutation. To assure that the negative charge introduced by the Asp substitutions was indeed responsible for the resulting effect, the protein with substitution of the corresponding Ser to alanine (Ser15,19Ala) was also assayed. Transient expression of bZIP53-GFP in an Arabidopsis protoplast system showed induced expression of P(ACTCAT)2x::GUS compared to the ‘no effector’ control (Fig. 1C). A similar induction of the reporter gene was observed upon transfection of protoplasts with bZIP53-GFP carrying the Ser15,19Ala mutations in its DBD. In contrast, no increase in GUS activity was observed when bZIP53-GFP bearing the Ser15,19Asp mutations was co-expressed with the reporter gene. These data highlight the functional importance of the conserved serines in bZIP DBD for in vivo activity of bZIP factors. Since ProDH1 is known as a target gene of the C/S1 bZIP network (Weltmeier et al., 2006), we tested the C-group members bZIP10 and 25 for their ability to activate the same reporter construct. In contrast to bZIP53, neither the wild-type nor the mutated versions could induce GUS expression (Fig. 1C). This is in accordance with previous results (Weltmeier et al., 2006), and indicates that bZIP10 and 25 are able to induce ProDH1 only as heterodimers with bZIP53. A high sucrose level (13.7%) in the protoplast incubation medium clearly leads to u-ORF-mediated translational repression of intrinsic S1 bZIP factors (Hummel et al., 2009) and thus a lack of dimerization partners of bZIP10 and 25.
We then generated transgenic Arabidopsis lines constitutively expressing bZIP53-GFP, bZIP10-GFP, or bZIP25-GFP. Similar to the results of ProDH1 transactivation, we were not able to find significant alterations in the lines overexpressing bZIP10 or 25 when compared to wild-type plants at optimal greenhouse conditions (Fig. 1D). These results indicate that post-transcriptional events, such as availability of heterodimerization partners and/or signal-induced protein modifications, are necessary for activation of bZIP10 and 25. In contrast, the overexpression of bZIP53-GFP led to a strongly dwarfed phenotype, reflecting its important role in the regulatory C/S1 network.
Transactivation properties of S1/C bZIP heterodimers depend on the particular residue at positions 15 and 19 in the DBD
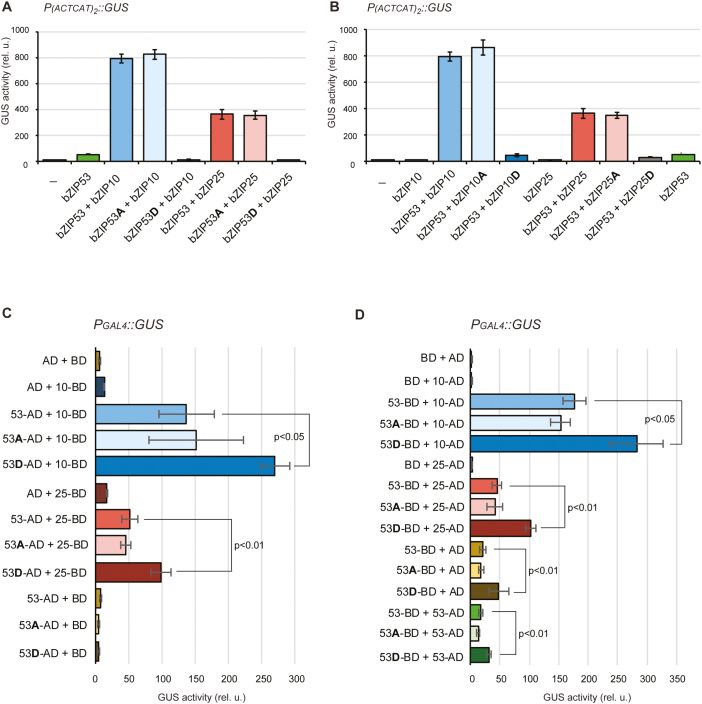
Since S1- and C-group members strongly tend to form inter-group heterodimers, we wondered how the introduction of a negative charge at positions 15 and 19 in the DBD of just one partner would influence the dimer transactivity. We therefore examined the activation of P(ACTCAT)2x::GUS upon co-expression of bZIP53 and its mutated versions either with bZIP10 or 25 in an Arabidopsis protoplast system. As compared to expression of bZIP53 alone, greatly induced GUS activity was observed upon co-expression of each C-group member with bZIP53 (Fig. 2A). Heterodimers of bZIP53 with bZIP10 induced stronger GUS activation than bZIP53/25 heterodimers. The activation of the GUS reporter was not affected by a Ser-to-Ala mutation in the bZIP53 DBD; however, it was completely abolished when the bZIP53 version contained a Ser-to-Asp mutation.
Fig. 2.
Analysis of the transcriptional activity and the heterodimerization of Arabidopsis bZIP factors. (A) Protoplast reporter gene assay of bZIP53, bZIP53Ser15,19Ala (bZIP53A) and bZIP53Ser15,19Asp (bZIP53D) co-expressed with bZIP10 or 25 as indicated. (B) Reciprocal assays with mutation of conserved Ser residues to Ala or Asp in bZIP10 and 25. P(ACTCAT)2x::GUS was used as the reporter. (C, D) Protoplast two-hybrid assays of bZIP53 with bZIP10 and 25. PGAL4::GUS was used as the reporter. (C) bZIP53 was fused to the GAL4 activation domain (AD), bZIP10 and 25 were fused to the GAL4 DNA-binding domain (BD). (D) Reciprocal assays applying bZIP53 and its mutated forms as BD-fusions and bZIP10 and 25 as AD-fusions. Data are means (±SD), n=3, of relative GUS/NAN activities. A, substitution of Ser15 and Ser19 to Ala; D, substitution of Ser15 and Ser19 to Asp. (This figure is available in colour at JXB online.)
