Although hypervirulent Klebsiella pneumoniae (hvKp) has been associated with severe community-acquired infections that occur among relatively healthy individuals, information about hvKp infections in health care settings remains limited. Here, we systematically analyzed the clinical and molecular characteristics of K. pneumoniae isolates causing bloodstream infections in a cross-sectional study.
KEYWORDS: bloodstream infection, disseminated infection, hypervirulent Klebsiella pneumoniae, whole-genome sequencing
ABSTRACT
Although hypervirulent Klebsiella pneumoniae (hvKp) has been associated with severe community-acquired infections that occur among relatively healthy individuals, information about hvKp infections in health care settings remains limited. Here, we systematically analyzed the clinical and molecular characteristics of K. pneumoniae isolates causing bloodstream infections in a cross-sectional study. Clinical characteristics of K. pneumoniae bloodstream infections from hospitals across Japan were analyzed by a review of the medical records. Whole-genome sequencing of the causative isolates was performed. Bacterial species were confirmed and hvKp were identified using whole-genome sequencing data. Clinical characteristics of hvKp infections were compared with those of non-hvKp infections by bivariate analyses. Of 140 cases of K. pneumoniae bloodstream infections, 26 cases (18.6%) were caused by various clones of hvKp defined by the carriage of cardinal virulence genes. Molecular identification revealed that 24 (17.1%) and 14 (10%) cases were caused by Klebsiella variicola and Klebsiella quasipneumoniae, respectively. Patients with hvKp infections had higher proportions of diabetes mellitus (risk ratio [RR], 1.75; 95% confidence interval [CI], 1.05 to 2.94), and their infections had significantly higher propensity to involve pneumonia (RR, 5.85; 95% CI, 1.39 to 24.6), liver abscess (RR, 5.85; 95% CI, 1.39 to 24.6), and disseminated infections (RR, 6.58; 95% CI, 1.16 to 37.4) than infections by other isolates. More than one-half of hvKp infections were health care associated or hospital acquired, and a probable event of health care-associated transmission of hvKp was documented. hvKp isolates, which are significantly associated with severe and disseminated infections, are frequently involved in health care-associated and hospital-acquired infections in Japan.
INTRODUCTION
Klebsiella pneumoniae causes infections mainly in hospitalized patients or patients with underlying medical conditions, but occurrence of severe infections by this organism in healthy community-dwelling persons has been well documented, especially in East and Southeast Asia. The typical presentation of this “invasive syndrome” is liver abscess occasionally accompanied by endophthalmitis or meningitis (1). Detailed analysis of the isolates causing community-acquired liver abscess in Taiwan has led to the first recognition of the presence of hypervirulent K. pneumoniae (hvKp) isolates. Molecular analysis revealed that most of these isolates produced K1 capsular polysaccharides and belong to sequence type (ST) 23 (2, 3).
Severe community-acquired infections caused by K. pneumoniae isolates other than capsular genotype K1 have been reported from a number of countries and have gained attention in recent years (4, 5). While most of these isolates have capsule types classically recognized as high risk for invasive infections (K1, K2, K5, K20, K54, and K57), some of them have capsules of other types (6). Furthermore, the genetic backgrounds are diverse, even among isolates producing the same capsule. For example, hvKp isolates of capsular genotype K2 belong to different STs (e.g., ST65, ST86, and ST380) (4, 7). In addition to liver abscess, clinical presentations of severe infections caused by hvKp include necrotizing fasciitis, osteomyelitis, and meningitis and are sometimes accompanied by disseminated infections (5, 8, 9). hvKp has also been associated with community-acquired pneumonia with bacteremia (10, 11).
A common feature of various clones of hvKp is carriage of virulence genes such as rmpA, rmpA2, and those encoding siderophore biosynthesis. Carriage of rmpA and rmpA2 has been associated with hypermucoviscosity of bacterial colonies on agar plates, a frequent feature of hvKp that originally triggered attention to the presence of K1-ST23 isolates in Taiwan (12, 13). Siderophores are small molecules with a high affinity for iron and are common virulence factors of pathogenic bacteria. Among the various siderophores K. pneumoniae produces, the production of aerobactin and salmochelin appears to be specific to hvKp (14). These cardinal virulence genes were found on the prototypical hvKp plasmid pLVPK, carried by the K. pneumoniae CG43 strain (15). Despite the diversity of genetic backgrounds among hvKp isolates, they commonly carry plasmids resembling pLVPK (16). Virulence genes, such as rmpA-rmpA2, genes for aerobactin biosynthesis, and genes for salmochelin biosynthesis, have been found to be sensitive and specific biomarkers to identify hvKp compared with the string test, which has been commonly employed to identify isolates with hypermucoviscosity (6).
