A comprehensive tissue type-specific transcriptional study identifies 45 genes consistently differentially expressed during Boechera sexual as compared with apomictic germline specification, suggesting their relevance for apomixis.
Keywords: Apomixis, Boechera, E3-ligase, F-box, laser-assisted microdissection, megasporogenesis, nucellus tissue, plant reproduction, RNA-seq, tissue-specific transcriptome
Abstract
Germline specification is the first step during sexual and apomictic plant reproduction, and takes place in the nucellus of the ovule, a specialized domain of the reproductive flower tissues. In each case, a sporophytic cell is determined to form the sexual megaspore mother cell (MMC) or an apomictic initial cell (AIC). These differ in their developmental fates: while the MMC undergoes meiosis, the AIC modifies or omits meiosis to form the female gametophyte. Despite great interest in these distinct developmental processes, little is known about their gene regulatory basis.
To elucidate the gene regulatory networks underlying germline specification, we conducted tissue-specific transcriptional profiling using laser-assisted microdissection and RNA sequencing to compare the transcriptomes of nucellar tissues between different sexual and apomictic Boechera accessions representing four species and two ploidy levels. This allowed us to distinguish between expression differences caused by genetic background or reproductive mode. Statistical data analysis revealed 45 genes that were significantly differentially expressed, and which potentially play a role for determination of the reproductive mode. Based on annotations, these included F-box genes and E3 ligases that most likely relate to genes previously described as regulators important for germline development. Our findings provide novel insights into the transcriptional basis of sexual and apomictic reproduction.
Introduction
The acquisition of reproductive or germline fate is a key step in the life cycle of higher plants, as it mediates the transition from the sporophytic to the gametophytic generation. The formation of the female germline in sexual plants initiates with the selection of a single sporophytic cell of the nucellus for reproductive fate, the megaspore mother cell (MMC). During megasporogenesis, the MMC undergoes meiosis to generate the functional megaspore (FMS) as the first cell of the gametophytic generation. Subsequently, three rounds of mitosis starting from the FMS followed by cellularization (gametogenesis) lead to the formation of the mature gametophyte. This consists of two female gametes (egg and central cell), and accessory synergid cells and antipodals (Sprunck and Gross-Hardt, 2011). Double-fertilization of the egg and central cell, which gives rise to the embryo and endosperm, respectively, completes the life cycle by starting a new sporophytic generation.
Apart from sexual reproduction through seeds, more than 400 angiosperm species produce seeds asexually in a process called gametophytic apomixis (hereafter referred to simply as apomixis; Asker and Jerling, 1992; Carman, 1995; Spillane et al., 2001). Apomixis comprises three consecutive developmental processes that differ from sexual reproduction. It begins with the formation of an unreduced female gametophyte (apomeiosis), which subsequently enables fertilization-independent embryogenesis (parthenogenesis), and fertilization-dependent or autonomous endosperm development (Nogler, 1984; Asker and Jerling, 1992). Generally, two major types of apomixis can be distinguished based on how the female germline is established, namely diplospory and apospory, (Nogler, 1984; Asker and Jerling, 1992; Koltunow, 1993; Bicknell and Koltunow, 2004). In diplospory, a single apomictic initial cell (AIC) is specified instead of the MMC. It typically omits meiosis, or enters a form of meiosis that is modified to proceed as a mitotic-like division, thus leading to the formation of an unreduced FMS. In contrast, apospory is characterized by an aposporous initial cell (also termed AIC), which specifies adjacent to a sexual MMC. This cell directly differentiates into an unreduced FMS without undergoing meiosis. In each case, the FMS develops further into a mature gametophyte as during sexual reproduction; however, gametes typically remain genetically identical to the mother plant. Subsequently, clonal offspring is formed by parthenogenesis.
As it is foreseen that complex genotypes from hybrid crops can be fixed over subsequent generations by harnessing apomixis, it holds great potential for use in agriculture. Hence, the understanding of germline specification in both sexual and apomictic reproduction is not only of particular scientific interest, but also can have practical implications in the future. Despite increasing interest in the developmental processes that lead to apomixis, knowledge about the underlying gene regulatory mechanisms is still scarce.
A widely accepted hypothesis states that apomixis results from asynchronous expression of genes of the sexual pathway (Carman, 1997). In accordance with this, there is increasing evidence to suggest that apomixis evolved from deregulation of the sexual reproductive pathway several times independently (Koltunow, 1993; Grimanelli et al., 2001; Grossniklaus et al., 2001; Koltunow and Grossniklaus, 2003; Tucker et al., 2003; Sharbel et al., 2009, 2010).
Studies in sexual model species such as Arabidopsis, Zea mays, and Oryza sativa have provided strong evidence that complex regulatory networks are required for megasporogenesis (Schmidt et al., 2015; Nakajima, 2018). Interestingly, mutations in a number of genes important for sexual germline development lead to aposporous- or diplosporous-like phenotypes in sexual species (Barcaccia and Albertini, 2013; Hand and Koltunow, 2014; Schmidt et al., 2015). These include regulatory components involved in restricting reproductive fate (Hand and Koltunow, 2014; Schmidt et al., 2015). In contrast to aposporous apomicts where the AIC typically forms adjacent to the sexual MMC, the formation of only one germline in each ovule is tightly controlled in sexual species (Albertini et al., 2005; Albrecht et al., 2005; Colcombet et al., 2005; Olmedo-Monfil et al., 2010; Schmidt et al., 2011; Singh et al., 2011, 2017; Wang et al., 2012; Zhao et al., 2017b; Cao et al., 2018). Developmental alterations resembling apospory have been described for mutations that disrupt members of epigenetic regulatory and small RNA pathways, for example Arabidopsis ARGONAUTE 9 (AGO9), RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), and SUPPRESSOR OF GENE SILENCING 3 (SGS3) (Ravi et al., 2008; Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010; Singh et al., 2017). In contrast, phenotypes resembling diplospory are induced in maize mutant lines of AGO104, a homologue of Arabidopsis AGO9, in addition to Arabidopsis DYAD, and in certain combinations of mutants in meiotic genes, for example in SPORYLATION 11-1 (SPO11-1), which is involved in pairing and recombination of homologous chromosomes, in addition to REC8, which takes part in chromatid segregation, and OMISSION OF SECOND DEVISION 1 (OSD1) (Ravi et al., 2008; d’Erfurth et al., 2009; Singh et al., 2011). However, it remains to be determined whether alterations in these genes and pathways underlie reproductive development in natural apomicts.
The genus Boechera is increasingly being appreciated as a model for studying apomictic reproduction (Sharbel et al., 2009, 2010; Aliyu et al., 2010, 2013; Corral et al., 2013; Lovell et al., 2013; Schmidt et al., 2014; Mau et al., 2015; Rojek et al., 2018). While apomixis is almost exclusively restricted to polyploids in most species, it occurs at low ploidy levels, including diploid and triploid, throughout Boechera (Bicknell and Koltunow, 2004; Windham and Al-Shehbaz, 2007; Lovell et al., 2013). Furthermore, both apospory and diplospory are represented (Mateo de Arias, 2015; Rojek et al., 2018). Most frequently diplospory is of the Taraxacum-type, which is characterized by first division restitution and normal meiosis II, but the Antennaria-type also occurs where the AIC directly adopts FMS identity without meiosis (Naumova et al., 2001; Mateo de Arias, 2015; Rojek et al., 2018).
Transcriptional profiling has previously been used to identify genes involved in apomictic development. Studies in Boechera have compared entire ovules of sexual and apomictic plants, while cell type-specific investigations have compared the sexual MMC of Arabidopsis and the apomictic AIC of triploid B. gunnisoniana (Sharbel et al., 2009, 2010; Schmidt et al., 2011, 2014). This has revealed several hundreds of genes that are differentially expressed in apomictic versus sexual germlines, which are involved in cell-cycle control, and epigenetic, transcriptional, and hormonal regulation (Schmidt et al., 2014).
To narrow down candidate genes of putative importance for apomixis and to minimize confounding effects of ploidy or species differences, we conducted a comprehensive investigation of two sexual and four apomictic Boechera accessions. We used laser-assisted microdissection combined with RNA sequencing to perform a comparative transcriptome analysis of nucellus tissues harbouring the MMC or AIC. This study therefore allowed us to examine germline specification in a tissue-specific manner. Particularly with regard to the female germline that develops inside of sporophytic flower tissues, cell or tissue type-specific investigations have great potential to identify important genes that might otherwise be overlooked due to an over-abundance of sporophytic tissues in the sample (Wuest et al., 2010; Schmid et al., 2012; Schmidt et al., 2012; Florez Rueda et al., 2016). Moreover, any relevant signalling processes involving cells of the surrounding nucellus tissue can simultaneously be detected. In this way, we identified 45 genes that were consistently differentially regulated between sexual and apomictic accessions, pointing to the involvement of genes and pathways previously described as regulators for germline development, namely cell-cycle regulation and stress responses.
Material and methods
Plant material
Seeds of Boechera Á.Löve & D.Löve were kindly provided by Timothy F. Sharbel (Global Institute for Food Security, University of Saskatchewan, Canada) and Thomas Mitchell-Olds (Duke University, USA), or were obtained from the laboratory of Ueli Grossniklaus (University of Zürich, Switzerland). Seeds were stratified for at least 1 week at 4 °C before surface-sterilization and germination as described previously (Wuest et al., 2010). About 2 weeks after germination, seedlings were transferred to a mixture of soil and sand as described previously (Schmidt et al., 2014). Plants were germinated and grown in a growth chamber with 55% relative humidity and under 16/8 h light/darkness at 20–22 °C. For each apomictic plant, ploidy was confirmed by flow cytometry using a CyStainUV Precise P Kit (Sysmex Partec, Görlitz', Germany) on a Partec CyFlow Space instrument following the manufacturer’s instructions.