Since bZIP53 binds directly to the ACTCAT core sequence in the reporter gene, we tested an alternative scenario, i.e. whether Ser-to-Asp mutation in the C-group member has a similar effect on the heterodimer-dependent P(ACTCAT)2x::GUS activation. In this case, the GUS activity was once again comparable when either the wild-type C-group factor or its Ser-to-Ala mutant version was co-expressed with bZIP53 (Fig. 2B). Conversely, the co-expression of bZIP53 with bZIP10 or 25 versions containing the Ser-to-Asp mutation drastically reduced activation of the reporter gene. It should, however, be noted that in this case the GUS activity was reduced only to a level comparable with the expression of bZIP53 alone (and not to the level of the ‘no effector’ control, as was observed upon expression of bZIP53Ser15,19Asp). This might indicate the existence of some intrinsic bZIP53 dimerization partners, which are probably able to outcompete bZIP10 or 25 under the tested conditions. Taken together, the results indicate that the substitution of the conserved Ser in the DBD with Asp in only one subunit is sufficient for the suppression of heterodimer transactivity.
In vivo interaction between bZIP53 and C-group bZIP factors
Directly adjacent to their DBD, bZIP factors contain the zipper domain that provides the hydrophobic surface for dimerization (Jakoby et al., 2002). As mentioned above, both C- and S1-group factors preferentially heterodimerize with each other (Ehlert et al., 2006; Llorca et al., 2015), which correlates with their in vivo functionality. To clarify whether the drastically reduced transactivity of bZIP factors with a Ser-to-Asp substitution in their DBD might be caused by their inability to form dimers, we carried out quantitative interaction studies using an Arabidopsis protoplast-based two-hybrid assay (Ehlert et al., 2006). The principle of the method is based on the fusion of one protein of interest with the GAL4 activation domain (AD) and another with the GAL4 binding domain (BD). Upon protein interaction, AD is recruited to the DNA sequence recognized by BD (PGAL4) and activates the PGAL4::GUS reporter gene. Although bZIP53-BD fusions alone could induce a certain level of GUS activation, no further GUS activation was detected after the co-expression of bZIP53-BD with bZIP53-AD. Instead, the co-expression of bZIP53 and its mutated forms with bZIP10 or 25 led to a strong increase of GUS activity compared to co-expression with BD or AD alone (Fig. 2C, D). As was observed in the transactivation assays, the GUS activity was higher in the case of bZIP53–bZIP10 dimers than of bZIP53–bZIP25 dimers. As shown in Fig. 2(C, D), we observed a comparable interaction of bZIP53 and 53Ser15,19Ala either with bZIP10 or 25 irrespective of AD- or BD-fusions. Interestingly, even higher GUS activity was detected when bZIP10 or 25 was co-expressed with bZIP53S15,19D.
We also examined the opposite situation, i.e. the interaction of bZIP53 with mutated bZIP10 or 25. Similar to the above results, Ser-to-Ala mutations in the DBD of bZIP10 or 25 did not affect their ability to form heterodimers with bZIP53, whereas Ser-to-Asp even enhanced the detected signal (Supplementary Fig. S1). These results indicate that the substitutions of Ser15 and Ser19 with Asp do not restrain the in vivo dimerization ability of bZIP factors.
The functionality of the nuclear localization signal is maintained in the mutated versions
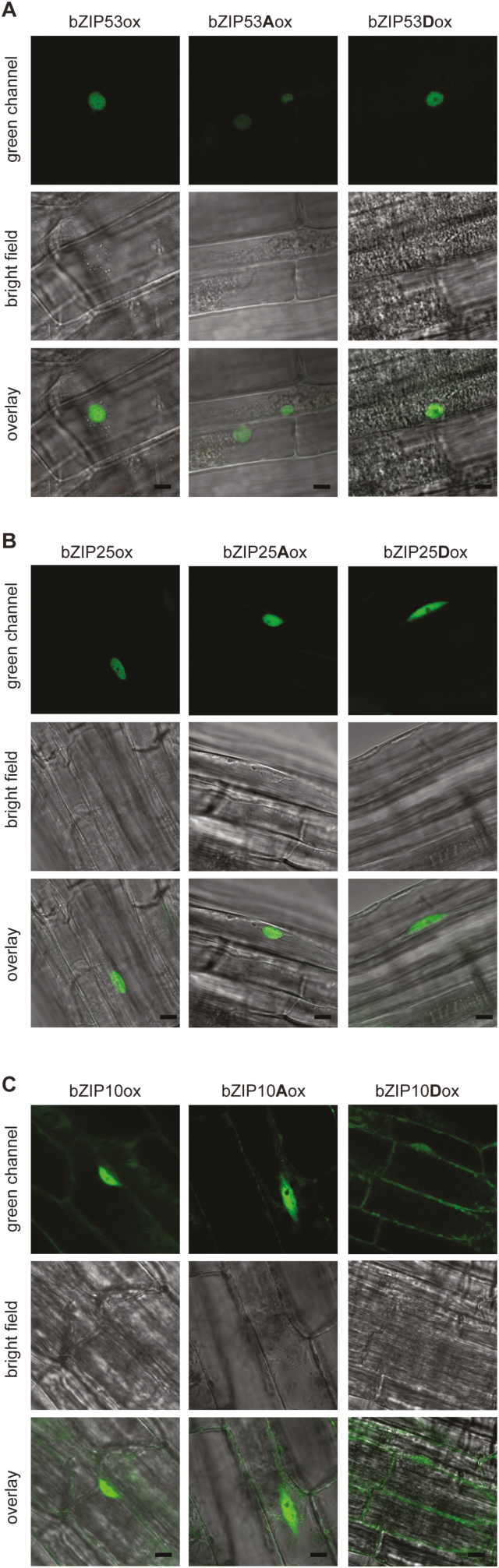
Since the basic region of bZIP factors also serves as a NLS, the phospho-mimicking mutations might disturb the correct localization of bZIPs, which could be the reason for their reduced transcriptional activity. To test this, we used both transiently transfected Arabidopsis leaf protoplasts and stably transformed lines. To visualize bZIP proteins, corresponding C-terminal GFP-fusions under the control of the 35S or UBI10 promoter were used. Upon expression of bZIP53-GFP, the GFP signal was detected only in the nucleus (Fig. 3A). Likewise, the bZIP25-GFP fusion was localized to the nucleus (Fig. 3B), which is in accordance with previous data for bZIP53 and 25 localization in transiently transformed Arabidopsis protoplasts (Llorca et al., 2015). The same nuclear localization of the GFP signal was observed when either Ser-to-Ala or Ser-to-Asp forms of bZIP53-GFP and bZIP25-GFP were examined (Fig. 3A, B). This demonstrated that mutations at positions 15 and 19 in the DBD do not influence the subcellular localization of bZIP53 or 25, and suggests that the functionality of the NLS is not disrupted.