While these studies have advanced our understanding of the pathogenesis and epidemiology of hvKp, data regarding health care-associated and hospital-acquired infections caused by hvKp remain limited. To address this knowledge gap, we systematically analyzed the clinical and molecular characteristics of K. pneumoniae isolates causing bloodstream infections at hospitals across Japan.
(This study was presented in part at the 55th Interscience Conference on Antimicrobial Agents and Chemotherapy, October 2015, San Diego, CA.)
MATERIALS AND METHODS
Ethics.
The protocols were approved by the institutional review boards of all participating hospitals, including the coordinating hospital (Cancer Institute Hospital, Japanese Foundation for Cancer Research; approval number 2013-1063). Informed consent to participate in the study was obtained from the patients or their proxies (if the patient had significant altered mental status or had expired).
Participants.
Patients 20 years of age or older at 23 acute care hospitals in Japan from December 2013 through March 2014 whose blood cultures grew K. pneumoniae were prospectively and sequentially recruited for this cross-sectional study. These hospitals were located in seven geographic regions across the country (see Table S1 in the supplemental material). K. pneumoniae was identified by automated systems at each hospital. For patients with multiple episodes of K. pneumoniae bloodstream infections during the study period, only the first episode was included.
Clinical data collection.
Infectious disease specialists at each participating hospital collected the following clinical information of the patients from medical records: age; sex; severity of underlying diseases according to the Charlson Comorbidity Index (17); immunocompromised conditions (see Table S2); source of bloodstream infections defined by culture results of relevant clinical samples (definite), specific radiographic or laboratory findings other than cultures (probable), or suggestive clinical signs combined with risk factors for urinary tract or biliary tract infections (possible); presence of disseminated infection defined by multiple organ involvement; presence of severe sepsis or septic shock according to sepsis-2 criteria (18); and 30-day crude mortality. The origins of infections were classified into community acquired, health care associated, or hospital acquired. Hospital-acquired infection was defined as infection documented by a positive blood culture obtained more than 48 h after hospital admission, health care-associated infection was defined as infection documented by a positive blood culture obtained on an outpatient basis or within 48 h of admission from a patient with recent history of medical care (e.g., hospitalization in an acute care hospital for 2 or more days within 90 days, receipt of intravenous chemotherapy within 30 days, or residence in a nursing home or long-term-care facility), and community-acquired infection was defined as infection documented by a positive blood culture obtained on an outpatient basis or within 48 h of admission from a patient who did not fulfill the criteria for a health care-associated infection (19).
Microbiological and molecular analyses.
K. pneumoniae isolates from each hospital were collected and analyzed at a central research laboratory for the study. The string test was performed with a standard inoculation loop to evaluate the hypermucoviscosity, and the formation of viscous strings >5 mm was regarded as positive (12).
The genomes of all study isolates were sequenced and assembled according to the methods detailed in the supplemental text. Identification of bacterial species and capsular genotyping were performed with KmerFinder and Kaptive web, respectively (supplemental text). Multilocus sequence typing and identification of β-lactamase genes and virulence genes were implemented on the Klebsiella locus/sequence definitions database (https://bigsdb.pasteur.fr/klebsiella/). Characteristics of β-lactamase genes were queried on the Comprehensive Antibiotic Research Database (20). Of the K. pneumoniae isolates confirmed by KmerFinder, those carrying any of the virulence genes, rmpA, rmpA2, iroBCDN (salmochelin siderophore biosynthesis), iucABCD (aerobactin siderophore biosynthesis), and iutA (aerobactin transporter), were defined as hvKp and others were defined as classic K. pneumoniae (6, 14). Single-nucleotide polymorphism (SNP) identification was performed for isolates of K1-ST23 and K62-ST36 with the methods detailed in the supplemental text.
Statistical analysis.
Continuous data are expressed as medians with interquartile ranges. Categorical variables are shown as proportions. Sensitivity, specificity, positive predictive value, and negative predictive value of the string test for identification of hvKp were computed with 95% confidence intervals (CIs). Bivariate analyses of the clinical characteristics of the patients with hvKp infection and those with infection by other isolates were performed, and risk ratios (RRs) and 95% CIs were calculated. EZR (ver. 1.37) was used for the statistical analysis (21).
Accession number(s).
All genome sequences have been deposited in the NCBI database under BioProject accession number PRJDB7163.
