Higher Pi acquisition efficiency in wheat was related to an improved root system under Pi starvation, allowing higher Pi uptake. This response correlated with faster modulation of the IPS1–miR399–PHO2 pathway and strigolactone levels.
Keywords: P signalling, phosphate starvation responses, phosphate transporters, root traits, strigolactones, wheat
Abstract
Inorganic phosphorus (Pi) fertilizers are expected to become scarce in the near future; so, breeding for improved Pi acquisition-related root traits would decrease the need for fertilizer application. This work aimed to decipher the physiological and molecular mechanisms underlying the differences between two commercial wheat cultivars (Crac and Tukan) with contrasting Pi acquisition efficiencies (PAE). For that, four independent experiments with different growth conditions were conducted. When grown under non-limiting Pi conditions, both cultivars performed similarly. Crac was less affected by Pi starvation than Tukan, presenting higher biomass production, and an enhanced root development, root:shoot ratio, and root efficiency for Pi uptake under this condition. Higher PAE in Crac correlated with enhanced expression of the Pi transporter genes TaPht1;2 and TaPht1;10. Crac also presented a faster and higher modulation of the IPS1–miR399–PHO2 pathway upon Pi starvation. Interestingly, Crac showed increased levels of strigolactones, suggesting a direct relationship between this phytohormone and plant P responses. Based on these findings, we propose that higher PAE of the cultivar Crac is associated with an improved P signalling through a fine-tuning modulation of PHO2 activity, which seems to be regulated by strigolactones. This knowledge will help to develop new strategies for improved plant performance under P stress conditions.
Introduction
Phosphorus (P) is the second most limiting nutrient in plants besides nitrogen, being involved in numerous cellular processes such as protein activation, energy transfer, signalling, and regulation of carbon metabolism (Lan et al., 2015; Xu et al., 2018). However, unlike nitrogen, which can be fixed by microorganisms, the amount of P available for agriculture is finite (Bovill et al., 2013). Moreover, when compared with other essential macronutrients, P is one of the less abundant elements in the lithosphere (0.1% of the total), highlighting the need for supplying P fertilizers to sustain modern agricultural production (Campos et al., 2018). As a consequence, consumption of P fertilizers has increased worldwide in the past decades. P fertilizers are made from non-renewable resources such as rock phosphates, which are expected to become scarce in the near future as few mining sites are discovered and demand is expected to increase further by 50–100% in the next 30 years (Cordell et al., 2009; Ulrich and Frossard, 2014). P is present in plants either as organic phosphate esters or as the free inorganic orthophosphate form (Pi). Remarkably, Pi has high affinity for both soil mineral particles and organic matter; therefore, its availability in agroecosystems is generally below a plant’s demand, even in fertilized sites, where up to ~90% of the applied P fertilizer is not taken up by the roots in the first year (Syers et al., 2008; López-Arredondo et al., 2014). Therefore, although a huge amount of P fertilizers are used, plants are normally subjected to stress due to the low availability of this essential nutrient. Nevertheless, when P fertilization exceeds soil holding capacity, environmental problems associated with eutrophication due to P leaching are likely to occur (Bennett et al., 2001). In addition, these fertilizers can contain heavy metals, such as cadmium, that may accumulate in arable soils as a result of the addition of rock phosphate (van de Wiel et al., 2016).
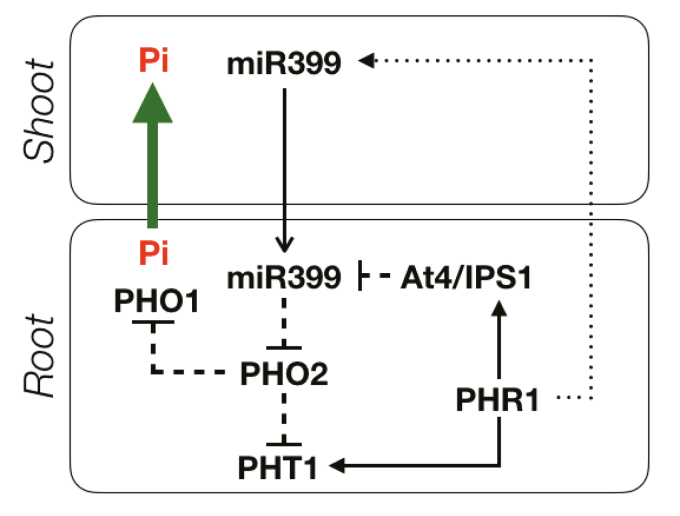
Plants have developed an array of complex regulatory mechanisms to adapt themselves to low Pi availability in the soil, known as P starvation responses (PSRs), aiming to optimize its external and internal use (Puga et al., 2017; Ham et al., 2018). These responses include changes at genetic, biochemical, physiological, morphological, and rhizospheric levels (Puga et al., 2017). PSRs include alterations in shoot and root morphology, growth and development, exudation of low molecular weight organic acid anions and Pi-releasing enzymes, modifications in lipids and carbohydrate metabolism, association with soil microorganisms, as well as the regulation of expression and activity of high-affinity Pi transporters (PHTs) (Lambers et al., 2015; Campos et al., 2018). Nevertheless, in order to respond accurately, plants need first to sense the P status both locally and systemically in order to orchestrate the appropriate responses (Lan et al., 2015; Scheible and Rojas-Triana, 2015). PSRs are themselves complex, with a large set of genes (>1000) being regulated. However, new genomic findings have contributed to shed light on some mechanisms of P sensing, signalling, and homeostasis, especially in the model plants Arabidopsis thaliana and rice (Oryza sativa) (Liu et al., 2012; Lan et al., 2015). It is well established that the transcriptional activator PHOSPHATE STARVATION RESPONSE 1 (PHR1) in Arabidopsis and its orthologous OsPHR2 in rice play a key role in regulating the expression of numerous Pi starvation-induced (PSI) genes (Rubio et al., 2001; Zhou et al., 2008). Among them, special attention has been paid to the miRNA miR399, whose expression is highly induced by Pi deprivation (Pant et al., 2008). This regulator has been shown to be a key systemic cue between plant tissues by modulating the activity of PHO2, which encodes a ubiquitin-conjugating E2 enzyme (UBC24) implicated in protein degradation (Lin et al., 2008). Down-regulation of PHO2 prevents the degradation of the Pi transporter PHO1, involved in Pi xylem loading, and some transporters of the PHT1 family, associated with Pi acquisition and translocation within the plant (Liu et al., 2012; Huang et al., 2013). Another key PSI gene family involved in P signalling and homeostasis is At4/IPS1 in Arabidopsis and rice, respectively. These genes affect the miR399–PHO2 interaction by sequestering free miR399 through a target mimicry mechanism, preventing its binding to PHO2 transcripts and, thus, its degradation (Fig. 1) (Franco-Zorrilla et al., 2007). Therefore, Pi acquisition and distribution within the plant are regulated mainly by the interaction of the triad IPS1–miR399–PHO2, which serves to fine-tune PSRs (Fig. 1).
Fig. 1.
Schematic summary of the P signalling and homeostasis pathway in plants. Upon Pi deficiency, expression of the miRNA miR399 is induced in the shoot. miR399 moves downwards, inactivating PHO2 in the roots. Regulation of PHO2 prevents the degradation of the Pi transporters PHO1 and PHT1, which increase Pi uptake and translocation. Pi deficiency also induces the expression of IPS1, which binds miR399, modulating this response. Based on Puga et al. (2017). (This figure is available in colour at JXB online.)
