Abstract
Potassium (K+) is a critical determinant of salinity tolerance, and H2O2 has been recognized as an important signaling molecule that mediates many physiological responses. However, the details of how H2O2 signaling regulates K+ uptake in the root under salt stress remain elusive. In this study, salt-sensitive cucumber and salt-tolerant pumpkin which belong to the same family, Cucurbitaceae, were used to answer the above question. We show that higher salt tolerance in pumpkin was related to its superior ability for K+ uptake and higher H2O2 accumulation in the root apex. Transcriptome analysis showed that salinity induced 5816 (3005 up- and 2811 down-) and 4679 (3965 up- and 714 down-) differentially expressed genes (DEGs) in cucumber and pumpkin, respectively. DEGs encoding NADPH oxidase (respiratory burst oxidase homolog D; RBOHD), 14-3-3 protein (GRF12), plasma membrane H+-ATPase (AHA1), and potassium transporter (HAK5) showed higher expression in pumpkin than in cucumber under salinity stress. Treatment with the NADPH oxidase inhibitor diphenylene iodonium resulted in lower RBOHD, GRF12, AHA1, and HAK5 expression, reduced plasma membrane H+-ATPase activity, and lower K+ uptake, leading to a loss of the salinity tolerance trait in pumpkin. The opposite results were obtained when the plants were pre-treated with exogenous H2O2. Knocking out of RBOHD in pumpkin by CRISPR/Cas9 [clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9] editing of coding sequences resulted in lower root apex H2O2 and K+ content and GRF12, AHA1, and HAK5 expression, ultimately resulting in a salt-sensitive phenotype. However, ectopic expression of pumpkin RBOHD in Arabidopsis led to the opposite effect. Taken together, this study shows that RBOHD-dependent H2O2 signaling in the root apex is important for pumpkin salt tolerance and suggests a novel mechanism that confers this trait, namely RBOHD-mediated transcriptional and post-translational activation of plasma membrane H+-ATPase operating upstream of HAK5 K+ uptake transporters.
Keywords: HAK5, potassium uptake, pumpkin, RBOH, ROS, salinity, transcriptome
RBOHD-mediated transcriptional and post-translational activation of plasma membrane H+-ATPase leads to improved K+ uptake and cell viability in the root apex, conferring salinity tolerance in pumpkin
Introduction
Soil salinity is one of the major environmental stresses affecting crop production worldwide. It is estimated that ~950 Mha of arable land in the world, including 250 Mha of irrigated land, are affected by salinity (Ruan et al., 2010). Although improving salt tolerance in major cultivated crops is of a great importance for global food security in the 21st century, the process is significantly handicapped due to the complexity of salinity tolerance (Ismail and Horie, 2017).
The main targeted trait of salinity tolerance is sodium (Na+) extrusion from the cytosol, either to external media or via intracellular Na+ sequestration in the vacuoles (Ismail and Horie, 2017; Yang and Guo, 2018). The importance of maintaining optimal potassium (K+)/Na+ ratios is also widely recognized (Lauchli and Wieneke, 1979; Koyro and Stelzer, 1988; Gassman et al., 1996; Zhu et al., 1998; Lauchli, 1999; Hasegawa et al., 2000). K+ is an important macronutrient that plays pivotal roles in many processes in living cells, including maintaining membrane potential, enzyme activation, and protein synthesis (Wang and Wu, 2017), as well as having an important signaling role (Anschütz et al., 2014; Shabala, 2019). Root K+ retention ability is associated with salt tolerance in a broad range of plant species (Bose et al., 2014; Chakraborty et al., 2016; Wu et al., 2018). K+ uptake is controlled by various types of channels and transporters (Véry et al., 2014); amongst these, AKT1 and HAK5 are primary players in K+ uptake into roots under saline conditions (Nieves-Cordones et al., 2010), while GORK channels are considered as a major pathway for K+ efflux under saline conditions (Wu et al., 2018).
The plasma membrane (PM) H+-ATPase is an important component of cell ionic homeostasis. By extruding protons from the cell and generating a membrane potential, this pump energizes the PM, which is essential for nutrient uptake and, hence, regulation of many physiological processes (Falhof et al., 2016). It was reported that cell type-specific H+-ATPase activity in root tissues enables K+ accumulation and mediates salinity tolerance of barley (Shabala et al., 2016).
Reactive oxygen species (ROS) have been characterized as important signaling molecules that mediate many developmental and physiological responses (He et al., 2018; Waszczak et al., 2018). Numerous pieces of evidence have suggested that ROS play an important role in acclimation-induced salt tolerance (Jiang et al., 2013; Peng et al., 2014; Hossain et al., 2017; Niu et al., 2018; Saini et al., 2018). Due to its relative stability and ability to diffuse through membranes, facilitated by aquaporins, H2O2 is often considered as a predominant ROS signaling molecule (Niu and Liao, 2016).
Previous studies found that H2O2 signaling can regulate K+ uptake by HAK5 under low K+ conditions (Shin and Schachtman, 2004; Jung et al., 2009), and a positive relationship between H2O2 production and root K+ accumulation under salt stress was also reported for Arabidopsis (Ma et al., 2012) and cucumber (Redwan et al., 2016). However, the details of how H2O2 signaling regulates K+ uptake in the root under salt stress remain elusive. One of the currently favored scenarios is that H2O2 interacts with transition metals (such as iron or copper) in the cell wall, forming highly reactive hydroxyl radicals (Demidchik et al., 2014). The latter then activate K+-efflux GORK channels, resulting in a K+ loss from the cytosol (Demidchik et al., 2014). However, this scenario cannot explain the beneficial effects of ROS production on root K+ uptake under salt stress mentioned above. This implies that some other mechanisms may operate in plant cells.
Plant roots are the first organs sensing and responding to salinity stress. These responses are manifested differentially at the tissue-specific level, as the longitudinal structure of plant roots contains specialized zones of development (Sun et al., 2009; Hill et al., 2016; Liu et al., 2019). It was shown that the sensitivity of the root apex to salt is higher than that of the mature zone (Hill et al., 2016; Shabala et al., 2016). This implies that analyses of whole-root tissue may be counterproductive as it can dilute information important to understand the complex physiological and molecular programs that define root response to salt stress. However, very few studies have accounted for this, with the bulk of reported results obtained at the whole-root level.
A genomics approach can be instrumental in identification of important genes involved in plant adaptation to salt stress. This can be done by comparing the transcriptome of tolerant and sensitive species in response to salinity (Gong et al., 2005). Transcriptomics approaches have been applied to identify salinity-induced differentially expressed genes (DEGs) in roots for a range of species (Senadheera et al., 2009; Cotsaftis et al., 2011; Zahaf et al., 2012; Geng et al., 2013; Xie et al., 2018). However, most of these results were conducted at the whole-root level, with only a few studies (Dinneny et al., 2008; Gruber et al., 2009; Hill et al., 2016) investigating the response of the transcriptome to salinity in different root zones. As a result, very few (if any) studies have attempted to link a comprehensive transcriptomic analysis of tissue-specific gene expression with the functional studies of their operation under saline conditions. Meanwhile, such integration is critically important for understanding of the mechanisms underlying differential salt tolerance, to facilitate the progress in breeding for plant salt tolerance.
Cucumber and pumpkin belong to the same family, the Cucurbitaceae, but differ drastically in salt tolerance, with cucumber being sensitive and pumpkin tolerant (Zhu et al., 2006). As a result, pumpkin is often used as a salt-tolerant rootstock for cucumber and other cucurbit crops (Edelstein et al., 2011; Huang et al., 2013; Lei et al., 2014). Previous studies have suggested that pumpkin roots exhibited a high efficiency in extruding Na+ (Edelstein et al., 2011; Lei et al., 2014; Niu et al., 2018). However, the role of K+ uptake as a component in the salt tolerance of pumpkin has not been investigated so far.
The present study attempts to investigate the physiological and molecular mechanisms underlying the differential salt tolerance between cucumber and pumpkin, with a specific focus on H2O2 signaling and K+ uptake, at the root tissue-specific level. The physiological responses were linked with changes in the transcriptional profile of several key DEGs. Our analyses yielded the new physiological and molecular mechanisms that are likely to contribute to the differential salt tolerance within the Cucurbitaceae family.
Materials and methods
Plant materials and growth conditions
A salt-sensitive cucumber (Cucumis sativus L.) cv. Jinchun No. 2 and a salt-tolerant pumpkin (Cucurbita moschata) cv. Chaojiquanwang were used. Seeds were surface sterilized with 1% HClO for 30 min and rinsed thoroughly with distilled water, and then incubated in the dark at 28 °C until germination. The germinated seeds were then grown hydroponically in an aerated basic salt medium (BSM) solution (0.5 mM KCl, 0.1 mM CaCl2, pH 5.7 non-buffered) in the dark at room temperature (24 °C). Five days later, 75 mM NaCl was added to BSM solution for either 24 h or 48 h, to assess effects of acute salinity stress on cell viability, root ion fluxes, K+ content, H2O2 content, PM H+-ATPase activity, transcriptome, and quantitative real-time PCR analysis. The main reason for using dark-grown seedlings was to maximize the species difference in physiological and molecular responses from the roots (Cuin et al., 2011).
Cell viability staining
Plants were treated with 75 mM NaCl for 48 h, then the cell viability of roots (apex and mature zone) was assessed using the fluorescein diacetate (FDA)–propidium iodide (PI) double staining method as described by Chakraborty et al. (2016). To determine the extent of cell death, the red and green channels of the fluorescent images were separated and the fluorescent intensity of the respective images was quantified using ImageJ software (version 1.48, US National Institutes of Health, Bethesda, MD, USA).