RESULTS
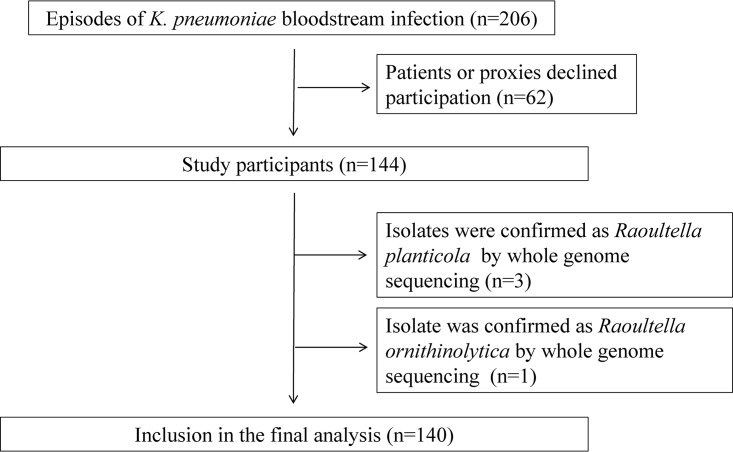
Of the 206 patients with K. pneumoniae bloodstream infections identified during the study period, 144 patients (69.9%) agreed to participate in this study (Fig. 1). The remaining 62 patients (30.1%) declined to participate and were not included in the analysis.
FIG 1.
Patient inclusion.
Species identification.
Assembled genomes of the K. pneumoniae isolates had an average of 75.2 contigs (standard deviation [SD], 66.9) and N50 value of 400,608 bp (SD, 301,862 bp). KmerFinder identified 102 K. pneumoniae, 24 Klebsiella variicola, and 14 Klebsiella quasipneumoniae isolates. Three isolates and one isolate were identified as Raoultella planticola and Raoultella ornithinolytica, respectively, and were excluded from the further analysis. Therefore, 140 cases were included in final analysis (Fig. 1).
Virulence gene profile and hypermucoviscosity.
Of the 102 K. pneumoniae isolates, 26 isolates (25.5%) were hvKp, and the remaining 76 isolates (74.5%) were classic K. pneumoniae. Of the 26 hvKp isolates, all carried iroBCDN, 23 (88.5%) carried both iucABCD and iutA, and 22 (84.6%) carried rmpA and/or rmpA2. In addition, two isolates of K. variicola carried rmpA and iroBCDN (see Table S3 in the supplemental material).
The string test was positive for 30 isolates, including 18 hvKp isolates, 9 classic K. pneumoniae isolates, and 3 K. variicola isolates (Table S3). Sensitivity, specificity, positive predictive value, and negative predictive value of string tests for identification of hvKp were 69.2% (95% CI, 48.2 to 85.7%), 89.5% (95% CI, 82.3 to 94.4%), 60.0% (95% CI, 40.6 to 77.3%), and 92.7% (95% CI, 86.2 to 96.8%), respectively.
Clinical characteristics.
The median age of the 140 patients was 74 years (interquartile range, 67 to 81 years), and 87 patients (62.1%) were male. Fifty-five cases (39.3%) were community acquired, 34 cases (24.3%) were health care associated, and 51 cases (36.4%) were hospital acquired. The sources of bloodstream infections were confirmed in 121 cases (86.4%), with definite diagnosis in 59 cases (42.1%), probable diagnosis in 43 cases (30.7%), and possible diagnosis in 19 cases (13.6%). Patient backgrounds and characteristics of infections with respect to the groups of isolates are presented in Tables 1 and 2, respectively. hvKp infections were community acquired in 10 cases (38.5%), health care associated in 8 cases (30.8%), and hospital acquired in 8 cases (30.8%). Classic K. pneumoniae infections were community acquired in 30 cases (39.4%), health care associated in 18 cases (23.7%), and hospital acquired in 28 cases (36.8%). K. variicola infections were community acquired in 9 cases (37.5%), health care associated in 3 cases (12.5%), and hospital acquired in 12 cases (50%). K. quasipneumoniae infections were community acquired in 6 cases (42.9%), health care associated in 5 cases (35.7%), and hospital acquired in 3 cases (21.4%). Detailed characteristics of hvKp bloodstream infections are presented in Table 3.
TABLE 1.