Wheat (Triticum aestivum L.) is one of the most important food crops in the world, with global grain production of 7.5×1014 g in 2016, making it the third most harvested crop worldwide, after sugarcane (Saccharum spp.) and maize (Zea mays L.) (FAO, http://faostat3.fao.org/home/E). However, wheat production is highly dependent on P fertilizers, leading to a higher consumption per area when compared with other major crops (Heffer, 2013). Therefore, improving P fertilization efficiency in wheat cropping is a major goal in order to achieve a more sustainable agricultural production. The last can be achieved by improving the availability of P fertilizers in soil, such as by avoiding Pi sorption to soil particles, and/or by the development of P use-/acquisition-efficient plants (Campos et al., 2018). While the first option increases operational costs and requires modern technology, often not accessible for producers, breeding for Pi acquisition-efficient root systems would provide benefits to both high- and low-input systems (Wissuwa et al., 2001; Rose and Wissuwa, 2012). In recent years, some molecular mechanisms underlying P signalling and homeostasis in wheat have been revealed. Wang and co-workers characterized TaPHR1, and showed that overexpressing lines had improved root system architecture, enhanced Pi uptake, and higher yield (Wang et al., 2013). In another breakthrough, the IPS1–miR399–PHO2 system was shown to be functional in wheat (Ouyang et al., 2016). In that study, TaPHO2 expression was found to be related to root and shoot growth, shoot Pi accumulation, and activity of some PHT1 transporters (Ouyang et al., 2016). In this context, the Pi transporter gene family PHT1 from wheat has been recently identified, consisting of 16 phylogenetically distinct transporters (Grün et al., 2017).
On the other hand, it is well known that phytohormones such as auxin, cytokinin, abscisic acid, ethylene, and in particular strigolactones (SLs) play synergistic roles in the regulation of P homeostasis when plants are subjected to P stress, through modulation of the P signalling- and homeostasis-associated pathways and ultimately root functioning (Waters et al., 2017; Chien et al., 2018). SLs are the latest class of phytohormones described, and have been shown to function as regulators of plant development/architecture and as signalling molecules in the rhizosphere to recruit arbuscular mycorrhizal fungi under Pi limitation (López-Ráez et al., 2017; Waters et al., 2017). Indeed, their biosynthesis is highly promoted under this stress condition (Yoneyama et al., 2007, 2012; López-Ráez et al., 2008). Recently, it has been shown that exogenous application of the synthetic SL analogue GR24 induced root hair elongation, anthocyanin accumulation, production of acid phosphatases, and reduced plant weight (Ito et al., 2015), which are characteristic PSRs, suggesting a potential overlap between these two signalling and homeostasis pathways in plants. Although the molecular mechanisms that regulate P signalling and homeostasis, and their associated plant morphological changes and Pi uptake capacity, are being established at the laboratory level, only a few studies have verified these findings in commercial cultivars so far. In the present work, we aimed to characterize at the physiological and molecular level two commercial wheat cultivars—Crac and Tukan—with different Pi acquisition efficiencies, and to relate these phenotypes to their ability to regulate P signalling and homeostasis under Pi-limited conditions. The results provide new insights into the regulation of P signalling and homeostasis in plants and suggest new potential targets for future breeding strategies in plant production.
Materials and methods
Plant material and growth conditions
Seeds from the wheat cultivars Crac and Tukan (formerly know as TCRB14 and STKI14, respectively) were surface-sterilized in 4% sodium hypochlorite, rinsed thoroughly with sterile distilled water, and germinated for 72 h on moistened filter paper at 25 °C in darkness. Precise phenotyping for optimal root system characteristics is difficult and time-consuming as root traits are hidden under the soil, making their extraction for observation difficult (Zhu et al., 2011). Therefore, different ‘artificial’ growing methods are used under laboratory conditions to facilitate their access, such as growing plants in liquid culture (hydroponics) or in transparent surfaces (rhizoboxes). These methods, although they do not fully represent the root growth in soil, give valuable clues to understand general features, and the physiological and genetic background behind them (Hargreaves et al., 2009). In order to access the effects of Pi deficiency on plant development and Pi acquisition of these cultivars, seedlings of each genotype were grown hydroponically (Supplementary Fig. S1a at JXB online) for 2 weeks with a standard nutrient solution (Taylor and Foy, 1985) containing 200 μM Pi in 1 litre containers and then half of the plants were submitted to Pi starvation (10 μM Pi in nutrient solution) for 3 weeks. In parallel, seedlings of each genotype were transferred to 0.5 litre plastic pots with a mixture of autoclaved substrate of sand and vermiculite (1:1) and were watered manually with standard nutrient solution low in Pi (10 μM) for 33 d (Supplementary Fig. S1b). In addition to the experiments previously mentioned, another set of plants were grown for 8 weeks in rhizoboxes (30 cm height, 20 cm width, and 0.7 cm depth) filled with an acidic high P-fixing soil without P fertilization (Supplementary Table S1; Supplementary Fig. S2a). Plants were grown under greenhouse conditions with temperatures ranging from 16 °C to 23 °C during the day and from 10 °C to 18 °C at night, and were harvested at Zadoks growth stage 23 (Zadoks et al., 1974).
For gene expression analysis and SL quantification, six seedlings of each cultivar were grown hydroponically in 3 litre containers, containing a modified Long Ashton nutrient solution with 150 μM Pi (Hewitt, 1966) in a greenhouse for a total of 5 weeks (Supplementary Fig. S1c). Nutrient solution was replaced twice a week. After 4 weeks, half of the plants were transferred to a modified nutrient solution without P and were left to grow for another week. For the time course Pi starvation experiment, plants were grown for 4 weeks under normal P conditions, and then half of the plants were subjected to 2, 4, and 7 d Pi deprivation. Six independent plants were grown per treatment and time point. Shoots, roots, and root exudates were collected, weighed, frozen with liquid nitrogen, and kept at –80 °C until use.
Root architecture measurements
For the phenotyping experiments, root systems were cleaned after harvest, arranged to minimize overlaps (Yao et al., 2009), placed in an A3-sized Perspex tray filled with water, and scanned in both grey-scale and colour in a Epson Expression 11000XL calibrated for Image Analysis. The images were then subjected to software analysis (WinRhizo; Regent Instruments, Quebec, Canada), and root length, specific area, and average diameter were assessed.
Pi acquisition
After root architecture determination, plants were separated into roots and shoots, and both parts were weighed and dried at 65 °C in a forced-air oven for 72 h. After drying, the root and shoot samples were weighed, crushed, ground, ashed in a furnace at 550 °C, and digested using an H2O:HCl:HNO3 mixture (8:1:1, v/v/v). Then, Pi content was determined using the vanadate–molybdate colorimetric method (Murphy and Riley, 1962).
RNA extraction and gene expression analysis by qPCR
Total RNA from roots was extracted using TRIsure™ (Bioline, Toronto, Canada) according to the manufacturer’s instructions. The RNA was treated with RQ1 DNase (Promega, Madison, WI, USA) and purified through a silica column using the RNA Clean & Concentrator™ (Zymo Research, irvine, CA, USA). Before storage at –80 °C, RNA was quantified using a Nanodrop 2000C spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and its integrity checked by gel electrophoresis. The first-strand cDNA was synthesized with 1 μg of purified total RNA using the PrimeScript™ RT Master Mix (Clontech, Fremont, CA, USA) according to the manufacturer’s instructions. The expression of marker genes for different P signalling and homeostasis pathways (Supplementary Table S1) was analysed by real-time quantitative PCR (qPCR). All reactions were performed using TB Green™ Premix Ex Taq™ (Kusatsu, Shiga, Japan) on an iCycler iQ5 system (Bio-Rad), with 5 µl of single-stranded cDNA (diluted 1:50) and specific primers for each gene, except for TaPht1;10 and tae-miR399b, where a dilution of 1:5 was used. In the case of the gene TaPht1:2, it is present in chromosomes A and B, showing high sequence similarities among them; therefore, it was not possible to design specific primers able to differentiate the expression of the two alleles (Grün et al., 2017). The amplification protocol included an initial denaturation at 95 °C for 3 min followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. The specificity of the different amplicons was checked by a melting curve analysis (from 65 °C to 100 °C) at the end of the amplification protocol. Five independent biological replicates were analysed per treatment and time point, and each PCR was done in duplicate. Relative quantification of specific mRNA levels was performed using the comparative 2−ΔCt method. Expression values were normalized using the housekeeping gene TahnRNPQ (Grün et al., 2017).