Determination of K+ content
Plants were grown and salt treated as described in the cell viability staining section. Roots were washed with distilled water. The apical (0–5 mm from the root tip) and mature (5–25 mm from the root tip) segments of root were collected and weighed. Root sap was mechanically extracted as described elsewhere (Cuin et al., 2009), and its K+ content was quantified using an atomic absorption spectrophotometer (Varian spectra AA 220, Varian, Palo Alto, CA, USA).
Determination of H2O2 levels
Confocal laser scanning microscopy (Leica TCS-SP2, Leica Microsystems GmbH, Wetzlar, Germany) was used to visualize H2O2 accumulation in roots in vivo, according to the method described by Niu et al. (2018). For spectrophotometric measurements, H2O2 was extracted from 0.5 g fresh root (apex, 0–5 mm from the tip) samples ground in 3 ml of 1 M HClO4. After centrifugation at 6000 g for 5 min, the supernatant was adjusted to pH 6.0–7.0 using KOH and filtered through a Sep-Pak C18 cartridge (Millipore, Milford, MA, USA). After elution with 4 ml of distilled water, an aliquot of the sample (800 μl) was mixed with 400 μl of reaction buffer containing 4 mM 2,2′-azino-di (3-ethylbenzthiazoline-6-sulfonic acid), 100 mM potassium acetate at pH 4.4, and 400 μl of deionized water. The reaction was started by the addition of 3 μl (0.5 U) of horseradish peroxidase, then the H2O2 content was measured at an optical density of 412 nm (Willekens et al., 1997). Relative H2O2 content (NaCl/control) in the root apex of pumpkin was calculated.
K+ flux measurements
Net K+ flux in the root apex (1 mm from the tip) was measured under the BSM background in the presence of NaCl, using the non-invasive microelectrode ion flux estimation (MIFE) technique (University of Tasmania, Hobart, Australia) as described previously (Bose et al., 2014; Chakraborty et al., 2016). The tips of the electrodes were front filled with K+-selective cocktails (Potassium ionophore I-Cocktail 99311, Sigma-Aldrich).
Pharmacology
In pharmacological experiments, plant roots were pre-treated for 1 h prior to application of 75 mM NaCl stress with either 20 µM diphenylene iodonium (DPI, an NADPH oxidase inhibitor) or 1 mM H2O2. Then net K+ flux, PM H+-ATPase activity, RBOHD, GRF (encoding a 14-3-3 protein), AHA (PM H+-ATPase genes), and HAK5 expression in the root apex (0–5 mm from the tip) was measured at 24 h after 75 mM NaCl stress. The H+-ATPase activity was determined by measuring the release of inorganic phosphate (Pi) (Kłobus and Janicka-Russak, 2004) and expressed as the difference between the activities measured in the absence and presence of Na3VO4.
Transcriptome analysis
Plants were subjected to 75 mM NaCl stress for 24 h. The root apex (0–5 mm from the tip) was then harvested. Total RNA was isolated using a TRIzol kit (Invitrogen, Carlsbad, CA, USA) with three biological replicates for each treatment. RNA concentration was measured in a Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit in the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Library construction and RNA sequencing (RNA-seq) were conducted by the Novogene Bioinformatics Institute (Beijing, China) on a HiSeq 4000 platform (Illumina, San Diego, CA, USA).
Clean data reads were obtained by removing low quality reads from the raw data and then mapped to the cucumber (Chinese Long) and Cucurbita moschata (Rifu) genome assembly using TopHat v2.0.12 (Trapnell et al., 2009). Fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) were calculated using HTSeq v0.6.1 to estimate gene expression levels. Differential expression analysis of RNA-seq between NaCl and control for cucumber and pumpkin was performed using the edgeR package (Robinson et al., 2010). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes with FDR <0.01 and |log2foldchange (log2FC)|>2 were assigned as DEGs. The identified DEGs were then subjected to GO (Gene Ontology) enrichment analyses by using the GO enrichment Tool from the Cucurbit Genomics Database website (http://cucurbitgenomics.org). GO terms with corrected P-value <0.05 were considered significantly enriched. RNA-seq data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA437579.
Quantitative real-time PCR (qRT-PCR)
Twenty-three genes were selected to carry out qRT-PCR for determination of the DEG results of RNA-seq. In addition, relative expression of RBOHD, GRF, AHA, and HAK5 was also determined after 24 h of exposure to 75 mM NaCl, with or without pre-treatment with DPI and H2O2. We amplified the PCR products in triplicate using 1× Top Green qPCR SuperMix (TransGen Biotech, Inc., Beijing, China) in 10 μl of qRT-PCR assays. The PCR was performed using the ABI 7000 machine (Applied Biosystems), and the cycling conditions consisted of denaturation at 94 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C for 15 s, and extension at 72 °C for 15 s. The specific primers (Supplementary Table S1 at JXB online) were designed based on the published mRNA of cucumber (Chinese Long) and Cucurbita moschata (Rifu) on the Cucurbit Genomics Database (http://cucurbitgenomics.org/) using Primer 5 software. The relative gene expression was determined as previously described by Livak and Schmittgen (2001).
Functional analysis of pumpkin RBOHD by CRISPR/Cas9 genome editing and ectopic expression in Arabidopsis
Generation of RBOHD mutation mediated by CRISPR/Cas9 [clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9] in the hairy root transformation system of pumpkin was conducted following the methods described by Hu et al. (2017) and Kirchner et al. (2017). The genome DNA sequence of RBOHD was downloaded from the Cucurbit Genomics Database (http://cucurbitgenomics.org/), and the small guide RNA (sgRNA) was designed by Geneious (Bioinformatics Software for Sequence Data Analysis). The construct (pKSE402) containing the Cas9 nuclease and the sgRNA were delivered by the Agrobacterium rhizogenes (Ar Qual)-mediated hairy root transformation technique, and a successful transformation was monitored by green fluorescent protein (GFP) fluorescence. The non-transgenic roots were cut off and plants with transgenic roots were cultivated further. For the verification of the gene editing, the gene region including the sgRNA region was amplified by PCR (RBOHD Forward,TTCGGACGCAGATGGAAGAA; RBOHD Reverse, TTAGAACTCGGCTGTCGCTC), and was sequenced and compared with the control (empty vector). The regenerated pumpkin plants at the fourth true leaf stage were treated with 75 mM NaCl as described by Niu et al. (2018). Relative plant dry weight (NaCl/control) and K+ content in the root apex (0–5 mm from the tip) were measured at day 12 after salt treatment. H2O2 content and relative gene expression (NaCl/control) in the root apex were measured at 24 h after salt treatment. The methods for K+ and H2O2 determination and gene (RBOHD, GRF12, AHA1/9/11, and HAK5) expression analysis by qRT-PCR were described above. The specific primers are listed in Supplementary Table S1.
For the ectopic expression of pumpkin RBOHD in wild-type (WT) Arabidopsis (Columbia-0), RBOHD was cloned, and transgenic Arabidopsis plants were generated following the method described by Cao et al. (2017). Arabidopsis T3 homozygous lines were used for the salt experiment. All the Arabidopsis plants were grown at 22–24 °C under a 16 h light (100 µmol m−2 s−1) and 8 h dark photoperiod. Seeds were imbibed at 4 °C for 2 d and geminated on Murashige and Skoog (MS) Petri dishes containing 0.5% phytagel and 3.0% sucrose in a light growth chamber. For salt tolerance assays, 10-day-old seedlings grown on MS medium were transferred to medium containing 75 mM NaCl. At 24 h after salt treatment, gene expression (NaCl/control) was analyzed in the root apex (0–5 mm from the tip) of RBOHD ectopic expression (OE-RBOHD) and WT plants. Plant fresh weight and K+ content of the whole plant were measured at day 7 after salt treatment. The methods for K+ and gene (RBOHD, GRF12, AHA1/5/9/11, and HAK5) expression analysis by qRT-PCR were described above. The specific primers are listed in Supplementary Table S1.
Statistical analysis
Data were presented as the means of 3–6 biological replicates. Duncan’s multiple range tests were used to evaluate significant differences between treatments at P<0.05. Student’s two-sample t-test was also used for comparing the means of two independent samples. Statistical analyses were performed using SAS version 8.0 (SAS Institute Inc., Cary, NC, USA).