Baseline characteristics of the patients with K. pneumoniae bloodstream infections divided into groups by molecular identification
| Characteristic | Value |
|||
|---|---|---|---|---|
| Hypervirulent K. pneumoniae (n = 26) | Classic K. pneumoniae (n = 76) | K. variicola (n = 24) | K. quasipneumoniae (n = 14) | |
| Age (yrs) (median [IQR]a) | 75.5 (68.5–83.5) | 73 (66.75–80) | 78 (68.5–81.5) | 72 (64.75–80.75) |
| Male sex (n [%]) | 14 (53.8) | 52 (68.4) | 16 (66.7) | 5 (35.7) |
| Isolation setting (n [%]) | ||||
| Community acquired | 10 (38.5) | 30 (39.4) | 9 (37.5) | 6 (42.9) |
| Healthcare associated | 8 (30.8) | 18 (23.7) | 3 (12.5) | 5 (35.7) |
| Hospital acquired | 8 (30.8) | 28 (36.8) | 12 (50) | 3 (21.4) |
| Diabetes mellitus (n [%]) | 12 (46.2) | 21 (27.6) | 6 (25) | 3 (21.4) |
| Solid tumors (n [%]) | 13 (50) | 35 (46.1) | 15 (62.5) | 5 (35.7) |
| Hematological malignancy (n [%]) | 1 (3.8) | 4 (5.3) | 0 (0) | 0 (0) |
| Immunocompromised conditions (n [%]) | ||||
| Neutropenia | 1 (3.8) | 4 (5.3) | 1 (4.2) | 0 (0) |
| Cellular immunodeficiency | 1 (3.8) | 6 (7.9) | 0 (0) | 2 (14.3) |
| Humoral immunodeficiency | 0 (0) | 1 (1.3) | 0 (0) | 0 (0) |
| Charlson score (n [%]) | ||||
| 0 | 3 (11.5) | 8 (10.5) | 2 (8.3) | 2 (14.3) |
| 1–2 | 7 (26.9) | 28 (36.8) | 8 (33.3) | 6 (42.9) |
| 3–4 | 6 (23.1) | 21 (27.6) | 8 (33.3) | 3 (21.4) |
| ≥5 | 10 (38.5) | 19 (25) | 6 (25) | 3 (21.4) |
IQR, interquartile range.
TABLE 2.
Clinical characteristics and outcomes of K. pneumoniae bloodstream infections divided into groups defined by molecular analysis
| Characteristic | No. (%) patients infected with: |
|||
|---|---|---|---|---|
| Hypervirulent K. pneumoniae (n = 26) |
Classic K. pneumoniae (n = 76) |
K. variicola (n = 24) | K. quasipneumoniae (n = 14) | |
| Source of bloodstream infectiona | ||||
| Biliary tract | 6 (23.1) | 32 (42.1) | 8 (33.3) | 8 (57.1) |
| Urinary tract | 8 (30.8) | 22 (28.9) | 6 (25) | 4 (28.6) |
| Intra-abdominal | 1 (3.8) | 3 (3.9) | 2 (8.3) | 1 (7.1) |
| Pneumonia | 4 (15.4) | 3 (3.9) | 0 (0) | 0 (0) |
| Intravenous catheter related | 0 (0) | 3 (3.9) | 2 (8.3) | 1 (7.1) |
| Liver abscess | 4 (15.4) | 2 (2.6) | 1 (4.2) | 0 (0) |
| Endophthalmitis | 2 (7.7) | 0 (0) | 0 (0) | 0 (0) |
| Meningitis | 1 (3.8) | 0 (0) | 0 (0) | 0 (0) |
| Unknown | 3 (11.5) | 11 (14.5) | 5 (20.8) | 0 (0) |
| Disseminated infection | 3 (11.5) | 2 (2.6) | 0 (0) | 0 (0) |
| Severity of infection | ||||
| Severe sepsis | 17 (65.4) | 46 (60.5) | 15 (62.5) | 13 (92.9) |
| Septic shock | 6 (23.1) | 12 (15.8) | 3 (12.5) | 2 (14.3) |
| Altered mental status | 7 (26.9) | 13 (17.1) | 5 (20.8) | 2 (14.3) |
| 30-day crude mortality | 2 (7.7) | 4 (5.3) | 1 (4.2) | 2 (14.3) |
If a patient had multiple infected organs (disseminated infection), all were counted separately.
TABLE 3.