Fig. 3.
Confocal images of Arabidopsis root epidermal cells expressing GFP-fused bZIP factors. The 2-week-old transgenic seedlings are expressing following sequences: (A) PUBI10::bZIP53-GFP (bZIP53ox), PUBI10::bZIP53Ser15,19Ala-GFP (bZIP53Aox), or PUBI10::bZIP53Ser15,19Asp-GFP (bZIP53Dox); (B) P35S::bZIP25-GFP (bZIP25ox), P35s::bZIP25Ser15,19Ala-GFP (bZIP25Aox), or P35S::bZIP25Ser15,19Asp-GFP (bZIP25Dox); and (C) P35S::bZIP10-GFP (bZIP10ox), P35s::bZIP10Ser15,19Ala-GFP (bZIP10Aox), or P35S::bZIP10Ser15,19Asp-GFP (bZIP10Dox). Scale bars are 5 μm.
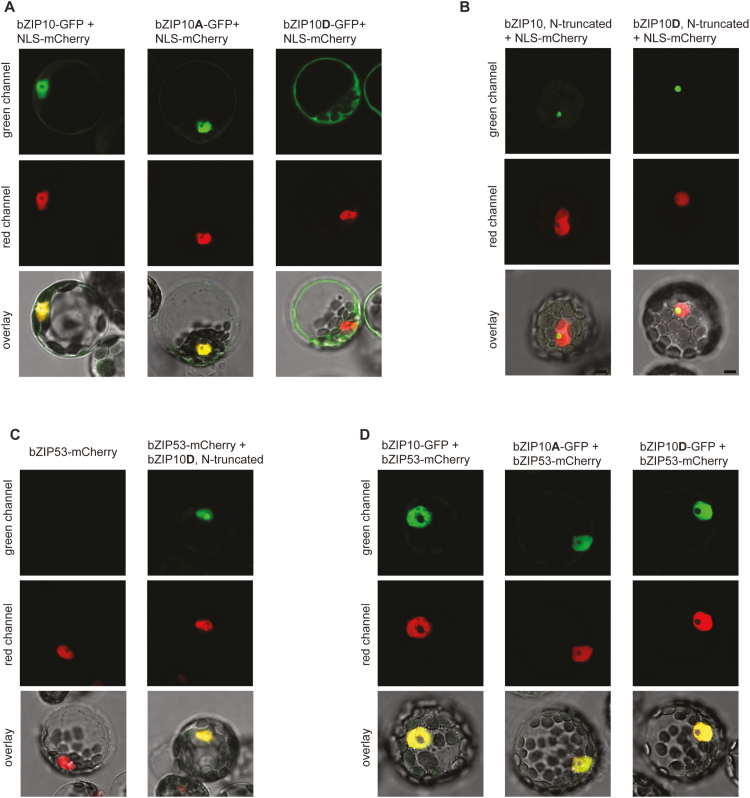
It has been previously shown that, due to the nuclear export signal in its N-terminus, bZIP10 can be detected in the nucleus and in the cytoplasm (Kaminaka et al., 2006). This nucleo-cytoplasmic localization was also observed in transgenic lines overexpressing bZIP10-GFP and bZIP10Ser15,19Ala-GFP (Fig. 3C); however, upon overexpression of the bZIP10Ser15,19Asp-GFP variant, the GFP signal was detected preferentially in the cytoplasm. A similar localization pattern was observed during transient expression of bZIP10 in Arabidopsis leaf protoplasts. In this case, however, Ser-to-Asp substitution led to the detection of the mutated variant exclusively in the cytoplasm (Fig. 4A). To determine whether such cytoplasmic localization was caused by the disruption of the NLS due to the Ser-to-Asp mutation, we used the GFP-bZIP10 version with a truncated N-terminus, which has previously been used to demonstrate the functionality of the NLS (Näke, 2001), and introduced the corresponding mutations. Both constructs were transiently expressed in Arabidopsis leaf protoplasts. Microscopic analysis of the protoplasts revealed similar nucleolar localization of the truncated bZIP10 peptides independently of the mutation in the DBD (Fig. 4B). These results clearly support the conclusion that Ser-to-Asp mutations in the DBD do not disrupt the NLS of the bZIP factors.
Fig. 4.
Confocal images of Arabidopsis protoplasts expressing GFP- and/or RFP-fused proteins. (A) Full-length bZIP10-GFP, bZIP10Ser15,19Ala-GFP, or bZIP10Ser15,19Asp-GFP fusions were co-expressed with nuclear-localization signal (NLS)-mCherry. (B) N-terminally truncated (lacking 215 amino acids) forms of GFP-bZIP10 and GFP-bZIP10Ser15,19Asp were co-expressed with NLS-mCherry. (C) N-terminally truncated form of GFP-bZIP10Ser15,19Asp was co-expressed with bZIP53-mCherry. (D) Full-length bZIP10-GFP, bZIP10Ser15,19Ala-GFP, or bZIP10Ser15,19Asp-GFP fusions were co-expressed with bZIP53-mCherry. A, substitution of Ser15 and Ser19 to Ala; D, substitution of Ser15 and Ser19 to Asp. The scale bar is 5 μm.
Given our findings of exclusively nuclear localization of bZIP53 (both wild-type and mutated forms) and cytoplasmic localization of bZIP10Ser15,19Asp, we investigated where their interaction occurs. To this end, we transiently expressed bZIP10Ser15,19Asp-GFP and bZIP53-mCherry in Arabidopsis leaf protoplasts. As shown in Fig. 4(C, D), co-expression of both proteins led to their exclusive nuclear localization, independent of the ΔN-truncated or full-length bZIP10 protein expression.