Results
The difference in salt tolerance between pumpkin and cucumber is conferred by the root apex
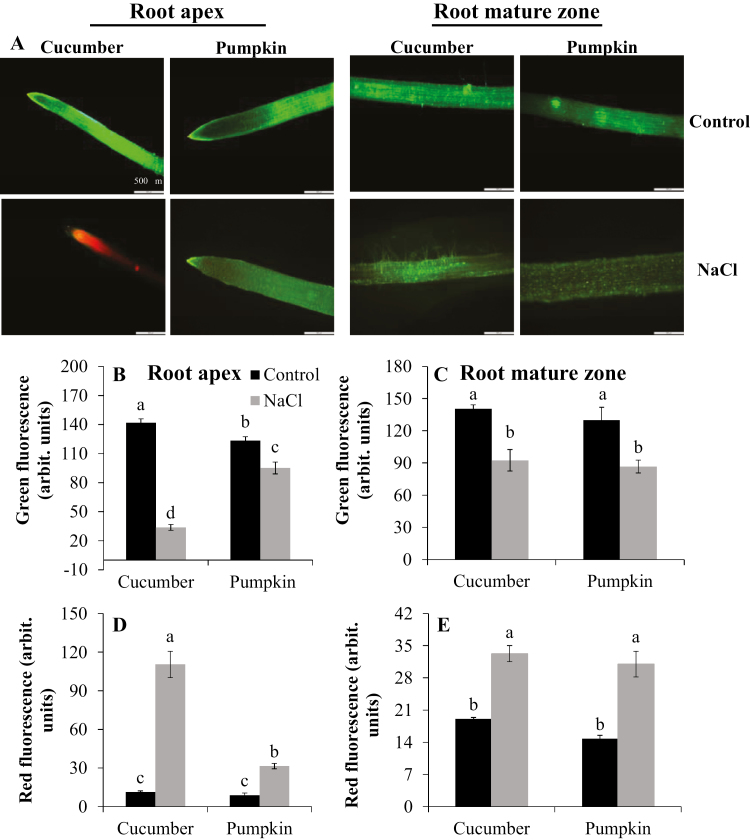
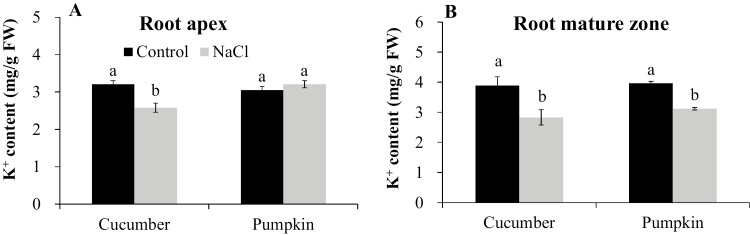
Pumpkin species have higher salt tolerance than cucumber (Supplementary Fig. S1; see also Zhu et al., 2006; Huang et al., 2013; Lei et al., 2014). This difference originates from the superior ability of pumpkin to maintain viability of the apical cells (Fig. 1A, B, D). Unlike the root apex, the mature zone of both species showed less loss of viability under salt stress, and there was no difference between cucumber and pumpkin (Fig. 1C, E). Salinity treatment significantly decreased K+ content in the root apex of cucumber (by ~20%) but it remained unchanged in pumpkin (Fig. 2A). At the same time, K+ content between the two species was similar in the mature zone under salt stress (Fig. 2B).
Fig. 1.
Cell viability staining of the root apex and mature zone of cucumber and pumpkin. Plants were treated with 75 mM NaCl for 48 h. (A) One (of six) typical image is shown for each treatment. Intensity of the green (B, apex; C, mature) and red (D, apex; E, mature) fluorescent signal is quantified. Values are the mean ±SE (n=6 plants); different letters indicate a significant difference (P<0.05) according to Duncan’s multiple range tests.
Fig. 2.
K+ content in the root apex (A; 0–5 mm from the tip) and root mature zone (B; 5–25 mm from the tip) of cucumber and pumpkin after 75 mM NaCl treatment for 48 h. Values are the mean ±SE (n=6). Different letters indicate a significant difference (P<0.05) according to Duncan’s multiple range tests.
Salinity induced a higher genome-wide expression level in pumpkin
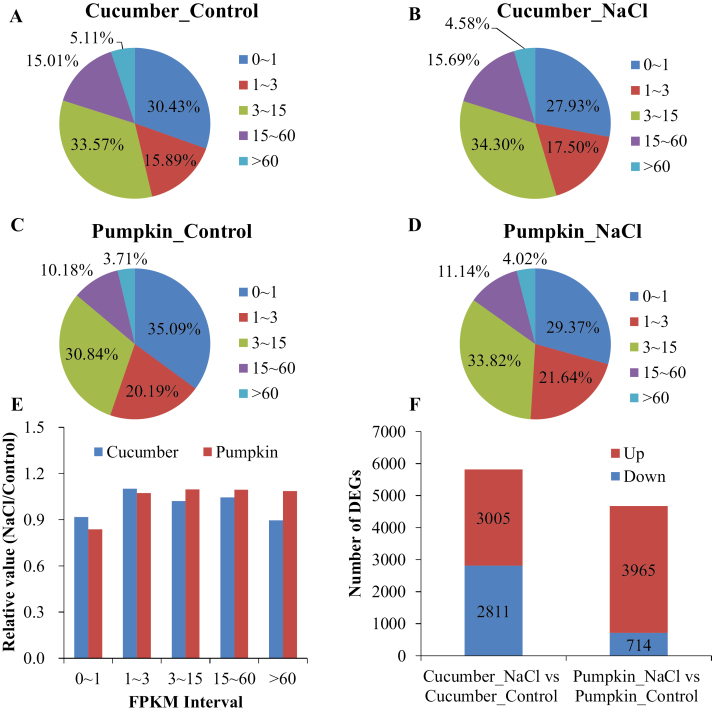
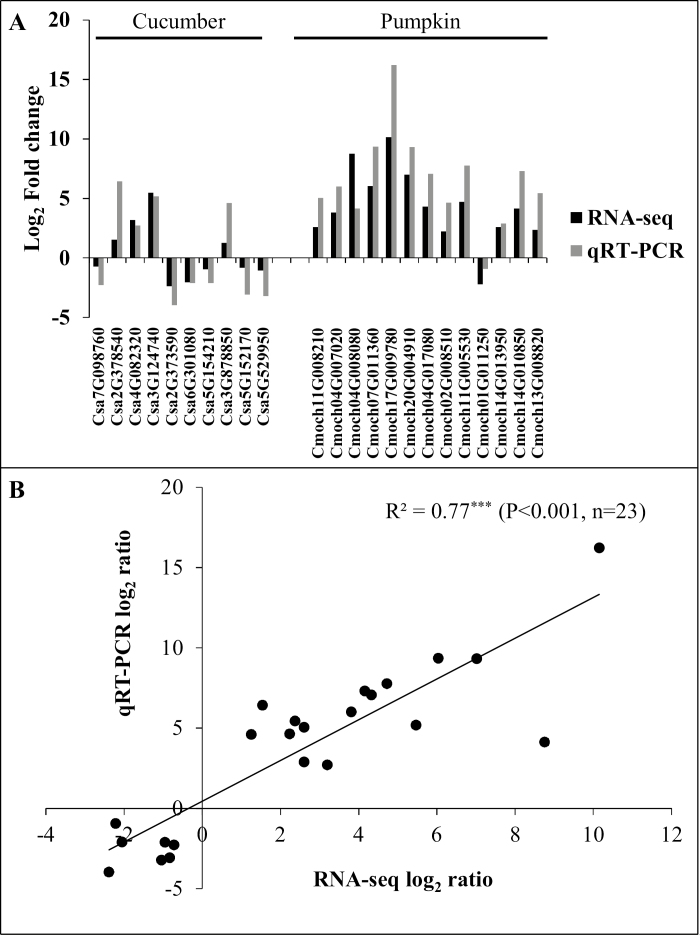
RNA-seq was used to investigate the molecular response of the two species to salinity (Fig. 3A–F). High-quality transcriptome readings were obtained, as shown by the sequencing data quality (Supplementary Table S2), transcriptome assembly evaluation (Supplementary Table S3), Pearson correlation between samples (Supplementary Table S4), and the validation by qRT-PCR (Fig. 4A, B). The genome-wide expression level was distinctly higher in pumpkin compared with cucumber, as revealed by a higher relative value of the FPKM interval of 3–15, 15–60, and >60 under NaCl stress, compared with control (Fig. 3A–E). Salinity decreased the percentage of ‘FPKM interval >60’ by 10% in cucumber, but increased it by 8% in pumpkin (Fig. 3A–E). Meanwhile, salinity induced 5816 (3005 up- and 2811 down-) and 4679 (3965 up- and 714 down-) DEGs in cucumber and pumpkin, respectively (Fig. 3F; Supplementary Tables S5, S6), suggesting that the percentage of up-regulated DEGs was higher in pumpkin (85%) than in cucumber (51%). In cucumber, DEGs related to membrane transport were significantly enriched in GO terms, namely transporter activity (GO:0005215) (Supplementary Table S7). In pumpkin, GO terms related to membrane transport including secondary active transmembrane transporter activity (GO:0015291), transporter activity (GO:0005215), transmembrane transporter activity (GO:0022857), antiporter activity (GO:0015297), and active transmembrane transporter activity (GO:0022804) were significantly enriched (Supplementary Table S8).
Fig. 3.
Gene expression analysis (n=3). Percentage of FPKM interval in the root apex (0–5 mm from the tip) transcriptome of cucumber and pumpkin under control (A, C) and NaCl (B, D) for 24 h. (E) The relative value of the FPKM interval under NaCl compared with control. (F) The number of differentially expressed genes (DEGs) in cucumber and pumpkin in response to NaCl stress.
Fig. 4.
Validation (A) and correlation analysis (B) of gene expression values obtained from transcriptome (RNA-seq) and data obtained using qRT-PCR of cucumber and pumpkin. All qRT-PCRs were performed in three biological replicates. Values are the log2 ratio (NaCl/control) for all genes.
Pumpkin species had higher expression of DEGs involved in K+ transport and cell division but lower expression of those involved in cell death
There were 9 and 11 DEGs involved in K+ transport in cucumber and pumpkin, respectively (Table 1). In cucumber, two HAK5 homologs (Csa4G001590 and Csa4G007060) and one AKT1 homolog (Csa3G878850) were found. In pumpkin, there were three HAK5 homologs (CmoCh14G009180, CmoCh17G010860, and CmoCh08G004000) (Table 1). The average log2FC of HAK5 homologs was –0.04 in cucumber, while the value was 4.36 in pumpkin (Table 1). Two of the three DEGs involved in cell division were down-regulated, and all eight DEGs involved in cell death were up-regulated in cucumber (Table 1). In pumpkin, one DEG (CmoCh14G001330) involved in cell division and two DEGs (CmoCh13G008030 and CmoCh13G008040) involved in cell death were up-regulated (Table 1).
Table 1.
