Capsidiol, a phytoalexin accumulated in Nicotiana attenuata in response to Alternaria alternata attack, plays an important role in pathogen resistance, and its biosynthesis is transcriptionally regulated by an ERF2-like transcription factor.
Keywords: 5-epi-Aristolochene hydroxylase (EAH), 5-epi-aristolochene synthase (EAS), ethylene responsive factor, Nicotiana attenuata, phytoalexin, sesquiterpene, virus induced gene silencing (VIGS)
Abstract
Capsidiol is a sesquiterpenoid phytoalexin produced in Nicotiana and Capsicum species in response to pathogen attack. Whether capsidiol plays a defensive role and how its biosynthesis is regulated in the wild tobacco Nicotiana attenuata when the plant is attacked by Alternaria alternata (tobacco pathotype), a notorious necrotrophic fungus causing brown spot disease, are unknown. Transcriptome analysis indicated that a metabolic switch to sesquiterpene biosynthesis occurred in young leaves of N. attenuata after A. alternata inoculation: many genes leading to sesquiterpene production were strongly up-regulated, including the capsidiol biosynthetic genes 5-epi-aristolochene synthase (EAS) and 5-epi-aristolochene hydroxylase (EAH). Consistently, the level of capsidiol was increased dramatically in young leaves after fungal inoculation, from not detectable in mock control to 50.68±3.10 µg g−1 fresh leaf at 3 d post-inoculation. Capsidiol-reduced or capsidiol-depleted plants, which were generated by silencing EAHs or EASs by virus-induced gene silencing, were more susceptible to the fungus. In addition, this sesquiterpene when purified from infected plants exhibited strong anti-fungal activities against A. alternata in vitro. Furthermore, an ERF2-like transcription factor was found to positively regulate capsidiol production and plant resistance through the direct transactivation of a capsidiol biosynthetic gene, EAS12. Taken together, our results demonstrate that capsidiol, a phytoalexin highly accumulated in N. attenuata plants in response to A. alternata infection, plays an important role in pathogen resistance independent of jasmonate and ethylene signaling pathways, and its biosynthesis is transcriptionally regulated by an ERF2-like transcription factor.
Introduction
Plants are constantly attacked by a wide variety of microbial pathogens. In response, they activate a large number of intricate defense mechanisms, including the formation of reactive oxygen species, physical reinforcement of cell walls, and production of phytohormones, antimicrobial proteins, and metabolites (Glazebrook, 2005; Ahuja et al., 2012; Mengiste, 2012). The class of small molecules known as phytoalexins is produced by plants de novo in response to pathogen attack, and is an important part of the plant defense repertoire (Ahuja et al., 2012). Arabidopsis mutants impaired in the production of the phytoalexin camalexin are more susceptible to infection by necrotrophic fungi, such as Alternaria brassicicola (Nafisi et al., 2007), Botrytis cinerea (Kliebenstein et al., 2005), and Pletosphaerella cucumerina (Sanchez-Vallet et al., 2010). In Nicotiana species, two phytoalexins have recently received attention: scopoletin (El Oirdi et al., 2010; Sun et al., 2014b) and capsidiol (Mialoundama et al., 2009; Shibata et al., 2010, 2016; Grosskinsky et al., 2011).
Capsidiol has been proposed to be an important ‘chemical weapon’ against pathogens in Nicotiana species. This bicyclic sesquiterpene is produced via cyclization of farnesyl pyrophosphate (FPP) to 5-epi-aristolochene by 5-epi-aristolochene synthase (EAS), followed by two hydroxylation reactions catalysed by 5-epi-aristolochene dihydroxylase (EAH) (Facchini and Chappell, 1992; Ralston et al., 2001). Capsidiol exhibits toxicity towards many pathogens, including Phytophthora capsici and B. cinerea (Stoessl et al., 1972; Ward et al., 1974). Recently, molecular evidence also supports its role in non-host resistance against P. infestans in N. benthamiana, as NbEAS- or NbEAH-silenced plants were highly susceptible (Shibata et al., 2010).
Ethylene response factors (ERFs) are transcription factors that play crucial roles in plant immunity (Huang et al., 2016). In Arabidopsis, several ERFs have been identified as important regulators in Botrytis resistance, such as ORA59, ERF1, and RAP2.2 (Solano et al., 1998; Berrocal-Lobo et al., 2002; Pré et al., 2008; Zhao et al., 2012). Most ERFs are able to bind specifically to DNA sequences containing a GCC (GCC box) and/or a dehydration-responsive element/C-repeat (DRE/CRT box, A/GCCGAC) (Hao et al., 1998, 2002). However, the ERF RAP2.2 binds the consensus sequence ATCTA in the promoter region of phytoene synthase and phytoene desaturase to regulate carotenoid biosynthesis (Welsch et al., 2007).
The necrotrophic fungal pathogen Alternaria alternata (tobacco pathotype) causes brown spot disease in Nicotiana tabacum (LaMondia, 2001) and many other Nicotiana species, including the wild tobacco Nicotiana attenuata (Schuck et al., 2014). Inoculation with A. alternata elicits the activation of both jasmonate (JA) and ethylene signaling pathways in N. attenuata plants, which subsequently lead to the accumulation of the phytoalexins scopoletin and its glycoside form, scopolin (Sun et al., 2014b; Li and Wu, 2016; Sun et al., 2017). Currently it is not known if fungus-inoculated N. attenuata plants produce other phytoalexins, such as capsidiol. If they do, how their biosynthesis is transcriptionally regulated is a significant question.
In this study, capsidiol was identified and confirmed to be an important phytoalexin produced in N. attenuata when challenged by A. alternata, and its regulation by an ERF2-like transcription factor was investigated in detail.
Materials and methods
Plant and fungal materials
Seeds of the 31st generation of an inbred line of Nicotiana attenuata were used as the wild-type (WT) genotype. Ethylene-deficient and -insensitive (irACO and Ov-etr1, respectively), and JA-deficient (irAOC) N. attenuata plants were generated previously (von Dahl et al., 2007; Kallenbach et al., 2012). Seed germination and plant growth were conducted as described by Krügel et al. (2002).
Alternaria alternata was grown and inoculated into leaves as described by Sun et al. (2014a).
RNA-seq data processing and analysis
Source–sink transition leaves of rosette-staged WT plants (35 d old) were detached and inoculated with A. alternata for 1 d, when only a few of fungal hyphae had penetrated into leaf tissues (Sun et al., 2014a). Total RNA of three biological replicates of mock (WT_0L_M, with sample names S716, S717, and S719) or inoculated leaf samples (WT_0L_Inf, with sample names S20, S722, and S726) were isolated with TRIzol reagent (Invitrogen). RNA sequencing was conducted by Shanghai OE-Biotech (http://www.oebiotech.com/) with the Illumina Hiseq 2000.
Sequencing was performed at 8 G depth, and mapped to the N. attenuata reference genome sequence. The relative abundance of the transcripts was measured with the FPKM (RPKM) method, which measures the transcript abundance as RPKM (reads per kilobase of exon model per million mapped reads). The differential expression between mock and 1 d post-inoculation (dpi) samples with a cutoff of 2-fold change and its significance were calculated.
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA001483 (publicly accessible at http://bigd.big.ac.cn/gsa).
Generation of NaEAS- and NaEAH-silenced virus-induced gene silencing plants
A 579 bp NaEAS cDNA fragment amplified with primers (Z003_F and Z004_R; Supplementary Table S1) and a 424 bp fragment of NaEAH cloned with primers (Z047_F and Z048_R; Supplementary Table S1) were individually inserted into pTV00 (Ratcliff et al., 2001) in reverse orientations. Agrobacterium tumefaciens (strain GV3101) harboring these constructs was mixed with the strain with pBINTRA and inoculated into N. attenuata leaves generating NaEASs- and NaEAHs-silenced plants (virus-induced gene silencing (VIGS) NaEASs and VIGS NaEAHs). The A. tumefaciens-mediated transformation procedure was performed as described previously (Saedler and Baldwin, 2004). To monitor the progress of VIGS, phytoene desaturase (PDS) was also silenced. Silencing PDS results in the visible bleaching of green tissues (Saedler and Baldwin, 2004; Wu et al., 2008) about 2–3 weeks after the inoculation. When the leaves of PDS-silenced plants began to bleach, the young leaves of VIGS plants and empty vector-inoculated plants (EV plants) were selected for further experiments. Around 20 plants were inoculated with each construct, and usually 10 biological replicates per construct exhibiting efficient silencing were used for each experiment, and all VIGS experiments were repeated twice.
Purification and quantification of capsidiol in N. attenuata after infection
Around 500 g leaves that had been inoculated with A. alternata for 3 d were collected for capsidiol extraction. Leaves were twice extracted with 70% acetone (2 liters) at room temperature. The solvent was evaporated and suspended in water, and then extracted with ethyl acetate. The ethyl acetate-soluble fraction (10 g) was decolorized on an MCI gel (www.gls.co.jp) with methanol: H2O (90:10) to obtain a yellow gum (7 g), which was subsequently purified with a silica gel column with a chloroform: acetone gradient system (from 10:0, 9:1, 8:2, 7:3, 6:4, to 1:1) to yield six main fractions (A–F). Fraction B (chloroform: acetone, 9:1; 2 g) was subjected to repeated chromatography over silica gel (petroleum ether: acetone, from 30:1 to 1:1) to yield fractions B1–B4. Fraction B3 (petroleum ether: acetone, 10:1) was separated further with an RP-18 column (acetonitrile: H2O, 30:70). The obtained crude capsidiol was further purified by semi-preparative HPLC (3 ml min−1, UV detection at λ max=202 nm, acetonitrile: H2O, 40:60; ZORBAX SB-C18 column (5 μm, 9.4×250 mm, Agilent 1200, USA) to yield capsidiol (30 mg, >99.5% purity). The purified capsidiol showed the same characteristic NMR data and HPLC retention times as the authentic standard provided by Prof. J. Chappell (University of Kentucky, USA).