Strigolactone quantification
Germination bioassays were performed using extracts from frozen roots from plants grown for molecular analysis. For SL extraction, root extracts were processed as described by López-Ráez et al. (2008). SLs are germination stimulants of root parasitic plants of the family Orobancheaceae (Bouwmeester et al., 2007; López-Ráez et al., 2017). Therefore, germination assays with seeds of these parasitic weeds is an indirect way to estimate SL levels. Germination bioassays with pre-conditioned seeds of the parasitic plants Phelipanche ramosa were performed as described by López-Ráez et al. (2008). The synthetic SL analogue 2-epi-GR24 and demineralized water were included as positive and negative controls, respectively. Extract dilutions of 1:10 and 1:20 were tested for seed germination. After 7 d, the germinated and non-germinated seeds were counted using a binocular loupe. In addition to the germination bioassays, SLs were quantified by UHPLC-MS/MS using GR24 as internal standard as described by Rial et al. (2019). Briefly, 0.1 g of ground root material was extracted with 1 ml of ethyl acetate in an ultrasonic bath for 10 min, centrifuged for 10 min at 5000 rpm, concentrated in a rotary evaporator, and stored at –80 °C. Before the chromatographic analysis, extracts were dissolved with MeOH (1:1, v/v), and GR24 was added to each sample to a final concentration of 10 µg l−1. Chromatographic analyses were carried in a Bruker EVOQ Triple Quadrupole Mass Spectrometer with an electrospray ionization (ESI) source in positive mode. The mobile phase consisted of solvent A (water, 0.1% formic acid) and solvent B (MeOH, 0.1% formic acid) and the flow rate was set to 0.3 ml min−1. The optimized linear gradient system was as follow: 0–0.5 min, 50% B; 0.5–5 min, to 100% B; 5–7 min, 100% B; 7–7.5 min, to 50% B; 7.5–10.5 min, 50% B. The injection volume was 5 μl. The instrument parameters were set as described by Rial et al. (2019)
Statistical analysis
Means for plant growth, root architecture measurements, Pi acquisition, gene expression analysis by qPCR, and SL production were obtained from the results of five replicates. Data were assessed for normality, transformed when necessary, and significant differences between means were analysed by independent Student’s t-test or ANOVA followed by Tukey LSD when suited. Correlations among the different variables were performed using the r Pearson coefficient. All statistical analyses were carried out with R software.
Results
Pi acquisition and root system architecture in Crac and Tukan
In acidic soils, including Andisols in Chile, Pi bioavailability is rather low, many times due to high levels of iron and aluminium, which greatly affects plant productivity. In a previous study, a screening of wheat cultivars commonly used in Chile revealed high variations in Pi acquisition and grain yield when grown in high Pi-fixing Andisol (Seguel et al., 2017). From that study, we selected two cultivars—Crac and Tukan—showing contrasting Pi acquisition efficiency (PAE) under Pi-deficient conditions. The most efficient cultivar, Crac, yielded almost three times more than the less efficient cultivar, Tukan, at low Pi fertilization levels (Seguel et al., 2017). In the present work, we aim to decipher the physiological and molecular mechanisms behind such phenotypes. For that, plants of these two wheat cultivars were grown in different substrates and under different P conditions, and their Pi acquisition capacity compared. Similar results were obtained with the different growing conditions. When grown in hydroponics (Supplementary Fig. S1a) with sufficient Pi, both cultivars accumulated and allocated P in a similar manner to shoot and roots. Pi starvation reduced Pi uptake in both cultivars. However, despite the loss of Pi accumulation in both organs, the loss was significantly lower in Crac than in Tukan (Table 1; Supplementary Table S2). Indeed, Pi-starved Crac plants accumulated 25% and 17% more Pi in the shoots and in the roots, respectively, than the less efficient cultivar Tukan. Differences in Pi uptake between the two cultivars were even higher when using pots with inert substrate, where Crac accumulated 40% and 60% more Pi in shoots and roots, respectively, than Tukan (Table 1). The same pattern was also observed in soil-grown plants in rhizoboxes, although to a lesser extent (Supplementary Fig. S2b). Interestingly, despite the different Pi uptake capacities of the two cultivars under Pi limitation, no significant differences were observed in their root Pi concentration (Table 1), suggesting that the increased P accumulation in Crac was associated with a larger root system.
Table 1.
Phosphate (Pi) uptake, concentration, and root efficiency in accumulating Pi in shoots, roots, and the whole plant in Crac and Tukan
| Tissue | cv | Hydroponic (+P) | Hydroponic (–P) | Pot (-P) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P uptake (μg) | P concentration (mg g−1) | Root efficiency (mg P m−2) | P uptake (μg) | P concentration (mg g−1) | Root efficiency (mg P m−2) | P uptake (μg) | P concentration (mg g−1) | Root efficiency (mg P m−2) | ||
| Shoot | Crac | 1060.±158 | 1.57±0.23 | 61.57±1.60 | 600±32* | 1.18±0.09* | 17.73±0.55 | 678±43** | 1.06±0.04** | 22.41±1.61* |
| Tukan | 1130±43 | 1.41±0.04 | 74.17±5.05 | 480±22 | 0.92±0.05 | 17.04±1.99 | 406±28 | 0.89±0.02 | 18.48±0.77 | |
| Roots | Crac | 180±16 | 1.01±0.08 | 10.51±1.02 | 120±02* | 0.44±0.03 | 03.65±0.11 | 120±18** | 0.40±0.06 | 3.61±0.51* |
| Tukan | 180±24 | 1.12±0.14 | 11.94±1.31 | 100±06 | 0.51±0.03 | 03.76±0.23 | 46±5 | 0.30±0.02 | 2.28±0.16 | |
| Plant | Crac | 1240±157 | 1.45±0.13 | 72.07±6.56 | 720±29** | 0.92±0.01** | 21.38±0.45 | 797±44*** | 0.88±0.04** | 26.46±2.05* |
| Tukan | 1310±47 | 1.36±0.02 | 86.10±5.17 | 580±15 | 0.81±0.02 | 20.80±1.81 | 421±41 | 0.73±0.01 | 20.76±0.71 |
Plants were grown in Pi-sufficient conditions (+P; 200 μM, hydroponically) and under Pi starvation (–P; 10 μM, hydroponically and in pots) and were harvested at Zadoks growth stage 23 (Zadoks et al., 1974).
Data represent the means of five independent replicates (±SE). Asterisks indicate the significance of differences between the cultivars in the same condition, as determined by Student’s t-test analysis: *P<0.05, **P<0.01, ***P<0.001.
Shoot and root development under Pi deficiency
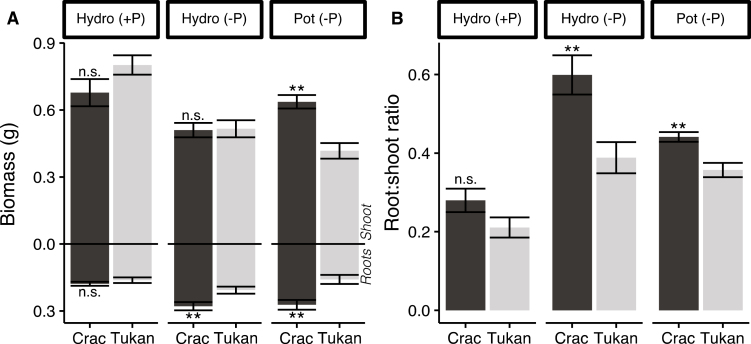
One of the main symptoms of Pi deficiency in plants is the enhanced root:shoot ratio; by either a reduction in shoot growth or an increase in root production, or both (Ericsson, 1995; Chien et al., 2018). To ascertain whether the differences observed in PAE between Crac and Tukan are associated with plant development, their shoot and root architecture were analysed. As expected, no significant differences in growth were observed under normal Pi conditions when grown hydroponically (Fig. 2a). However, under Pi starvation, plants showed a clear reduction in shoot growth (P<0.01), with a concomitant increase of root production compared with plants growing under normal Pi conditions (Fig. 2a; Supplementary Table S3). The effect was more severe in the less efficient cultivar Tukan, with a 37% reduction in shoot biomass, while the reduction was only 23% in Crac. The opposite effect was observed in the roots, where an increase in biomass of 30% was found in Tukan under Pi starvation, while the increase was up to 59% in Crac (Fig. 2a; Supplementary Table S3). Taken together, hydroponically grown plants displayed an average increase of the root:shoot ratio of 128% and 87% for Crac and Tukan, respectively (Fig. 2b; Supplementary Table S3). The same trend was observed in plants grown in pots with substrate, where Crac showed more shoot (34%) and root (42%) biomass than Tukan under Pi starvation, giving rise to a 20% higher root:shoot ratio in Crac (Fig. 2b). A similar pattern was also observed in plants grown in soil (Supplementary Fig. S2d, e). Therefore, a positive correlation between Pi accumulation and root:shoot ratio (R2=0.87, R2=0.95, and R2=0.85, P<0.01, for hydroponic, pot, and rhizobox experiments, respectively) under Pi starvation was observed in all growing conditions.