A summary of the differentially expressed genes (DEGs) (significant at FDR <0.01, |log2FC|>2) involved in potassium transport, cell division, and cell death in the transcriptome of cucumber and pumpkin
| ID | Log2FC | Annotation | Arabidopsis homologs |
|---|---|---|---|
| Cucumber | |||
| Csa4G001590 | 2.78 | Potassium transporter | AT4G13420/HAK5 |
| Csa4G007060 | –2.86 | Potassium transporter | AT4G13420/HAK5 |
| Csa3G878850 | 4.60 | Voltage-dependent potassium channel | AT2G26650/AKT1 |
| Csa5G220400 | –2.03 | Potassium transporter | AT2G40540/KUP2 |
| Csa1G532340 | 2.98 | Potassium transporter | AT2G30070/KUP1 |
| Csa3G835810 | –3.88 | Potassium transporter | AT1G70300/KUP6 |
| Csa4G107490 | 3.15 | Potassium transporter | AT5G09400/KUP7 |
| Csa5G070180 | –2.28 | Potassium transporter | AT5G09400/KUP7 |
| Csa7G098760 | –2.28 | Outward rectifying potassium channel | AT5G55630/TPK1 |
| Csa7G343310 | –3.48 | Cell division control protein 45 | AT3G25100/CDC45 |
| Csa1G571790 | 2.13 | Cell division cycle protein | AT4G05440/EDA35 |
| Csa4G095030 | –2.43 | Cell division cycle-associated 7-like protein | AT5G38690/unknown |
| Csa3G121010 | 2.04 | Programmed cell death protein | AT2G46200/unknown |
| Csa6G014680 | 3.18 | Programmed cell death protein 2-like protein | AT5G64830/unknown |
| Csa1G050020 | 2.03 | Programmed cell death protein | AT5G63190/MRF1 |
| Csa5G608410 | 2.32 | Programmed cell death 6 interacting protein | AT1G13310/unknown |
| Csa1G701950 | 3.06 | Development and Cell Death domain protein | AT2G35140/DCD |
| Csa4G052730 | 3.06 | Development and Cell Death domain protein | AT5G42050/DCD |
| CsaUNG029290 | 2.41 | Development and Cell Death domain protein | AT2G32910/DCD |
| Csa4G007630 | 2.90 | Development and Cell Death domain protein | AT5G61910/DCD |
| Pumpkin | |||
| CmoCh14G009180 | 2.96 | Potassium transporter | AT4G13420/HAK5 |
| CmoCh17G010860 | 4.19 | Potassium transporter | AT4G13420/HAK5 |
| CmoCh08G004000 | 5.94 | Potassium transporter | AT4G13420/HAK5 |
| CmoCh11G017020 | –2.18 | Potassium transporter | AT1G70300/KUP6 |
| CmoCh13G008810 | 2.45 | Potassium transporter | AT2G35060/KUP11 |
| CmoCh13G008820 | 2.37 | Potassium transporter | AT1G31120/KUP10 |
| CmoCh15G007530 | 2.25 | Potassium voltage-gated channel | AT3G02850/SKOR |
| CmoCh04G017080 | 4.32 | Potassium voltage-gated channel | At2G02710/TLP1 |
| CmoCh11G014470 | 2.59 | Glutathione-regulated potassium-efflux protein | AT4G04850/KEA3 |
| CmoCh01G011250 | –2.22 | Outward rectifying potassium channel | AT5G55630/TPK1 |
| CmoCh10G005870 | 2.24 | Outward-rectifying potassium channel | AT3G15760/unknown |
| CmoCh14G001330 | 6.05 | FtsJ cell division protein-like | AT5G02220/unknown |
| CmoCh13G008030 | 2.31 | Programmed cell death protein | AT5G63190/MRF1 |
| CmoCh13G008040 | 2.14 | Programmed cell death protein | AT5G63190/MRF1 |
Measurements were taken in the root apex (0–5 mm from the tip) in plants exposed to 75 mM NaCl for 24 h.
Salt-tolerant pumpkin up-regulates DEGs encoding respiratory burst oxidase (NADPH oxidase), 14-3-3 protein, and PM H+-ATPase under salt stress
There were two and three DEGs encoding respiratory burst oxidase (NADPH oxidase) in cucumber and pumpkin, respectively (Table 2). Among them, Csa5G529950 (RBOHE) and Csa5G152170 (RBOHF) were down-regulated in cucumber and CmoCh06G017360 and CmoCh14G010850 (RBOHD) were up-regulated in pumpkin (Table 2). The average log2FC of RBOH homologs was –3.15 in cucumber, while the value was 2.45 in pumpkin (Table 2). There were two DEGs (Csa3G890040 and Csa2G372150) encoding 14-3-3 protein in cucumber; both were down-regulated (Table 2). One DEG encoding 14-3-3 protein was observed in pumpkin, and it was up-regulated (Table 2). In cucumber, two out of five DEGs encoding PM H+-ATPase were down-regulated (Csa1G423270 and Csa1G045600); while three others were up-regulated (Table 2). In pumpkin, all three DEGs (CmoCh11G003690, CmoCh14G013950, and CmoCh04G028780) were up-regulated (Table 2). The average log2FC of AHA homologs was 1.0 in cucumber, while the value was 3.71 in pumpkin (Table 2).
Table 2.
A summary of the differentially expressed genes (DEGs) (significant at FDR<0.01, |log2FC|>2) encoding NADPH oxidase (respiratory burst oxidase), 14-3-3 protein, and PM H+-ATPase in the transcriptome of cucumber and pumpkin
| ID | Log2FC | Annotation | Arabidopsis homologs |
|---|---|---|---|
| Cucumber | |||
| Csa5G529950 | –3.22 | Respiratory burst oxidase | AT1G19230/RBOHE |
| Csa5G152170 | –3.07 | Respiratory burst oxidase | AT1G64060/RBOHF |
| Csa3G890040 | –2.28 | 14-3-3 protein | AT5G65430/GRF8 |
| Csa2G372150 | –2.70 | 14-3-3 protein | AT5G65430/GRF8 |
| Csa2G378540 | 6.43 | Plasma membrane H+-ATPase | AT2G24520/AHA5 |
| Csa1G423270 | –3.08 | Plasma membrane H+-ATPase | AT2G24520/AHA5 |
| Csa4G006220 | 2.96 | Plasma membrane H+-ATPase | AT1G80660/AHA9 |
| Csa1G045600 | –3.80 | Plasma membrane H+-ATPase | AT5G62670/AHA11 |
| Csa5G635370 | 2.48 | Plasma membrane H+-ATPase | AT5G62670/AHA11 |
| Pumpkin | |||
| CmoCh06G017360 | 5.20 | Respiratory burst oxidase | AT5G47910/RBOHD |
| CmoCh14G010850 | 4.16 | Respiratory burst oxidase | AT5G47910/RBOHD |
| CmoCh11G002090 | –2.01 | Respiratory burst oxidase | AT1G64060/RBOHF |
| CmoCh01G016540 | 4.25 | 14-3-3 protein | AT1G26480/GRF12 |
| CmoCh11G003690 | 4.30 | Plasma membrane H+-ATPase | AT2G18960/AHA1 |
| CmoCh14G013950 | 2.60 | Plasma membrane H+-ATPase | AT1G80660/AHA9 |
| CmoCh04G028780 | 4.22 | Plasma membrane H+-ATPase | AT5G62670/AHA11 |
Measurements were taken in the root apex (0–5 mm from the tip) in plants exposed to 75 mM NaCl for 24 h.