Preparation of plant material for laser-assisted microdissection
For laser-assisted microdissection (LAM), flower buds with ovules were harvested that harboured either a MMC or AIC, predominantly before meiosis or apomeiosis, respectively, but covering a range of developmental stages until approximately prophase of meiosis I. Tissue fixation and manual embedding was done as described by Wuest et al. (2010), except that we applied a xylene gradient with 25%, 50%, 75%, and twice 100% xylene in EtOH. After blocking in Paraplast X-tra (Leica Biosystems, Nussloch, Germany), samples were stored at 4 °C until further use. Thin sections of 7 µm were prepared from embedded tissues using a RM2255 microtome (Leica Biosystems), mounted on 1.4 µm PET-membrane frame slides (Leica Microsystems, Wetzlar, Germany), and de-waxed according to Schmidt et al. (2011).
Laser-assisted microdissection
LAM was performed using an Eclipse Ti microscope (Nikon Instruments) equipped with a mmiCellCut instrument (Molecular Machines & Industries, Eching, Germany). Nucellus tissue was isolated from 7 μm thin sections using a 40× SPF Ph2 ELWD objective. As controls for RNA quality corresponding to the samples, ~10 ovary sections from the same slides as the nucelli sections were isolated afterwards. All samples were stored at –80 °C until RNA extraction.
RNA extraction and amplification
RNA extraction from LAM samples including DNA digest was done using PicoPure RNA isolation kits (ThermoFisher Scientific) following the manufacturer’s instructions, except that 15 µl extraction buffer was used. Each sample contained pooled individuals from the same accession to obtain 38–226 nucelli sections per sample (Supplementary Table S1 at JXB online). The RNA integrity of controls was determined using Agilent 2100 Bioanalyzer RNA Pico Chips. Good and reproducible RNA integrity was consistently achieved (Supplementary Fig. S1A).
A SMARTseq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio USA) was used for linear amplification of mRNA derived from nucelli samples following the manufacturer’s instructions (Supplementary Fig. S1B, C). Amplified cDNA was purified using AMPure Sample Purification Beads (Beckman Coulter, Brea, USA) and eluted in nuclease-free H2O and stored at –20 °C until subsequent library preparation.
RNA sequencing
A Nextera XT DNA Library Prep Kit (Illumina) was used to prepare libraries for RNA-seq of the nine samples (Supplementary Table S1) according to the manufacturer’s instructions. The quality of individual libraries and equimolar library pool was determined using Qubit (ThermoFisher Scientific) and Bioanalyzer High Sensitivity DNA assays (Agilent) (Supplementary Fig. S1D).
RNA-seq was conducted on one lane of a flow cell on a NextSeq 500 platform (Illumina) by the Deep Sequencing Core Facility of Heidelberg University at the EMBL Genomics Core Facility (EMBL, Heidelberg). The 75-bp single-end protocol was applied with 94 cycles (including index read). Original data files are deposited in the NCBI SRA database with the accession number SRP159014.
Data analysis
Pre-analysis and processing of raw reads
The quality of raw reads was assessed with FastQC version 0.11.2. For trimming with cutadapt version 1.14 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc;Martin, 2011) parameters were set to hard trimming of final base, and quality trimming at the 3´-end with phred score threshold 20. Adapter sequences were provided as fasta files. Trimmed reads shorter than 30 bases were filtered out. The B. stricta genome assembly and annotations from Lee et al. (2017) were used for read-mapping using STAR version 2.5.3a_modified (Dobin et al., 2013). The number of reads mapped per gene was determined with featureCounts as described previously (Liao et al., 2014; Schmid, 2017).
As quality controls, mapping statistics and gene body coverage were assessed with the modules bam_stat.py and geneBody_coverage.py from the RseQC package version 2.6.4 (Supplementary Fig. S2) (Wang et al., 2012).
Differential gene expression analysis
Read counts derived from featureCounts were used to analyse differential gene expression with the Bioconductor package edgeR version 3.12.1 (Robinson et al., 2010). Samples were grouped by accession and reproductive mode, low-expressed genes were filtered out (>1 counts per million bases in ≥2 libraries). Normalization factors for libraries were calculated with the TMM-method (Robinson and Oshlack, 2010). Normalized read counts were used for statistical analysis. For pairwise comparisons between apomictic and sexual accessions, the biological coefficient of variation was set to 0.8 as previously described (Schmidt et al., 2014). For comparisons treating groups of sexual or apomictic accessions as biological replicates, common and tagwise dispersions were estimated by edgeR. To identify genes differentially expressed between any of the accessions, dispersions were estimated using edgeR and a generalized linear model was applied to model all contrasts by grouping samples per accessions. All comparisons were performed with exact test and Benjamini–Hochberg adjustment of the false discovery rate (FDR). Differential expressed genes (DEGs) with FDR≤0.05 were considered significant. To identify genes that were differentially expressed in all comparisons of sexual against apomictic accessions, an intersection of gene lists was calculated in R. DEGs between any of the accessions were afterwards searched for core cell-cycle genes and meiotic genes (Vandepoele et al., 2002; Gutierrez, 2009; Mercier et al., 2015).
Validation of gene expression by comparison to published datasets
Our datasets were compared with published cell- and tissue type-specific transcriptome analyses of megasporogenesis in Arabidopsis and the triploid apomict B. gunnisoniana (Schmidt et al., 2011, 2014). Genes were defined to be expressed if they had either ≥10 mapped reads or present calls in ≥3 of 4 microarray replicates (Schmidt et al., 2011, 2014; this study).
Gene ontology analysis.
For gene ontology (GO) analysis we used the Bioconductor package topGO v.2.36.0 (http://bioconductor.org/packages/release/bioc/html/topGO.html). Over-representation of biological processes was done using Fisher’s exact test combined with the function ‘weight’. Genes with annotations for biological processes in the B. stricta genome were used as the gene universe (Lee et al., 2017).
Data visualization
Venn diagrams of overlapping gene expression were constructed using Venny 2.1 or Biovenn for comparison of four or fewer datasets, respectively (http://bioinfogp.cnb.csic.es/tools/venny/index.html;Hulsen et al., 2008).
Heatmaps showing expression levels of DEGs were generated using the R package gplots version 3.0.1. (https://CRAN.R-project.org/package=gplots). Heatmaps were based on log2-transformed, TMM-normalized read counts generated by NOISeq (Tarazona et al., 2011).
In situ hybridizations
Flower tissues for in situ hybridizations were prepared as for LAM, except that 8 µm thin sections were mounted on glass slides and dewaxed with Histoclear (Carl Roth, Karlsruhe, Germany). For cloning of in situ probes, RNA was extracted from B. stricta acc. LTM buds using an RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions and reverse-transcribed to cDNA using oligo dT12-dT18 primers and Superscript IV (ThermoFisher Scientific) following the manufacturer’s instructions. Sequences for probe synthesis were amplified using the primers listed in Supplementary Table S2. Probe synthesis, labelling with T7 RNA polymerase, and hybridizations were performed as described by Medzihradszky et al. (2014). An Axio Imager M1 (Zeiss) was used to capture images, which were cropped and processed in Adobe Photoshop CS2 Version 9.0.
Results
Transcriptome profiling identifies genes expressed in sexual and apomictic nucelli
To gain new insights into the gene regulatory networks that underlie germline specification during sexual and apomictic reproduction, tissue-specific RNA-seq was performed. The study design comprised four obligatory apomictic and two sexually reproducing accessions of the genus Boechera (Table 1) (Schranz et al., 2005; Aliyu et al., 2010; Mau et al., 2015; Lee et al., 2017). We selected accessions based on reproductive mode but also to include four different species at two ploidy levels to account for part of the genetic diversity of Boechera (Mau et al., 2015). Specifically, we included the sequenced accession B. stricta LTM, B. divaricarpa as a hybrid of B. stricta, together with B. williamsii and B. pallidifolia (Table 1) (Dobeš et al., 2004a; Lee, 2017). The origin of apomixis in Boechera is typically associated with hybridization or intraspecific crosses (Lovell et al., 2013), which might result in broader changes in gene regulation. Therefore, our study design enabled us to distinguish differential expression based on reproductive mode from effects of ploidy or genetic background.
Table 1.
List of Boechera accessions included in this study
| Boechera Species | Accession no. | Reproductive mode | Ploidy | Reference |
|---|---|---|---|---|
| B. divaricarpa | ES517 | Apomictic | 2x | Aliyu et al. (2010) |
| B. pallidifolia | B12-1578 | Apomictic | 2x | Mau et al. (2015) |
| B. pallidifolia | B12-1599 | Apomictic | 3x | Mau et al. (2015) |
| B. williamsii | B12-1524 | Apomictic | 2x | Mau et al. (2015) |
| B. williamsii | B12-558 | Sexual | 2x | Mau et al. (2015) |
| B. stricta | LTM | Sexual | 2x | Schranz et al. (2005), Lee et al. (2017) |
All accessions have been previously described as indicated, and represent different reproductive modes (sexual, apomictic), ploidy levels (2x, 3x), and species.