The dwarf phenotype of bZIP53-overexpressing plants is not observed for bZIP53S15,19D overexpression
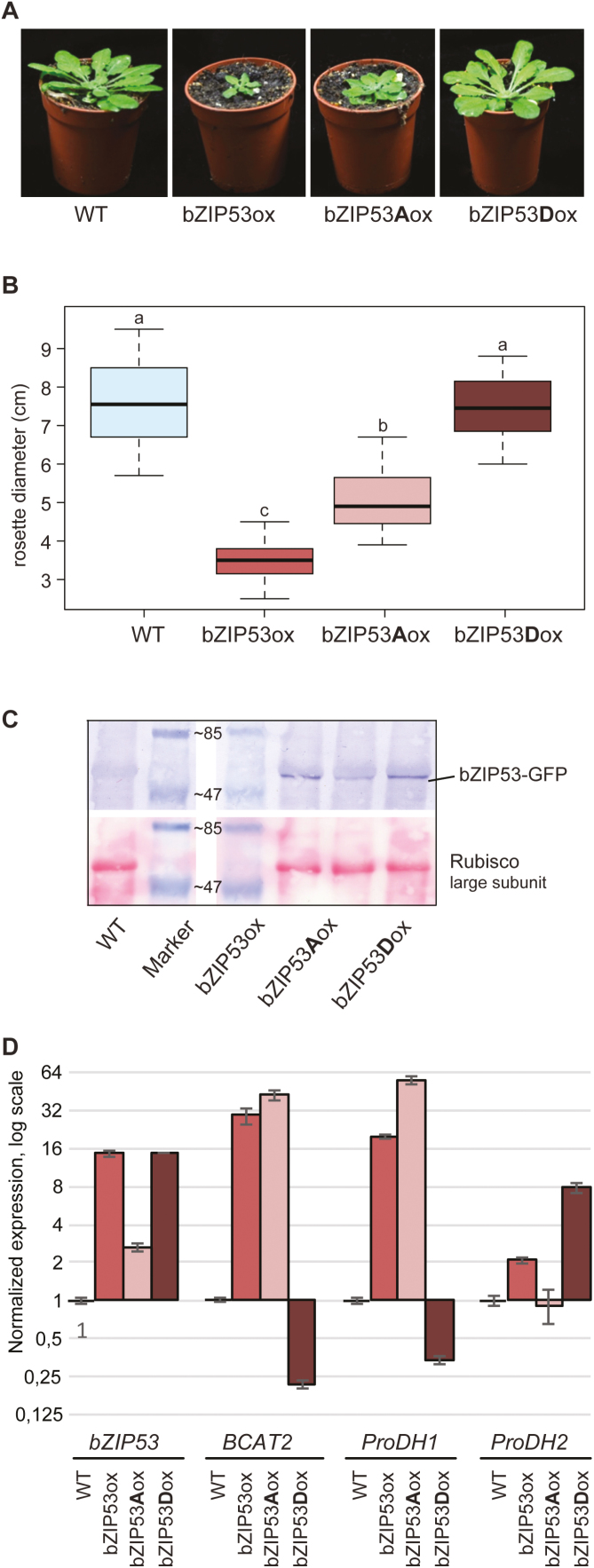
As shown above, the mutations of conserved DBD Ser to Asp disrupted the DNA binding and transactivity of bZIP factors in protoplasts. To address the possibility of targeted bZIP inactivation in plants, we generated transgenic Arabidopsis lines constitutively expressing the mutated variants of bZIP53-GFP. As already noted above, the overexpression of wild-type bZIP53 caused an extreme dwarf phenotype (Fig. 5A, B), which is associated with reduced seed production (Alonso et al., 2009; Dietrich et al., 2011). Overexpression of bZIP53Ser15,19Ala resulted in a slightly weaker dwarf phenotype compared to that of plants overexpressing bZIP53. This was probably due to the strong tendency of gene-silencing in bZIP53Ser15,19Ala-overexpressing plants, which was not observed for bZIP53 overexpressors (Fig. 5C). In sharp contrast, plants overexpressing bZIP53Ser15,19Asp did not show any phenotypic alterations compared to non-transformed Col-0 plants (Fig. 5A, B), despite the high level of the transgene expression (Fig. 5C).
Fig. 5.
Characterization of Arabidopsis lines overexpressing bZIP53 and its mutant forms. (A) Representative images of each transgenic line expressing coding sequences of bZIP53-GFP (bZIP53ox), bZIP53Ser15,19Ala-GFP (bZIP53Aox), or bZIP53Ser15,19Asp-GFP (bZIP53Dox). (B) Box-plot of leaf rosette diameter in the transgenic plants overexpressing bZIP53 and its mutant forms. Different letters indicate significantly different diameters (ANOVA, P<0.05); 12 plants from three independent lines for each construct were used. The seeds from the transgenic lines and corresponding wild-type belonged to the same seed batch. (C) Western blot analysis of the transgenic lines using anti-GFP antibody (upper panel). Ponceau staining was used to confirm the equal transfer of proteins onto the membrane (lower panel). The correct size of the bZIP53-GFP protein (48 kDa) was confirmed with peqGOLD Prestained Protein-Marker (PEQLAB, Lot No.1422). (D) RT-qPCR analysis of transgenic lines. For each gene, the expression level in the wild-type was set to 1, and different lines are presented as the mean of relative fold-changes compared to the control. Data are means (±SD), n=3. A, substitution of Ser15 and Ser19 to Ala; D, substitution of Ser15 and Ser19 to Asp.
To examine the physiological importance of bZIP53 Ser-to-Ala and Ser-to-Asp substitutions on the molecular level, we performed qPCR analysis using leaf RNA derived from transgenic and wild-type plants. The transcript levels of BRANCHED-CHAIN AMINO ACID TRANSFERASE 2 (BCAT2) and ProDH1, which are genes known to be regulated by S1-group bZIPs (Pedrotti et al., 2018), were up-regulated in plants overexpressing bZIP53 and 53Ser15,19Ala, whereas they did show down-regulation in plants overexpressing bZIP53Ser15,19Asp (Fig. 5D). In contrast, ProDH2, the known target gene of bZIP11 (Hanson et al., 2008), did not correlate with the expression of the active or inactive form of bZIP53. Using DPI-ELISA, we directly confirmed the disruption of the interaction with the cognate DNA sequence of bZIP heterodimers that contained just one subunit with the Ser15,19Asp substitution (Supplementary Fig. S2). Thus, these results indicated that the Ser-to-Asp mutation in the DBD of the bZIP factor creates a non-functional and apparently dominant-negative form of the transcription factor due to the perturbation of its DNA-binding capacity.