NADPH oxidase-dependent H2O2 signaling is essential for the salt tolerance of pumpkin and is upstream of the PM H+-ATPase-mediated K+ uptake
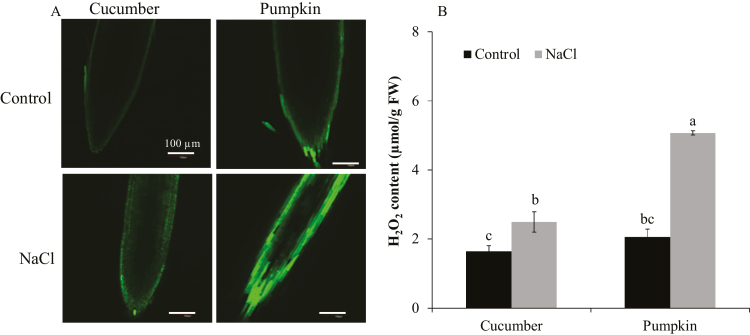
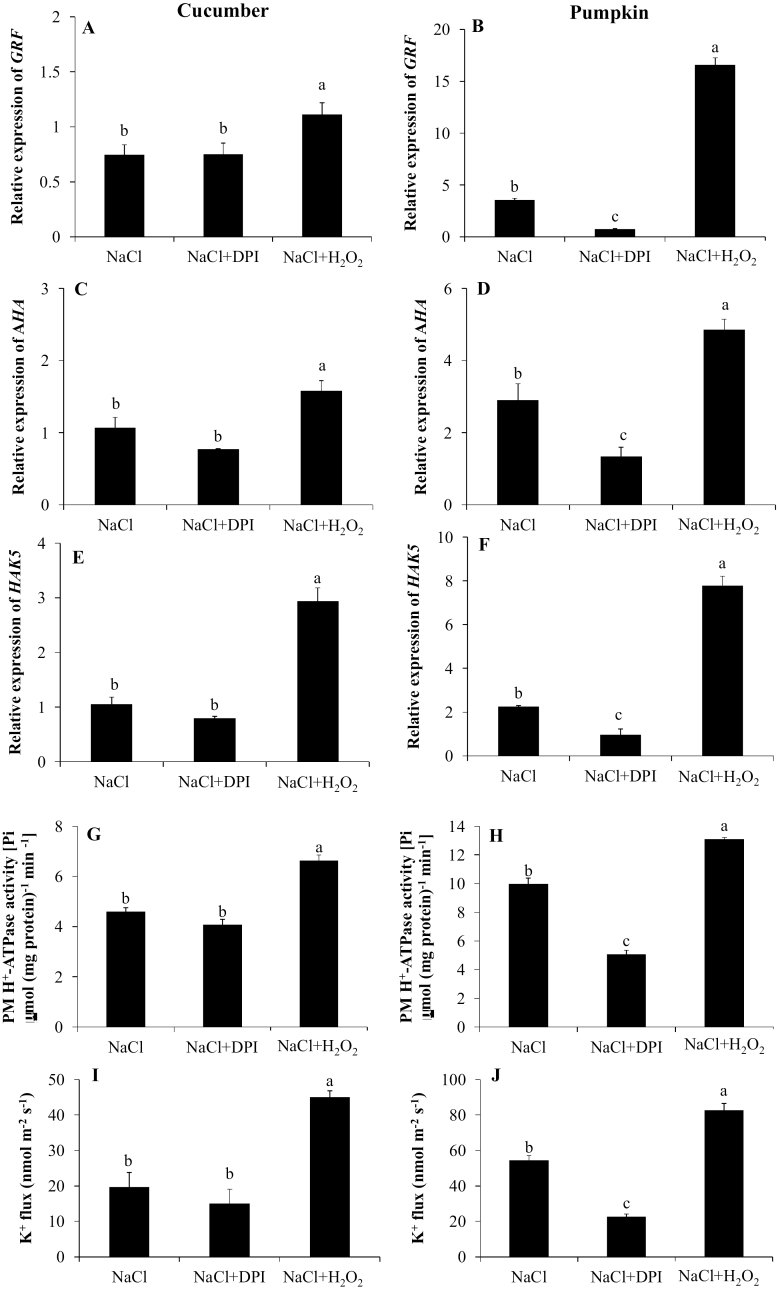
To determine the possible involvement of H2O2 signaling in salt tolerance, the levels of H2O2 in cucumber and pumpkin were measured (Fig. 5A, B). The H2O2 level was significantly higher in the roots of pumpkin than in those of cucumber, when visualized using the H2DCF-DA fluorescence probe (Fig. 5A), and quantified by spectrophotometry tools (Fig. 5B). An NADPH oxidase inhibitor, DPI, was used to investigate the potential role of the RBOH-dependent H2O2 production in regulating root K+ uptake (Figs 6A–D, 7A–J). Inhibition of the NADPH oxidase resulted in a significantly lower expression of RBOHD, GRF12 (encoding 14-3-3 protein), AHA1 (encoding PM H+-ATPase), and HAK5, reduced PM H+-ATPase activity, and smaller K+ influx in pumpkin (Figs 6C, D, 7B, D, F, H, J). In contrast, in cucumber, NADPH oxidase inhibitor did not result in a significant difference in GRF8, AHA11, and HAK5 expression, PM H+-ATPase activity, and K+ flux (Fig. 7A, C, E, G, I). In addition, exogenous application of H2O2 significantly increased the above parameters in both species (Fig. 7A–J). Taken together, these data suggest that RBOHD-dependent H2O2 operates upstream of H+-ATPase and 14-3-3 protein, and affects K+ uptake via regulation of H+-ATPase in pumpkin.
Fig. 5.
Endogenous H2O2 level in the root apex of cucumber and pumpkin exposed to 75 mM NaCl for 24 h. H2O2 levels were measured by using confocal fluorescence imaging from roots stained with H2DCF-DA. (A) Typical (one of six) images for each treatment. Scale bar=100 μm. Higher green fluorescence intensity means higher H2O2 content. (B) Quantifying H2O2 content in root samples by spectrophotometry. Values are the mean ±SE (n=4). Different letters indicate a significant difference (P<0.05) according to Duncan’s multiple range tests.
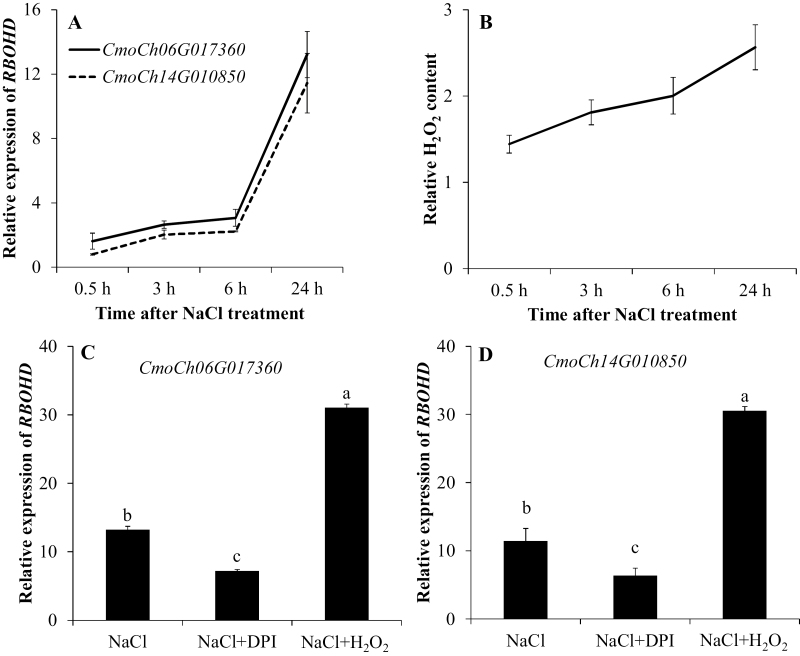
Fig. 6.
Relative expression of RBOHD (A) and relative H2O2 content (B, NaCl/control) in the root apex of pumpkin after 75 mM NaCl stress for 0.5, 3, 6, and 24 h. RBOHD expression (C, D) measured after 24 h of exposure to 75 mM NaCl stress from the pumpkin root apex pre-treated for 1 h in solutions containing specific chemicals (DPI, an NADPH oxidase inhibitor, and H2O2). Values are the mean ±SE (n=4). Different letters indicate a significant difference (P<0.05) according to Duncan’s multiple range tests.
Fig. 7.
Relative expression of GRF (14-3-3 protein), AHA (plasma membrane H+-ATPase), and HAK5 (high affinity K+ transporter) in the root apex of cucumber (A, C, E) and pumpkin (B, D, F) exposed to 75 mM NaCl for 24 h. (G–J) Plasma membrane H+-ATPase activity and net K+ flux measured after 24 h of exposure to 75 mM NaCl stress from the root apex pre-treated for 1 h in solutions containing specific chemicals (DPI, an NADPH oxidase inhibitor, and H2O2) in cucumber (G, I) and pumpkin (H, J). Values are the mean ±SE (n=4). Different letters indicate a significant difference (P<0.05) according to Duncan’s multiple range tests. The gene IDs for are GRF, AHA, and HAK5 are Csa3G890040 (GRF8), Csa1G045600 (AHA11), and Csa3G835810 (HAK5) for cucumber, and CmoCh01G016540 (GRF12), CmoCh11G003690 (AHA1), and CmoCh08G004000 (HAK5) for pumpkin.