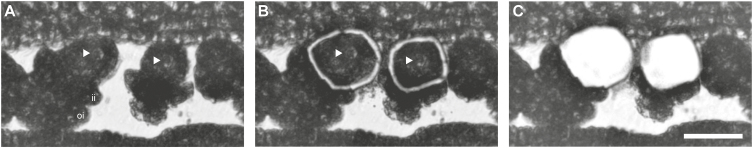
LAM was used to isolate nucelli from developing ovules at the onset of megasporogenesis. Tissues were precisely dissected from dry sections of young flower buds (Fig. 1B); however, minor cross-contamination with surrounding tissue could not be avoided completely. In order to identify genes consistently differentially regulated among all sexual and all apomictic accessions included in the analysis, a total of nine samples was prepared (Supplementary Table S1).
Fig. 1.
Precise sampling of nucelli using laser-assisted microdissection (LAM). Nucelli harbouring apomictic/aposporous initial cells (AICs) in ovary of apomictic Boechera pallidifolia (B12-1578) (A) before and (B) after LAM, which allows precise isolation with only ~2 µm cutting width of the laser beam. (C) After sample collection. Nuclei of AICs are marked with arrowheads. ii, inner integument; oi, outer integument. The scale bar is 30 µm.
For transcriptional profiling, RNA was extracted, amplified, and subjected to library preparation (Supplementary Fig. S1). RNA-seq was then performed and resulted in individual libraries that comprised 28 396 884–99 921 406 reads of overall good quality (Supplementary Table S1).
After quality controls and trimming, reads were mapped to the B. stricta reference genome using STAR (Dobin et al., 2013; Lee et al., 2017). On average, ~98% of the reads could be mapped per library (Supplementary Table S1). Unique reads constituted 95% of total reads on average, and gene bodies were covered homogenously by mapped reads (Supplementary Fig. S2). Further, 85% of total reads mapped to exonic regions, when counted with featureCounts (Liao et al., 2014). Taken together, the quality controls performed during processing of the RNA-seq reads, the general composition of read sequences, and the library statistics demonstrated an overall good quality of the data obtained (Table S1, Supplementary Figs S1–S3).
Global gene expression in Boechera nucelli widely overlaps
Nucellus tissues at similar developmental stages from related sexual and apomictic Boechera accessions were profiled. This provided a good basis to identify genes consistently transcribed and of general importance independent of the reproductive mode.
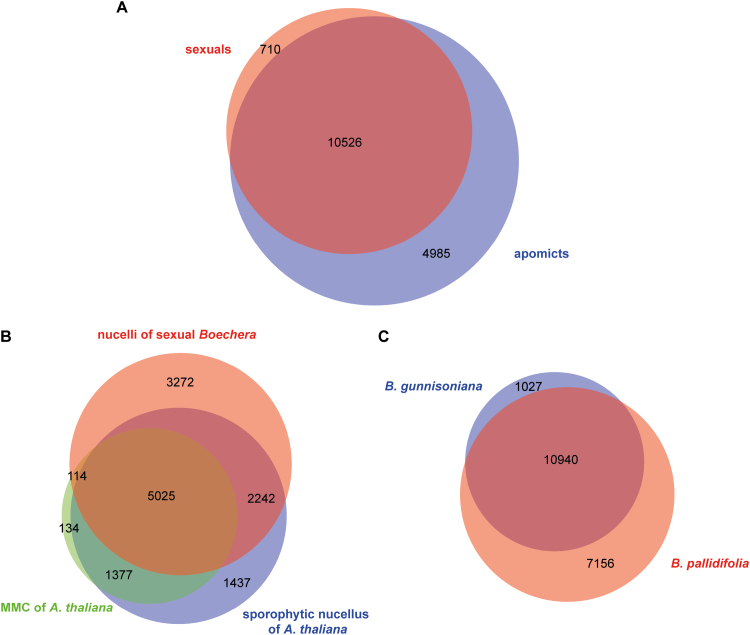
In total, we identified 24 197 genes that are expressed (≥10 normalized read counts) in at least one sample, thus representing 88% of the 27 416 genes in B. stricta (Supplementary Fig. S4) (Lee et al., 2017). Expression of 15 511 genes was shared among all the samples from apomicts, while 11 236 genes were expressed in all samples from sexual accessions (Fig. 2, Supplementary Fig. S4). Overlapping expression in all samples was detected for 10 526 genes, representing 38% of the genes in the genome (Fig. 2A).
Fig. 2.
Comparison of gene expression between different reproductive modes. (A) Venn diagram representing the overlap of genes expressed commonly in either all samples from apomictic or all samples from sexual accessions of Boechera (≥10 normalized read counts). (B) Venn diagram representing the intersections of genes expressed in isolated megaspore mother cells (MMCs) or sporophytic nucellar tissue of Arabidopsis (present calls in at least three of four of microarray replicates; Schmidt et al., 2011) and nucelli of all analysed samples of sexual Boechera accessions in the current study (LTM, B12.558). (C) Venn diagram representing the overlap of expressed genes (≥10 reads) in apomictic initial cells (AICs) of triploid B. gunnisoniana (Schmidt et al., 2014) and nucelli of B. pallidifolia (B12.1599, this study). Each comparison was based on annotated Arabidopsis homologues of corresponding Boechera genes, with available data having been generated on Affymetrix ATH1 microarrays for Arabidopsis or mapped using the reference transcriptome of B. gunnisoniana (B, C).
To classify biological processes that were up-regulated during germline specification independently of the reproductive mode, we applied a GO enrichment analysis on the consistently active set of 10 526 genes using topGO. Based on the available annotations for 33% of the genes of the genome, 37 GO terms were significantly enriched (P<0.01, Supplementary Table S3) (Lee et al., 2017). In agreement with previous findings for Arabidopsis MMCs (Schmidt et al., 2011), terms related to ‘translation’, ‘ubiquitin dependent protein catabolism’, and ‘ribosome biogenesis’ were amongst the significantly enriched processes, in addition to terms pointing towards the importance of redox homeostasis, signal transduction, cell-cycle, and different metabolic processes.
Despite similar regulation for a large number of genes, variation of expression between accessions of the same reproductive mode was also observed. However, only seven genes were identified to be expressed in all sexual accessions and not in any apomict, while 82 genes were exclusively active in all samples from apomicts (Supplementary Fig. S4A, B).
Overall, our analyses revealed widely overlapping gene expression patterns in nucelli of apomictic and sexually reproducing Boechera. Nevertheless, the comparisons indicated distinct regulation of a subset of genes according to the reproductive mode.
Comparisons to the transcriptomes of Arabidopsis MMCs and B. gunnisoniana AICs indicate the overall accuracy of the dataset
Cell and tissue type-specific transcriptome analyses have previously been described for Arabidopsis MMCs and surrounding nucellus tissues isolated separately, and the AICs of the triploid apomict B. gunnisoniana (Schmidt et al., 2011, 2014). To determine how much these datasets conform with ours, the transcriptomes were compared. Generally, comparisons were restricted to annotated Arabidopsis homologues (Fig. 2).
Overlapping expression in nucelli from all analysed sexual Boechera accessions (B12-558, LTM) compared with the MMCs and nucellar tissue without the MMCs of Arabidopsis was determined to comprise 5139 of 6650 (77%) and 7267 of 10 081 (72%) genes, respectively (Fig. 2B). In addition, ~91% (10 940) of 11 967 genes expressed (≥10 reads) in the AICs of B. gunnisoniana were also identified to be active in the triploid B. pallidifolia included in this study (B12-1599; Fig. 2C) (Schmidt et al., 2014). This high overlap of gene expression indicated an overall high accuracy of the dataset presented in this study.
We also checked the activity of the homologue of KNUCKLES, a marker gene for MMC identity in Arabidopsis (Tucker et al., 2012), and APOLLO, which shows allelic variants linked to apomixis in Boechera (Corral et al., 2013). In contrast to APOLLO, KNUCKLES was either expressed at low levels or was absent in all samples (Supplementary Fig. S5, Supplementary Table S4).
Few core cell-cycle and meiotic genes are differentially regulated between any accessions
As the sampled tissues harboured MMCs or AICs, we analysed transcription of meiotic genes and core cell-cycle regulators (Vandepoele et al., 2002; Gutierrez, 2009; Mercier et al., 2015). Consistent with previous findings (Schmidt et al., 2014), homologues of 16 out of 24 selected meiotic genes were active in samples from all accessions (Supplementary Fig. S5, Supplementary Table S4). To identify which of the genes were differentially regulated between accessions, we searched DEGs obtained using edgeR to apply a generalized linear model to compare all accessions (Supplementary Table S5) (Robinson et al., 2010). Interestingly, this identified SPO11-1 and REC8 in addition to the cell-cycle regulators PROLIFERATING CELL NUCLEAR ANTIGEN, CYCLIN D3;3, CYCLIN A2;3, CYCLIN B2;1, and E2F TRANSCRIPTION FACTOR 3 (Supplementary Fig. S5). However, the differences observed were not consistent between all sexual and all apomictic accessions.
Statistical analysis reveals genes consistently differentially expressed between apomictic and sexual nucelli
Despite the large number of genes active in all samples, differential expression would be expected for genes important for the determination of the reproductive mode or for processes specific to apomictic or sexual development. To identify such genes, statistical analysis of differential gene expression between apomictic and sexual nucelli samples was applied using edgeR (Robinson et al., 2010).