Microbiological, molecular, and clinical characteristics of hypervirulent K. pneumoniae bloodstream infectionsa
|
Strain no. |
Area | Hospital | Age groupb | Settingc |
String test |
K typed | STe | Source of BSIf |
Septic shock |
Disturbed consciousness | 30-day death | Charlson score | Malignancy |
Diabetes mellitus |
Immunosuppression |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14133 | Kanto | C | 80 | CA | + | 1 | 23 | Liver abscess | + | + | − | 1–2 | − | + | − |
| 14056 | Kanto | D | 50 | HC | + | 1 | 23 | Unknown | − | − | − | 1–2 | Endometrial cancer | − | − |
| 14005 | Chubu | I | 60 | HA | − | 1 | 23 | Biliary tract infection | − | − | − | 3–4 | Bile duct cancer | + | − |
| 14036 | Kinki | M | 60 | CA | + | 1 | 23 | Liver abscess | − | − | − | 0 | − | − | − |
| 14041 | Kinki | M | 70 | CA | + | 1 | 23 | Liver abscess, endophthalmitis | − | − | − | 1–2 | − | − | − |
| 14030 | Chugoku | R | 80 | CA | + | 1 | 23 | Biliary tract infection | − | − | − | ≥5 | Bile duct cancer | + | − |
| 14089 | Shikoku | T | 90 | HC | − | 1 | 23 | Urinary tract infection | − | − | − | ≥5 | Renal cell carcinoma | + | Neutropenia |
| 14087 | Tohoku | A | 90 | CA | + | 2 | 65 | Pneumonia | − | + | − | 3–4 | Gastric cancer, bladder cancer | + | − |
| 14106 | Kinki | L | 70 | CA | − | 2 | 65 | Unknown | + | − | − | 3–4 | − | − | − |
| 14139 | Kanto | C | 90 | CA | + | 2 | 86 | Urinary tract infection | − | − | − | 0 | − | − | − |
| 14111 | Kanto | F | 70 | CA | + | 2 | 86 | Biliary tract infection | − | − | − | ≥5 | Pancreatic cancer | + | − |
| 14023 | Kinki | O | 40 | HA | + | 2 | 86 | Pneumonia | − | + | − | 1–2 | Intravascular lymphoma | − | − |
| 14090 | Kyushu | V | 70 | HA | + | 2 | 86 | Urinary tract infection, endocarditis | + | − | − | 3–4 | − | − | − |
| 14123 | Kyushu | U | 80 | HA | + | 20 | 268 | Urinary tract infection | − | − | − | ≥5 | − | + | − |
| 14128 | Kyushu | U | 60 | HA | − | 20 | 268 |
Intra-abdominal infection |
+ | − | − | ≥5 |
Hepatocellular carcinoma |
− | − |
| 14120 | Kanto | H | 70 | CA | − | 20 | 1544 | Urinary tract infection | − | + | − | ≥5 | Colon cancer | + | − |
| 14081 | Tohoku | A | 80 | CA | + | 35 | 1266 | Urinary tract infection | − | − | − | 1–2 | − | + | − |
| 14062 | Kinki | N | 80 | HC | − | 54 | 29 | Urinary tract infection | + | − | − | 1–2 | − | − | − |
| 14059 | Kanto | D | 80 | HA | + | 57 | 218 | Pneumonia | − | + | + | ≥5 | Oropharynx cancer | + | − |
| 14017 | Kanto | G | 60 | HC | + | 57 | 218 | Pneumonia | + | + | + | 1–2 | − | + | − |
| 14098 | Kinki | L | 70 | HC | + | 57 | 218 | Biliary tract infection | − | − | − | 0 | − | − | − |
| 14013 | Chubu | I | 70 | HA | − | 57 | 412 | Liver abscess, meningitis, endophthalmitis |
− | + | − | 3–4 | Bile duct cancer | + | Cellular immunodeficiency |
| 14001 | Chubu | I | 70 | HC | + | 62 | 36 | Unknown | − | − | − | ≥5 | Pancreatic cancer | − | − |
| 14002 | Chubu | I | 70 | HC | + | 62 | 36 | Biliary tract infection | − | − | − | ≥5 | Pancreatic cancer | − | − |
| 14009 | Chubu | I | 60 | HC | + | 62 | 36 | Biliary tract infection | − | − | − | ≥5 | Pancreatic cancer | − | − |
| 14126 | Kyushu | U | 70 | HA | − | 62 | 36 | Urinary tract infection | − | − | − | 3–4 | − | − | − |
Isolates were aligned according to capsular genotype and sequence type.
Ages of the patients were described as groups within a 10-year range (e.g., age group 80 represents ages from 80 through 89).
CA, community acquired; HC, health care associated; HA, hospital acquired.
K type, capsular genotype.
ST, sequence type.
BSI, bloodstream infection.
When comparing patients with hvKp infections and those with infections by other isolates, the hvKp group had a significantly higher rate of diabetes mellitus and had significantly higher propensity to present with pneumonia, liver abscess, and disseminated infections (Table 4). The 30-day crude mortality rates were not significantly different between the two groups.
TABLE 4.