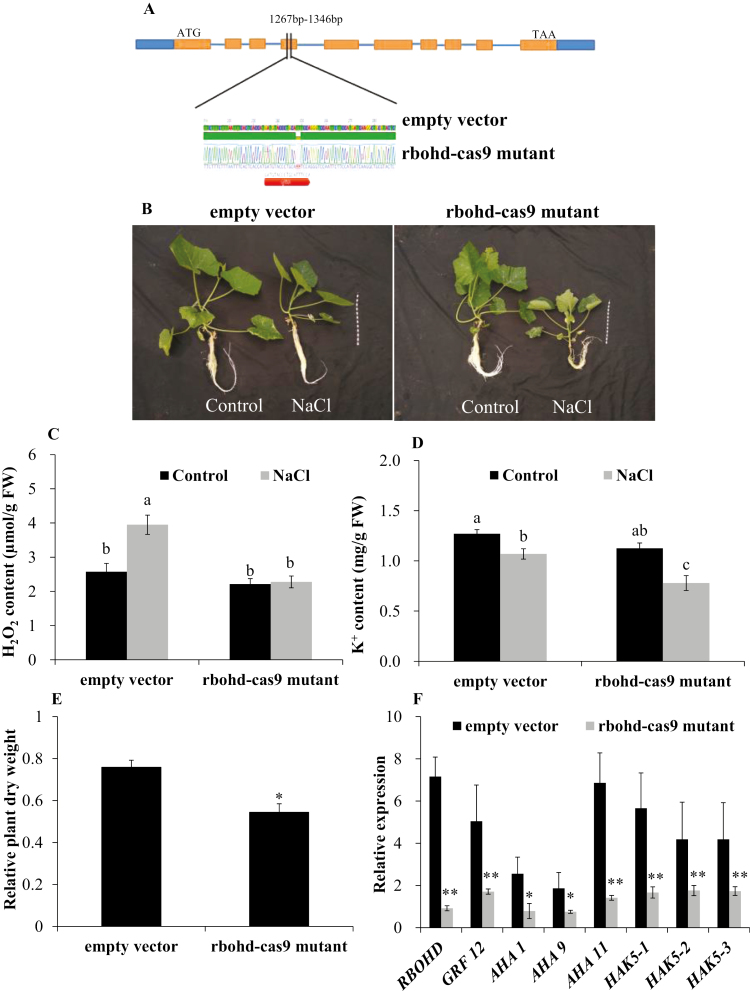
Functional analysis of pumpkin RBOHD showed that H2O2 signaling is essential for K+ uptake and salt tolerance at the genetic level
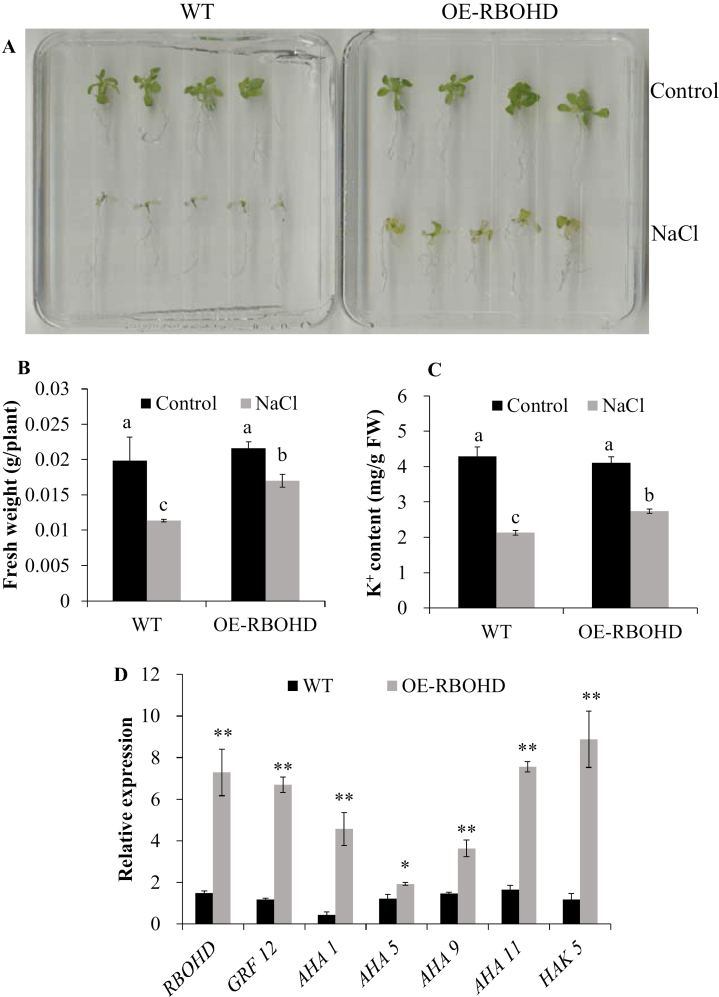
The CRISPR/Cas9 system has emerged as a robust technology for efficient genome editing, and has been successfully applied in many crops (Hu et al., 2017; Kirchner et al., 2017). Since stable transgenic pumpkin plants require a relatively long time (~12 months) to develop, we opted to use hairy roots as a model system which only take ~2 months (Supplementary Fig. S2A–F). In this study, we set out to test the function of pumpkin RBOHD by hairy root transformation using A. rhizogenes combined with the CRISPR/Cas9 system. Compared with control (empty vector), deletions of two nucleotides (TT) were observed in rbohd-cas9 mutant pumpkin plants (Fig. 8A), resulting in a loss-of-function mutation in RBOHD. The rbohd-cas9 mutant had significantly lower salt tolerance, as indicated by the relative plant dry weight (Fig. 8B, E), and H2O2 and K+ content, as well as expression of RBOHD, GRF12, AHA1/9/11, and HAK5 (Fig. 8C, D, F). In this study, the function of pumpkin RBOHD was also investigated by the ectopic expression system in Arabidopsis (Supplementary Fig. S3). Compared with the WT, ectopic expression of RBOHD (OE-RBOHD) significantly increased plant fresh weight (Fig. 9A, B), K+ content, and RBOHD, GRF12, AHA1/5/9/11, and HAK5 expression (Fig. 9C, D). These genetic results strongly demonstrated that RBOHD-dependent H2O2 signaling is essential for K+ uptake and salt tolerance of pumpkin.
Fig. 8.
CRISPR/Cas9-induced nucleotide deletion (TT) in pumpkin RBOHD (A, rbohd-cas9 mutant) results in a salt-sensitive phenotype (B). (C, D) K+ and H2O2 content of empty vector-transformed and rbohd-cas9 mutant pumpkin plants under control and NaCl stress conditions, respectively. (E) Relative plant dry weight (NaCl/control); (F) relative expression of RBOHD, GRF12, AHA1/9/11, and HAK5 genes (NaCl/control). Values are the mean ±SE (n=4). Different letters indicate a significant difference at P<0.05 according to Duncan’s multiple range tests. * and ** indicate a significant difference at P<0.05 and 0.01 levels between empty vector-transformed and rbohd-cas9 mutant plants by Student’s t-tests. Plant pictures were taken and relative plant dry weight and K+ content were measured at day 12 after onset of 75 mM NaCl treatment. H2O2 content and relative gene expression in the root apex was measured 24 h after 75 mM NaCl treatment. The gene IDs are CmoCh14G010850 (RBOHD), CmoCh01G016540 (GRF12), CmoCh11G003690 (AHA1), CmoCh14G013950 (AHA9), CmoCh04G028780 (AHA11), Cmoch08G004000 (HAK5-1), CmoCh14G009180 (HAK5-2), and CmoCh17G010860 (HAK5-3).
Fig. 9.
Plant phenotype (A), fresh weight (B), and K+ content (C) of Arabidopsis WT (Columbia-0) and plants ectopically expressing pumpkin RBOHD (OE-RBOHD) under control and NaCl stress conditions. (D) Relative (NaCl/control) expression of RBOHD, GRF12, AHA1/5/9/11, and HAK5 genes in WT and ectopic plants. Values are the mean ±SE (n=4). Different letters indicate a significant difference at P<0.05 according to Duncan’s multiple range tests. * and ** indicate a significant difference at P<0.05 and 0.01 levels between WT and OE-RBOHD plants by Student’s t-tests. Plant pictures were taken and relative plant dry weight and whole-plant K+ content were measured at day 7 after onset of 75 mM NaCl treatment. Relative gene expression in the root apex was measured 24 h after 75 mM NaCl treatment. The gene IDs are AT5G47910 (RBOHD), AT1G26480 (GRF12), AT2G18960 (AHA1), AT2G24520 (AHA5), AT1G80660 (AHA9), AT5G62670 (AHA11), and AT4G13420 (HAK5).
Discussion
In this work, we describe a novel mechanism that contributes to differential salinity tolerance amongst Cucurbitaceae species, namely RBOHD-dependent H2O2 signaling to the PM H+-ATPase that results in improved HAK5-mediated K+ uptake and a salt-tolerant phenotype. The supporting arguments are given below.
Maintenance of K+ homeostasis in the root apex is critical for salt tolerance in pumpkin
Maintenance of K+ homeostasis is essential for enzyme activities, ionic and pH homeostasis, and charge balance (Dreyer and Uozumi, 2011), and salinity stress results in a K+ loss from plant roots (Shabala and Pottosin, 2014). A strong correlation between the root’s K+ retention ability and plant salinity stress tolerance was reported for several species including wheat (Cuin et al., 2008), barley (Chen et al., 2007), poplar (Sun et al., 2009), and cucumber (Redwan et al., 2016). Our present study demonstrated that K+ retention in the root apex, but not the mature zone, is important for the salt tolerance of pumpkin, possibly due to the higher cell division and metabolic demand for K+ in the root tip.
HAK5 contributes to the K+ accumulation in the root apex of pumpkin
HAK5 is one of the primary players in K+ uptake into roots under low K+ concentrations (Shin and Schachtman, 2004; Jung et al., 2009; Véry et al., 2014). In this study, no DEGs encoding AKT1 were observed in pumpkin, and pumpkin had significantly higher HAK5 expression under salt stress than cucumber. This is consistent with higher K+ influx (Fig. 7I, J), suggesting that HAK5 contributes to the K+ accumulation in salt-tolerant pumpkin species. Jiang et al. (2013) found that ethylene promotes the accumulation of K+ via an AtrbohF-independent mechanism, possibly via an increase in the level of transcripts encoding AtHAK5 in saline conditions.
Higher AHA expression and PM H+-ATPase activity are responsible for the K+ uptake in the roots of pumpkin during salt stress
HAK5 is a K+/H+ symporter, so a higher rate of H+ efflux via H+-ATPase may create steeper H+ gradients, hence being a driving force for K+ uptake (Scherzer et al., 2015). Pre-treating pumpkin roots with 20 µM DPI significantly decreased AHA expression and PM H+-ATPase activity, decreasing K+ influx (Fig. 7B, D, F, H, J). This indicates that pumpkin roots rely heavily on PM H+-ATPase for the regulation of K+ uptake during salt stress, at both the transcriptional and protein (enzyme activity) level. The H+-ATPase activity can be affected by the cytosolic K+ concentration, and vice versa, as it was shown that the plant PM H+-ATPase is regulated by K+ bound to the proton pump at a site involving Asp617 in the cytoplasmic phosphorylation domain, suggesting a role for K+ as an intrinsic uncoupler (Buch-Pedersen et al., 2006).
H2O2 probably regulates PM H+-ATPase through 14-3-3 protein binding in pumpkin under salt stress
Despite the wealth of evidence indicating that H+-ATPase activity confers regulation of K+ homeostasis (Shabala et al., 2016), the exact molecular mechanism whereby H2O2 regulates H+-ATPase remains largely unknown (Majumdar and Kar, 2018). In addition to the transcriptional control, phosphorylation of the PM H+-ATPase plays a major role in its regulation (Falhof et al., 2016). Our study revealed the presence of 7 and 14 AHA members in cucumber and pumpkin, respectively (Supplementary Table S9). Only 3 of 14 AHA members in pumpkin were regulated at the transcriptional level, implying that post-transcriptional control may be more critical for other AHA members in this species. Phosphorylation of several Thr and Ser residues in the C-terminal region is crucial for the regulation of PM H+-ATPase activity (Rudashevskaya et al., 2012). 14-3-3 proteins interact directly with the C-terminal region of the Thr947 phosphorylation site to activate PM H+-ATPase (Jahn et al., 1997; Svennelid et al., 1999). Salt stress induces phosphorylation and 14-3-3 protein binding to PM H+-ATPase (Janicka-Russak et al., 2013). In pumpkin, salinity up-regulated the expression of GRF12 which encodes 14-3-3 protein (Table 2), and DPI treatment decreased the expression of GRF12 (Fig. 7B), suggesting that H2O2 regulates PM H+ ATPase through 14-3-3 protein.