Alternaria alternata-elicited capsidiol levels were determined by HPLC by reference to the authentic capsidiol standard. Each leaf was inoculated with four agar plugs with fungal mycelium, and a 1.5×1.5 cm2 area of the leaf lamina was harvested at 1 or 3 dpi (around 200 mg fresh mass). Samples were ground in liquid nitrogen and twice extracted with 1 ml dichloromethane; 2 ml extracts were dried in a SpeedVac concentrator (Eppendorf), and finally dissolved in 1 ml methanol for HPLC. At a flow rate of 1 ml min−1, 10 μl of each sample was injected onto a ZORBAX SB-C18 column (5 μm, 4.6×250 mm) (Agilent 1260). The mobile phase was composed of solvent A (water), and solvent B (acetonitrile) was used in a isocratic elution (40% of B). Capsidiol was detected at 202 nm, with a retention time of 10.38 min. The standard capsidiol was dissolved in methanol at six concentrations (6.25, 12.5, 25, 50, 100, and 200 μg ml−1) to create an external standard curve that was used to calculate fungus-induced capsidiol levels.
Bioassays for the inhibition of A. alternata growth by capsidiol in vitro
The inhibition of A. alternata mycelium growth by capsidiol in vitro was tested in Petri dishes by sub-culturing a 3 mm-diameter mycelium plug on potato dextrose agar (PDA) medium containing various concentrations of capsidiol for 6 d in the dark at 25 ℃. Twenty milligrams of capsidiol was dissolved in 10 ml methanol, and added to the PDA medium at final concentrations of 0, 50, 100, and 200 µg ml−1. PDA plates with 1% methanol served as controls. Photos were taken every 2 d, and the area of mycelium growth was calculated with ImageJ (http://imagej.nih.gov/ij/).
Quantification of scopoletin and scopolin
Around 0.2 g leaf samples at 6 dpi of EV, VIGS NaEAS and VIGS NaEAH plants were harvested and ground to fine powder in liquid nitrogen. The levels of scopoletin and scopolin were determined by HPLC-MS/MS as described in Sun et al. (2014b).
Real-time PCR
Total RNA was extracted from a 1.5×1.5 cm2 area of leaf lamina that encompassed the inoculation site with TRI reagent (Invitrogen). For each treatment, four to five replicate biological samples were collected. cDNA was synthesized from 500 ng total RNA with reverse transcriptases (Thermo Fisher Scientific). Real-time PCR was performed on a CFX Connect qPCR System (Bio-Rad) with iTaq Universal SYBR Green Supermix (Bio-Rad) and gene-specific primers as described (Wu et al., 2013). For each analysis, a linear standard curve (obtained from threshold cycle number versus log DNA quantity) was constructed by using a dilution series of a specific cDNA sample, and the transcript levels of unknown samples were calculated according to the standard curve. Finally, the relative transcript levels of target genes were obtained by dividing the extrapolated transcript levels of the target genes by the levels of a housekeeping gene, NaActin 2, as an internal standard from the same sample. The transcript abundance of NaActin 2 was not altered in leaves inoculated with A. alternata at 1 and 3 dpi (Xu et al., 2018). All primers are listed in Supplementary Table S1 at JXB online.
Transient expression assays
To determine the subcellular localization of NaERF2-like, a 35S::ERF2-eGFP construct was used for transient expression in N. attenuata protoplasts. The full-length coding sequence of NaERF2-like was amplified by primers Z141_F and Z142_R (Supplementary Table S1) and inserted into vector pM999 via SacI and XhoI. The method of protoplast isolation and transient transformation was adopted from (Yoo et al., 2007) with some modifications. In brief, mesophyll protoplasts were isolated from source–sink transition leaves, and 20 µg of plasmids was transfected into 2×105 protoplasts with polyethylene glycol (PEG) solution (0.4 g ml−1 PEG 4000, 0.2 M mannitol, 0.1 M CaCl2). Transformed cells were cultured in solution (4 mM MES, 0.5 M mannitol, 20 mM KCl) for 18 h in the dark and images were obtained with a fluorescence microscope (Leica DM5500 B) with excitation at 488 nm for the green fluorescent protein (GFP) signal.
For a transient transactivation assay, the promoter region of NaEAS12 (−1671 to −1; numbered from the first ATG) and firefly luciferase (LUC) were amplified and cloned into pCAMBIA1301. Next, the PCR fragment including the promoter region of NaEAS12, LUC, and Nos terminator was subcloned into pMD18 via HindIII and SacI to reduce the size of the vector and increase the transformation efficiency. Similarly, 35S::NaERF2-like-2HA-eGFP with Nos terminator was also subcloned into pMD18 vector. Nicotiana attenuata protoplasts were prepared as describe above, and transformed with both NaEAS12Promoter::LUC and 35S::NaERF2-like-2HA-eGFP, or with NaEAS12Promoter::LUC and 35S::2HA-eGFP as control. After transformation, the protoplasts were subjected to RNA extraction with a PrimeScript™ RT reagent kit with gDNA Eraser followed by a gene expression assay. All the experiments were repeated twice with similar results.
Yeast-one-hybrid assay
The Matchmaker yeast one-hybrid system (Clontech) was used to test the binding of NaERF2-like and the NaEAS12 promoter in vitro according to the user manual. The promoter region of NaEAS12 (pEAS12-b; −926 to −699; numbered from the first ATG), which contained the candidate binding motif of EM13 (5′-tagattATCTaattctact-3′), was inserted into pAbAi vector with HindIII and XhoI. The bait construct was linearized and integrated into the genome of yeast strain Y1HGold. The full-length coding sequence of NaERF2-like was introduced into pGADT7 AD vector via ClaI and XhoI, and then the construct was transformed into the yeast cells containing the bait. The positive clones were analysed on SD/−His/−Leu medium supplied with 200 ng ml−1 (final concentration) 3-amino-1,2,4-trazole (3-AT). Y1HGold [pGADT7/pEAS12-b-AbAi] was used as a negative control and Y1HGold [pGADT7 Rec-p53/p53-AbAi] was used as positive control.
Electromobillity shift assays
The full-length coding sequence of NaERF2-like was cloned in-frame into the EcoRI–XhoI sites of the pET28a (+), and His-NaERF2-like was expressed and purified with Ni-NTA agarose (Qiagen). A biotin-labeled probe, EM13 (5′-tagattATCTaattctact-3′), and a mutant probe (5′-tagattAATTaattctact-3′) were synthesized by Sangon Biotech (Shanghai). The detection of the binding of the recombinant protein and the probes (300 ng of recombinant protein and 30 ng labeled probe) was carried out with a chemiluminescence electrophoretic mobility shift assay (EMSA) kit (Beyotime Biotechnology) according to the protocol suggested by the manufacturer.
Generation of Ov-NaERF2-like plants and chromatin immune-precipitation assay
The full-length cDNA of NaERF2-like with two hemagglutinin (HA: YPYDVPDYA) epitopes at its C-terminus was cloned into pCAMBIA1301 vector after the 35S promoter with the in-fusion technique (Clontech). Nicotiana attenuata plants (31st generation of an inbred line) were transformed by Agrobacterium tumefaciens with this construct according to Krügel et al. (2002). T1 seeds were screened for single T-DNA inserts (1:3 segregation of hygromycin resistance), and two lines of T2 (Ov-NaERF2-like line 1 and 2) with HA signals (see Results, Supplementary Fig. S2) were selected and used in this study.
Chromatin immunoprecipitation was performed with EpiQuik Plant ChIP Kit (EPIGENTEK) according to the user manual. The source–sink transition leaves of Ov-NaERF2-like line 2 at 1 dpi (1 g) were used for ChIP assays. A ChIP grade anti-HA antibody (Abcam) was used to immunoprecipitate the protein–DNA complex, and the precipitated DNA was further purified for real time PCR by using primer sets from NaEAS12 promoter (5′-CACTTTAACCCCCGGGTAACT-3′ and 5′-CACTTCTCAGATTCTCCAGTTTGG-3′) and NaActin 2 gene (5′-GGTCGTACCACCGGTATTGTG-3′ and 5′-GTCAAGACGGAGAATGGCATG-3′) for negative control. The ChIP experiments were performed twice with similar results. Chromatins that were precipitated without antibody served as the negative controls.