Altered amino acid metabolism in bZIP53 transgenic plants
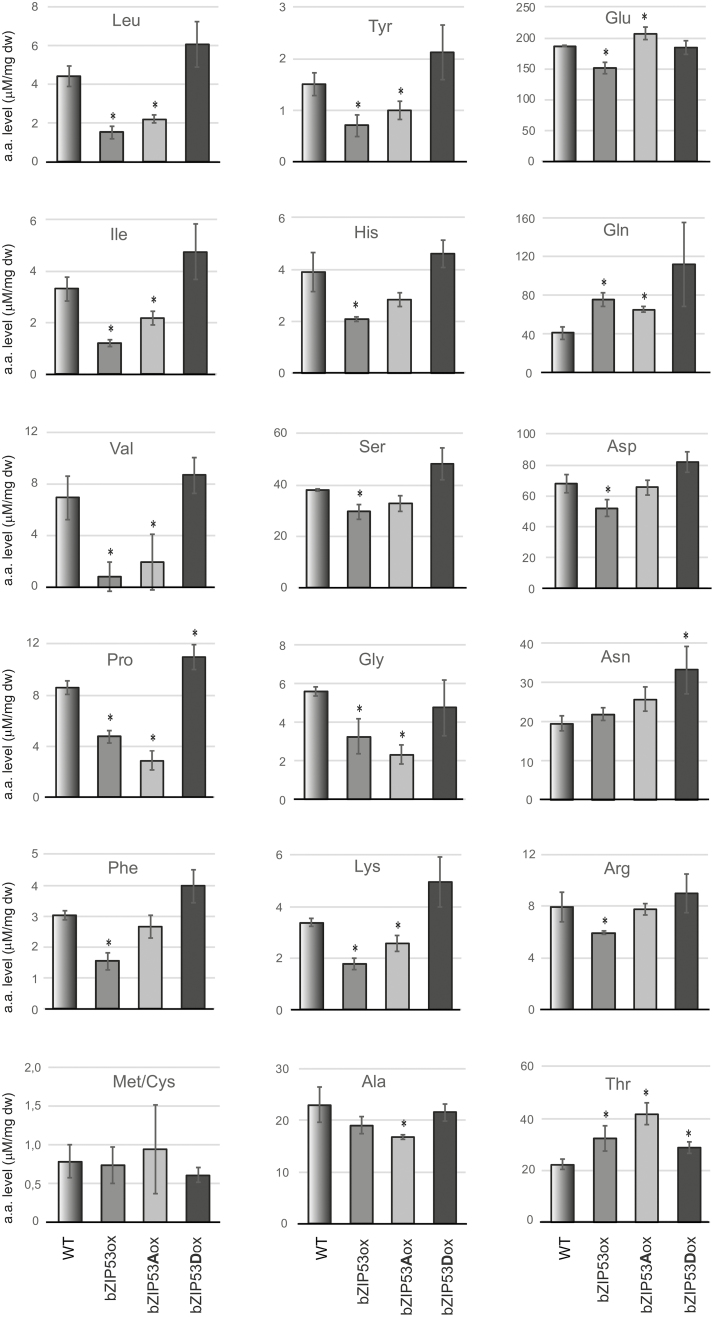
To date, the role of bZIP53 as a key regulator of metabolic reprogramming has mostly been shown in plants under energy starvation conditions, where it is mainly involved in the regulation of amino acid metabolism (Dietrich et al., 2011; Pedrotti et al., 2018). We decided to test whether bZIP53 might also be functioning under non-stressed conditions. Since it is known that S1-group bZIPs, including bZIP53, are post-transcriptionally repressed by sucrose (Hummel et al., 2009), and several C-group members are transcriptionally up-regulated during night periods (Weltmeier et al., 2009), the plants for the amino acid analysis were harvested at dawn, i.e. before light-induced activation of photosynthesis. Under these conditions, there should be a higher probability of expression of intrinsic bZIP53 and its heterodimerization partners. The amount of free amino acids in 7-week-old plants is shown in (Fig. 6). In transgenic lines with ectopic expression of either native bZIP53 or 53Ser15,19Ala, the concentration of several amino acids was significantly different from the wild-type, with decreased levels in both the lines. The strongest decrease was observed for proline (Pro) and all three branched-chain amino acids, whereas the level of these amino acids was moderately higher in transgenic plants overexpressing bZIP53Ser15,19Asp. However, due to considerable variation between samples (Supplementary Table S5), the difference between bZIP53Ser15,19Asp-overexpression (ox) and wild-type plants was significant only in the case of Pro. The levels of five further amino acids, Tyr, His, Ser, Gly, and Lys, were also substantially lower in bZIP53-ox and in bZIP53Ser15,19Alaox. At the same time, Tyr, Ser, and Lys showed a tendency to be increased in bZIP53Ser15,19Aspox plants as compared to the wild-type (Fig. 6). In addition, the total amount of free amino acids was higher in these plants compared to the others (Supplementary Table S5). Thus, the metabolism of these amino acids in plants at dawn showed a bZIP53-dependent pattern. Although the impact of the ectopically expressed active forms of bZIP53 was more pronounced, our approach allowed us to detect bZIP53-dependent differences in the plants overexpressing non-DNA-binding bZIP53Ser15,19Asp, supporting its dominant negative nature.
Fig. 6.
The amount of free amino acids in 7-week-old Arabidopsis plants expressing the following transgenes: bZIP53-GFP (bZIP53ox), bZIP53Ser15,19Ala-GFP (bZIP53Aox), or bZIP53Ser15,19Asp-GFP (bZIP53Dox). Data are means (±SD) from either three (wild-type, WT) or five (two independent homozygous lines per transgene) biological replicates. Significant differences compared to the WT were determined using Student’s t-test: *P<0.05.
It should be noted that we did not detect a significant increase in Asn levels in bZIP53- and 53Ser15,19Ala-overexpressing plants, as we had expected given previously published results (see Discussion). The amount of Asn seemed to be slightly decreased in the wild-type compared to all the transgenic lines. The same tendency was observed for Gln, where the difference was even larger and was significant (Fig. 6, Supplementary Table S5).