Fig. 2.
Growth rate of wheat cultivars Crac (dark bars) and Tukan (light bars) in sufficient Pi conditions (+P; 200 μM, hydroponically) and under Pi starvation (–P; 10 μM, hydroponically and in pots) harvested at Zadoks growth stage 23 (Zadoks et al., 1974). Graphics represent shoot and root biomass (A) and root:shoot ratio (B). Data represent the means of five independent replicates (±SE). n.s, non-significant differences. Asterisks indicate the significance of the differences between the cultivars in the same condition as determined by Student’s t-test: **P<0.01.
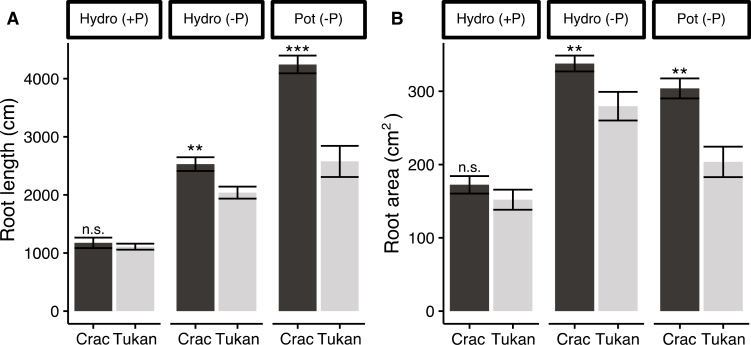
Additionally, parameters associated with root system architecture, such as total root length and root area, were evaluated (Fig. 3). In hydroponics, Pi deprivation increased root length in both cultivars by 113% and 80% in Crac and Tukan, respectively (Fig. 3a; Supplementary Table S3). Root surface area also increased under Pi starvation, by 98% and 73%, respectively (Fig. 3b; Supplementary Table S3). In the pot experiment, root length (~65%) and surface area (~50%) were also greater in the most efficient cultivar, Crac, than in Tukan (Fig. 3). The same behaviour was observed in plants growing in rhizoboxes, with Crac showing wider root systems (Supplementary Fig. S2f, g). Differences in the diameter of the roots were also observed, roots of the cultivar Crac being significantly thinner (2.27 mm) than those of Tukan (2.41 mm) (P<0.01). These differences in average diameter were also detected in plants growing in rhizoboxes with soil (Supplementary Fig. S2h). In order to assess the root system efficiency in acquiring Pi, root efficiency (Pi uptake per root area) was calculated. Although few differences were observed among cultivars in the hydroponic experiment, the losses of efficiency from Pi-sufficient to Pi-deficient conditions in Tukan were significantly higher for all the experimental variables compared with those observed in Crac (Supplementary Table S3). Nevertheless, Crac plants growing in pots and rhizoboxes acquired significantly more Pi per root area compared with Tukan (Table 1; Supplementary Fig. S2i). Together, these results suggest a more developed and efficient root system for this genotype under P deficiency.
Fig. 3.
Root system architecture of wheat cultivars Crac (dark bars) and Tukan (light bars) grown in sufficient Pi condition (+P; 200 μM, hydroponically) and under Pi starvation (–P; 10 μM, hydroponically and in pots) harvested at Zadoks growth stage 23 (Zadoks et al., 1974). Graphics show total root length (A) and root surface area (B). Data represent the means of five independent replicates (±SE). n.s, non-significant differences. Asterisks indicate the significance of differences between the cultivars in the same condition as determined by Student’s t-test: **P<0.01, ***P<0.001.
Gene expression of Pi transporters
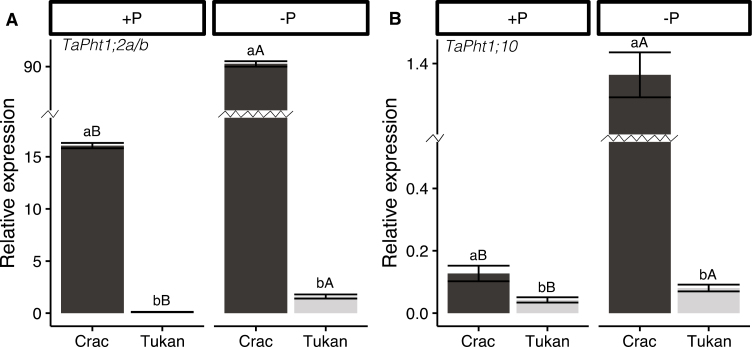
We investigated whether the higher root efficiency observed for Crac was related to a higher induction of Pi transporters. For that, a new experiment under normal and Pi-deficient conditions was carried out in hydroponics, and gene expression levels of different Pi transporters were analysed by qPCR. Wheat has 16 phylogenetically distinct Pi transporters, seven of them being induced by Pi deprivation (Grün et al., 2017). We analysed the expression profile of two of them, TaPht1;2a/b and TaPht1;10. The first one is the most hightly expressed, and it was described as a fast responsive Pi marker. The other shows low expression levels, and its expression is increased with time (Grün et al., 2017). The remaining Pi-inducible TaPHT1 shows similar expression patterns either to TaPht1;2a/b or to TaPht1;10, but with lower expression levels (Grün et al., 2017; Teng et al., 2017). For that reason, they were not analysed in this work. Expression analysis revealed major differences among the two cultivars and their responses to Pi deprivation, with Crac showing higher basal levels of both transporters. The expression of TaPht1;2a/b was ~140 times higher in Crac than in Tukan, and Pi starvation induced its expression levels 6- and 13-fold in Crac and Tukan, respectively (Fig. 4a). Therefore, the levels of TaPht1;2a/b upon Pi stress were ~50-fold higher in Crac. A similar pattern was observed for TaPht1;10, although its expression levels were much lower than that of TaPht1;2a/b (>70 times in Crac). In this case, basal transcripts levels of TaPht1;10 in Crac were three times higher than in Tukan, and the induction by Pi deficiency was 10- and 2-fold in Crac and Tukan, respectively, giving rise to a 15-fold greater expression in the former under stress conditions (Fig. 4b).
Fig. 4.
Gene expression analysis of two TaPHT1s Pi transporters in roots of Crac (dark bars) and Tukan (light bars) plants grown in nutrient solution with Pi (+P; 150 µM) and without Pi for the last week (–P) harvested at Zadoks growth stage 24 (Zadoks et al., 1974). Graphics represent expression of TaPht1;2a/b (A) and TaPht1;10 (B). Expression levels were referenced to the expression of the housekeeping gene TahnRNPQ. Bars represent the means of five independent replicates (±SE). Lower case letters indicate differences between cultivars in the same condition, and upper case letters indicate differences within the same cultivar under normal and deficient Pi conditions as determined by Student’s t-test analysis (P<0.05).