Results
Transcriptome analysis reveals the strong regulation of sesquiterpene biosynthetic genes in N. attenuata after A. alternata inoculation
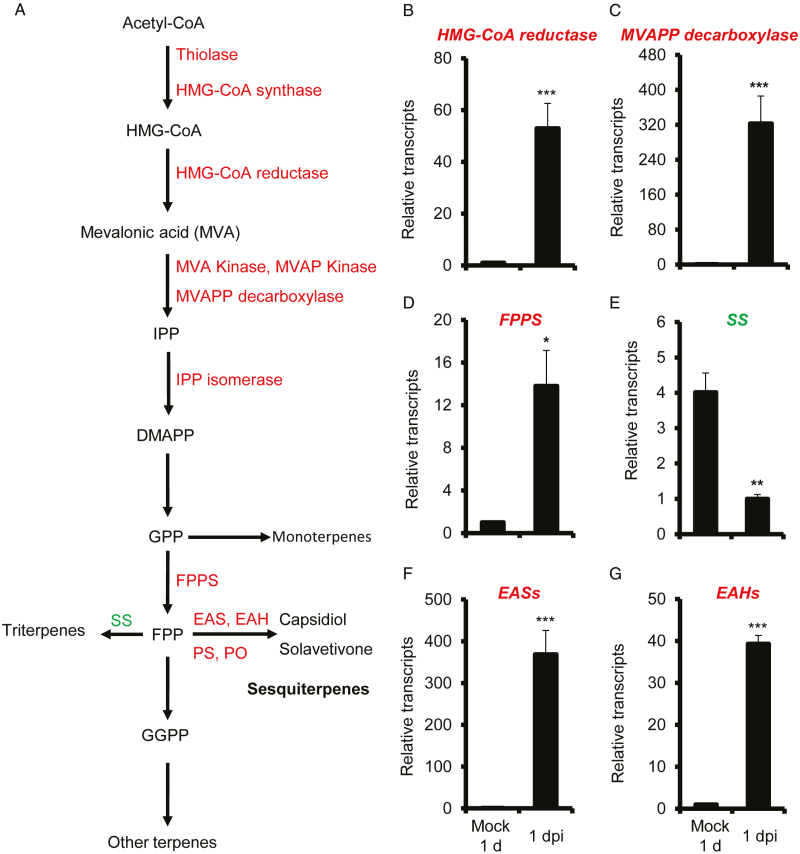
Previously, we have demonstrated that, in response to A. alternata inoculation, N. attenuata plants activate both JA and ethylene signaling pathways to regulate the biosynthesis of the phytoalexins scopoletin and its β-glycoside form, scopolin (Sun et al., 2014b, 2017; Li and Wu, 2016). As feruloyl-CoA 6′-hydroxylase 1 (NaF6′H1), the key enzyme gene for scopoletin biosynthesis, was highly elicited at 1 d post-inoculation (dpi) when the infection was at an early stage and only a few fungal hyphae had been observed to penetrate into leaf tissues via stomata (Sun et al., 2014a), transcriptome analysis was performed in A. alternata-inoculated N. attenuata leaves at 1 dpi to identify secondary metabolites that could potentially act similarly to scopoletin. Notably, a set of genes involved in terpene synthesis were strongly up-regulated, including thiolase, HMG-CoA synthase, HMG-CoA reductase, mevalonate kinase, phosphomevalonate kinase, diphosphomevalonate decarboxylase (MVPP decarboxylase), isopentenyl-diphosphate delta-isomerase, FPP synthase, the capsidiol biosynthetic genes 5-epi-aristolochene synthases (EASs) and 5-epi-aristolochene 1,3-dihydroxylases (EAHs), and the solavetivone biosynthetic genes premnaspirodiene oxygenase and premnaspirodiene synthase (Fig. 1A; Supplementary Table S2). The squalene synthase involved in triterpene biosynthesis was down-regulated (Fig. 1A; Supplementary Table S2). These results strongly indicate that the sesquiterpene biosynthetic pathway is activated during the inoculation of A. alternata.
Fig. 1.
The regulation of terpene biosynthetic genes in N. attenuata in response to A. alternata inoculation at 1 d post-inoculation (dpi). Genes involved in terpene synthesis were strongly regulated during transcriptome analysis in three biological replicate samples of mock and 1 dpi. (A) Transcripts of all enzymes marked in red font were up-regulated at 1 dpi, while squalene synthase (SS), shown with green font, was down-regulated. (B–G) To validate this regulation, relative mean transcripts (±SE) of HMG-CoA reductase (B), MVAPP decarboxylase (C), FPPS (D), squalene synthase (E), EASs (F), and EAHs (G) were measured by real-time PCR in four biological replicate 0 leaves at 1 dpi. Leaves without inoculation were collected as controls (Mock 1 d). Both EASs and EAHs were detected by primers conserved in gene family members. Asterisks indicate level of significant difference between Mock 1 d and 1 dpi samples (Student’s t-test: *P<0.05, **P<0.01, ***P<0.005). DMAPP, 3′3-dimethylallyl diphosphate; EA, 5-epi-aristolochene; EAH, 5-epi-aristolochene hydroxylase; EAS, 5-epi-aristolochene synthase; FPP, farnesyl diphosphate; FPPS, farnesyl pyrophosphate synthase; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate; HMG-CoA, 3-hydroxy-3-methyl-glutaryl-CoA; IPP, isopentenyl diphosphate; MVA, mevalonic acid; MVAP, mevalonic acid 5-phosphate; MVAPP, mevalonic acid 5-diphosphate; PO, premnaspirodiene oxidase; PS, premnaspirodiene synthase.
To confirm the regulation of terpene biosynthesis genes in response to A. alternata, we performed quantitative real time PCR on N. attenuata samples collected at 1 dpi. Compared with mock infection controls, the transcripts of MVPP decarboxylase, HMG-CoA reductase, and FPP synthase at 1 dpi were increased 322-, 53-, and 14-fold, respectively (Fig. 1B–D), while the transcripts of squalene synthase were reduced by 75% (Fig. 1E). More importantly, transcripts of the capsidiol biosynthetic genes NaEASs and NaEAHs, which are encoded by a multi-gene family (Supplementary Table S2) and thus were detected by primers designed at the consensus region, increased 368- and 40-fold compared with control (Fig. 1F, G). These data fully confirmed the transcriptomics result of strong activation of the sesquiterpene biosynthesis pathway in N. attenuata after inoculation.
Accumulation of capsidiol in N. attenuata in response to A. alternata inoculation
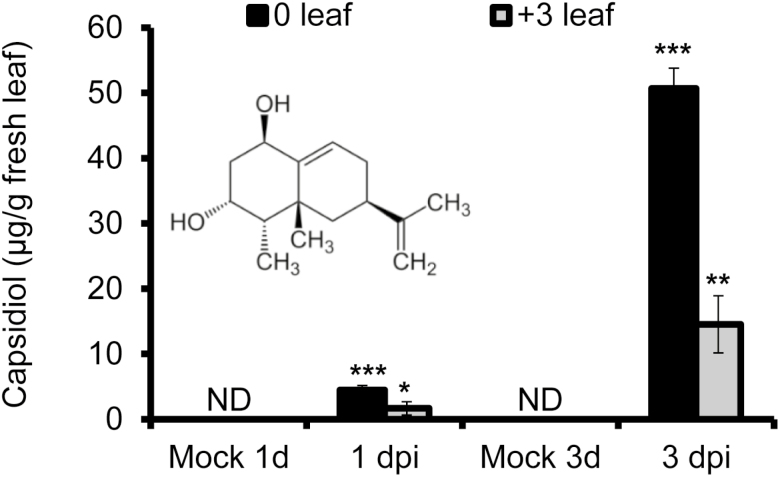
Since NaEAS and NaEAH are key genes in the capsidiol biosynthesis pathway, we expected to see an increase in capsidiol production after A. alternata inoculation. The levels of capsidiol at 1 and 3 dpi in the young source–sink transition leaves (0 leaves) and mature leaves (+3 leaves) were determined by HPLC. The +3 leaves are three phyllotaxic positions older than 0 leaves, and are more susceptible to A. alternata (Sun et al., 2014a). Our results indicated that capsidiol was not detectable in the 0 leaves of mock control, but its level was increased to 4.45±0.72 µg g−1 fresh leaf in 0 leaves at 1 dpi, and to 50.68±3.10 µg g−1 at 3 dpi (Fig. 2). Interestingly, this compound was also detected in the susceptible +3 leaves, but its level was only about one-third of that of 0 leaves, with 1.66±1.00 µg g−1 fresh leaf at 1 dpi, and 14.54±4.39 µg g−1 at 3 dpi (Fig. 2). These results indicate that (i) capsidiol is highly elicited in N. attenuata leaves after inoculation, and (ii) the lower accumulation of capsidiol in mature leaves may account for their susceptibility to the fungus.
Fig. 2.
Accumulation of capsidiol in 0 (young) and +3 (mature) leaves after A. alternata inoculation. Mean (±SE) capsidiol levels were determined by HPLC in five biological replicate 0 and +3 leaves at 1 and 3 dpi. Asterisks indicate level of significant difference between mock and infected samples (Student’s t-test: *P<0.05, **P<0.01, ***P<0.005). ND, not detectable.
Capsidiol accumulation is essential for A. alternata resistance in N. attenuata
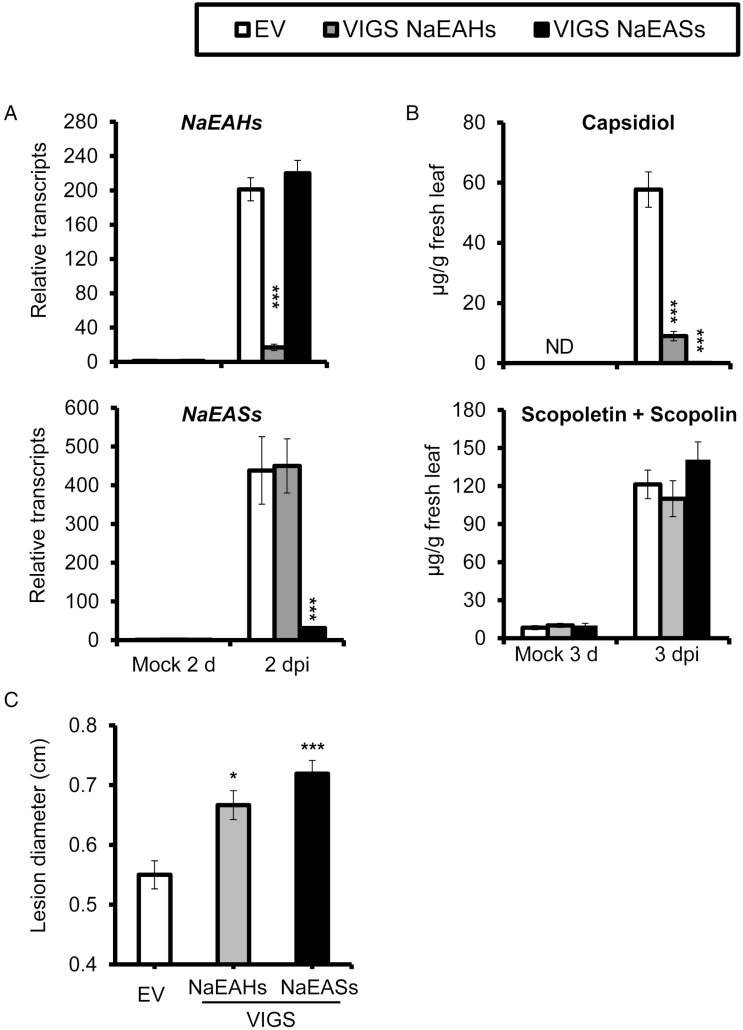
To further evaluate the role of capsidiol in N. attenuata resistance against A. alternata, we silenced NaEASs and NaEAHs separately with their conserved sequences, as both are members of a multi-gene family. Compared with mock controls, transcripts of NaEAHs were dramatically induced in young leaves of N. attenuata plants transformed with empty vector (EV) at 2 dpi; however, plants transformed with the NaEAHs-silencing construct (VIGS NaEAHs) showed a 92% reduction in NaEAHs transcripts compared with EV plants with the same treatments, indicating effective silencing of the NaEAHs (Fig. 3A). We also investigated the capsidiol level in EV and VIGS NaEAHs plants at 3 dpi. A capsidiol level of 57.72±5.88 µg g−1 fresh leaf was detected in EV plants, while only 8.99±1.56 µg g−1 fresh leaf was found in VIGS NaEAHs plants (Fig. 3B).