Comparison of clinical characteristics of hypervirulent K. pneumoniae infections and those caused by other isolates
| Clinical characteristic | No. (%) of patients infected with: |
Risk ratio (95% confidence interval)a | |
|---|---|---|---|
| Hypervirulent K. pneumoniae (n = 26) | Other isolates (n = 114) | ||
| Diabetes mellitus as an underlying condition | 12 (46.2) | 30 (26.3) | 1.75 (1.05–2.94) |
| Pneumonia | 4 (15.4) | 3 (2.6) | 5.85 (1.39–24.6) |
| Liver abscess | 4 (15.4) | 3 (2.6) | 5.85 (1.39–24.6) |
| Disseminated infections | 3 (11.5) | 2 (1.8) | 6.58 (1.16–37.4) |
| Septic shock | 6 (23.1) | 17 (14.9) | 1.55 (0.68–3.54) |
| Altered mental status | 7 (26.9) | 20 (17.5) | 1.54 (0.73–3.24) |
| 30-day crude mortality | 2 (7.7) | 7 (6.1) | 1.25 (0.28–5.69) |
Risk ratio of hypervirulent K. pneumoniae infections to have each characteristic compared with those caused by other isolates.
Two K. variicola isolates carrying rmpA and iroBCDN caused intravenous catheter-associated bloodstream infections with septic shock and bacteremic urinary tract infection, both hospital acquired (Table 5).
TABLE 5.
Microbiological, molecular, and clinical characteristics of K. variicola bloodstream infectionsa
| Strain no. | Area | Hospital | Age groupb | Settingc | String test | K typed | STe | Source of BSIf | Septic shock | Disturbed consciousness | 30-day death | Charlson score | Malignancy |
Diabetes mellitus |
Immunosuppression |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14028 | Chugoku | R | 80 | HA | − | 2 | 919 | Intravenous catheter-related infection |
+ | − | + | 3–4 | − | − | − |
| 14085 | Tohoku | A | 70 | HA | + | 2 | 919 | Urinary tract infection | − | − | − | ≥5 | Colon cancer | − | − |
| 14037 | Kinki | M | 80 | CA | + | 10 | 3954 | Biliary tract infection | − | − | − | 12 | − | − | − |
| 14011 | Chubu | I | 80 | HA | − | 16 | 3950 | Intravenous catheter-related infection |
− | − | − | 3–4 | Oral cavity cancer | − | − |
| 14012 | Chubu | I | 80 | CA | − | 16 | 3950 | Biliary tract infection | + | + | − | 3–4 | Pancreatic cancer | − | − |
| 14006 | Chubu | I | 50 | HC | − | 18 | 1903 | Biliary tract infection | − | − | − | 1–2 | Pancreatic cancer | − | − |
| 14007 | Chubu | I | 60 | HA | − | 18 | 1903 | Intra-abdominal infection | − | − | − | 1 − 2 | Gastric cancer | − | − |
| 14078 | Tohoku | B | 60 | HA | − | 19 | 3956 | Unknown | − | + | − | 3 − 4 | Lung cancer | − | Neutropenia |
| 14134 | Kanto | C | 80 | HC | − | 24 | 3961 | Urinary tract infection | − | + | − | ≥5 | − | − | − |
| 14035 | Kinki | M | 80 | CA | − | 30 | 3953 | Biliary tract infection | − | − | − | 1 − 2 | − | − | − |
| 14025 | Kinki | O | 70 | HA | − | 34 | 363 | Urinary tract infection | − | − | − | ≥5 | Colon cancer | + | − |
| 14136 | Kanto | C | 70 | HA | − | 47 | 3962 | Urinary tract infection | − | − | − | 1 − 2 | Lung cancer | − | − |
| 14077 | Tohoku | B | 70 | HA | − | 57 | 3955 | Biliary tract infection | − | − | − | 3 − 4 | Colon cancer | − | − |
| 14010 | Chubu | I | 60 | HA | − | 60 | 697 | Unknown | − | − | − | 1–2 | Bile duct cancer | − | − |
| 14021 | Kyushu | W | 80 | CA | − | 64 | 3951 | Urinary tract infection | − | + | + | 0 | − | − | − |
| 14109 | Kanto | F | 70 | CA | − | 64 | 3959 | Intra-abdominal infection | − | − | − | ≥5 | Colon cancer | − | − |
| 14103 | Kinki | L | 70 | HC | − | 105 | 1142 | Urinary tract infection | − | − | − | ≥5 | − | + | − |
| 14033 | Kinki | M | 30 | CA | − | 114 | 3952 | Biliary tract infection | − | − | − | 1 − 2 | − | − | − |
| 14096 | Kinki | L | 70 | CA | − | 125 | 355 | Liver abscess | − | − | − | 3 − 4 | Thyroid cancer | + | − |
| 14079 | Tohoku | B | 70 | HA | − | 125 | 3957 | Unknown | − | − | − | 3 − 4 | Lung cancer, oral cavity cancer | + | − |
| 14115 | Chubu | J | 80 | CA | + | 125 | 3960 | Biliary tract infection | − | − | − | ≥5 | − | + | − |
| 14058 | Kanto | D | 60 | HA | − | 127 | 3239 | Unknown | + | + | − | 3 − 4 | Gastric cancer | + | − |
| 14086 | Tohoku | A | 70 | HA | − | UD | 3958 | Unknown | − | − | − | 1 − 2 | Gastric cancer | − | − |
| 14095 | Kinki | L | 60 | CA | − | UD | 919 | Biliary tract infection | − | − | − | 0 | − | − | − |
Isolates were aligned according to capsular genotype and sequence type.