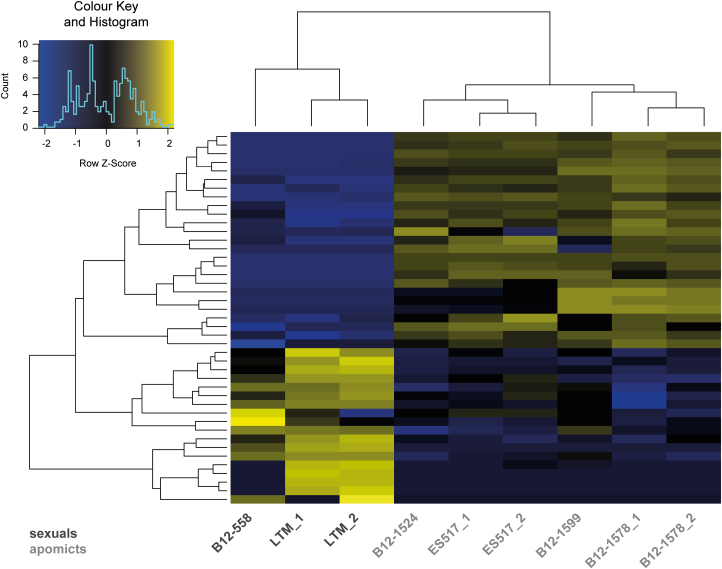
First, we compared the transcriptomes of all apomictic against all sexual samples, treating samples of same reproductive mode as biological replicates. Using this approach, 43 genes were identified to be significantly differentially expressed (FDR≤0.05 after Benjamini–Hochberg adjustment; Fig. 3, Supplementary Tables S6, S7). Of these DEGs, 28 were significantly up-regulated in apomictic and 18 in sexual samples. Hierarchical clustering and heatmap representation of the expression levels demonstrated a clear distinction between reproductive modes and a close relation between biological replicates of the same accession (Fig. 3). Moreover, the expression patterns of samples from the B. pallidifolia accessions (B12-1578 and B12-1599) clustered together, as did those of the apomictic B. divaricarpa (ES517) and apomictic B. williamsii (B12-1524) accessions.
Fig. 3.
Heatmap of log2-transformed read counts of differentially expressed genes (DEGs) in Boechera accessions. Expression levels are shown for 43 DEGs in all sexual as compared to all apomictic samples. The heatmap is based on TMM-normalized and log2-transformed read counts. The hierarchical clustering of samples and genes was based on Euclidean distance and hierarchical agglomerative clustering. The colours within the heatmap are scaled per row with blue indicating low expression and yellow high expression.
In a second approach, the ploidy and genetic backgrounds of the analysed accessions were taken into account. To this end, DEGs were identified by individual pairwise comparisons of sexual and apomictic accessions (FDR≤0.05 after Benjamini–Hochberg adjustment; Supplementary Table S7). Relevant candidate genes for germline specification were narrowed down by taking the intersection of all DEGs identified in individual pairwise comparisons. This resulted in the identification of seven common DEGs, which were all more highly expressed in apomictic than in sexual accessions (Supplementary Fig. S6). Five of these were consistently identified in both analyses (Table 2, Supplementary Table S8).
Table 2.
List of 43 genes differentially expressed between all samples originating from all apomictic versus sexual nucellar tissues isolated by laser-assisted microdissection
| Gene locus* | GO terms | Arabidopsis gene locus | Arabidopsis gene name | Arabidopsis gene/ protein description |
|---|---|---|---|---|
| (A) DEGs up-regulated in apomictic accessions | ||||
| Bostr.26527s0134 | GO:0030246, GO:0005975, GO:0004553 | At5G20710 | BGAL7 | Beta-galactosidase 7 |
| Bostr.7867s0569 | GO:0046872 | At4G27470 | AtRMA3, RMA3 | RING membrane-anchor 3 |
| Bostr.25375s0042 | NA | At1G64290 | NA | F-box protein-related |
| Bostr.15697s0319 | NA | At1G30790 | NA | F-box and associated interaction domains-containing protein |
| Bostr.15697s0321 | NA | At1G32660 | NA | F-box and associated interaction domains-containing protein |
| Bostr.13129s0275 | NA | NA | NA | NA |
| Bostr.29044s0001 | NA | At5G36710 | NA | NA |
| Bostr.3288s0214 | GO:0033177, GO:0015991, GO:0015078 | At1G19910 | AtVHA-C2, AVA-2PE, AVA-P2 | ATPase, F0/V0 complex, subunit C protein |
| Bostr.2983s0066 | GO:0005515 | At4G05310 | NA | Ubiquitin-like superfamily protein |
| Bostr.15697s0461 | GO:0046872 | At1G31480 | SGR2 | Shoot gravitropism 2 (SGR2) |
| Bostr.0568s0078 | GO:0008270 | At5G17890 | CHS3, DAR4 | DA1-related protein 4 |
| Bostr.0697s0107 | NA | At4G10400 | NA | F-box/RNI-like/FBD-like domains-containing protein |
| Bostr.26833s0971 | NA | NA | NA | NA |
| Bostr.19424s0775 | NA | NA | NA | NA |
| Bostr.10058s0023 | GO:0008168 | At4G00740 | NA | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| Bostr.13158s0243 | GO:0055114, GO:0009396, GO:0004488, GO:0003824 | At4G00620 | NA | Amino acid dehydrogenase family protein |
| Bostr.26959s0357 | GO:0055114, GO:0009396, GO:0004488, GO:0003824 | At4G00620 | NA | Amino acid dehydrogenase family protein |
| Bostr.7305s0030 | NA | At5G52390 | NA | PAR1 protein |
| Bostr.18351s0192 | GO:0005515 | At2G17310 | SON1 | F-box and associated interaction domains-containing protein |
| Bostr.3279s0010 | GO:0010468, GO:0005777 | At4G30760 | NA | Putative endonuclease or glycosyl hydrolase |
| Bostr.26833s0821 | NA | At5G54510 | DFL1, GH3.6 | Auxin-responsive GH3 family protein |
| Bostr.5022s0132 | NA | At2G21140 | AtPRP2, PRP2 | Proline-rich protein 2 |
| Bostr.25993s0534 | NA | At2G42480 | NA | TRAF-like family protein |
| Bostr.13158s0246 | GO:0055114, GO:0009396, GO:0004488, GO:0003824 | At4G00620 | NA | Amino acid dehydrogenase family protein |
| Bostr.13671s0276 | GO:0000062 | At1G31812 | ACBP, ACBP6 | Acyl-CoA-binding protein 6 |
| (B) DEGs up-regulated in sexual accessions: | ||||
| Bostr.13129s0114 | GO:0005515, GO:0003677 | At5G08430 | NA | SWIB/MDM2 domain; Plus-3; GYF |
| Bostr.26959s0127 | NA | At1G67800 | NA | Copine (Calcium-dependent phospholipid-binding protein) family |
| Bostr.7867s0507 | GO:0005975, GO:0004553 | At4G26830 | NA | O-Glycosyl hydrolases family 17 protein |
| Bostr.12396s0001 | GO:0055114, GO:0051287, GO:0016616 | At1G26570 | ATUGD1, UGD1 | UDP-glucose dehydrogenase 1 |
| Bostr.7867s1023 | GO:0003677 | At4G31680 | NA | Transcriptional factor B3 family protein |
| Bostr.18351s0250 | GO:0007165, GO:0000155, GO:0000160 | At2G17820 | AHK1, ATHK1, HK1 | Histidine kinase 1 |
| Bostr.7867s1594 | GO:0055114, GO:0020037, GO:0016705, GO:0005506 | At4G37330 | CYP81D4 | Cytochrome P450, family 81, subfamily D, polypeptide 4 |
| Bostr.0697s0084 | GO:0055085, GO:0016021 | At3G54700 | PHT1;7 | Phosphate transporter 1;7 |
| Bostr.26833s0862 | GO:0016758, GO:0008152 | At3G21790 | NA | UDP-Glycosyltransferase superfamily protein |
| Bostr.15774s0342 | GO:0055114, GO:0016614, GO:0050660 | At5G51950 | NA | Glucose-methanol-choline (GMC) oxidoreductase family protein |
| Bostr.2021s0098 | NA | NA | NA | NA |
| Bostr.29223s0097 | NA | At1G64230 | UBC28 | Ubiquitin-conjugating enzyme 28 |
| Bostr.2618s0047 | GO:0005975, GO:0004553 | At5G17500 | NA | Lycosyl hydrolase superfamily protein |
| Bostr.15697s0040 | NA | NA | NA | NA |
| Bostr.7867s0172 | NA | At1G25290 | AtRBL10, RBL10 | RHOMBOID-like protein 10 |
| Bostr.26833s0734 | NA | At5G55240 | AtPXG2 | ARABIDOPSIS THALIANA PEROXYGENASE 2 |
| Bostr.25993s0569 | NA | At3G53750 | ACT3 | Actin 3 |
| Bostr.3751s0034 | NA | At5G40680 | NA | Galactose oxidase/kelch repeat superfamily protein |
*Boechera stricta gene locus., Arabidopsis homologues are based on genome annotations by Lee et al. (2017). DEGs are ordered according to Supplementary Table S7 (sheet 1) and those identified consistently in both analyses are highlighted in bold. NA, no available information.