RBOHD is essential for H2O2 production in pumpkin under salt stress
NADPH oxidases represent an important source of apoplastic ROS production. They transfer electrons across the PM from the cytoplasmic NADPH to molecular oxygen to produce O2· −, which can be dismutated to H2O2 either spontaneously or by the apoplastic superoxide dismutases (Waszczak et al., 2018). The Arabidopsis genome contains 10 genes for the RBOH isoforms (Suzuki et al., 2011). Two specific isoforms, RBOHD and RBOHF, seem to be critical to salt stress responses (Xie et al., 2011; Ma et al., 2012). There are 8 and 12 genes for the RBOH isoforms in cucumber and pumpkin (Supplementary Table S10) genomes, respectively. This study showed that RBOHD is the key member for H2O2 production in pumpkin, as only RBOHD was found to be up-regulated under salt stress (Table 1). In addition, RBOHD expression was consistent with H2O2 accumulation in the root apex of pumpkin at different times of salt stress (Fig. 6A, B). In Arabidopsis, it was shown that a ROS-assisted calcium wave, which was dependent on the AtRBOHD NADPH oxidase, propagates the systemic response to salt stress (Evans et al., 2016). In this study, the importance of RBOHD in the salt tolerance of pumpkin was also verified at the genetic level. Using pumpkin CRISPR/Cas9-mediated genome editing and ectopic expression of pumpkin RBOHD in Arabidopsis, our study explicitly demonstrated that RBOHD-mediated activation of PM H+-ATPase results in a salt-tolerant phenotype, as a result of enhanced K+ uptake mediated by HAK5 transporters.
Compared with other cucurbit genera, such as cucumber (2n=14), Cucurbita has a higher chromosome number (2n=40) (Bisognin, 2002). A recent study provided evidence supporting an allotetraploidization event in Cucurbita (Sun et al., 2017). The ploidy level was shown to be causally related to salinity tolerance (Yang et al., 2014), and both K+ uptake (Chao et al., 2013) and operation of RBOH (Liu et al., 2019; Shabala, 2019) were found to be more efficient in polyploids. This work is fully consistent with these recent findings, demonstrating that the larger number of genes in the AHA and RBOH family in pumpkin lead to its superior salinity tolerance, as compared with cucumber.
Conclusions
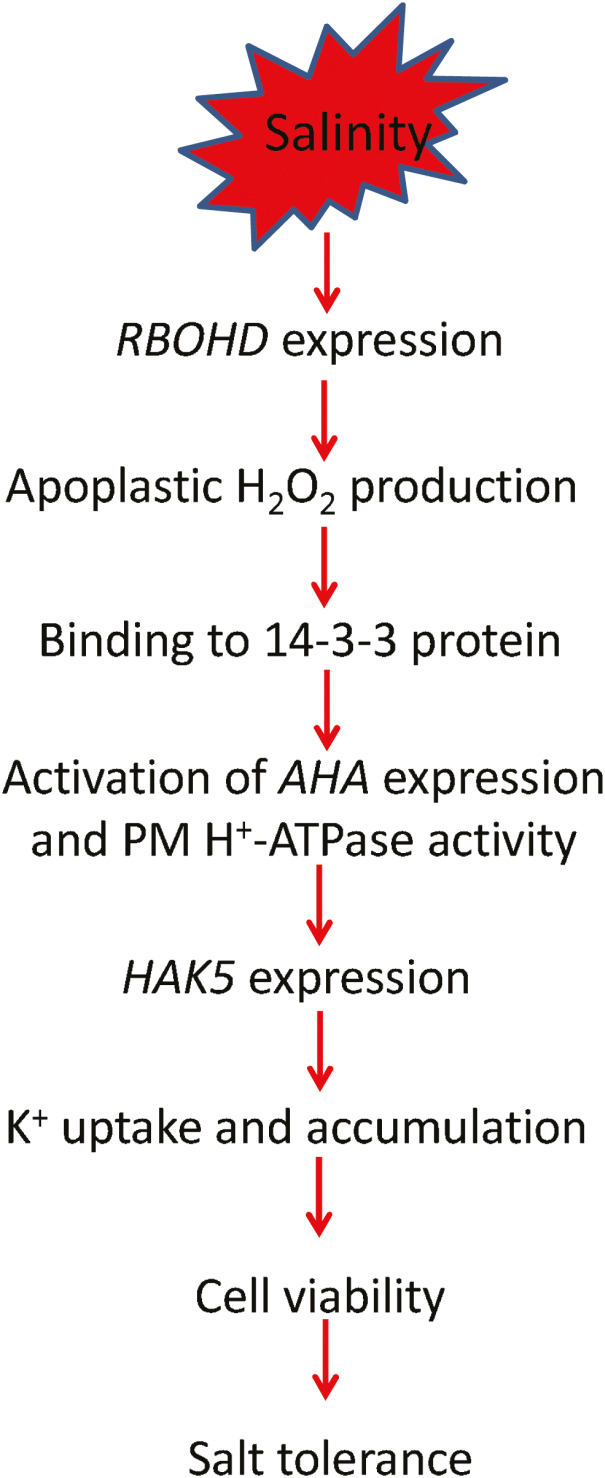
The results reported here demonstrate that the superior salinity stress tolerance in pumpkin was conferred by its better ability to take up K+ in the root apex (Fig. 10). This trait was mediated by a HAK5 transporter that operated downstream of RBOH-generated H2O2 signaling that resulted in increased AHA expression and post-translational regulation of the PM H+-ATPase (most probably via 14-3-3 proteins) (Fig. 10). As a result, pumpkin can accumulate more K+ in the root apex, thus preventing programmed cell death in this tissue and hence displaying superior salinity stress tolerance.
Fig. 10.
A suggested model depicting RBOHD-dependent H2O2 signaling to H+-ATPase and its impact on K+ uptake and accumulation in the root apex (thus, overall salt tolerance) in pumpkin compared with cucumber.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Effect of NaCl stress on the root morphological parameters in cucumber and pumpkin.
Fig. S2. Generation of the pumpkin rbohd-cas9 mutant by CRISPR/Cas9 in the hairy root transformation system.
Fig. S3. Ectopic expression of pumpkin RBOHD in Arabidopsis plants (OE-RBOHD) under control and 75 mM NaCl stress (24 h) conditions.
Table S1. List of primer sequences used for qRT-PCR and RT-PCR analysis.
Table S2. Sequencing data quality.
Table S3. Transcriptome assembly evaluation.
Table S4. Pearson correlation between samples.
Table S5. Significant differentially expressed (FDR<0.01, |log2FC|>2) genes in the transcriptome of cucumber under NaCl stress compared with control.
Table S6. Significant differentially expressed (FDR<0.01, |log2FC|>2) genes in the transcriptome of pumpkin under NaCl stress compared with control.
Table S7. Summary of Gene Ontology biological process, cellular component, and molecular function terms significantly enriched in differentially expressed genes in the cucumber transcriptome in response to NaCl stress.
Table S8. Summary of Gene Ontology biological process, cellular component, and molecular function terms significantly enriched in differentially expressed genes in the pumpkin transcriptome in response to NaCl stress.
Table S9. The characteristics of AHA family members in cucumber and pumpkin.
Table S10. The characteristics of RBOH family members in cucumber and pumpkin.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD1000800) and the National Natural Science Foundation of China (31572168, 31772357) to ZB, the National Natural Science Foundation of China (31471919), the Fundamental Research Funds for the Central Universities (2662018JC037, 2662018PY039), China Scholarship Council (CSC) (grant no. 201606765073) to YH, and funding from the Australian Research Council and Grain Research and Development Corporation to SS. The authors have declared that no competing interests exist.
Author contributions
YH, SS, and ZLB designed the experiments; YH, HSC, LY, CC, LS, MX, MLN, JL, ZHZ, LJZ, and ZWP performed the experiments; YH, SS, and ZLB analyzed the data. YH, SS, LS, and ZLB wrote the article. All authors read and approved the final manuscript.