Ages of the patients were described as groups within a 10-year range (e.g., age group 80 represents ages from 80 through 89).
CA, community acquired; HC, health care associated; HA, hospital acquired.
K type, capsular genotype; UD, undetermined with Kaptive database.
ST, sequence type.
BSI, bloodstream infection.
β-Lactamase genes.
All except one K. pneumoniae isolate carried blaSHV, all K. variicola isolates carried blaLEN, and all K. quasipneumoniae isolates carried blaOKP (Table S3). Twelve K. pneumoniae isolates carried broad-spectrum β-lactamase genes, including those encoding SHV-type extended-spectrum β-lactamase (ESBL) (n = 5), CTX-M-type ESBL (n = 4), CMY-type cephalosporinase (n = 1), and both carbapenemase and CTX-M-type ESBL genes (n = 2). The carbapenemase genes were blaIMP-6 (n = 1) and blaGES-4 (n = 1). While one hvKp isolate carried blaCTX-M-2, all other isolates with β-lactamase genes for expanded spectrum of activity were classic K. pneumoniae.
Capsular genotype and clonality of the isolates.
The isolates were classified into a number of clones defined by capsular genotypes and STs, and isolates of a given ST had a corresponding specific capsular genotype with a few exceptions (Table S3). Of the hvKp clones, K1-ST23 (n = 7), K2-ST65 (n = 2), K2-ST86 (n = 4), K20-ST268 (n = 2), K57-ST218 (n = 3), and K62-ST36 (n = 4) were the common combinations (Table 3). Three sets of distinct capsular genotype and ST combinations were recovered from the patients at the same hospitals: three K62-ST36 isolates from health care-associated infections, two K1-ST23 isolates from community-acquired infections, and two K20-ST268 isolates from hospital-acquired infections. The K62-ST36 isolates and K1-ST23 shared identical profiles of β-lactamase genes, whereas the two K20-ST268 isolates had different profiles. Notably, a pair of K62-ST36 isolates (strain numbers 14001 and 14002) were genetically almost identical with only one core genome SNP, while other pairs had greater core genome SNPs ranging from 93 to 108. The core genome SNP numbers of seven K1-ST23 isolates, including those recovered in the same hospital, ranged between 165 and 399.
Classic K. pneumoniae, K. variicola, and K. quasipneumoniae had diverse capsular genotypes and STs (Table S3). Of interest, the two K. variicola isolates carrying rmpA and iroBCDN both belonged to ST919 and had capsular genotype K2.
DISCUSSION
In this study, we analyzed the clinical and molecular characteristics of K. pneumoniae bloodstream infections in Japan. Twenty-six of the 140 K. pneumoniae bloodstream infections were caused by hvKp identified by several molecular biomarkers. hvKp isolates consisting of various clones were distributed across the country.
Of the hvKp isolates identified, K1-ST23 isolates were most common, which was consistent with previous studies in Japan and other Asian countries (22, 23). Nevertheless, various other clones were also observed, including established hvKp clones such as K2-ST86 and previously unrecognized clones such as K62-ST36. Minor differences in the patterns of carriage of virulence genes could be explained by probable variations in the pLVPK-like virulence plasmid due to insertions and deletions, which are known to occur (16, 24). Higher risk of hvKp infection in patients with diabetes and its propensity to cause pneumonia, liver abscess, and disseminated infections have been reported in infections caused by K1 isolates, hypermucoviscous isolates, and isolates carrying rmpA (10, 25, 26). These clinical characteristics were preserved in our hvKp isolates that were defined by a more comprehensive set of biomarkers. This may suggest that identification of hvKp among blood isolates using biomarkers may be able to guide comprehensive screening for disseminated infection and implementation of antibiotic therapy with good tissue and central nervous system penetration. While the string test is a popular method to identify hvKp due to its simplicity to perform in the clinical microbiology laboratory, data from this report and others demonstrate that its sensitivity and specificity are not optimal, and the test would not be adequate for standalone use in the identification of hvKp (6).