Fig. 3.
Silencing expression of NaEAHs or NaEASs dramatically reduces A. alternata-induced capsidiol levels and plant resistance without affecting scopoletin. (A) Mean (±SE) relative A. alternata-induced transcripts of NaEAHs and NaEASs as measured by real-time PCR in five replicate young leaves of EV, VIGS NaEAHs, and VIGS NaEASs plants at 2 dpi. (B) Mean (±SE) capsidiol and scopoletin (including scopolin) levels were determined by HPLC in five replicate young leaves of EV, VIGS NaEAHs, and VIGS NaEASs plants at 3 dpi. (C) Mean (±SE) diameter of necrotic lesions in eight replicate young leaves of EV, VIGS NaEAHs, and VIGS NaEASs plants infected with A. alternata for 6 d. As both NaEAH and NaEAS are encoded by large gene families, conserved cDNA regions were used for silencing and real time PCR. Asterisks indicate level of significant difference between EV and VIGS leaves (Student’s t-test: *P<0.05, ***P<0.005). ND, not detectable.
Because a small amount of capsidiol was still present in VIGS NaEAHs plants, we attempted to generate additional capsidiol-depleted N. attenuata plants by silencing NaEASs. Expression of NaEASs was successfully silenced, as only 7% of the transcripts of NaEASs were detected at 2 dpi in VIGS NaEASs plants compared with EV plants (Fig. 3A). More importantly, the A. alternata-elicited capsidiol level at 3 dpi was abolished in VIGS NaEASs plants, with only 0.2% of the level detected in EV plants, which was comparable to the levels quantified in the mock controls of EV plants (Fig. 3B). From these data, we infer that NaEASs genes are crucial for A. alternata-elicited capsidiol production.
To test whether capsidiol-reduced or -depleted plants are more susceptible to A. alternata, young leaves of EV, VIGS NaEAHs, and VIGS NaEASs plants were inoculated with the fungus. Two independent VIGS experiments showed significantly increased lesion diameters in VIGS NaEAHs (121% of EV plants) and VIGS NaEASs plants (131% of EV plants) (Fig. 3C). We did not observe changes in scopoletin and scopolin, two important phytoalexins involved in A. alternata resistance (Fig. 3B). These results strongly indicate that capsidiol plays an important role in defending against A. alternata.
Capsidiol exhibits anti-fungal activity against A. alternata in vitro
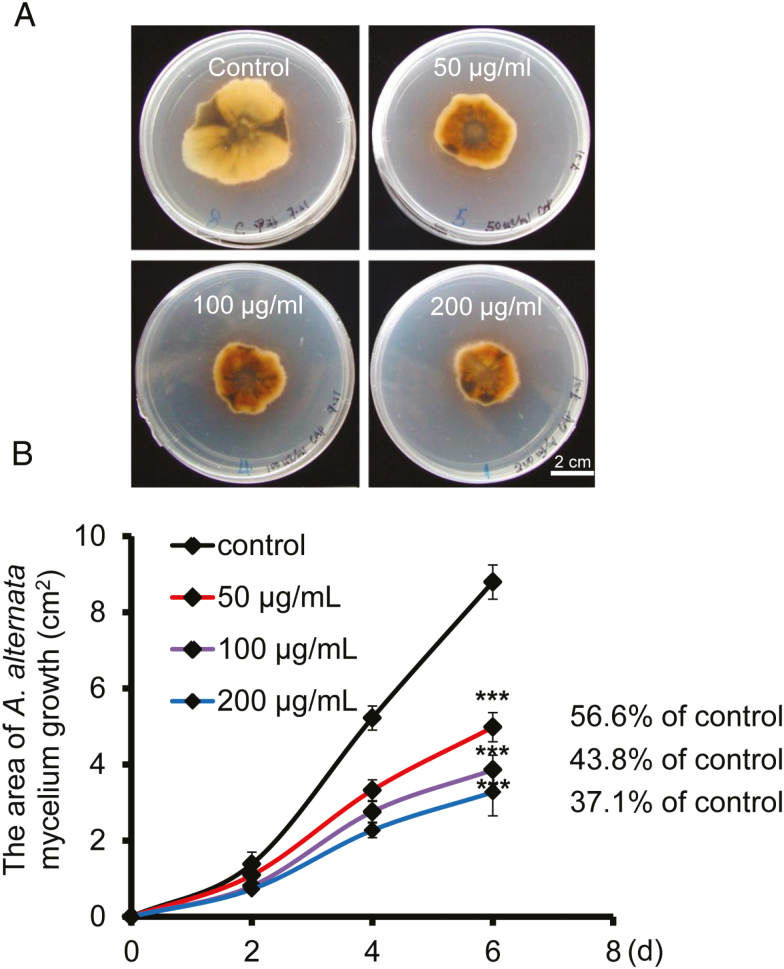
To test whether capsidiol has a direct impact on fungal growth or not, we purified 30 mg capsidiol from 500 g of A. alternata-inoculated leaves, and applied this compound in various concentrations to the growth medium to evaluate its inhibitory activity on fungal growth in vitro. The fungi were grown on PDA plates containing 50 or 100 µg ml−1 capsidiol, and we observed that fungal growth was reduced to 56.6% or 43.8%, respectively, of that of controls. Application of 200 µg ml−1 capsidiol resulted in further reduction in growth to 37.1% of control (Fig. 4A, B). These results suggest that capsidiol at a concentration observed in planta has a direct impact on the fungal growth in vitro.
Fig. 4.
Capsidiol exhibits anti-fungal activity in vitro. (A) Growth of A. alternata mycelium at day 6 in PDA with capsidiol at final concentration of 0, 50, 100, and 200 µg ml−1. PDA plates with 1% methanol served as controls. (B) The area of A. alternata mycelium growth in PDA with different concentrations of capsidiol was determined by ImageJ. Data were collected every 2 d. Asterisks indicate level of significant difference between mock and infected samples (Student’s t-test: ***P<0.005).
Alternaria alternata-induced capsidiol accumulation is not dependent on JA and ethylene signaling
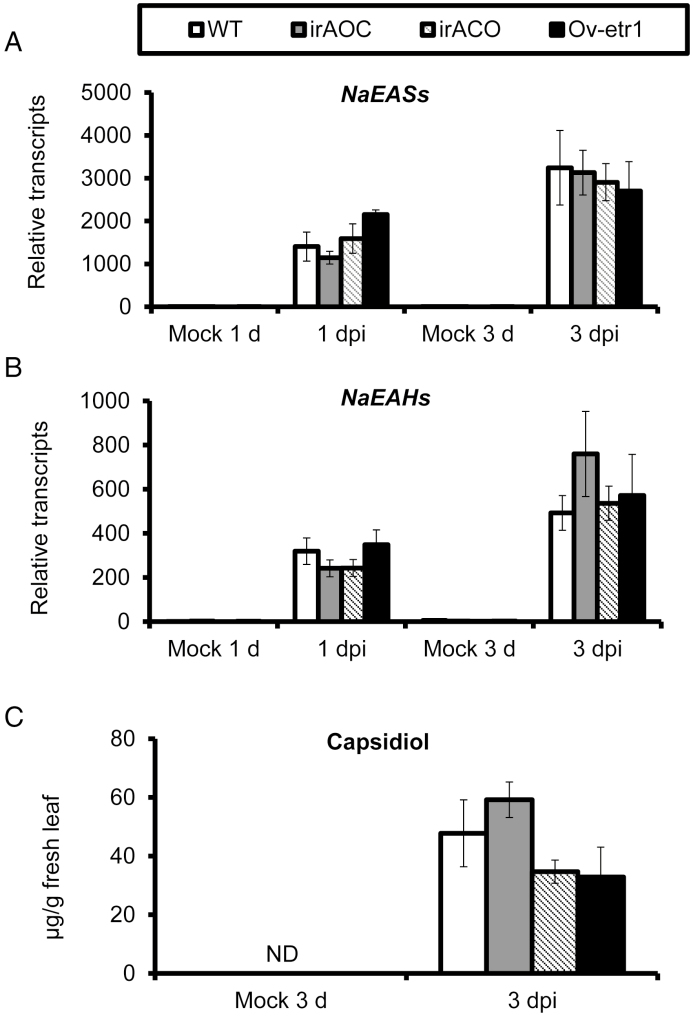
JA and ethylene signaling pathways are crucial for biosynthesis of the phytoalexin scopoletin. To investigate the roles of these two signaling pathways in capsidiol biosynthesis, we measured the levels of capsidiol and transcripts of NaEASs and NaEAHs after A. alternata inoculation in WT, JA-deficient (irAOC), ethylene-deficient (irACO), and ethylene-insensitive (Ov-etr1) plants generated previously (von Dahl et al., 2007; Kallenbach et al., 2012). We found that the induction levels of NaEASs and NaEAHs by A. alternata were similar in WT, irAOC, irACO, and Ov-etr1 plants at both 1 and 3 dpi (Fig. 5A, B). In addition, no significant differences of capsidiol production were observed in WT, irAOC, irACO, and Ov-etr1 plants at 3 dpi (Fig. 5C). Thus, our data indicated that A. alternata-induced capsidiol accumulation is not dependent on JA and ethylene signaling.
Fig. 5.