Despite the large overlap of global gene expression, our statistical data analysis identified 45 genes that were significantly differentially expressed in all sexual versus all apomictic samples (Supplementary Tables S6, S7). By comparing closely related Boechera accessions we reduced the influence of genetic backgrounds on our ability to identify promising new candidate genes with enriched expression in sexual or apomictic reproduction.
Annotations of DEGs suggest roles in protein degradation, transcriptional regulation and stress responses
Because it has been used as a model only relatively recently, knowledge about specific gene functions is scarce in the genus Boechera. However, its close relation to Arabidopsis and genome annotations of B. stricta provide a useful basis for investigations (Lee et al., 2017).
Arabidopsis homologues have been annotated for 21 890 genes in the B. stricta genome, equal to ~80% of the total of 27 416 genes (Lee et al., 2017). Based on this, DEGs could be attributed to the following different functional categories: protein degradation, transcriptional and post-transcriptional regulation of gene expression, redox processes and stress responses, and phytohormones and cell signalling (Table 2, Supplementary Tables S7, S8).
The first group, functionally related to protein degradation, in particular comprised genes coding for proteins with F-box and associated interaction domains (e.g. cyclin-like and Skp2-like domains), an F-box/RNI-like superfamily protein, or a protein related to F-box proteins. These genes were mostly up-regulated in apomictic nucelli (Table 2A, Supplementary Table S7). In addition, two E3 ligases were identified as DEGs: Bostr.7867s0569, the homologue of the E3 ligase RING MEMBRANE-ANCHOR 3 (RMA3), was consistently identified in both comparisons, and a gene encoding a Tumor Necrosis Factor Receptor Associated Factors-like (TRAF-like) family protein, Bostr.25993s0534, was found to have significantly higher expression levels in apomictic nucelli. In addition, two genes related to ubiquitination were represented in the set of DEGs: Bostr.29223s0097 and Bostr.2983s0066, the homologues of UBIQUITIN-CONJUGATING ENZYME 28 (UBC28), and of a ubiquitin-like superfamily protein, respectively.
In the second group, two DEGs related to transcriptional and post-transcriptional regulation processes were identified. A putative endonuclease, Bostr.3279s0010, was significantly up-regulated in apomictic nucelli in both analyses, whilst a B3 family transcription factor, Bostr.7867s1023, was identified as significantly lower expressed in apomictic than sexual nucelli (Table 2A, Supplementary Table S7).
The third group included DEGs related to redox processes and stress responses. Most prominently, a gene encoding a vacuolar ATPase subunit protein, Bostr.3288s0214, was found to have significantly higher expression levels in apomicts in both comparisons. Its homologue has previously been described to be related to abiotic stress responses (Kreps et al., 2002) (Table 2A, Supplementary Tables S7, S8). In contrast, several genes of this group had significantly lower levels of expression in apomictic nucelli. This subset was comprised of the homologue of Arabidopsis PEROXYGENASE 2 (AtPXG2), Bostr.26833s0734, a gene coding for a glucose-methanol-choline (GMC) oxidoreductase family protein, Bostr.15774s0342, and another oxidoreductase, a member of the cytochrome P450 superfamily CYP81D4, Bostr.7867s1594 (Table 2B, Supplementary Table S7).
In the fourth group, Arabidopsis homologues are involved in phytohormone-mediated cell signalling, or cell signalling per se. This group was comprised of a gene coding for an auxin-responsive GRETCHEN HAGEN 3 (GH3) family protein (GH3.6), Bostr.26833s0821, and the non-ethylene receptor HISTIDINE KINASE 1 (HK1), Bostr.18351s0250, which is a positive regulator in ABA signal transduction and drought responses (Tran et al., 2007) (Table 2, Supplementary Table S7).
Interestingly, apart from RMA3, the putative endonuclease and the gene encoding the vacuolar ATPase subunit protein, two genes with unknown function lacking any annotation, Bostr.13129s0275 and Bostr.19424s0775, were consistently detected with both approaches (Table 2A, Supplementary Tables S7, S8).
In summary, our dataset indicated consistent differential expression related to sexual and apomictic megasporogenesis for a rather small number of genes. The differences identified mainly pointed towards involvement of protein degradation mediated by poly-ubiquitination, transcriptional and post-transcriptional regulatory mechanisms, regulation by plant hormones, signal transduction, and stress responses. In addition, a few genes sharing no significant homology to Arabidopsis genes were identified as DEGs.
Independent validation of expression supports evidence for the candidate genes
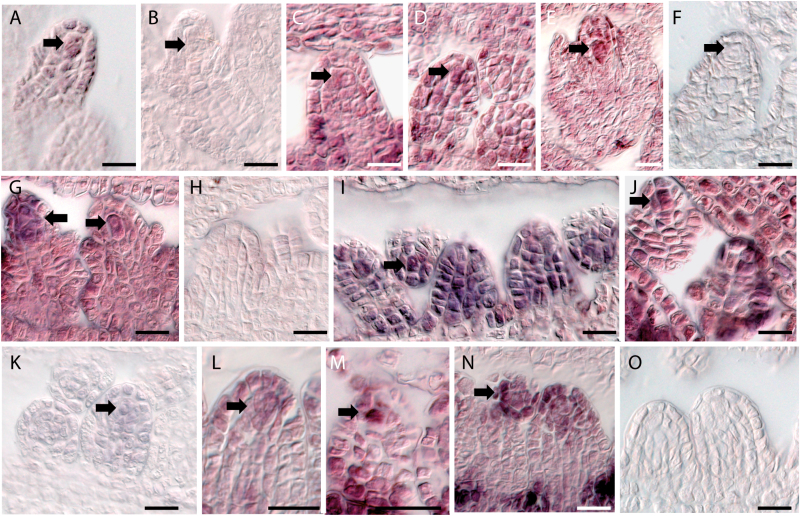
To independently confirm the expression in reproductive nucellus tissues we selected four DEGs for in situ hybridizations on B. stricta LTM or B. divaricarpa ES517 buds (Fig. 4). Probes were based on B. stricta LTM cDNA for the putative endonuclease Bostr.3279s0010 (Fig. 4A, B), the B3 transcription factor B3 TF (Bostr.7867s1023; Fig. 4C–F), CYP81D4 (Bostr.7867s1594; Fig. 4G–K), and for Bostr.13129s0275 (Fig. 4L–O). All probes were designed to have significant homology only to the gene of interest based on the B. stricta reference genome (Lee et al., 2017). Activity in the young reproductive nucellus tissues could be confirmed for the genes.
Fig. 4.
Independent data validation by in situ hybridization. Expression in reproductive nucellus tissues of young flower buds was confirmed for Bostr.3279s0010 in B. divaricarpa ES517 (A, B), for Bostr.7867s1023 in B. stricta LTM (C–F), for Bostr.7867s1594 in B. divaricarpa ES517 (G, H) and in B. stricta LTM (I–K), and for Bostr.13129s0275 in B. stricta LTM (L–O). In situ hybridizations were performed with antisense probes (A, C–E, G, I, J, L–N) or with sense probes as controls (B, F, H, K, O). Arrows indicate the specifying megaspore mother cells (MMCs) or apomictic/aposporous initial cells (AICs), with exception of (E) where it indicates the functional megaspore. Scale bars are 50 µm.
Discussion
Genes involved in cell-cycle regulation, transcriptional control, and stress responses are differentially regulated during sexual and apomictic germline specification
The identification of genes of putative relevance in determining the reproductive mode or in sustaining either sexual or apomictic development remains challenging. This is mainly due to the small size of the reproductive tissues. A previous comparison of gene activity between apomictic B. gunnisoniana and Arabidopsis identified almost 900 genes with evidence of expression that was different in the AIC compared with the MMC (Schmidt et al., 2014). This study provided important insights into the gene regulatory processes that govern germline specification in apomicts as compared with sexual plants; however, species and ploidy effects probably contributed to the differential regulation of such a large number of genes, thus complicating the identification of genes important for apomixis.
In our present study, different apomictic and sexual Boechera accessions were compared and our design allowed us to set apart differences due to ploidy or genetic background from those relevant for reproduction (Table 1, Supplementary Table S7). This allowed the number of DEGs to be narrowed down to 45 (Table 2, Supplementary Tables S6, S7, S8). Their consistent differential regulation in all sexual versus all apomictic samples indicated putative functional importance for discrimination of sexual and apomictic development. Interestingly, the genes identified could be functionally attributed to different regulatory mechanisms, including transcriptional and post-transcriptional regulatory processes, protein degradation, cell signalling, and stress responses. This is in agreement with previous findings (Schmidt et al., 2014, 2015; Mateo de Arias, 2015; Tang et al., 2017).