Crac and Tukan show different regulation of P signalling and homeostasis
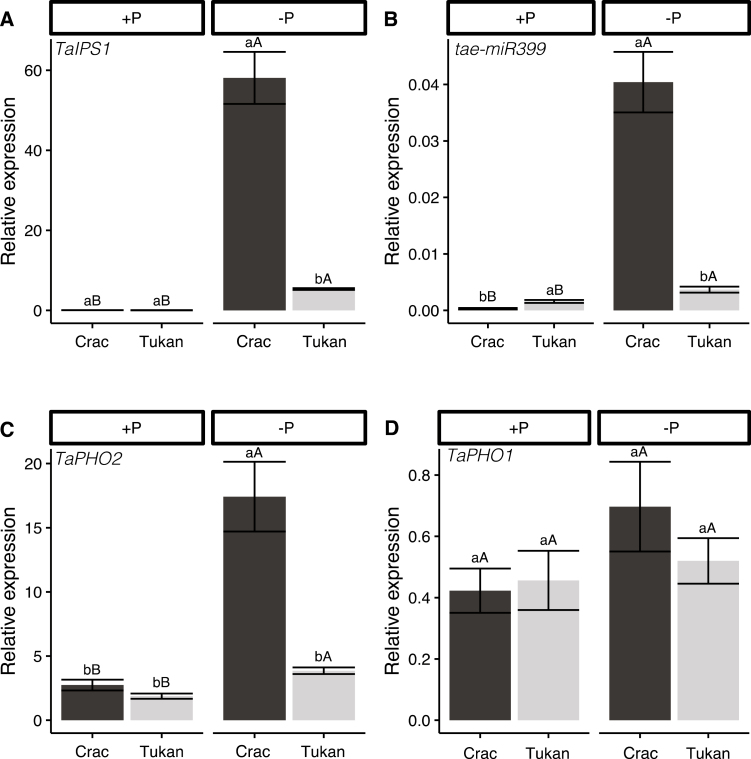
All adaptive responses that plants have evolved to cope with Pi deficiency are regulated through P signalling and homeostasis mechanisms, which begin with the integration of the information of the extracellular Pi concentration and its levels in the different organs (Puga et al., 2017). Here, the IPS1-mediated signalling cascade, including PHR1–IPS1–miR399–PHO2, plays a pivotal role in P homeostasis regulation by coordinating the activities of Pi uptake and its root–shoot translocation through the transporters PHT1s and PHO1 (Fig. 1) (Ouyang et al., 2016; Ham et al., 2018). To investigate whether Crac and Tukan presented differences in the P signalling and homeostasis pathway, we analysed the gene expression of TaIPS1, tae-miR399 [specifically from tae-miR399b family members, with confirmed expression and regulation activity in wheat roots under Pi starvation (Ouyang et al., 2016)], TaPHO2, and TaPHO1 in roots of the two cultivars after 7 d of Pi deprivation and under normal conditions. No differences in the transcript levels of TaIPS1 were found under optimal Pi conditions, but a clear induction by Pi starvation was observed in both cultivars (Fig. 5a). Interestingly, a 10-fold higher induction was observed in Crac compared with Tukan. As for TaIPS1, Pi starvation promoted tae-miR399b expression in both cultivars, this increase being much higher in Crac (128-fold) than in Tukan (~2-fold). Thus, the number of transcripts under stress conditions was >10 times higher in the most efficient cultivar Crac (Fig. 5b). The same behaviour was observed for TaPHO2 under Pi limitation. Here, an increase of 6- and 2-fold was observed for Crac and Tukan, respectively, resulting in almost 5-fold higher transcript levels in Crac (Fig. 5c). We further assessed the expression of the three TaPHO2 alleles present in the wheat genome (1A, 1B, and 1D), using specific primers (Supplementary Table S2). Different patterns were observed for the three alleles regarding P responses (Supplementary Fig. S3a–c). Overall, Crac TaPHO2 alleles presented a higher induction in Pi starvation, especially TaPHO2 1B, with an increase of 25-fold. Tukan TaPHO2 1B and 1D showed a small, but significant increase in its expression under Pi deprivation (2.8- and 2-fold, respectively), while no differences were observed for TaPHO2 1A expression. No differences in gene expression were observed for the transporter TaPHO1 (Fig. 5d).
Fig. 5.
Expression levels of genes involved in P signalling and homeostasis in roots of Crac (dark bars) and Tukan (light bars) plants grown in nutrient solution with Pi (+P; 150 µM) and without Pi for the last week (–P) harvested at Zadoks growth stage 24 (Zadoks et al., 1974). Graphics represent gene expression of TaIPS1 (A), tae-miR399 (B), TaPHO2 (C), and TaPHO1 (D). Expression levels were referenced to the expression of the housekeeping gene TahnRNPQ. Bars represent the means of five independent replicates (±SE). Lower case letters indicate differences between cultivars in the same condition, and upper case letters indicate differences within the same cultivar under normal and deficient Pi conditions, as determined by Student’s t-test (P<0.05).
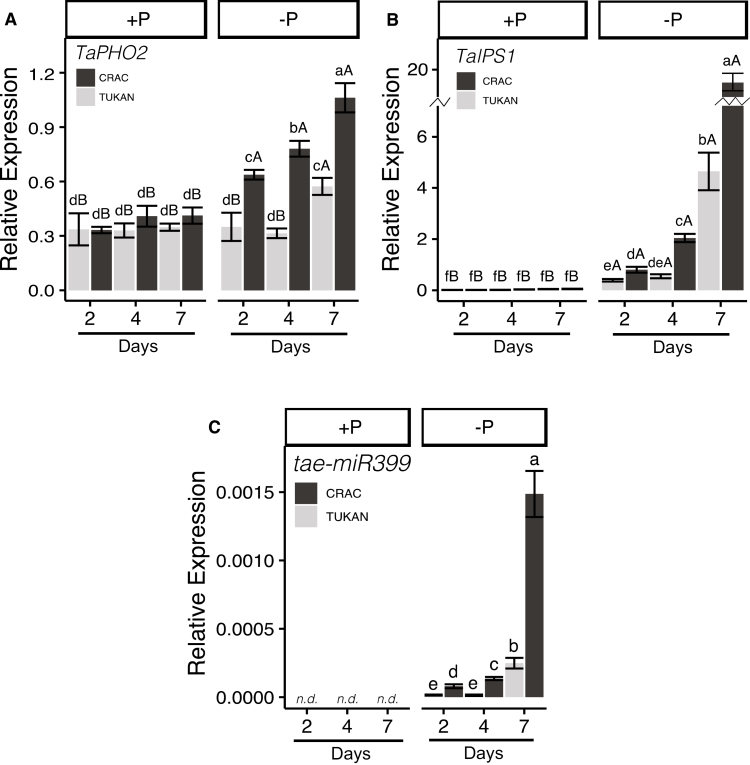
In order to assess the dynamics of Pi signalling in the two cultivars, a time-course experiment was performed under the same conditions as described before, but harvesting plants after 2, 4, and 7 d of Pi deprivation. Interestingly, Crac presented an induction of TaPHO2, TaIPS1, and tae-miR399b from day 2, which was steadily increased over time. However, in Tukan, the increasing response of these genes to Pi starvation was only observed at day 7 (Fig. 6), indicating a faster response by Crac to Pi deprivation.
Fig. 6.
Expression levels of genes involved in P signalling and homeostasis in roots of Crac (dark bars) and Tukan (light bars) plants grown in nutrient solution with Pi (+P; 150 µM) and without Pi (–P) harvested after 2, 4, and 7 d of Pi deprivation. Graphics represent the expression of genes TaPHO2, TaIPS1, and tae-miR399. Expression levels were referenced to the expression of the housekeeping gene TahnRNPQ. Bars represent the means of five independent replicates (±SE). Lower case letters indicate differences between cultivars in the same condition, and upper case letters indicate differences within the same cultivar and day between +P and –P conditions, as determined by Student’s t-test (P<0.05). n.d, non-detected.