Discussion
Transcription factors containing a highly conserved bZIP domain were identified as regulators of gene expression a long time ago. Since then, extensive studies have been carried out to decipher the regulatory circuits involving the bZIP factors. The degree of complexity is especially high in plants when compared to other eukaryotes, due to a much larger protein family and a broad spectrum of homo- and heterodimerization (Dröge-Laser et al., 2018). An additional step toward complexity is caused by functional redundancy, which necessitates the generation of higher-order knockout mutants when trying to resolve functions (Dietrich et al., 2011; Pedrotti et al., 2018). Consequently, alternative approaches that implement different types of dominant negative mutants have been proposed (Hiratsu et al., 2003; Jain et al., 2017). In this study, we made use of highly conserved residues at positions 15 and 19 in the DBD of bZIP factors that can be potentially phosphorylated in order to achieve a highly specific inactivation of particular bZIP proteins in a dominant negative manner.
bZIP inactivation via phospho-mimicking amino acid substitution
As has been demonstrated through co-crystallization of bZIP–DNA complexes and in silico modeling, the residues at position 15 and 19 in the DBD are in direct contact with the DNA phosphodiester bonds (Sibéril et al., 2001; Miller et al., 2003; Kirchler et al., 2010). Interestingly, in plant bZIPs these positions contain conserved residues potentially capable of phosphorylation (Sibéril et al., 2001; Kirchler et al., 2010). We have shown that phospho-mimicking mutations lead to the inability of the bZIP53 factor to bind to the DNA, and consequently transcriptional activation of its target gene was completely impaired (Fig. 1). This makes the mutated form an excellent candidate for the in vivo inactivation of the intrinsic counterpart, provided that its other features, such as dimer formation and intracellular localization, remain unaltered.
Using complementary approaches, we demonstrated that the NLS, which overlaps with DNA-binding domain, remained functional in the Ser-to-Asp mutants of the bZIP factors that we tested (Fig. 4): the nuclear localization of both bZIP53 and 25 was not altered by the Ser-to-Asp substitution. Although bZIP10 localization was mostly shifted to the cytoplasm after Ser-to-Asp substitutions, the truncation of the N-terminal nuclear export signal completely eliminated bZIP10 from the cytoplasm, demonstrating the functionality of the NLS. It has previously been proposed that the cytoplasmic localization of bZIP10 is controlled by two mechanisms: XPO1-dependent export from the nucleus and LSD1-dependent cytoplasmic retention (Kaminaka et al., 2006). Our study revealed the impact of additional mechanisms, such as DNA-binding and dimerization. It appeared that dimerization with bZIP53 prevented the export of bZIP10 from the nucleus, independently of the interaction with the cognate DNA (Fig. 4D). In cases where bZIP10 is forced to form homodimers, for example due to very high expression levels in transient protoplast systems at high sucrose content in the medium (Hummel et al., 2009), its nuclear export may be restricted by the binding to the DNA. This is supported by the fact that impairment of DNA binding led to the exclusive cytoplasmic localization of bZIP10Ser15,19Asp in protoplasts (Fig. 4).
In order to maintain a high specificity of the generated mutant, a minimal alteration of dimerization preferences is desired. The dimerization specificity of bZIP factors is thought to depend on particular amino acid residues in the leucine zipper domain, especially at positions a, e, and g in the corresponding heptad repeats (Sibéril et al., 2001; Deppmann et al., 2004; Jain et al., 2017). For the C/S1 network, heterodimerization specificity is proposed to be facilitated by the exceptionally long zipper domain, consisting of up to eight repeats (Ehlert et al., 2006; Llorca et al., 2015; Dröge-Laser and Weiste, 2018). Therefore, the substitution of two residues in the DBD (i.e. positioned outside of the leucine zipper) theoretically should not influence the bZIP dimerization. In their attempt to create a dominant negative form of bZIP53, Jain et al. (2017) replaced the complete basic region with the peptide containing preferentially acidic residues. The resulting so-called A-ZIP53 demonstrated significantly altered affinity to its native counterpart and to its C-group partners bZIP10 and 25, as demonstrated by in vitro circular dichroism measurements. In our case, protoplast two-hybrid analysis showed stronger activation of PGAL4::GUS when one bZIP partner contained Ser-to-Asp substitutions (Fig. 2). This enhanced PGAL4::GUS activation, however, was also observed in the negative control when bZIP53Ser15,19Asp was used as bait (compared to BD-bZIP53). Since enhancement was observed in the absence of the dimerization partner, it therefore seems to be rather unspecific. A simple explanation might be that a portion of bZIP proteins expressed in the protoplasts, either as AD- or BD-fusions, might be bound to cognate DNA sequences on the Arabidopsis chromosomes and thus be less available for binding at the PGAL4::GUS. The non-DNA-binding mutant form, in contrast, is completely available. An alternative quantitative method, such as circular dichroism-based analysis, might help to verify this possibility. More importantly, Ser-to-Asp substitutions neither diminished the specific heterodimerization of bZIP53 with 10 or 25, nor enhanced the unspecific homodimerization of bZIP53 (Fig. 2, Supplementary Fig. S1).
Taken together, our data showed that a dominant negative mutant can be generated by ectopic expression of a particular bZIP factor that bears negatively charged residues at positions 15 and 19 in the DBD. As bZIP factors exist throughout all eukaryotes, this strategy may also be used in other organisms in addition to plants. The clear advantage of this approach is that it does not require tagging of the proteins, which otherwise may influence protein folding, localization, or its interactions. In contrast to the implementation of the EAR motif repression domain (Ohta et al., 2001; Hiratsu et al., 2003), Ser-to-Asp substitution does not reverse the function of the transcription factor from transactivating to repressing, it simply causes its inactivation by substituting it in corresponding dimers and hence blocks their binding to cognate DNA sequences. This would be also helpful when working with bZIP factors that function as transcriptional repressors. Furthermore, exploiting DBD Ser15,19Asp protein forms might prove to be especially useful in the case of bZIP factors with partially overlapping dimerization properties. Due to redundancy, knocking out a single gene usually does not lead to noticeable phenotypes. Saturation of such a bZIP network with a dominant-negative form—for example, through varying the gene doses using corresponding promoters—would lead to specific inactivation of the hub-dependent dimerization network through competition for the dimerization partner(s).
bZIP53 contributes to amino acid metabolism in non-stressed plants
In the last decade, great progress has been made in our understanding of low-energy signaling in plants. bZIP factors of C/S1 networks have been identified as key players that mediate transcriptional responses, both SnRK1-dependent and -independent, and lead to metabolic reprogramming during low-energy stress (reviewed by Dröge-Laser and Weiste, 2018). Although C- and S1-group factors generally work as heterodimers, S1 bZIPs, and especially bZIP11 and 53, seem to be major limiting factors as their ectopic expression leads to strongly dwarfed phenotypes (Hanson et al., 2008; Alonso et al., 2009). Due to a sugar-induced co-translational repression mechanism (Hummel et al., 2009), these proteins apparently occur at very low levels in photosynthesizing wild-type plants and hence are not available as dimerization partners for the C-group members. This may at least partially explain why the overexpression of either bZIP10, 25, or 63 caused no visible phenotype in plants at ambient conditions (Fig. 1; Kaminaka et al., 2006; Veerabagu et al., 2014; Mair et al., 2015).