Cell-cycle control involving protein degradation is probably of crucial importance during germline specification
Interestingly, based on homology and annotations, some of the DEGs that we identified are potentially involved in pathways that are described to have regulatory roles either during germline development or in the restriction of reproductive fate (Kim et al., 2008; Gusti et al., 2009; Schallau et al., 2010; Schmidt et al., 2014; Singh et al., 2017; Tang et al., 2017; Zhao et al., 2017a, 2017b; Cao et al., 2018). For example, six of the identified candidates encode for proteins containing F-box and/or associated interaction domains (Table 2, Supplementary Table S8). F-box proteins are involved in a multitude of biological processes in plants (Lechner et al., 2006; Stefanowicz et al., 2015). They are part of Skp1-Cullin1-F-box protein (SCF) ligase complexes that act in 26S proteasomal degradation mediated by polyubiquitination (Skowyra et al., 1997). As an essential component of SCF complexes, they facilitate the regulation of many core cell-cycle genes and are thus responsible for the proper progression of the cycle, including the G1/S transition (Krek, 1998; Vodermaier, 2004). For example, Arabidopsis SCFSKP2A complex, which contains the F-box protein SKP2A, positively regulates cell division (del Pozo et al., 2002, 2006; Jurado et al., 2008). One of the F-box candidate genes, Bostr.15697s0319 (Table 2), even contains a cyclin-like and Skp2-like domain. Based on this homology of the interaction domains, it is probably part of an SCF complex that guides specific protein degradation during the progression of the cell-cycle. Interestingly, the Arabidopsis SCFFBL17 complex, which contains the F-box-like 17 protein, has been described to act specifically in male germ cells, where it targets the cyclin-dependent kinase inhibitors KIP-RELATED PROTEIN 6 (KRP6) and KRP7 to control germ cell proliferation (Kim et al., 2008; Gusti et al., 2009). KRPs in turn are also required to restrict female reproductive fate to only one MMC per ovule since they repress the cell-cycle regulator RETINOBLASTOMA-RELATED 1 (RBR1) via the cyclin-dependent kinase A;1 (CDKA;1) (Zhao et al., 2017b; Cao et al., 2018).
These interrelated findings demonstrate that F-box proteins are essential for cell-cycle regulation during germline development. In line with this, the F-box genes that we identified as being differentially regulated, particularly Bostr.15697s0319, might be part of the SCF complex(es) governing cell-cycle mechanisms that are distinct between apomeiosis and meiosis.
Distinct regulation of cell-cycle and meiotic genes was also observed between the Boechera accessions (Supplementary Fig. S5, Supplementary Table S4). Interestingly, triple-mutants in SPO11-1, REC8, and OSD1 lead to mitotic instead of meiotic division (d’Erfurth et al., 2009). However, the lack of consistent regulation observed in apomicts might relate to differences in apomeiotic development (Supplementary Fig. S5, Supplementary Table S4), and also reflect the developmental stage of sampled tissues that are predominantly before (apo)meiosis. Further studies focusing on the proteins encoded are needed to shed more light on their roles and regulation in Boechera.
Another DEG, Bostr.25993s0534 (Table 2), encodes a TRAF-like family protein. TRAFs comprise a class of E3 ubiquitin ligases with characteristic RING finger, TRAF, and Meprine And TRAF Homology (MATH) domains, which enable them to mediate interactions between other TRAF members, receptors, and several different intracellular signalling molecules (Ye et al., 1999; Zapata, 2003; Alvarez et al., 2010). In Arabidopsis, TRAF Mediated Gametogenesis Progression (TRAMGaP), which shares interaction domains with Bostr.25993s0534, is an important regulator of germline development involved in restricting reproductive fate (Singh et al., 2017). The expression of several genes that act in germline specification and regulation of sporophyte-to-gametophyte transition has been shown to be dependent on TRAMGaP. Importantly, these not only include RBR1 and the core meiotic gene DYAD, but also AGO9, SGS3, and RDR6 (Singh et al., 2017). While dyad mutants largely lead to sterility, formation of triploid offspring that fully retain parental heterozygosity has been observed at low frequencies (Ravi et al., 2008). Furthermore, apospory-like phenotypes have been described for ago9, sgs3, and rdr6 mutants (Ravi et al., 2008; Olmedo-Monfil et al., 2010). It is tempting to speculate that Bostr.25993s0534 might have similar functions to TRAMGaP. However, future functional studies are required to determine whether this gene mediates apospory or diplospory by targeting DYAD or genes active in the small RNA pathway, including AGO9.
Apart from this TRAF-like gene, additional DEGs with higher expression in the apomicts than in the sexual plants may be related to E3 ligases for which Arabidopsis homologs have previously been functionally described. In particular, Bostr.2983s0066 (Table 2) encodes for a ubiquitin-like superfamily protein homologous to At4G05310, for which a Cullin3A (CUL3A)-dependent expression has been reported (Dieterle et al., 2005). CUL3 proteins, which form E3 ligase complexes with BTB and MATH domain proteins among others, are required for female gametogenesis and cul3a cul3b plants show maternal effect embryo lethality (Dieterle et al., 2005; Thomann et al., 2005). Bostr.7867s0569 (Table 2, Supplementary Table S8) encodes a RING E3 ligase homologous to AtRMA3. Notably, AtRMA3 is expressed during pollen germination and pollen tube growth, and is co-expressed with UBC28 (Wang et al., 2008). UBC28 in turn belongs to the UBC8 group of proteins that are known to interact with RING E3 ligases (Kraft et al., 2005; Stone et al., 2005). Interestingly, the Boechera homologue of UBC28, Bostr.29223s0097, was also identified as a DEG in our study; however, it was more highly expressed in the sexual accessions, unlike the homologue of AtRMA3. HpARI7 completes the set of RING E3 ligases that have been described as potentially related to cell-cycle regulation during megasporogenesis, since it is a gene located on the apospory-linked Hypericum APOSPORY locus in H. perforatum (Schallau et al., 2010).
Taken together, our study identified a number of candidate genes that code for F-box proteins, E3 ligases, or associated factors. Their Arabidopsis homologues are directly or indirectly interrelated with previously described proteasomal degradation-mediated control of cell-cycle regulators and other targets, which influence reproductive fate decisions and germline development. This provides further evidence that tightly regulated protein degradation that affects cell-cycle progression may be crucial for governing the distinct specification and differentiation of apomictic and sexual germlines.
Involvement of stress responses and cell signalling in early megasporogenesis
In addition to those considered above, other regulatory mechanisms have previously been described to be involved in megasporogenesis, including stress responses and hormonal pathways (Schmidt et al., 2015), and some DEGs identified in our study were related to such pathways.
For example, oxidative stress and signalling, including reactive oxygen species (ROS), have been described to play roles in germline specification and possibly in meiosis (Schmidt et al., 2015). In accordance with this, members of the cytochrome P450 (CYP450) superfamily of oxidoreductases are enriched in the nucellar tissue of Arabidopsis compared to other tissues (Schmidt et al., 2011). Furthermore, they show cell cycle-dependent expression (Menges et al., 2002). Interestingly, recent findings have demonstrated that the Arabidopsis cytochrome P450 gene KLU (CYP78A5) is involved in restricting female germline fate to only one MMC per ovule (Zhao et al., 2017a). Likewise, Bostr.7867s1594 is a homologue of Arabidopsis CYP81D4 and a member of the CYP450 superfamily, which potentially functions in response to ROS (Table 2B). It had significantly higher expression levels in sexual than in apomictic nucelli. Thus, the detection of CYP81D4 and other DEGs related to redox processes (Bostr.26833s0734 and Bostr.15774s0342) provides further evidence that regulation of redox homeostasis and response to oxidative stress might contribute to the determination and differentiation of female sexual and apomictic germlines.
Beside their damaging properties and the consequent triggering of oxidative stress responses, ROS are also known to function as signalling cues between and within cells, for example during male germline development (Kelliher and Walbot, 2012; De Storme and Geelen, 2014). Similarly, cell-to-cell signalling mechanisms are also required during female germline specification and differentiation (Grossniklaus and Schneitz, 1998; Koltunow and Grossniklaus, 2003; Schmidt et al., 2015; Zhou et al., 2017). In accordance with this, some of the DEGs that we found could be attributed to cell signalling mechanisms, such as the homologue of AtHK1 (Table 2B), which potentially acts as a non-ethylene phytohormone receptor (Tran et al., 2007). Other DEGs that we found might be involved in auxin signalling in ovules, which is an important component of sporo- and gametogenesis (Li et al., 2008; Pagnussat et al., 2009; Schmidt et al., 2011; Lituiev et al., 2013; Freire Rios et al., 2014; Schaller et al., 2015). These were Bostr.7867s1023, a transcription factor of the B3 family, and Bostr.26833s0821, the homologue of AtGH3.6, for which auxin-responsiveness has been demonstrated previously (Nakazawa et al., 2001). Interestingly, AUXIN RESPONSE FACTORs belong to the same B3 superfamily of transcription factors and are well known as key regulators of auxin signalling (Chandler, 2016).
In summary, the putative functions of various candidate genes that we identified are consistent with the not yet fully understood complex regulatory network that governs germline specification and differentiation in plants. Notably, Tang et al. (2017) found DEGs with similar functional annotations in a comparative transcriptome analysis of entire flower buds of sexual and apomictic Boehmeria tricuspis. Moreover, based on a transcriptome analysis in Hieracium praeltum, some homologues of the DEGs that we identified were also differentially expressed between AICs as compared to the early stages of gametophyte development, or somatic cells of the ovule, or both, including homologues of AtPXG2, CYP81D4, GH3, and AtHK1 (Supplementary Table S9) (Juranić et al., 2018). In contrast to our study, the work of Juranić et al. (2018) included an investigation of different cell and tissue types in one aposporous apomict, further supporting evidence for roles of the genes in reproductive development.
Taken together, the DEGs that we identified might be involved in the different apomictic and sexual fate decisions by facilitating oxidative stress responses, auxin-driven cell-signalling, and in particular ubiquitination-mediated control of the cell-cycle.