Strigolactone levels in Crac and Tukan
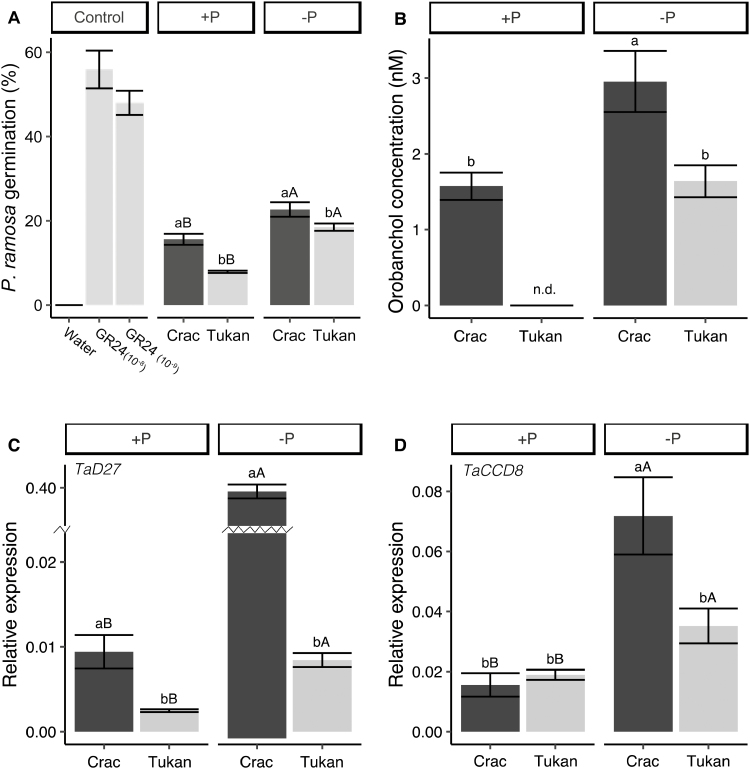
Since the root architecture of Crac and Tukan was different under Pi starvation, we assessed whether SLs were involved in such changes. We first performed a germination bioassay with P. ramosa seeds using root extracts from the two cultivars. GR24 (10–8 M and 10–9 M), used as positive control, induced high germination, while water (negative control) did not induce any germination. Under Pi-sufficient conditions, Crac induced twice the germination of P. ramosa comparted with Tukan (Fig. 7a), suggesting a higher basal level of SLs in that cultivar. Pi starvation increased germination in both cultivars: 7% and 10% for Crac and Tukan, respectively (Fig. 7a). Orobanchol was detected in the root extracts of both cultivars. This SL was reported as the main SL present in wheat exudates (Yoneyama et al., 2012). Orobanchol levels were significantly increased ~80% in Crac plants subjected to Pi starvation (P<0.01; Fig. 7b). This stress also increased the amount of orobanchol in Tukan, which was not detected under Pi-sufficient conditions. Here, orobanchol levels under Pi deficiency were similar to those in Crac in the absence of stress, indicating lower levels of SLs in Tukan, as previously observed in the P. ramosa germination bioassays (Fig. 7a). In addition to orobanchol, the SL fabacyl acetate was also detected in trace amounts in some samples. It was only detected in extracts from plants grown under Pi limitation, indicating that its biosynthesis was also promoted by Pi starvation, and also showing higher contents in Crac (data not shown).
Fig. 7.
Analysis of SL levels in roots of Crac and Tukan plants grown in nutrient solution with Pi (+P; 150 µM) and without Pi for the last week (–P) harvested at Zadoks growth stage 24 (Zadoks et al., 1974). (A) Germination of P. ramosa seeds induced by extracts from Crac (dark bars) and Tukan (light bars). (B) Orobanchol levels in root extracts determined by UHPLC-MS-MS. Data represent the means of three independent replicates (±SE). Different letters indicate significant differences between means, as determined by Tukey LSD test analysis. n.d, not detected. Expression levels of two SL biosynthesis genes: (C) TaD27 and (D) TaCCD8. Bars represent the means of five independent replicates (±SE). Lower case letters indicate differences between cultivars in the same condition, and upper case letters indicate differences within the same cultivar under normal and deficient Pi conditions, as determined by Student’s t-test (P<0.05).
To explore whether the elevated SL levels are related to a higher activity of the SL biosynthetic pathway in the Crac cultivar, the expression of two key genes involved in SL biosynthesis—TaD27, and TaCCD8—was analysed. The sequential action of these two enzymes gives rise to carlactone, the precursor of all the canonical SLs, including strigol- and orobanchol-type SLs (Al-Babili and Bouwmeester, 2015; Zhang et al., 2018). The search for the wheat D27 gene was conducted using BLAST against its orthologue sequence from rice (LOC107276001). Two complete sequences were found for copies in chromosomes 7A and 7D (accession numbers: KX168420.1 and KX168421.1), showing 52% and 54% homology, respectively, with rice D27 (Supplementary Fig. S4). The same strategy was applied for CCD8; however, no direct match was found. Therefore, the sequence encoding the putative wheat CCD8 was searched in the wheat genome database (IWGSC database), using BLAST against its orthologue sequence from Zea mays (ZmCCD8). One sequence for each chromosome was found (3A, 3B, and 3D) (Supplementary Fig. S5). The sequences obtained for TaD27 and TaCCD8 were checked for the presence of the functional domain DUF4033 and RPE65, respectively, and their homology with wheat close relatives was assessed (Supplementary Figs S4a, S5b). Specific primers for TaD27 and TaCCD8 were designed to perform qPCR (Supplementary Table S1). The basal expression of TaD27 under Pi-sufficient conditions was almost 4-fold higher in Crac roots than in Tukan (Fig. 7c). However, no differences in basal expression levels of TaCCD8 were found between the two cultivars (Fig. 7d). Pi deprivation induced TaD27 transcript levels ~40-fold in Crac, but only 3-fold in Tukan (Fig. 7c). A similar trend was observed for TaCCD8, with an increase in the expression levels of ~5-fold in Crac and only ~2-fold in Tukan (Fig. 7d). These results confirm that Crac produces higher basal levels of SLs than Tukan, and responds more efficiently to P starvation through a stronger promotion of SL biosynthesis, which would favour a greater and faster development of the root system in response to Pi-limiting conditions.
Discussion
Crac has an improved root system development and a higher Pi acquisition capacity
In the present work, we analysed the physiological and molecular traits of two commercial wheat cultivars—Crac and Tukan—showing differential PAE under Pi-limiting conditions (Seguel et al., 2017). Interestingly, under that stress condition, yield parameters correlated with root biomass and Pi acquisition (Seguel et al., 2017). Here, when plants were grown under Pi-sufficient conditions, no differences in Pi acquisition and biomass production/partitioning were observed, highlighting that the differences further discussed are specifically related to contrasting responses to Pi limitation. In this sense, our results are in agreement with a previous study in plants grown in an acidic high Pi-fixing soil (Seguel et al., 2017). We show that Crac also presents a higher Pi accumulation in shoots and roots when subjected to Pi deprivation (Table 1). Alterations in shoot and root growth and/or architecture are the most widespread plant adaptations to Pi starvation, affecting the root:shoot ratio (Haling et al., 2016; Chien et al., 2018). Interestingly, under these conditions, Crac showed smaller losses of shoot biomass production and greater increments in root growth, giving rise to higher root:shoot ratios, which correlated with total Pi acquisition in all experiments. In addition to greater root:shoot ratios, Crac showed larger and thinner roots than Tukan (Supplementary Figs S2, S6). Altogether, these phenotypic differences would allow Crac to have a higher soil exploration capacity in search of Pi patches under limiting conditions.
Improved PAE is associated with higher expression of PHT1 Pi transporter genes
Several studies correlate higher Pi accumulation and, in most cases, plant growth with higher expression of Pi transporters of the PHT1 family (Liu et al., 2013; Wang et al., 2013; Ham et al., 2018). Recently, it has been shown that under Pi starvation, transcript levels of the gene TaPht1;2 were the most abundant of all the PHT1 transporter geness described in wheat (Grün et al., 2017). This transporter is the orthologue of the rice OsPht1;2, which is also highly expressed in Pi-deprived roots, and it is characterized as a low-affinity Pi transporter, mainly involved in internal Pi translocation (Ai et al., 2009). The other Pi transporter gene analysed was TaPht1;10, whose expression levels were lower (~25-fold) than those of TaPht1;2. TaPht1;10 and its orthologues in rice—OsPht1;9 and OsPht1.10—are considered as high-affinity Pi transporters, and are mainly induced under long-term Pi starvation (Ai et al., 2009; Grün et al., 2017). As expected, both genes were highly induced by Pi deficiency in both cultivars, confirming their role in Pi uptake and distribution under nutritional stress. In agreement with this, it has been shown that transgenic Nicotiana tabacum plants overexpressing a Pht1;2 gene displayed a higher Pi content and better growth than the corresponding wild type (Cao et al., 2018). These plants also showed enhanced Pi in the xylem sap, indicating that this transporter is involved in root:shoot Pi translocation. No differences in gene expression were observed for TaPHO1, a component of the other family of Pi transporters and involved in Pi xylem loading (Franco-Zorrilla et al., 2007). However, this was not surprising since it was previously shown that this transporter is regulated post-transcriptionally by the action of PHO2 (Fig. 1) (Lin et al., 2008; Huang et al., 2013). Notably, basal levels of the two genes encoding PHT1 transporters under sufficient Pi conditions were higher in the most efficient cultivar Crac, and the final levels under Pi starvation were much higher than in Tukan (Fig. 4). In this sense, these differences could be related to the internal SL levels, which are directly linked to PHT1 expression and other PSI genes in Arabidopsis (Ito et al., 2015). These results, together with the enhanced root system of Crac, indicate that this cultivar is better suited to respond readily to this type of nutritional stress by an improved Pi uptake and root:shoot translocation capacity.