JA and ethylene pathways play a minor role in A. alternata-induced transcription of NaEASs and NaEAHs, and capsidiol biosynthesis. (A, B) Mean (±SE) relative A. alternata-induced NaEASs (A) and NaEAHs (B) transcripts as measured by real-time PCR in five biological replicates of young leaves (0 leaves) in WT, irAOC (JA deficient), irACO (ethylene deficient), and Ov-etr1 (ethylene insensitive) plants at 1 and 3 dpi. (C) Mean (±SE) capsidiol levels determined by HPLC in five replicate 0 leaves of WT, irAOC, irACO, and Ov-etr1 plants at 3 dpi. ND, not detectable.
NaERF2-like, a transcription factor highly induced in young leaves, is required to mount a capsidiol-based defense
Since we observed increases in both capsidiol production and NaEASs and NaEAHs transcripts in N. attenuata leaves after fungal inoculation, we hypothesized that the transcription factors regulating expression of NaEASs and NaEAHs were also increased. Thus, we silenced the expression of the six fungus-elicited transcription factor genes with the most abundant transcripts after fungal inoculation (Supplementary Table S3), namely ethylene-responsive transcription factor ABR1-like (NaERF ABR1-like; gene accession no.: XM_019374371), zinc finger protein ZAT12-like (NaZAT12; gene accession no.: XM_019368773.1), probable WRKY transcription factor 40 (NaWRKY40; gene accession no.: XM_019402562.1), probable WRKY transcription factor 43 (NaWRKY43; gene accession no.: XM_019375046.1), probable WRKY transcription factor 61 (NaWRKY61; gene accession no.: XM_019371308.1), and ethylene-responsive transcription factor 2-like(NaERF2-like; gene accession no.: XM_019399671.1), to identify the regulator(s) responsible for capsidiol biosynthesis. Alternaria alternata-elicited NaEAS12 was not altered in plants silenced with NaERF ABR1-like, NaZAT12, NaWRKY40, NaWRKY43, or NaWRKY61 (Supplementary Fig. S1).
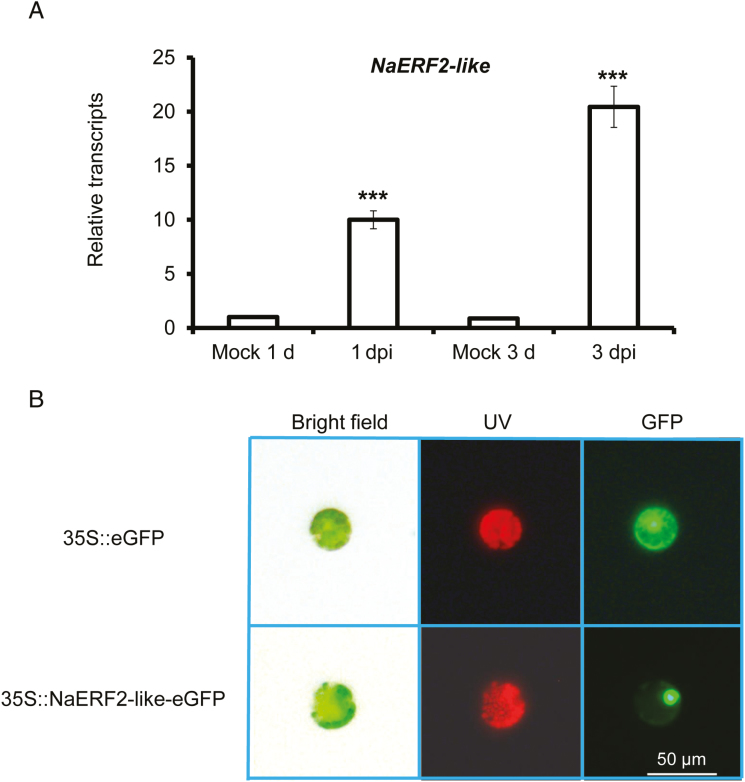
NaERF2-like (gene accession no.: XM_019399671.1), one of the top six transcription factors strongly up-regulated in response to fungal inoculation in our transcriptome analysis, was highly induced in N. attenuata 0 leaves at both 1 and 3 dpi (Fig. 6A; Supplementary Table S3). The NaERF2-like protein exhibited nuclear localization in the protoplast of N. attenuata when its eGFP fusion protein was driven by a constitutive 35S promoter (Fig. 6B).
Fig. 6.
NaERF2-like is highly elicited after A. alternata inoculation and is targeted to the nucleus. (A) Mean (±SE) relative A. alternata-induced NaERF2-like transcripts as measured by real-time PCR in four replicate 0 leaves at 1 and 3 dpi. Asterisks indicate level of significant difference between mock and infected samples (Student’s t-test: ***P<0.005). (B) Two fusion proteins, 35S::eGFP and 35S::NaERF2-like-eGFP, were expressed transiently in N. attenuata protoplasts. NaERF2-like was found to be targeted to the nucleus. Images were taken in bright field (left), UV (middle) and GFP channels (right).
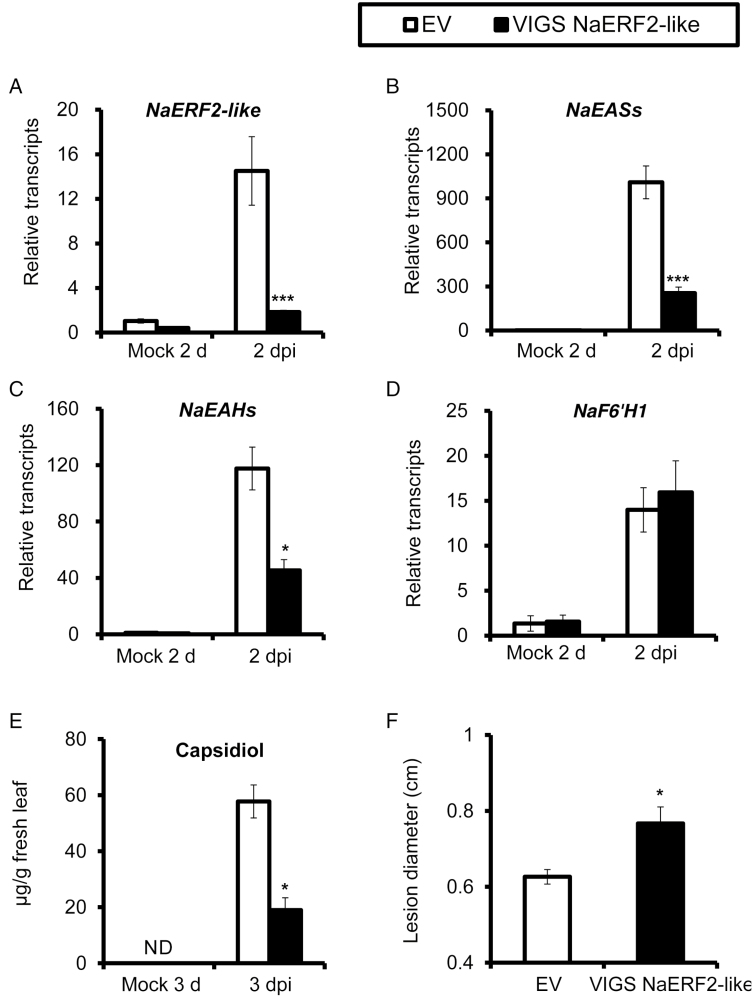
To investigate the role of the NaERF2-like transcription factor in capsidiol biosynthesis in detail, we silenced the gene by VIGS, and then measured the levels of capsidiol and transcripts of NaEASs and NaEAHs after fungal inoculation. NaERF2-like transcripts were highly elicited at 2 dpi in EV plants; in contrast, VIGS NaERF2-like plants showed an 87% reduction in NaERF2-like transcripts after the same treatment (Fig. 7A). Compared with EV plants at 2 dpi, the transcripts of NaEASs and NaEAHs in VIGS NaERF2-like plants were reduced by 75% and 62%, respectively (Fig. 7B, C). As expected, A. alternata-induced capsidiol level in VIGS NaERF2-like plants at 3 dpi was reduced by 68% when compared with EV plants (Fig. 7E). However, the fungus-elicited transcriptional levels of NaF6′H1 were not affected in VIGS NaERF2-like plants (Fig. 7D). These results indicate that silencing NaERF2-like substantially decreases A. alternata-elicited transcription of NaEASs and NaEAHs, and consequently, capsidiol level, without affecting scopoletin-based defense.
Fig. 7.
Silencing NaERF2-like impairs A. alternata-induced transcripts of NaEASs and NaEAHs, capsidiol levels and plant resistance without affecting NaF6′H1 transcript accumulation. (A–D) Mean (±SE) relative A. alternata-induced NaERF2-like (A), NaEASs (B), NaEAHs (C), and NaF6′H1 (D) transcript abundance as measured by real-time PCR in five replicate young leaves of EV and VIGS NaERF2-like plants at 2 dpi. (E) Capsidiol levels determined by HPLC in five replicate young leaves of EV and VIGS NaERF2-like plants at 3 dpi. (F) Mean (±SE) diameter of necrotic lesions recorded in eight replicate young leaves of EV and VIGS NaERF2-like plants infected with A. alternata for 6 d. Two independent VIGS experiments returned similar results. Asterisks indicate level of significant difference between EV and VIGS leaves (Student’s t-test: *P<0.05, ***P<0.005). ND, not detectable.
In addition, silencing NaERF2-like led to plants being more susceptible to A. alternata, as significantly larger lesions were observed in VIGS NaERF2-like plants at 6 dpi in two independent VIGS experiments (Fig. 7F).