Commonly and differentially regulated genes in apomictic and sexual nucelli
In accordance with the finding that hybrid origin or polyploidy are associated with most apomicts (Bicknell and Koltunow, 2004; Lovell et al., 2013), some variation of expression was observed between the apomictic accessions that we analysed (Supplementary Fig. S4, Supplementary Table S7). This might have been caused by the genomic effects of polyploidy and hybridization and is consistent with the hypothesis that apomixis derived several times independently (Sharbel and Mitchell-Olds, 2001; Koch et al., 2003; Dobeš et al., 2004a, 2004b; Kiefer et al., 2009; Kiefer and Koch, 2012; Lovell et al., 2013). However, it is still not fully understood whether broad deregulation of the sexual regulatory programme is the cause of or only correlated with apomixis. The rather small number of DEGs consistently identified in this study tends to suggest that a small number of genes is sufficient to sustain the switch from sexual reproduction to apomixis. This is in agreement with apomixis being genetically linked to only a few loci in most species (Barcaccia and Albertini, 2013).
Despite differential regulation of a subset of genes, almost 40% of all Boechera genes shared expression in all the samples that we analysed, regardless of the reproductive mode (Fig. 2, Supplementary Fig. S4). These genes probably include common regulators of reproductive development during megasporogenesis. However, gene expression for either reproductive mode appeared imbalanced, with more genes being expressed commonly in apomicts than in sexual nucelli. But fewer genes were identified to be active in sample LTM_2 compared to the other samples from sexual accessions because only 38 nucelli were dissected, representing the lowest input amount used. Thus, this apparent difference is probably not of biological relevance. In addition, for certain genes marked by ‘sexalleles’ and ‘apoalleles’, such as APOLLO (Corral et al., 2013), read-mapping to the genome of sexual B. stricta (Lee et al., 2017) possibly results in underestimation of expression levels in apomicts. Overall, the comparison of transcriptomes of Boechera nucelli with datasets from MMCs or nucellus tissue of Arabidopsis and B. gunnisoniana AICs (Schmidt et al., 2011, 2014), in addition to independent validations of selected genes by in situ hybridizations (Fig. 4), indicated overall good accuracy of our data.
In conclusion, our in-depth comparative analysis identified 45 genes that were differentially expressed between sexual and apomictic Boechera accessions. Most interestingly, the newly identified genes could be associated to the control of genes and pathways previously described to relate to regulation of apomixis. The identification of these genes will provide a very good basis for future functional investigations. Thus, our study contributes to a detailed understanding of the regulatory mechanisms that govern apomixis and mediate the specification of the female germline in apomictic and sexually reproducing plants in general.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Summary of samples and mapping statistics.
Table S2. Cloning of constructs for in situ probes.
Table S3. Gene ontology analysis.
Table S4. Expression of selected meiotic genes, core cell-cycle regulators, APOLLO, and KNUCKLES.
Table S5. Genes differentially expressed between any accessions.
Table S6. Transcriptome analysis of nucelli of apomictic and sexual Boechera.
Table S7. Differential expression analysis of nucelli of apomictic versus sexual Boechera.
Table S8. Table of common differentially expressed genes between apomictic and sexual nucelli.
Table S9. List of candidate genes, homologues of which are differentially expressed in Hieracium praealtum cells and tissue types.
Fig. S1. Quality controls of sample preparation for RNA-seq.
Fig. S2. Coverage of gene bodies by mapped reads.
Fig. S3. Assessment of read duplication rates.
Fig. S4. Visualization of overlap of gene expression.
Fig. S5. Heatmap of log2-transformed mean read counts of selected genes.
Fig. S6. Heatmap of log2-transformed read counts of common differentially expressed genes.
Author contributions
AS conceived the project; AS and LZ planned the experiments; LZ conducted the experiments with the support of DI and AS; LZ conducted the data analysis; CV performed the in situ hybridizations; LZ, AS, and DI interpreted the data; LZ and AS wrote the manuscript; all authors approved the manuscript.
Acknowledgements
We thank Timothy Sharbel (Global Institute for Food Security, Canada), Thomas Mitchell-Olds (Duke University, USA), and Ueli Grossniklaus (University of Zurich, Switzerland) for providing Boechera seeds, Joachim P. Spatz (Max Planck Institute for Medical Research, Germany), Marcus A. Koch and Jan U. Lohmann [Centre for Organismal Studies Heidelberg (COS), Germany] for contributing instruments and facilities, Christian Wenzl (COS Heidelberg, Germany) for an introduction to in situ hybridizations, Markus Kiefer (COS Heidelberg, Germany) for bioinformatics support, and Marc W. Schmid (MWSchmid GmbH, Switzerland) for helpful discussions and critical comments on the manuscript. We also acknowledge DFG for funding (SCHM2448/2-1 and SCHM2448/2-2) to AS.
Glossary
Abbreviations
- AGO9
ARGONAUTE 9
- AIC
apomictic/aposporous initial cell
- RMA3
RING MEMBRANE-ANCHOR 3
- AtPXG2
Arabidopsis thaliana PEROXYGENASE 2
- CUL3
CULLIN3
- CYP
cytochrome P450
- DEG
differentially expressed gene
- FMS
functional megaspore
- GH3
GRETCHEN HAGEN 3
- GO
, Gene Ontology;
- KRP
KIP-RELATED PROTEIN
- LAM
laser-assisted microdissection
- MMC
megaspore mother cell
- OSD1
OMISSION OF SECOND DEVISION 1
- RDR6
RNA-DEPENDENT RNA POLYMERASE 6
- SCF
Skp1-Cullin1-F-box protein ligase complex
- SGS3
SUPPRESSOR OF GENE SILENCING 3
- SPO11-1
SPORULATION 11-1
- TRAF-like
Tumor Necrosis Factor Receptor (TNF-R) Associated Factor-like
- TRAMGaP
TRAF Mediated Gametogenesis Progression
- UBC28
UBIQUITIN-CONJUGATING ENZYME 28
References
- Albertini E, Marconi G, Reale L, Barcaccia G, Porceddu A, Ferranti F, Falcinelli M. 2005. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiology 138, 2185–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. 2005. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. The Plant Cell 17, 3337–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliyu OM, Schranz ME, Sharbel TF. 2010. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). American Journal of Botany 97, 1719–1731. [DOI] [PubMed] [Google Scholar]
- Aliyu OM, Seifert M, Corral JM, Fuchs J, Sharbel TF. 2013. Copy number variation in transcriptionally active regions of sexual and apomictic Boechera demonstrates independently derived apomictic lineages. The Plant Cell 25, 3808–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez SE, Harikumar KB, Hait NC, et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465, 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker SE, Jerling L. 1992. Apomixis in plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Barcaccia G, Albertini E. 2013. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reproduction 26, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell RA, Koltunow AM. 2004. Understanding apomixis: recent advances and remaining conundrums. The Plant Cell 16, S228–S245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Wang S, Venglat P, Zhao L, Cheng Y, Ye S, Qin Y, Datla R, Zhou Y, Wang H. 2018. Arabidopsis ICK/KRP cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLoS Genetics 14, e1007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman JG. 1995. Gametophytic angiosperm apomicts and the occurrence of polyspory and polyembryony among their relatives. Apomixis Newsletter 8, 39–53. [Google Scholar]
- Carman JG. 1997. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biological Journal of the Linnean Society 61, 51–94. [Google Scholar]
- Chandler JW. 2016. Auxin response factors. Plant, Cell & Environment 39, 1014–1028. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. 2005. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. The Plant Cell 17, 3350–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral JM, Vogel H, Aliyu OM, Hensel G, Thiel T, Kumlehn J, Sharbel TF. 2013. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiology 163, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. 2009. Turning meiosis into mitosis. PLoS Biology 7, e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Geelen D. 2014. The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant, Cell & Environment 37, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C. 2002. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. The Plant Cell 14, 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. 2006. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. The Plant Cell 18, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Thomann A, Renou JP, Parmentier Y, Cognat V, Lemonnier G, Müller R, Shen WH, Kretsch T, Genschik P. 2005. Molecular and functional characterization of Arabidopsis Cullin 3A. The Plant Journal 41, 386–399. [DOI] [PubMed] [Google Scholar]
- Dobeš C, Mitchell-Olds T, Koch MA. 2004a. Intraspecific diversification in North American Boechera stricta (=Arabis drummondii), Boechera ×divaricarpa, and Boechera holboellii (Brassicaceae) inferred from nuclear and chloroplast molecular markers—an integrative approach. American Journal of Botany 91, 2087–2101. [DOI] [PubMed] [Google Scholar]
- Dobeš CH, Mitchell-Olds T, Koch MA. 2004b. Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. × divaricarpa, and A. holboellii (Brassicaceae). Molecular Ecology 13, 349–370. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez Rueda AM, Grossniklaus U, Schmidt A. 2016. Laser-assisted microdissection (LAM) as a tool for transcriptional profiling of individual cell types. Journal of Visual Experiments 10, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire Rios A, Yoshida S, Weijers D. 2014. Auxin regulation of embryo development. In: Zažímalová E, Petrášek J,Benková E, eds. Auxin and its role in plant development. Vienna: Springer, 171–189. [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. 2010. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. The Plant Cell 22, 3249–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Perotti E, Grossniklaus U. 2001. Developmental genetics of gametophytic apomixis. Trends in Genetics 17, 597–604. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Nogler GA, van Dijk PJ. 2001. How to avoid sex: the genetic control of gametophytic apomixis. The Plant Cell 13, 1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K. 1998. The molecular and genetic basis of ovule and megagametophyte development. Seminars in Cell & Developmental Biology 9, 227–238. [DOI] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P. 2009. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4, e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. 2009. The Arabidopsis cell division cycle. The Arabidopsis Book 7, e0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand ML, Koltunow AM. 2014. The genetic control of apomixis: asexual seed formation. Genetics 197, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Díaz-Triviño S, Abraham Z, Manzano C, Gutierrez C, del Pozo C. 2008. SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. The Plant Journal 53, 828–841. [DOI] [PubMed] [Google Scholar]