In agreement with previous studies (Dietrich et al., 2011; Pedrotti et al., 2018), overexpression of bZIP53 caused a decrease in proline and branched-chain amino acids (Fig. 6) with concomitant transcriptional activation of ProDH1 and BCAT2, which encode the enzymes involved in the degradation of these amino acids (Fig. 5D). Similar changes were also observed in bZIP53Ser15,19Alaox plants. The regulation of these amino acids was, however, distinct: whereas the lower proline level in bZIP53ox was independent of the growth/harvesting conditions, differences for the branched-chain amino acids have so far been observed only during extended dark treatments (Dietrich et al., 2011). It appears that bZIP53 is important for the regulation of their catabolism during nocturnal growth periods as well, since we not only observed decreased levels in bZIP53ox and bZIP5353Ser15,19Alaox, but also a tendency to increased levels in bZIP53Ser15,19Aspox. Interestingly, amino acid analysis of the bzip1 bzip53 double-mutant has not shown an increase of branch-chained amino acids either in light-grown or in dark-grown plants, and only the quintuple-mutant with down-regulation of all five S1-group members shows increased levels of leucine and isoleucine after a 6-h extension of the night period (Dietrich et al., 2011; Pedrotti et al., 2018). The transcript level of BCAT2 has been reported to increase following the diurnal transition from light to dark, and to decrease during the light period (Peng et al., 2015). Thus, decreased BCAT2 expression and a tendency for increased levels of branched-chain amino acids in bZIP53Ser15,19Aspox at the end of the night period (Figs 5, 6) correspond well to a role of bZIP53 (and its dimerization partners) in the diurnally regulated catabolism of these amino acids. Previous studies on amino acid levels in the leaves of bcat2 mutants have shown partially contradictory results: whereas Angelovici et al. (2013) did not see any changes in the amino acid levels compared with the wild-type, Peng et al. (2015) found significant increases in Leu, Val, Trp, and His concentrations; they also found that Arg, Ile, Ser, Lys, and Tyr showed a clear tendency to increase, although this was not significant (P>0.05). These discrepancies might be caused by the harvesting time of the material. Unfortunately, this information was not provided by Angelovici et al. (2013), although harvesting was performed at the end of the night by Peng et al. (2015). Therefore, a similar tendency to increased levels of these amino acids that we observed in bZIP53Ser15,19Aspox (Fig. 6) might have been brought about, directly and indirectly, by impeded transcriptional activation of BCAT2. Taking into consideration a recently published study on S1-group quintuple-mutants (Pedrotti et al., 2018), the down-regulation of further catabolic genes involved in branched-chain amino acid degradation in the dominant negative bZIP53Ser15,19Aspox cannot be ruled out.
In addition to the regulation of ProDH1 and BCAT2, bZIP53 has previously been shown to activate the promoter of ASN1. This led to increased Asn levels in plants ectopically expressing bZIP53, with the increase being quite considerable in both light-grown and dark-grown plants, and a substantial decrease in Asn has been shown for the bzip1 bzip53 double-mutant (Alonso et al., 2009; Dietrich et al., 2011). On the other hand, compared to the wild-type no difference was observed either in ASN1 expression or the asparagine level in the S1-group quintuple-mutant under low-energy stress (a 6-h extension of the dark period), whereas ASN1 expression was down-regulated in the snrk1α1 snrk1α2 double-mutant under these conditions (Pedrotti et al., 2018). Dietrich et al. (2011) used 3-week-old plants grown under long-day conditions (16/8 h light/dark) whilst Pedrotti et al. (2018) used 3-week-old plants grown under short-day conditions (12/12 h). Our results from the analysis of 7-week-old plants growing under short-day conditions and harvested at the end of the dark period showed an insignificant increase of Asn level in bZIP53ox and bZIP53Ser15,19Alaox, as well as a significant increase in bZIP53Ser15,19Aspox plants. There might be several explanations for these different results. First of all, the harvesting time is of the great importance. In their kinetic analysis of ASN1 expression, Dietrich et al. (2011) detected an increase in bZIP53ox at all time-points except the one at dawn, and hence their findings are consistent with our data and explain the minimal increase in Asn level in bZIP53ox. On the other hand, Dietrich et al. (2011) found that the expression of ASPARAGINASE, which is involved in Asn degradation, was up-regulated in plants overexpressing bZIP1. Taking into account the overlapping dimerization partners for bZIP1 and 53, overexpression of the non-DNA-binding bZIP53 form could lead to interaction with, and removal of, the intrinsic C-group bZIPs from potential dimerization with bZIP1, thus restricting the activation capacity of bZIP1. As a consequence, higher Asn levels might be accumulated in the bZIP53Ser15,19Aspox plants. In addition, we used much older soil-grown plants in our experiment. Since Arabidopsis plants develop big rosettes under short-day conditions, the availability of nutrients, in particular nitrogen, may differ for 3-week-old and 7-week-old plants, leading to different gene regulation. Finally, the developmental stage per se is significant as well and influences the expression level of numerous genes, including bZIPs themselves and their modifying enzymes. In this context, crossing the bZIP53Ser15,19Alaox and bZIP53Ser15,19Aspox lines with bzip53 or other bzip mutants may help to shed light on the contribution of different bZIPs to the regulation of plant amino acid metabolism and development in Arabidopsis. Furthermore, the use of phospho-mimicking dominant negative bZIP mutants can help to further clarify the functional interconnections not only in the C/S1 network, but in other bZIP groups as well.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Analysis of the heterodimerization of bZIP53 with bZIP10 and 25 by protoplast two-hybrid assays.