NaERF2-like directly regulates the capsidiol biosynthetic gene NaEAS12
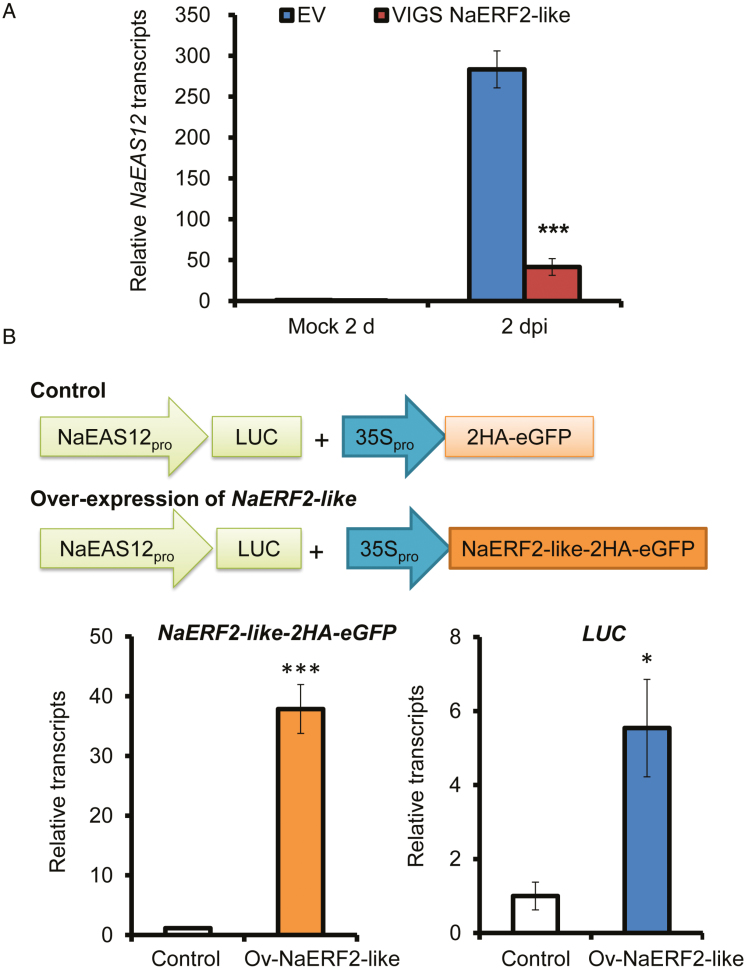
Next, we explored the mechanism by which NaERF2-like regulates capsidiol-based resistance. We hypothesized that NaERF2-like might directly regulate genes in the capsidiol biosynthetic pathway. Since NaEAS12 expression was greatly reduced in NaERF2-like-silenced plants (Fig. 8A), we selected this gene to test whether its promoter could be directly activated by NaERF2-like.
Fig. 8.
Alternaria alternata-elicited NaEAS12 transcripts are largely dependent on NaERF2-like, and transient overexpression of NaERF2-like leads to transactivation of NaEAS12 promoter. (A) Mean (±SE) relative A. alternata-induced NaEAS12 transcripts as measured by real-time PCR in five replicate young leaves of EV and VIGS NaERF2-like plants at 2 dpi. Asterisks indicate level of significant difference between EV and VIGS leaves at 2 dpi (Student’s t-test: ***P<0.005). (B) Transcript abundance of NaERF2-like-2HA-eGFP and LUC in N. attenuata protoplasts co-expressing NaEAS12 promoter::LUC and 35S::2HA-eGFP or 35S::NaERF2-like-2HA-eGFP measured in three biological samples. Experiments were repeated twice with similar results. Asterisks indicate level of significant difference between control and NaERF2-like overexpressing protoplasts (Student’s t-test: *P<0.05; ***P<0.005).
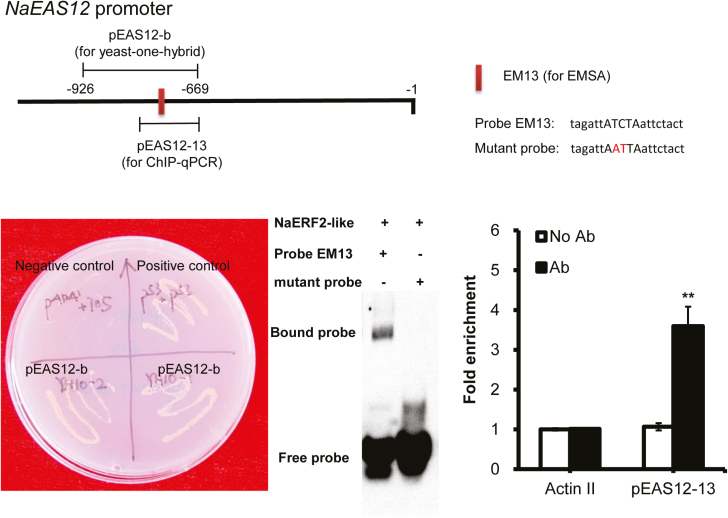
Several lines of evidence were consistent with the idea that NaERF2-like directly regulates the capsidiol biosynthetic gene NaEAS12. When the NaERF2-like gene was overexpressed in the protoplasts of N. attenuata, the luciferase gene (LUC) driven by the NaEAS12 promoter showed a 5.5-fold increase in expression (Fig. 8B), indicating that the overexpression of NaERF2-like enhanced the transcriptional activity of NaEAS12 promoter. From the promoter region of NaEAS12, an ATCTA motif, previously shown to be the binding site of RAP2.2 in Arabidopsis (Welsch et al., 2007), was identified and confirmed as the binding site for the ERF2-like by yeast one-hybrid, electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation-based qPCR (ChIP-qPCR). Yeast one-hybrid experiments revealed that the ERF2-like protein could bind to the NaEAS12 promoter fragment EAS12-b (located from −699 to −926 bp upstream of the starting codon), which contained the ATCTA motif (Fig. 9). Further EMSA experiments indicated that NaERF2-like could directly bind to the biotin labeled probe EM13 (5′-tagattATCTAattctact-3′), but not to the mutated one (5′-tagattAATTAattctact-3′) (Fig. 9). To further confirm these results in vivo, we generated a transgenic line ectopically expressing the NaERF2-like protein fused with two HA tags at the C-terminus (35S::NaERF2-like-2HA) for use in ChIP-qPCR. The fusion protein could be detected by a commercial HA antibody (Supplemental Fig. S2), suggesting that the protein was successfully overexpressed in N. attenuata plants. Transgenic plants were inoculated with A. alternata and sampled at 1 dpi. We found that the HA-tagged NaERF2-like protein bound the NaEAS12 promoter at a site that encompassed the ACTCA motif (Fig. 9).
Fig. 9.
The binding of NaERF2-like and NaEAS12 promoter as demonstrated by yeast one-hybrid, EMSA and chromatin immunoprecipitation. The NaEAS12 promoter structure indicated the pEAS12-b (−669 to −926 numbered from the ATG) for yeast one-hybrid, the probe EM13 (5′-tagattATCTaattctact-3′) for EMSA, and the pEAS12-13 (−669 to −781 numbered from the ATG) for ChIP assays. The red letters ‘AT’ indicate the mutated positions. Yeast one-hybrid analysis revealed that NaERF2-like could bind to EAS12-b as the yeast cells could grow on the SD/−His/−Leu medium supplied with 200 ng ml−1 (final concentration) 3-AT; Y1HGold [pGADT7/pEAS12-b-AbAi] was used as a negative control, and Y1HGold [pGADT7 Rec-p53/p53-AbAi] was used as a positive control. EMSA demonstrated that His tagged NaERF2-like could bind to probe EM13 but not to the mutated one. The mutant probe (5′-tagattAATTaattctact-3′) served as a negative control in EMSA. ChIP-real time PCR data indicated NaERF2-like bound to the promoter of NaEAS12. Negative controls were without antibody (no Ab) and with HA antibody but using primers detecting NaActin 2. Asterisks indicate level of significant difference between no Ab and with Ab in pEAS12-13 (Student’s t-test: **P<0.01).
Overexpression of NaERF2-like does not alter plant resistance, but increases A. alternata-induced NaEAS12 gene expression and capsidiol levels
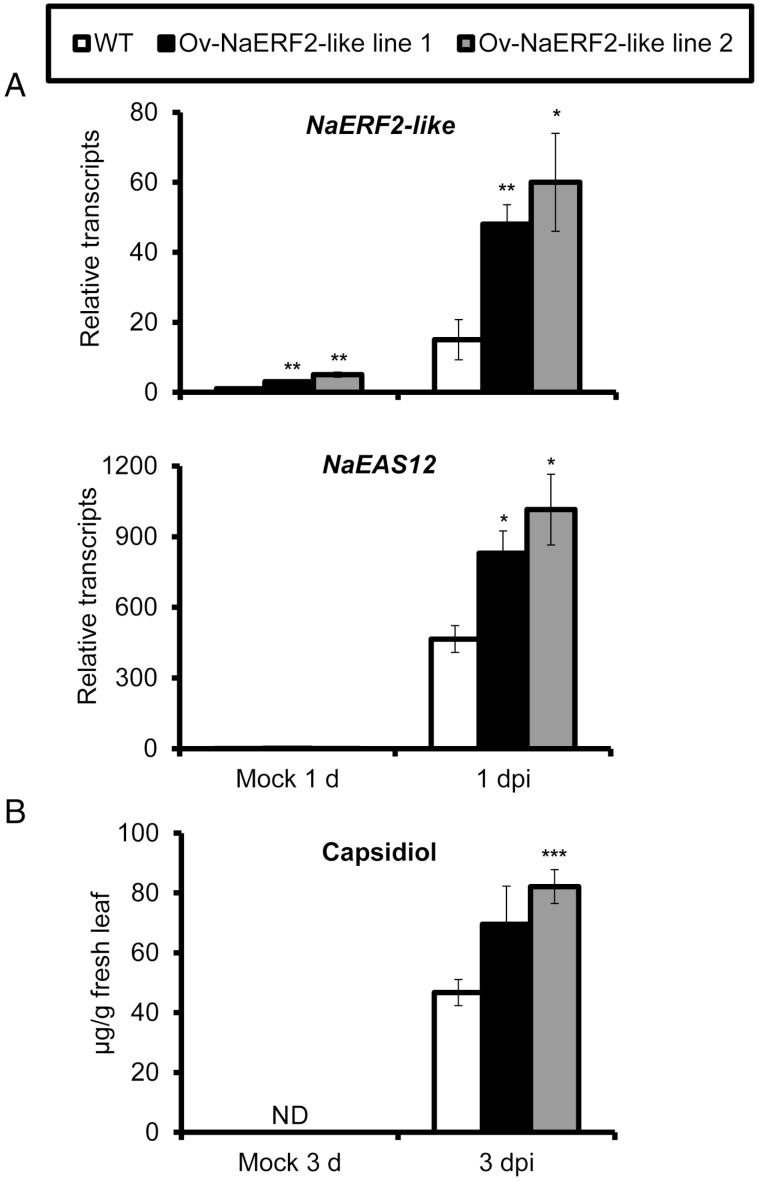
To further understand the role of NaERF2-like in pathogen defense, we investigated NaEAS12 gene expression, capsidiol level, and plant resistance in WT and two NaERF2-like overexpression (Ov-NaERF2-like) lines. The ectopic overexpression of NaERF2-like fused with two HA tags substantially increased the basal and induced transcriptional levels of NaERF2-like (Fig. 10A) without affecting the plants’ morphology and size. As expected, Ov-NaERF2-like line 1 and Ov-NaERF2-like line 2 showed 178% and 219% of the NaEAS12 expression of WT when 0 leaves were inoculated at 1 d (Fig. 10A). Consistently, capsidiol levels attained values 149% and 175% of that of WT in Ov-NaERF2-like lines 1 and 2 at 3 dpi, respectively (Fig. 10B). However, we found no difference in lesion diameters in both overexpression lines compared with WT (Supplementary Fig. S2), indicating that overexpression of NaERF2-like increased the gene expression of NaEAS12, and subsequently capsidiol biosynthesis, but had only a minor effect on the plants’ resistance.