- Juranić M, Tucker MR, Schultz CJ, Shirley NJ, Taylor JM, Spriggs A, Johnson SD, Bulone V, Koltunow AM. 2018. Asexual female gametogenesis involves contact with a sexually-fated megaspore in apomictic Hieracium. Plant Physiology 177, 1027–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V. 2012. Hypoxia triggers meiotic fate acquisition in maize. Science 337, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Dobeš C, Sharbel TF, Koch MA. 2009. Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera – a genus and continental-wide perspective. Molecular Phylogenetics and Evolution 52, 303–311. [DOI] [PubMed] [Google Scholar]
- Kiefer C, Koch MA. 2012. A continental-wide perspective: the genepool of nuclear encoded ribosomal DNA and single-copy gene sequences in North American Boechera (Brassicaceae). PLoS ONE 7, e36491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG. 2008. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455, 1134–1137. [DOI] [PubMed] [Google Scholar]
- Koch MA, Dobeš C, Mitchell-Olds T. 2003. Multiple hybrid formation in natural populations: concerted evolution of the internal transcribed spacer of nuclear ribosomal DNA (ITS) in North American Arabis divaricarpa (Brassicaceae). Molecular Biology and Evolution 20, 338–350. [DOI] [PubMed] [Google Scholar]
- Koltunow AM. 1993. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. The Plant Cell 5, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. 2003. Apomixis: a developmental perspective. Annual Review of Plant Biology 54, 547–574. [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. 2005. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiology 139, 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W. 1998. Proteolysis and the G1–S transition: the SCF connection. Current Opinion in Genetics & Development 8, 36–42. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. 2002. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology 130, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. 2006. F-box proteins everywhere. Current Opinion in Plant Biology 9, 631–638. [DOI] [PubMed] [Google Scholar]
- Lee CR, Wang B, Mojica JP, et al. 2017. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nature Ecology & Evolution 1, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Qin GJ, Tsuge T, et al. 2008. SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytologist 179, 751–764. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lituiev DS, Krohn NG, Müller B, Jackson D, Hellriegel B, Dresselhaus T, Grossniklaus U. 2013. Theoretical and experimental evidence indicates that there is no detectable auxin gradient in the angiosperm female gametophyte. Development 140, 4544–4553. [DOI] [PubMed] [Google Scholar]
- Lovell JT, Aliyu OM, Mau M, Schranz ME, Koch M, Kiefer C, Song BH, Mitchell-Olds T, Sharbel TF. 2013. On the origin and evolution of apomixis in Boechera. Plant Reproduction 26, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Mateo de Arias M. 2015. Effects of plant stress on facultative apomixis in Boechera (Brassicaceae). PhD thesis, Utah State University. [Google Scholar]
- Mau M, Lovell JT, Corral JM, Kiefer C, Koch MA, Aliyu OM, Sharbel TF. 2015. Hybrid apomicts trapped in the ecological niches of their sexual ancestors. Proceedings of the National Academy of Sciences, USA 112, E2357–E2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky A, Schneitz K, Lohmann JU. 2014. Detection of mRNA expression patterns by nonradioactive in situ hybridization on histological sections of floral tissue. Methods in Molecular Biology 1110, 275–293. [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. 2002. Cell cycle-regulated gene expression in Arabidopsis. The Journal of Biological Chemistry 277, 41987–42002. [DOI] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annual Review of Plant Biology 66, 297–327. [DOI] [PubMed] [Google Scholar]
- Nakajima K. 2018. Be my baby: patterning toward plant germ cells. Current Opinion in Plant Biology 41, 110–115. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. 2001. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. The Plant Journal 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Naumova TN, van der Laak J, Osadtchiy J, Matzk F, Kravtchenko A, Bergervoet J, Ramulu KS, Boutilier K. 2001. Reproductive development in apomictic populations of Arabis holboellii (Brassicaceae). Sexual Plant Reproduction 14, 195–200. [DOI] [PubMed] [Google Scholar]
- Nogler GA. 1984. Gametophytic apomixis. In: Johri BM, ed. Embryology of angiosperms. Berlin, Heidelberg: Springer. [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. 2010. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. 2009. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Ravi M, Marimuthu MP, Siddiqi I. 2008. Gamete formation without meiosis in Arabidopsis. Nature 451, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek J, Kapusta M, Kozieradzka-Kiszkurno M, Majcher D, Gorniak M, Sliwinska E, Sharbel TF, Bohdanowicz J. 2018. Establishing the cell biology of apomictic reproduction in diploid Boechera stricta (Brassicaceae). Annals of Botany 20, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallau A, Arzenton F, Johnston AJ, Hähnel U, Koszegi D, Blattner FR, Altschmied L, Haberer G, Barcaccia G, Bäumlein H. 2010. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. The Plant Journal 62, 773–784. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW. 2017. RNA-seq data analysis protocol: combining in-house and publicly available data. Methods in Molecular Biology 1669, 309–335. [DOI] [PubMed] [Google Scholar]
- Schmid MW, Schmidt A, Klostermeier UC, Barann M, Rosenstiel P, Grossniklaus U. 2012. A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS ONE 7, e29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Grossniklaus U. 2012. Analysis of plant germline development by high-throughput RNA profiling: technical advances and new insights. The Plant Journal 70, 18–29. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Grossniklaus U. 2015. Plant germline formation: common concepts and developmental flexibility in sexual and asexual reproduction. Development 142, 229–241. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Schmid MW, Klostermeier UC, Qi W, Guthörl D, Sailer C, Waller M, Rosenstiel P, Grossniklaus U. 2014. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genetics 10, e1004476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U. 2011. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biology 9, e1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Dobeš C, Koch MA, Mitchell-Olds T. 2005. Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). American Journal of Botany 92, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Mitchell-Olds T. 2001. Recurrent polyploid origins and chloroplast phylogeography in the Arabis holboellii complex (Brassicaceae). Heredity 87, 59–68. [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Galla G, Kumlehn J, Klukas C, Schreiber F, Vogel H, Rotter B. 2010. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. The Plant Cell 22, 655–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Thiel T, Varshney A, Kumlehn J, Vogel H, Rotter B. 2009. Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. The Plant Journal 58, 870–882. [DOI] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. 2011. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. The Plant Cell 23, 443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Kumar V, Srinivasan R, Ahuja PS, Bhat SR, Sreenivasulu Y. 2017. The TRAF mediated gametogenesis progression (TRAMGaP) gene is required for megaspore mother cell specification and gametophyte development. Plant Physiology 175, 1220–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Spillane C, Steimer A, Grossniklaus U. 2001. Apomixis in agriculture: the quest for clonal seeds. Sexual Plant Reproduction 14, 179–187. [DOI] [PubMed] [Google Scholar]
- Sprunck S, Gross-Hardt R. 2011. Nuclear behavior, cell polarity, and cell specification in the female gametophyte. Sexual Plant Reproduction 24, 123–136. [DOI] [PubMed] [Google Scholar]
- Stefanowicz K, Lannoo N, Van Damme EJM. 2015. Plant F-box proteins – judges between life and death. Critical Reviews in Plant Sciences 34, 523–552. [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. 2005. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiology 137, 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Zang G, Cheng C, Luan M, Dai Z, Xu Y, Yang Z, Zhao L, Su J. 2017. Diplosporous development in Boehmeria tricuspis: insights from de novo transcriptome assembly and comprehensive expression profiling. Scientific Reports 7, 46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Genome Research 21, 2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, Genschik P. 2005. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. The Plant Journal 43, 437–448. [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 20623–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Araujo AC, Paech NA, Hecht V, Schmidt ED, Rossell JB, De Vries SC, Koltunow AM. 2003. Sexual and apomictic reproduction in Hieracium subgenus pilosella are closely interrelated developmental pathways. The Plant Cell 15, 1524–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. 2012. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. 2002. Genome-wide analysis of core cell cycle genes in Arabidopsis. The Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodermaier HC. 2004. APC/C and SCF: controlling each other and the cell cycle. Current Biology 14, R787–R796. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ. 2012. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. 2008. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiology 148, 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windham MD, Al-Shehbaz IA. 2007. New and noteworthy species of Boechera (Brassicaceae) II: apomictic hybrids. Harvard Papers in Botany 11, 257–274. [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U. 2010. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Current Biology 20, 506–512. [DOI] [PubMed] [Google Scholar]
- Ye H, Park YC, Kreishman M, Kieff E, Wu H. 1999. The structural basis for the recognition of diverse receptor sequences by TRAF2. Molecular Cell 4, 321–330. [DOI] [PubMed] [Google Scholar]
- Zapata JM. 2003. TNF-receptor-associated factors as targets for drug development. Expert Opinion on Therapeutic Targets 7, 411–425. [DOI] [PubMed] [Google Scholar]
- Zhao L, Cai H, Su Z, et al. 2017a. KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115, E526–E535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, et al. 2017b. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356, 6336. [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Juranić M, Dresselhaus T. 2017. Germline development and fertilization mechanisms in maize. Molecular Plant 10, 389–401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.