Fig. S2. Binding of bZIP heterodimers to the cognate DNA sequences.
Table S1. Primer sequences used for Gateway cloning and mutagenesis.
Table S2. Primer sequences used for qPCR.
Table S3. Oligonucleotides used for the protein–DNA binding assay.
Table S4. Accession numbers of gene sequences used in this study.
Table S5. Summary of amino acid analysis in 7-week-old Arabidopsis leaf rosettes.
Acknowledgements
We thank Wolfgang Dröge-Laser and Niko Tintor for providing plasmids, Christel Kulibaba, Kerstin Haible, and Caterina Brancato for their excellent technical assistance, and the members of the MERIT Consortium for their helpful suggestions. This work was supported by a DFG grant to FW (SFB1101, Project D02), and by Marie Curie Actions FP7- People-2010-ITN (MERIT) to AG and CC.
Glossary
Abbreviations
- ASN1
asparagine synthetase 1
- BCAT2
branched-chain amino acid transferase 2
- DBD
DNA-binding domain
- GUS
glucuronidase
- NLS
nuclear localization signal
- ProDH
proline dehydrogenase
- SnRK1
SNF1-related protein kinase 1
References
- Alber T. 1992. Structure of the leucine zipper. Current Opinion in Genetics & Development 2, 205–210. [DOI] [PubMed] [Google Scholar]
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. 2009. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. The Plant Cell 21, 1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Bornberg-Bauer E, Oliver SG, Robertson DL. 2006. Reduction/oxidation-phosphorylation control of DNA binding in the bZIP dimerization network. BMC Genomics 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. 2013. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. The Plant Cell 25, 4827–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Hanson J. 2017. Shaping plant development through the SnRK1-TOR metabolic regulators. Current Opinion in Plant Biology 35, 152–157. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. 2008. Convergent energy and stress signaling. Trends in Plant Science 13, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D. 2010. DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C. 2004. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Research 32, 3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. 2011. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. The Plant Cell 23, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W, Snoek BL, Snel B, Weiste C. 2018. The Arabidopsis bZIP transcription factor family-an update. Current Opinion in Plant Biology 45, 36–49. [DOI] [PubMed] [Google Scholar]
- Dröge-Laser W, Weiste C. 2018. The C/S1 bZIP network: a regulatory hub orchestrating plant energy homeostasis. Trends in Plant Science 23, 422–433. [DOI] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W. 2006. Two-hybrid protein–protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. The Plant Journal 46, 890–900. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. 2000. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nature Structural Biology 7, 889–893. [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. The Plant Journal 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. 2008. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. The Plant Journal 53, 935–949. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W. 2006. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. The Plant Cell 18, 1931–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biometrical Journal 50, 346–363. [DOI] [PubMed] [Google Scholar]
- Hummel M, Rahmani F, Smeekens S, Hanson J. 2009. Sucrose-mediated translational control. Annals of Botany 104, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P, Shah K, Sharma N, Kaur R, Singh J, Vinson C, Rishi V. 2017. A-ZIP53, a dominant negative reveals the molecular mechanism of heterodimerization between bZIP53, bZIP10 and bZIP25 involved in Arabidopsis seed maturation. Scientific Reports 7, 14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F; bZIP Research Group. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Kaminaka H, Näke C, Epple P, et al. 2006. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. The EMBO Journal 25, 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kirchler T, Briesemeister S, Singer M, Schütze K, Keinath M, Kohlbacher O, Vicente-Carbajosa J, Teige M, Harter K, Chaban C. 2010. The role of phosphorylatable serine residues in the DNA-binding domain of Arabidopsis bZIP transcription factors. European Journal of Cell Biology 89, 175–183. [DOI] [PubMed] [Google Scholar]
- Llorca CM, Berendzen KW, Malik WA, Mahn S, Piepho HP, Zentgraf U. 2015. The elucidation of the interactome of 16 Arabidopsis bZIP factors reveals three independent functional networks. PLoS ONE 10, e0139884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. 1996. Coiled coils: new structures and new functions. Trends in Biochemical Sciences 21, 375–382. [PubMed] [Google Scholar]
- Mair A, Pedrotti L, Wurzinger B, et al. 2015. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLIFE 4, e05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. 2003. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. The Journal of Biological Chemistry 278, 15178–15184. [DOI] [PubMed] [Google Scholar]
- Näke C. 2001. Charakterisierung von CPRF2-homologen bZIP-Proteinen aus Arabidopsis thaliana unter besonderer Berücksichtigung ihrer intrazellulären Verteilung. PhD thesis, Freiburg University, Germany. [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti L, Weiste C, Nägele T, et al. 2018. Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. The Plant Cell 30, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL. 2015. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiology 169, 1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. 2009. Bimolecular fluorescence complementation (BiFC) to study protein–protein interactions in living plant cells. Methods in Molecular Biology 479, 189–202. [DOI] [PubMed] [Google Scholar]
- Shi L, Wu Y, Sheen J. 2018. TOR signaling in plants: conservation and innovation. Development 145, dev160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibéril Y, Doireau P, Gantet P. 2001. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. European Journal of Biochemistry 268, 5655–5666. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Hellmann HA. 2014. Sugar sensing and signaling in plants. Frontiers in Plant Science 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykowski A, Fischer SM, Zentgraf U. 2015. Phosphorylation affects DNA-binding of the senescence-regulating bZIP transcription factor GBF1. Plants 4, 691–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. 2012. Starch turnover: pathways, regulation and role in growth. Current Opinion in Plant Biology 15, 282–292. [DOI] [PubMed] [Google Scholar]
- Veerabagu M, Kirchler T, Elgass K, Stadelhofer B, Stahl M, Harter K, Mira-Rodado V, Chaban C. 2014. The interaction of the Arabidopsis response regulator ARR18 with bZIP63 mediates the regulation of PROLINE DEHYDROGENASE expression. Molecular Plant 7, 1560–1577. [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W. 2006. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. The EMBO Journal 25, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, et al. 2009. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Molecular Biology 69, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.