References
- Anschütz U, Becker D, Shabala S. 2014. Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. Journal of Plant Physiology 171, 670–687. [DOI] [PubMed] [Google Scholar]
- Bisognin DA. 2002. Origin and evolution of cultivated cucurbits. Ciencia Rural 32, 715–723. [Google Scholar]
- Bose J, Shabala L, Pottosin I, Zeng F, Velarde-Buendía AM, Massart A, Poschenrieder C, Hariadi Y, Shabala S. 2014. Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: physiological traits that differentiate salinity tolerance between pea and barley. Plant, Cell & Environment 37, 589–600. [DOI] [PubMed] [Google Scholar]
- Buch-Pedersen MJ, Rudashevskaya EL, Berner TS, Venema K, Palmgren MG. 2006. Potassium as an intrinsic uncoupler of the plasma membrane H+-ATPase. Journal of Biological Chemistry 281, 38285–38292. [DOI] [PubMed] [Google Scholar]
- Cao H, Wang L, Nawaz MA, Niu M, Sun J, Xie J, Kong Q, Huang Y, Cheng F, Bie Z. 2017. Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Frontiers in Plant Science 8, 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K, Bose J, Shabala L, Shabala S. 2016. Difference in root K+ retention ability and reduced sensitivity of K+-permeable channels to reactive oxygen species confer differential salt tolerance in three Brassica species. Journal of Experimental Botany 67, 4611–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE. 2013. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Pottosin II, Cuin TA, et al. 2007. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiology 145, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Johnson AA, Walia H, Wilson C, Ismail AM, Close TJ, Tester M, Baumann U. 2011. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Molecular Plant 4, 25–41. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Betts SA, Chalmandrier R, Shabala S. 2008. A root’s ability to retain K+ correlates with salt tolerance in wheat. Journal of Experimental Botany 59, 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S. 2011. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant, Cell & Environment 34, 947–961. [DOI] [PubMed] [Google Scholar]
- Cuin TA, Tian Y, Betts SA, Chalmandrier R, Shabala S. 2009. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Functional Plant Biology 36, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Straltsova D, Medvedev SS, Pozhvanov GA, Sokolik A, Yurin V. 2014. Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. Journal of Experimental Botany 65, 1259–1270. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. 2008. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Dreyer I, Uozumi N. 2011. Potassium channels in plant cells. FEBS Journal 278, 4293–4303. [DOI] [PubMed] [Google Scholar]
- Edelstein M, Plaut Z, Ben-Hur M. 2011. Sodium and chloride exclusion and retention by non-grafted and grafted melon and Cucurbita plants. Journal of Experimental Botany 62, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Choi WG, Gilroy S, Morris RJ. 2016. A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiology 171, 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J, Pedersen JT, Fuglsang AT, Palmgren M. 2016. Plasma membrane H+-ATPase regulation in the center of plant physiology. Molecular Plant 9, 323–337. [DOI] [PubMed] [Google Scholar]
- Gassman W, Rubio F, Schroeder JI. 1996. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. The Plant Journal 10, 869–852. [DOI] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR. 2013. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. The Plant Cell 25, 2132–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. 2005. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. The Plant Journal 44, 826–839. [DOI] [PubMed] [Google Scholar]
- Gruber V, Blanchet S, Diet A, Zahaf O, Boualem A, Kakar K, Alunni B, Udvardi M, Frugier F, Crespi M. 2009. Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Molecular Genetics and Genomics 281, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51, 463–499. [DOI] [PubMed] [Google Scholar]
- He H, Van Breusegem F, Mhamdi A. 2018. Redox-dependent control of nuclear transcription in plants. Journal of Experimental Botany 69, 3359–3372. [DOI] [PubMed] [Google Scholar]
- Hill CB, Cassin A, Keeble-Gagnère G, Doblin MS, Bacic A, Roessner U. 2016. De novo transcriptome assembly and analysis of differentially expressed genes of two barley genotypes reveal root-zone-specific responses to salt exposure. Scientific Reports 6, 31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, ElSayed AI, Moore M, Dietz KJ. 2017. Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. Journal of Experimental Botany 68, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Li D, Liu X, Qi J, Gao D, Zhao S, Huang S, Sun J, Yang L. 2017. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Molecular Plant 10, 1575–1578. [DOI] [PubMed] [Google Scholar]
- Huang Y, Bie ZL, Liu PY, Niu ML, Zhen A, Liu ZX, Lei B, Gu DJ, Lu C, Wang BT. 2013. Reciprocal grafting between cucumber and pumpkin demonstrates the roles of the rootstock in the determination of cucumber salt tolerance and sodium accumulation. Scientia Horticulturae 149, 47–54. [Google Scholar]
- Ismail AM, Horie T. 2017. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annual Review of Plant Biology 68, 405–434. [DOI] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. 1997. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. The Plant Cell 9, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicka-Russak M, Kabała K, Wdowikowska A, Kłobus G. 2013. Modification of plasma membrane proton pumps in cucumber roots as an adaptation mechanism to salt stress. Journal of Plant Physiology 170, 915–922. [DOI] [PubMed] [Google Scholar]
- Jiang C, Belfield EJ, Cao Y, Smith JA, Harberd NP. 2013. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. The Plant Cell 25, 3535–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP. 2009. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. The Plant Cell 21, 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner TW, Niehaus M, Debener T, Schenk MK, Herde M. 2017. Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PLoS One 12, e0185429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kłobus G, Janicka-Russak M. 2004. Modulation by cytosolic components of proton pump activities in plasma membrane and tonoplast from Cucumis sativus roots during salt stress. Physiologia Plantarum 121, 84–92. [DOI] [PubMed] [Google Scholar]
- Koyro HW, Stelzer R. 1988. Ion concentrations in the cytoplasm and vacuoles of rhizodermis cells from NaCl treated Sorghum, Spartina and Puccinellia plants. Journal of Plant Physiology 133, 441–446. [Google Scholar]
- Lauchli A. 1999. Salinity–potassium interactions in crop plants. In: Oosterhuis DM, Berkowitz GA, eds. Frontiers in potassium nutrition: new perspectives on the effects of potassium on physiology of plants. Norcross, GA: Potash & Phosphate Institute, 71–76. [Google Scholar]
- Lauchli A, Wieneke J. 1979. Studies on growth and distribution of Na+, K+ and Cl– in soybean varieties differing in salt tolerance. Zeitschrift für Pflanzenernährung und Bodenkunde 142, 3–13. [Google Scholar]
- Lei B, Huang Y, Sun J, Xie J, Niu M, Liu Z, Fan M, Bie Z. 2014. Scanning ion-selective electrode technique and X-ray microanalysis provide direct evidence of contrasting Na+ transport ability from root to shoot in salt-sensitive cucumber and salt-tolerant pumpkin under NaCl stress. Physiologia Plantarum 152, 738–748. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu Y, Sun J, Cao Q, Tang Z, Liu M, Xu T, Ma D, Li Z, Sun J. 2019. Root-zone-specific sensitivity of K+- and Ca2+-permeable channels to H2O2 determines ion homeostasis in salinized diploid and hexaploid Ipomoea trifida. Journal of Experimental Botany 70, 1389–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. 2012. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. Journal of Experimental Botany 63, 305–317. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Kar RK. 2018. Congruence between PM H+-ATPase and NADPH oxidase during root growth: a necessary probability. Protoplasma 255, 1129–1137. [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Alemán F, Martínez V, Rubio F. 2010. The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Molecular Plant 3, 326–333. [DOI] [PubMed] [Google Scholar]
- Niu L, Liao W. 2016. Hydrogen peroxide signaling in plant development and abiotic responses: crosstalk with nitric oxide and calcium. Frontiers in Plant Science 7, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M, Huang Y, Sun S, Sun J, Cao H, Shabala S, Bie Z. 2018. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. Journal of Experimental Botany 69, 3465–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Li Z, Wen X, Li W, Shi H, Yang L, Zhu H, Guo H. 2014. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genetics 10, e1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwan M, Spinelli F, Marti L, Weiland M, Palm E, Azzarello E, Mancuso S. 2016. Potassium fluxes and reactive oxygen species production as potential indicators of salt tolerance in Cucumis sativus. Functional Plant Biology 43, 1016–1027. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan CJ, da Silva JAT, Mopper S, Qin P, Lutts S. 2010. Halophyte improvement for a salinized world. Critical Reviews in Plant Sciences 29, 329–359. [Google Scholar]
- Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG. 2012. Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. Journal of Biological Chemistry 287, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Kaur N, Pati PK. 2018. Reactive oxygen species dynamics in roots of salt sensitive and salt tolerant cultivars of rice. Analytical Biochemistry 550, 99–108. [DOI] [PubMed] [Google Scholar]
- Scherzer S, Böhm J, Krol E, et al. 2015. Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proceedings of the National Academy of Sciences, USA 112, 7309–7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera P, Singh RK, Maathuis FJ. 2009. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. Journal of Experimental Botany 60, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala L, Zhang J, Pottosin I, et al. 2016. Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity stress. Plant Physiology 172, 2445–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S. 2019. Linking ploidy level with salinity tolerance: NADPH-dependent ‘ROS–Ca2+ hub’ in the spotlight. Journal of Experimental Botany 70, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Pottosin I. 2014. Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiologia Plantarum 151, 257–279. [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. 2004. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proceedings of the National Academy of Sciences, USA 101, 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Wu S, Zhang G, et al. 2017. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Molecular Plant 10, 1293–1306. [DOI] [PubMed] [Google Scholar]
- Sun J, Chen S, Dai S, et al. 2009. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiology 149, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. The Plant Cell 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H. 2014. Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? Journal of Plant Physiology 171, 748–769. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu WH. 2017. Regulation of potassium transport and signaling in plants. Current Opinion in Plant Biology 39, 123–128. [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J. 2018. Reactive oxygen species in plant signaling. Annual Review of Plant Biology 69, 209–236. [DOI] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W. 1997. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. The EMBO Journal 16, 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang X, Giraldo JP, Shabala S. 2018. It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant and Soil 431, 1–17. [Google Scholar]
- Xie R, Pan X, Zhang J, Ma Y, He S, Zheng Y, Ma Y. 2018. Effect of salt-stress on gene expression in citrus roots revealed by RNA-seq. Functional & Integrative Genomics 18, 155–173. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB. 2011. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. The Plant Journal 66, 280–292. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhao L, Zhang H, Yang ZZ, Wang H, Wen SS, Zhang CY, Rustgi S, von Wettstein D, Liu B. 2014. Evolution of physiological responses to salt stress in hexaploid wheat. Proceedings of the National Academy of Sciences, USA 111, 11882–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y. 2018. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytologist 217, 523–539. [DOI] [PubMed] [Google Scholar]
- Zahaf O, Blanchet S, de Zélicourt A, et al. 2012. Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes. Molecular Plant 5, 1068–1081. [DOI] [PubMed] [Google Scholar]
- Zhu J, Bie ZL, Li YN. 2006. Evaluation of salt resistance of cucumber at seed germination and rootstock-seedling stages. Scientia Agricultura Sinica 39, 772–778. [Google Scholar]
- Zhu JK, Liu J, Xiong L. 1998. Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. The Plant Cell 10, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.