Fig. 10.
Overexpression of NaERF2-like enhances NaEAS12 expression and capsidiol production in stable transformation plants. (A) Mean (±SE) relative A. alternata-induced NaERF2-like and NaEAS12 transcripts as measured by real-time PCR in five replicate 0 leaves of WT, Ov-NaERF2-like line 1, and Ov-NaERF2-like line 2 plants at 1 dpi. (B) Mean (±SE) capsidiol levels were determined by HPLC in five replicate 0 leaves of WT, Ov-NaERF2-like line 1, and Ov-NaERF2-like line 2 plants at 3 dpi. Asterisks indicate level of significant difference between WT and Ov-NaERF2-like lines after inoculation (Student’s t-test: *P<0.05, **P<0.01, ***P<0.005). ND, not detectable.
Discussion
Phytoalexins are important ‘chemical weapons’ employed by plants in defending against pathogens. In addition to scopoletin and scopolin, two phytoalexins regulated by JA and ethylene signaling pathways in response to A. alternata infection (Sun et al., 2014b; Li and Wu, 2016), we demonstrate in this study that capsidiol is another important phytoalexin produced by N. attenuata, and its biosynthesis is not dependent on JA and ethylene signaling pathways, but is transcriptionally regulated by a transcription factor, NaERF2-like.
Capsidiol is an important phytoalexin produced in N. attenuata in response to A. alternata infection
Capsidiol was initially isolated from pepper fruit after treatments with various pathogens, including Phytophthora capsici, Botrytis cinerea, and Fusarium oxysporum (Stoessl et al., 1972). Later, this compound was also identified from infected Nicotiana species (Bailey et al., 1975; Guedes et al., 1982; Mialoundama et al., 2009; Shibata et al., 2010; Grosskinsky et al., 2011; Shibata et al., 2016). In our N. attenuata–A. alternata pathosystem, we found that a large number of genes were strongly regulated in response to A. alternata inoculation at 1 dpi during transcriptome analysis. Many of these genes were involved in the biosynthesis of sesquiterpenes, capsidiol, and solavetivone (Fig. 1; Supplementary Table S2). Indeed, the levels of capsidiol were dramatically induced to 50.68±3.10 µg g−1 fresh leaf at 3 dpi in young 0 leaves (Fig. 2). These finding suggest that capsidiol is involved in the resistance of N. attenuata to A. alternata infection.
Ideally, the benefits of a putative resistant trait should be determined in plants differing only in a single gene that controls the defense trait and are otherwise identical (Bergelson et al., 1996). In this study, virus-induced gene silencing of NaEAHs or NaEASs was used to manipulate the production of capsidiol, and the results revealed that capsidiol-reduced or -depleted plants were more susceptible to A. alternata (Fig. 3). In response to A. alternata inoculation, capsidiol level was increased to 50.68±3.10 µg g−1 in young 0 leaves at 3 dpi (Fig. 2). Considering that this capsidiol level was averaged from leaf laminae of 1.5×1.5 cm2 after inoculation, we speculated that the concentration of capsidiol in planta at the interface of fungal infection would be higher than 50 µg g−1 fresh leaf. Therefore, we tested the inhibition rate of fungal growth in vitro by supplement of capsidiol at concentrations of 50, 100, and 200 µg ml−1. The fungal growth was reduced by 43.4% in PDA plates after 6 d with 50 µg ml−1 of capsidiol (Fig. 4), indicating that capsidiol had strong anti-fungal activities against A. alternata when it was applied in vitro at a concentration similar to those in planta.
Thus, our results demonstrate that capsidiol is an important phytoalexin involved in the defense mechanism of N. attenuata against A. alternata, and the high level of capsidiol accumulated in the young 0 leaves accounts for their high resistance to A. alternata infection. Whether solavetivone plays a role in resistance is unknown. Transcriptome data, especially the strong up-regulation of premnaspirodiene oxygenases and premnaspirodiene synthases at 1 dpi (Fig. 1 and Supplementary Table S2), are consistent with a defensive role of solavetivone. However, this question needs further investigation.
Regulation of capsidiol biosynthesis
Several reports indicate that the phytoalexin production is influenced by endogenous plant hormones. Increasing cytokinin (CK) levels in N. tabacum plants by exogenous CK application or overexpression of the bacterial isopentenyl transferase gene enhanced their resistance to the hemibiotrophic bacterium Psudomonas syringae by increasing levels of capsidiol and scopoletin (Grosskinsky et al., 2011). This CK-mediated resistance is independent of the salicylic acid, JA, and ethylene signaling pathways (Grosskinsky et al., 2011). Abscisic acid (ABA) negatively regulates elicitor-induced biosynthesis of capsidiol in N. plumbaginifolia; a 2-fold increase in capsidiol synthesis was observed in ABA-deficient mutants compared with WT plants when exposed to cellulose or B. cinerea (Mialoundama et al., 2009). In addition, when N. benthamiana was inoculated with Phytophthora infestans, pathogen-induced capsidiol and NbEAS and NbEAH expression were abolished in plants with ethylene insensitive 2 (NbEIN2) silenced, suggesting that the ethylene signaling pathway is essential for capsidiol production (Shibata et al., 2010; Ohtsu et al., 2014).
In contrast to the observation in N. benthamiana inoculated with P. infestans, our experiments did not support the role of ethylene signaling in capsidiol elicitation in the N. attenuata–A. alternata pathosystem. Compared with WT plants, both capsidiol production and transcripts of NaEAHs and NaEASs after A. alternata challenge were induced to the same high levels in irACO (ethylene-deficient) and Ov-etr1 (ethylene-insensitive) plants (Fig. 5), two transgenic plants generated previously by von Dahl et al (2007). To further support this idea, we also silenced NaEIN2 by VIGS. Alternaria alternata-elicited NaF6′H1 was dramatically reduced in VIGS NaEIN2 plants, which was consistent with our previous finding that ethylene signaling was essential for scopoletin biosynthesis (Sun et al., 2017), but NaEAHs and NaEASs transcripts were induced to levels similar to those of plants transformed with empty vector (Supplementary Fig. S3). This result also indicated that blocking the ethylene signaling pathway had little effect on the expression of these two key capsidiol biosynthesis genes. Thus, we concluded that the ethylene signaling pathway did not play a critical role in capsidiol production and gene expression of NaEAHs and NaEASs in N. attenuata after A. alternata challenge. Whether or not ethylene is involved in capsidiol biosynthesis is likely dependent on the pathosystem. Additionally, our experiments did not indicate a role for JA signaling in the regulation of capsidiol biosynthesis, as transcripts of NaEAHs and NaEASs were equivalent in WT and JA-deficient irAOC plants at 1 and 3 dpi (Fig. 5).
Transcription factors integrate developmental and environmental signals by interacting with the promoter regions of enzyme genes, thus enhancing or suppressing their expression to regulate the biosynthesis of secondary metabolites (Yang et al., 2012). For example, the sesquiterpene phytoalexin gossypol in cotton is regulated by GaWRKY1 (Xu et al., 2004), and EREB58 mediates the JA-induced production of sesquiterpene volatiles in maize (Li et al., 2015); artemisinin, a sesquiterpene lactone found exclusively in Artemisia annua, is controlled by bZIP1 and glandular trichome-specific WRKY1 (Zhang et al., 2015; Chen et al., 2017).
Despite the great role of capsidiol in resistance, the transcriptional regulation of its biosynthesis is still not clear. Due to the high level of gene expression seen in the capsidiol biosynthetic pathway, we hypothesized that the transcription factors regulating these genes must also be highly expressed during the initial infection period of the fungus. We performed a screen of the six most up-regulated transcription factors. Indeed, NaERF2-like was identified as being a positive regulator of plant resistance and capsidiol production. When compared with EV plants, those with a silenced NaERF2-like gene accumulated fewer transcripts of NaEASs and NaEAHs, as well as lower levels of capsidiol (Fig. 7). Consistently, NaERF2-like-silenced plants were susceptible to the fungus (Fig. 7).
Both NaEAS and NaEAH are members of a multi-gene family. Since NaEAS12 expression is greatly reduced in NaERF2-like-silenced plants (Fig. 8), we selected this gene to test whether its promoter could be directly activated by NaERF2-like. Several lines of evidence support that NaERF2-like directly regulates the capsidiol biosynthetic gene NaEAS12, including (i) the binding of NaERF2-like protein to the promoter region of NaEAS12, which was supported by yeast one-hybrid, EMSA and Chip-qPCR experiments (Fig. 9), and (ii) the NaEAS12 promoter was activated in response to transient NaERF2-like overexpression (Fig. 8); this result is further confirmed by stable transgenic lines of Ov-NaERF2-like, which exhibit increased NaEAS12 transcripts and higher levels of capsidiol accumulation (Fig. 10). Currently, it is not known how other NaEASs and NaEAHs are regulated by NaERF2-like. Further experiments are needed to test whether or not they are regulated in a way similar to NaEAS12.