P signalling and homeostasis, and its relationship to SLs
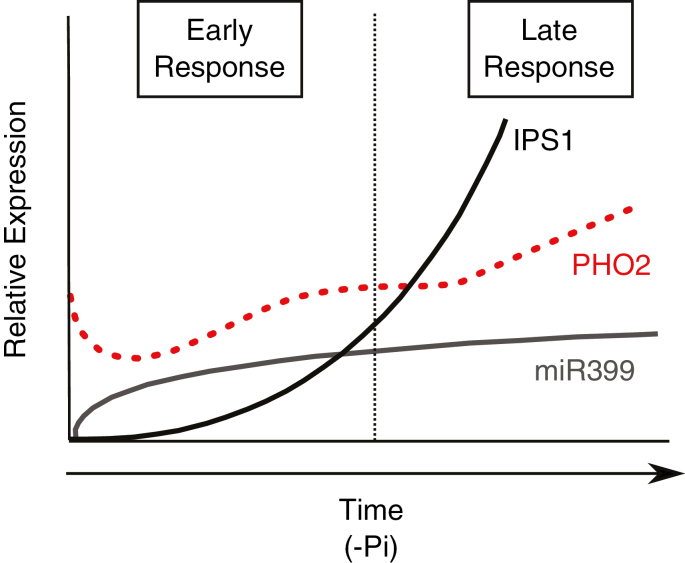
It is generally accepted that PHO2 activity is rapidly reduced under Pi deficiency due to high induction of miR399 in the first hours of stress, which overcomes protection by IPS1 (Ajmera et al., 2018). However, there are only a few studies evaluating the effect of long-term Pi deprivation on the regulation of this pathway. In a time-course study in rice, Ajmera et al. (2018) found that the number of IPS1 transcripts increased slowly, but more strongly than those of miR399, leading to a relative increase of PHO2 levels after 1 week of stress. A similar pattern was observed in barley plants after 16 d of Pi starvation, where cultivars showing high levels of HvIPS2 matched with those with higher HvPHO2 expression (Huang et al., 2011). In the present study, an induction of tae-miR399b was observed after 1 week of Pi deficiency. However, the total number of transcripts at this time point was much lower than those of TaIPS1 (>1000 times), leading to a complete sequestration of tae-miR399 transcripts. This blockage would explain the high induction of TaPHO2 observed in our system (Fig. 5). Arabidopsis thaliana IPS1-overexpressing lines also presented a enhanced PHO2 accumulation due to higher IPS1-mediated miR399 sequestration (Franco-Zorrilla et al., 2007). Therefore, it seems that the regulation of P signalling and homeostasis by the triad IPS1–miR399–PHO2 is dynamic, showing a different regulation over time. Based on the responses observed in wheat seedlings and other model plants at different developmental stages (Huang et al., 2011; Ouyang et al., 2016; Ajmera et al., 2018), we propose a model to explain the behaviour of these three regulators during early and late responses upon Pi starvation (Fig. 8). According to this, during the first hours of stress, there might be a rapid induction of miR399 levels, which mediates the cleavage of PHO2 transcripts, probably to increase the relative amount of PHT1 members to promote Pi uptake from the soil, with the corresponding translocation to the shoots. In the case where Pi limitation continues over time, transcripts of IPS1 would increase greatly to lock miR399, and probably to exert other regulatory functions as well, with the concomitant increase in PHO2 levels. This increase will trigger late Pi responses related to the improvement of Pi uptake and modification of root architecture, probably to search for new Pi ‘hotspots’, among others. In agreement with this hypothesis, higher PHO2 levels under Pi starvation in barley correlated with a higher root:shoot ratio (Huang et al., 2011). Conversely, wheat plants blocked at TaPHO2 showed a lower root:shoot ratio (Ouyang et al., 2016). The initial down-regulation of PHO2 proposed in our model was not observed in the time-course experiment, probably because this regulation may occur within hours after Pi deprivation at this developmental stage. On the other hand, the IPS1–miR399 relationship did not fully explain the variation in PHO2 levels, which was in accordance with the responses observed in rice (Ajmera et al., 2018). We suggest two possibilities to explain this fact: (i) there must be more genes involved in this complex regulation; and/or (ii) when IPS1 transcripts reach a certain level, they are sufficient to sequester miR399. Thus, further increases of these transcripts would not affect PHO2 degradation. Further studies are required to fully decipher how this signalling and homeostasis pathway works, especially under longer periods of Pi starvation and at different plant developmental stages.
Fig. 8.
Proposed model for the regulation of P signalling and homeostasis by the module IPS1–miR399–PHO2. At early stages of Pi starvation, the high miR399 induction favours PHO2 transcript degradation and triggers early phosphate starvation responses (PSRs). As Pi starvation follows, the miR399:IPS1 ratio diminishes due to sustained high expression of IPS1, increasing PHO2 degradation, and initiating late PSRs. Black and grey lines represent IPS1 and miR399 expression, respectively, and the dotted line represent PHO2 expression. (This figure is available in colour at JXB online.)
As for the TaPht1;2 and TaPht1;10 transporters, the levels of the three regulators IPS1–miR399b–PHO2 under Pi limitation were much higher in the cultivar Crac, with enhanced PAE, than in the less efficient cultivar Tukan. However, the basal transcript levels of these genes were similar under P-sufficient conditions. Again, these results show that Crac is more efficient in the response to Pi deficiency, and that this enhanced efficiency is due to a better and faster Pi starvation signalling and homeostasis regulation. In agreement with this, Crac showed higher basal levels of SLs than Tukan (Fig. 6). SLs, together with other phytohormones, are involved in the regulation of P homeostasis under Pi limitation by modulating P signalling-associated pathways and root development (Waters et al., 2017; Chien et al., 2018). Therefore, it might be the case that the higher SL levels in Crac would act as a priming signal under Pi starvation to boost plant responses to the stress. However, further research is needed to clarify the connection between SLs and P signalling.
Understanding the physiological mechanisms of improved PAE and the genetic basis therein will allow breeders to select more P-efficient cultivars. This knowledge will help to diminish the use of P fertilizers in agriculture, thus reducing costs and alleviating the excessive consumption of this non-renewable resource. However, traits associated with PAE are complex and context dependent. In the present study, we suggest that the higher PAE of the commercial cultivar Crac might be related to the SLs–P signalling relationship and homeostasis through fine-tuning modulation of PHO2 activity. This modulation, in the long term, would relatively reduce shoot Pi loading, favouring the development of an enhanced root system and giving rise to an increased soil exploration capacity in search of Pi patches under limiting conditions and to increase Pi acquisition in high Pi-fixing soils. Further research is needed to fully understand the SLs–P signalling relationship to develop new strategies for improved plant performance under P stress conditions.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Selected chemical properties of the soil used in the rhizobox experiment.
Table S2. Primer sequences used in the qPCR analysis.
Table S3. Effects (%) of Pi starvation on plants grown in hydroponics.
Fig. S1. Example of the different experimental conditions.
Fig. S2. Main results of the effects of Pi starvation in the rhizobox experiment.
Fig. S3. Expression levels of TaPHO2 alleles.
Fig. S4. Phylogenetic analysis of D27 sequences and amino acid sequences.
Fig. S5. Phylogenetic analysis of partial CCD8 sequences and amino acid sequences.
Fig. S6. Example of Crac and Tukan root systems.
Acknowledgements
This work was supported by funding from FONDECYT grants 11160385 and 1170264 from Comisión Nacional de Investigación Científica y Tecnológica (CONICYT-Chile), CONICYT DOCTORADO NACIONAL 2016 (21161474), CONICYT+PAI/CONCURSO NACIONAL TESIS DE DOCTORADO EN EL SECTOR PRODUCTIVO 2017 (T7817120011), CONICYT/FONDAP/15130015, and AGL2015-64990-C2-1R (MINECO-Spain). We also acknowledge Dr Koichi Yoneyama and Dr Xiaoan Xie for kindly providing the natural strigolactones used to set up their analytical quantification, Dr Gustavo Curaqueo for the analysis of the root scanning system, Campex Baer® for their agronomical support, BIOREN-UFRO for providing their facilities, and Dr María J. Pozo and Dr Francisco Macías for critical reading of the manuscript and support.