The ectopic overexpression of NaERF2-like increased the gene expression of NaEAS12 and capsidiol production after fungal inoculation, but did not result a higher resistance in Ov-NaERF2-like line 1 and Ov-NaERF2-like line 2 (Fig. 10). This was likely due to the reduced inhibition efficiency of fungal growth when capsidiol levels exceeded 50 µg g−1 fresh leaf. The fungal growth was reduced by 43.4% in PDA plates after 6 d with 50 µg ml−1 of capsidiol (Fig. 4). When 2-fold capsidiol (100 µg ml−1) was added into the PDA plates, the inhibition effect was only increased from 43.4 to 56.2% (Fig. 4). These results indicated that the inhibition rate of fungal growth by capsidiol was not linear with concentration, but rather decreased when capsidiol concentration exceeded 50 µg ml−1, and this could be the reason why NaERF2-like overexpression plants had higher levels of capsidiol but not higher resistance.
Accession numbers
Sequence data from this article can be found in the GenBank data library under accession numbers: XM_019375732.1 (HMG-CoA synthase), XM_019375278.1 (MVAPP decarboxylase), XM_019403732.1 (FPP synthase), XM_019409657.1 (squalene synthase), XM_019408556.1 (EAS12), and XM_019399671.1 (NaERF2-like).
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Alternaria alternata-elicited NaEAS12 transcripts in EV and plants individually silenced with the top five up-regulated transcription factors.
Fig. S2. Overexpression of NaERF2-like does not affect plant resistance.
Fig. S3. Silencing NaEIN2 has a great impact on A. alternata-induced transcripts of NaF6′H1 but does not affect transcripts of NaEAHs and NaEASs.
Table S1. Primers used in this study.
Table S2. Transcriptome analysis revealed regulation of genes involved in sesquiterpene biosynthesis in three biological replicate N. attenuata leaves after A. alternata inoculation at 1 d.
Table S3. Transcriptome analysis revealed top six highly elicited transcriptional factor genes in three biological replicate A. alternata-inoculated N. attenuata leaves at 1 d.
Acknowledgements
We thank Prof. Joe Chappell (University of Kentucky, USA) for the capsidiol standard, the Service Center for Experimental Biotechnology of Kunming Institute of Botany, the Chinese Academy of Sciences (CAS), for plant growth support. This project was supported by the National Science Foundation of China (NSFC Grant No. 31670262) and the Key Project of Applied Basic Research Program of Yunnan (2014FA040) to JW, NSFC grant (No. 31700231), the Applied Basic Research Program of Yunnan (2017FB048) and CAS “Light of West China” Program to LM.
References
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Burden RS, Vincent GG. 1975. Capsidiol: antifungal compound produced in Nicotiana tabacum and Nicotiana clevelandii following infection with tobacco necrosis virus. Phytochemistry 14, 597–597. [Google Scholar]
- Bergelson J, Purrington CB, Palm CJ, Lopez-Gutierrez JC. 1996. Costs of resistance: a test using transgenic Arabidopsis thaliana. Proceedings of the Royal Society B: Biological Sciences 263, 1659–1663. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. 2002. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Chen M, Yan T, Shen Q, et al. . 2017. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytologist 214, 304–316. [DOI] [PubMed] [Google Scholar]
- El Oirdi M, Trapani A, Bouarab K. 2010. The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environmental Microbiology 12, 239–253. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J. 1992. Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proceedings of the National Academy of Sciences, USA 89, 11088–11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Grosskinsky DK, Naseem M, Abdelmohsen UR, et al. . 2011. Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiology 157, 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes MEM, Kuc J, Hammerschmidt R, Bostock R. 1982. Accumulation of 6 sesquiterpenoid phytoalexins in tobacco leaves infiltrated with Pseudomonas lachrymans. Phytochemistry 21, 2987–2988. [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. 1998. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. The Journal of Biological Chemistry 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hao D, Yamasaki K, Sarai A, Ohme-Takagi M. 2002. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41, 4202–4208. [DOI] [PubMed] [Google Scholar]
- Huang PY, Catinot J, Zimmerli L. 2016. Ethylene response factors in Arabidopsis immunity. Journal of Experimental Botany 67, 1231–1241. [DOI] [PubMed] [Google Scholar]
- Kallenbach M, Bonaventure G, Gilardoni PA, Wissgott A, Baldwin IT. 2012. Empoasca leafhoppers attack wild tobacco plants in a jasmonate-dependent manner and identify jasmonate mutants in natural populations. Proceedings of the National Academy of Sciences, USA 109, E1548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Rowe HC, Denby KJ. 2005. Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. The Plant Journal 44, 25–36. [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. 2002. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12, 177–183 [Google Scholar]
- LaMondia JA. 2001. Outbreak of brown spot of tobacco caused by Alternaria alternata in connecticut and massachusetts. Plant Disease 85, 230. [DOI] [PubMed] [Google Scholar]
- Li J, Wu J. 2016. Scopolin, a glycoside form of the phytoalexin scopoletin, is likely involved in the resistance of Nicotiana attenuata against Alternaria alternata. Journal of Plant Pathology 98, 641–644. [Google Scholar]
- Li S, Wang H, Li F, Chen Z, Li X, Zhu L, Wang G, Yu J, Huang D, Lang Z. 2015. The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. The Plant Journal 84, 296–308. [DOI] [PubMed] [Google Scholar]
- Mengiste T. 2012. Plant immunity to necrotrophs. Annual Review of Phytopathology 50, 267–294. [DOI] [PubMed] [Google Scholar]
- Mialoundama AS, Heintz D, Debayle D, Rahier A, Camara B, Bouvier F. 2009. Abscisic acid negatively regulates elicitor-induced synthesis of capsidiol in wild tobacco. Plant Physiology 150, 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. 2007. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. The Plant Cell 19, 2039–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu M, Shibata Y, Ojika M, Tamura K, Hara-Nishimura I, Mori H, Kawakita K, Takemoto D. 2014. Nucleoporin 75 is involved in the ethylene-mediated production of phytoalexin for the resistance of Nicotiana benthamiana to Phytophthora infestans. Molecular Plant-Microbe Interactions 27, 1318–1330. [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J. 2001. Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Archives of Biochemistry and Biophysics 393, 222–235. [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. 2001. Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. The Plant Journal 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. 2004. Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. Journal of Experimental Botany 55, 151–157. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A, Ramos B, Bednarek P, Lopez G, Pislewska-Bednarek M, Schulze-Lefert P, Molina A. 2010. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. The Plant Journal 63, 115–127. [DOI] [PubMed] [Google Scholar]
- Schuck S, Weinhold A, Luu VT, Baldwin IT. 2014. Isolating fungal pathogens from a dynamic disease outbreak in a native plant population to establish plant-pathogen bioassays for the ecological model plant Nicotiana attenuata. PLoS One 9, e102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Kawakita K, Takemoto D. 2010. Age-related resistance of Nicotiana benthamiana against hemibiotrophic pathogen Phytophthora infestans requires both ethylene- and salicylic acid-mediated signaling pathways. Molecular Plant-Microbe Interactions 23, 1130–1142. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Ojika M, Sugiyama A, Yazaki K, Jones DA, Kawakita K, Takemoto D. 2016. The full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and postinvasion defense against Phytophthora infestans in Nicotiana benthamiana. The Plant Cell 28, 1163–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. 1998. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Development 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoessl A, Unwin CH, Ward EWB. 1972. Postinfectional inhibitors from plants I. Capsidiol, an antifungal compound from Capsicum frutescens. Phytopathology 74, 141–152. [Google Scholar]
- Sun H, Hu X, Ma J, Hettenhausen C, Wang L, Sun G, Wu J, Wu J. 2014a Requirement of ABA signalling-mediated stomatal closure for resistance of wild tobacco to Alternaria alternata. Plant Pathology 63, 1070–1077. [Google Scholar]
- Sun H, Song N, Ma L, Li J, Ma L, Wu J, Wu J. 2017. Ethylene signalling is essential for the resistance of Nicotiana attenuata against Alternaria alternata and phytoalexin scopoletin biosynthesis. Plant Pathology 66, 277–284. [Google Scholar]
- Sun H, Wang L, Zhang B, Ma J, Hettenhausen C, Cao G, Sun G, Wu J, Wu J. 2014b Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. Journal of Experimental Botany 65, 4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dahl CC, Winz RA, Halitschke R, Kuhnemann F, Gase K, Baldwin IT. 2007. Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. The Plant Journal 51, 293–307. [DOI] [PubMed] [Google Scholar]
- Ward EWB, Unwin CH, Stoessl A. 1974. Postinfectional inhibitors from plants 13. Fungitoxicity of phytoalexin, capsidiol, and related sesquiterpenes. Canadian Journal of Botany – Revue Canadienne De Botanique 52, 2481–2488. [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. 2007. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiology 145, 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang L, Baldwin IT. 2008. Methyl jasmonate-elicited herbivore resistance: does MeJA function as a signal without being hydrolyzed to JA? Planta 227, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang L, Wünsche H, Baldwin IT. 2013. Narboh D, a respiratory burst oxidase homolog in Nicotiana attenuata, is required for late defense responses after herbivore attack. Journal of Integrative Plant Biology 55, 187–198. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang J, Wang S, Wang J, Chen X. 2004. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiology 135, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Song N, Ma L, Fang D, Wu J. 2018. NaPDR1 and NaPDR1-like are essential for the resistance of Nicotiana attenuata against fungal pathogen Alternaria alternata. Plant Diversity 40, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Fang X, Wu X, Mao Y, Wang L, Chen X. 2012. Transcriptional regulation of plant secondary metabolism. Journal of Integrative Plant Biology 54, 703–712. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang F, Fu X, Lv Z, et al. . 2015. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Molecular Plant 8, 163–175. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wei T, Yin KQ, Chen Z, Gu H, Qu LJ, Qin G. 2012. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytologist 195, 450–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.