References
- Ai P, Sun S, Zhao J, et al. 2009. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. The Plant Journal 57, 798–809. [DOI] [PubMed] [Google Scholar]
- Ajmera I, Shi J, Giri J, Wu P, Stekel DJ, Lu C, Hodgman TC. 2018. Regulatory feedback response mechanisms to phosphate starvation in rice. NPJ Systems Biology and Applications 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Bennett EM, Carpenter SR, Caraco NF. 2001. Human impact on erodable phosphorus and eutrophication: a global perspective. BioScience 51, 227–234. [Google Scholar]
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. 2007. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends in Plant Science 12, 224–230. [DOI] [PubMed] [Google Scholar]
- Bovill W, Huang C, Mcdonald G. 2013. Genetic approaches to enhancing phosphorus-use efficiency (PUE) in crops: challenges and directions. Crop & Pasture Science 64, 179–198. [Google Scholar]
- Campos P, Borie F, Cornejo P, López-Ráez JA, López-García Á, Seguel A. 2018. Phosphorus acquisition efficiency related to root traits: is mycorrhizal symbiosis a key factor to wheat and barley cropping? Frontiers in Plant Science 9, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Sun D, Chen JX, Mei H, Ai H, Xu G, Chen Y, Ma LQ. 2018. Phosphate transporter PvPht1;2 enhances phosphorus accumulation and plant growth without impacting arsenic uptake in plants. Environmental Science & Technology 52, 3975–3981. [DOI] [PubMed] [Google Scholar]
- Chien P-S, Chiang C-P, Leong SJ, Chiou T-J. 2018. Sensing and signaling of phosphate starvation: from local to long distance. Plant & Cell Physiology 59, 1714–1722. [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. 2009. The story of phosphorus: global food security and food for thought. Global Environmental Change 19, 292–305. [Google Scholar]
- Ericsson T. 1995. Growth and shoot:root ratio of seedlings in relation to nutrient availability. Plant and Soil 168, 205–214. [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Grün A, Buchner P, Broadley MR, Hawkesford MJ. 2017. Identification and expression profiling of Pht1 phosphate transporters in wheat in controlled environments and in the field. Plant Biology 20, 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haling RE, Yang Z, Shadwell N, Culvenor RA, Stefanski A, Ryan MH, Sandral GA, Kidd DR, Lambers H, Simpson RJ. 2016. Root morphological traits that determine phosphorus-acquisition efficiency and critical external phosphorus requirement in pasture species. Functional Plant Biology 43, 815–826. [DOI] [PubMed] [Google Scholar]
- Ham BK, Chen J, Yan Y, Lucas WJ. 2018. Insights into plant phosphate sensing and signaling. Current Opinion in Biotechnology 49, 1–9. [DOI] [PubMed] [Google Scholar]
- Hargreaves CE, Gregory PJ, Bengough AG. 2009. Measuring root traits in barley (Hordeum vulgare ssp. vulgare and ssp. spontaneum) seedlings using gel chambers, soil sacs and X-ray microtomography. Plant and Soil 316, 285–297. [Google Scholar]
- Heffer P. 2013. Assessment of fertilizer use by crop at the global level. Paris: International Fertilizer Industry Association. [Google Scholar]
- Hewitt EJ. 1966. Sand and water culture methods used in the study of plant nutrition. Maidstone, UK: Commonwealth Bureau of Horticulture and Plantation Crops. [Google Scholar]
- Huang CY, Shirley N, Genc Y, Shi B, Langridge P. 2011. Phosphate utilization efficiency correlates with expression of low-affinity phosphate transporters and noncoding RNA, IPS1, in barley. Plant Physiology 156, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, et al. 2013. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. The Plant Cell 25, 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nozoye T, Sasaki E, et al. 2015. Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS One 10, e0119724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Martinoia E, Renton M. 2015. Plant adaptations to severely phosphorus-impoverished soils. Current Opinion in Plant Biology 25, 23–31. [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Schmidt W. 2015. ‘Omics’ approaches towards understanding plant phosphorus acquisition and use. Annual Plant Reviews 48, 65–98. [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ. 2008. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiology 147, 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ. 2012. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. The Plant Cell 24, 2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao X, Zhang L, Lu W, Li X, Xiao K. 2013. TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Functional Plant Biology 40, 329–341. [DOI] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. 2014. Phosphate nutrition: improving low-phosphate tolerance in crops. Annual Review of Plant Biology 65, 95–123. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, et al. 2008. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytologist 178, 863–874. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Shirasu K, Foo E. 2017. Strigolactones in plant interactions with beneficial and detrimental organisms: the Yin and Yang. Trends in Plant Science 22, 527–537. [DOI] [PubMed] [Google Scholar]
- Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27, 31–36. [Google Scholar]
- Ouyang X, Hong X, Zhao X, Zhang W, He X, Ma W, Teng W, Tong Y. 2016. Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Scientific Reports 6, 29850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. 2008. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. The Plant Journal 53, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J. 2017. Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Current Opinion in Plant Biology 39, 40–49. [DOI] [PubMed] [Google Scholar]
- Rial C, Varela RM, Molinillo JMG, López-Ráez JA, Macías FA. 2019. A new UHPLC-MS/MS method for the direct determination of strigolactones in root exudates and extracts. Phytochemical Analysis 30, 110–116. [DOI] [PubMed] [Google Scholar]
- Rose TJ, Wissuwa M. 2012. Rethinking internal phosphorus utilization efficiency; a new approach is needed to improve PUE in grain crops. Advances in Agronomy 116, 185–217. [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes & Development 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W-R, Rojas-Triana M. 2015. Sensing, signalling, and control of phosphate starvation in plants: molecular players and applications. Annual Plant Reviews 48, 23–63. [Google Scholar]
- Seguel A, Cornejo P, Ramos A, Von Baer E, Cumming J, Borie F. 2017. Phosphorus acquisition by three wheat cultivars contrasting in aluminium tolerance growing in an aluminium-rich volcanic soil. Crop and Pasture Science 68, 305–316. [Google Scholar]
- Syers J, Johnston A, Curtin D. 2008. Efficiency of soil and fertilizer phosphorus use: reconciling changing concepts of soil phosphorus behaviour with agronomic information. Rome: FAO. [Google Scholar]
- Taylor GJ, Foy CD. 1985. Mechanisms of aluminum tolerance in Triticum aestivum L. (wheat). I. Differential pH induced by winter cultivars in nutrient solutions. American Journal of Botany 72, 695–701. [Google Scholar]
- Teng W, Zhao Y-Y, Zhao X-Q, He X, Ma W-Y, Deng Y, Chen X-P, Tong Y-P. 2017. Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat. Frontiers in Plant Science 8, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich AE, Frossard E. 2014. On the history of a reoccurring concept: phosphorus scarcity. The Science of the Total Environment 490, 694–707. [DOI] [PubMed] [Google Scholar]
- van de Wiel CCM, van der Linden CG, Scholten OE. 2016. Improving phosphorus use efficiency in agriculture: opportunities for breeding. Euphytica 207, 1–22. [Google Scholar]
- Wang J, Sun J, Miao J, et al. 2013. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Annals of Botany 111, 1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC. 2017. Strigolactone signaling and evolution. Annual Review of Plant Biology 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Ae N, Jones SS. 2001. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breeding 120, 43–48. [Google Scholar]
- Xu Y, Liu F, Han G, Cheng B. 2018. Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Reports 37, 711–726. [DOI] [PubMed] [Google Scholar]
- Yao Q, Wang LR, Zhu HH, Chen JZ. 2009. Effect of arbuscular mycorrhizal fungal inoculation on root system architecture of trifoliate orange (Poncirus trifoliata L. Raf.) seedlings. Scientia Horticulturae 121, 458–461. [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K. 2012. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235, 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K. 2007. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227, 125–132. [DOI] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. 1974. A decimal code for the growth stages of cereals. Weed Research 14, 415–421. [Google Scholar]
- Zhang Y, Lv S, Wang G. 2018. Strigolactones are common regulators in induction of stomatal closure in planta. Plant Signaling & Behavior 13, e1444322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Ingram PA, Benfey PN, Elich T. 2011. From lab to field, new approaches to phenotyping root system architecture. Current Opinion in Plant Biology 14, 310–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.