Healthcare-associated and hospital-acquired hvKp infections were common in our study. Previous studies have presented severe community-acquired infections in relatively healthy persons as the typical feature of hvKp infections (10, 11). Nevertheless, it appears conceivable that hvKp isolates would also pose a high risk for severe infections among hospitalized patients or patients with comorbidities. It is also possible that hvKp infections occur as health care-associated and hospital-acquired infections commonly in countries with demographic and public health characteristics similar to those of Japan.
K62-ST36 was newly recognized as an hvKp clone in this study. A pair of K62-ST36 isolates, which were isolated from the same hospital as health care-associated infections, had only single SNPs in their core genomes. While the route of transmission was unclear, transmission during hospitalization was a distinct possibility, since both patients had pancreatic cancer and were cared for at the same hospital. Considering only clinical isolates from blood were studied in our study, this possible in-hospital transmission might suggest ongoing transmission of hvKp in health care settings that are currently unrecognized. Since transmission and subsequent infection caused by hvKp could pose significant risk of adverse outcome in debilitated inpatients, aggressive prevention measures, such as implementation of contact precaution for patients colonized with hvKp, may be considered.
Although spread of multidrug-resistant hvKp is an emerging problem globally, only one hvKp isolate carried an ESBL gene, and none carried a carbapenemase gene in this study (24, 27). Nevertheless, carbapenemase-producing hvKp was recently reported in Japan, and the future trend of antimicrobial resistance among hvKp isolates requires close monitoring (28).
Of 140 isolates identified as K. pneumoniae by automated methods, 24 and 14 were molecularly identified as K. variicola and K. quasipneumoniae, respectively. These species are difficult to differentiate from K. pneumoniae by conventional biochemical methods (29). Carriage of blaLEN by K. variicola isolates and blaOKP by K. quasipneumoniae isolates was in accordance with previous reports and supports the results of KmerFinder (29). Clinical significance of the carriage of virulence genes, rmpA and iroBCDN, found in two K. variicola ST919 isolates is unknown thus far. A single-hospital study showed a higher mortality rate of bloodstream infections caused by K. variicola than those caused by K. pneumoniae, and together with our findings might suggest the clinical significance of K. variicola as a distinct virulent species within the genus Klebsiella (30).
Our study has several limitations. First, this study was conducted in a single country. hvKp infections in other countries may have different clinical characteristics, especially if hvKp clones with distinct characteristics were involved. Nevertheless, the consistency of clinical characteristics of hvKp infections in our study with those reported in previous studies suggests the generalizability of the findings. Second, the limited number of hvKp isolates may have resulted in missed characteristics. Furthermore, clinical characteristics of K. variicola and K. quasipneumoniae infections could not be adequately assessed in our analysis due to the limited number of cases. Third, the use of whole-genome sequencing to identify hvKp applied in this study is difficult to employ in general clinical settings.
In conclusion, hvKp bloodstream infections, which were identified by molecular biomarkers, showed distinct clinical characteristics. Notably, health care-associated and hospital-acquired infections accounted for the majority of hvKp infections. The predominance of health care-associated and hospital-acquired hvKp infections observed in this study challenges the current paradigm of hvKp focused on community-acquired infections and suggests the need for further studies on diagnosis, treatment, and infection prevention of hvKp infections in health care settings.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research of Ministry of Education, Culture, Sports, Science and Technology, Japan (grant number 23791143).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We declare no conflict of interest relevant to the study.
We thank Koh Okamoto for helpful discussion and Noboru Hagino, Yoshiro Hayashi, and Ryosuke Osawa for their advice on the design of the research. We also thank Ayumi Yoshizumi, Junko Maeda, Hiroko Suzuki, Hisakazu Yano, Michi Shoji, Norio Ohmagari, Akihiko Hayashi, Norio Ikeda, and Kenichi Hayashi for their assistance in collecting clinical information and the isolates.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01206-19.
REFERENCES
- 1.Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wang JH, Wann SR, Lin HH. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 2.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 3.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decré D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, Offenstadt G, Maury E, Brisse S, Arlet G. 2011. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 49:3012–3014. doi: 10.1128/JCM.00676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki Y, Inokuchi R, Harada S, Aoki K, Ishii Y, Shinohara K. 2017. Bacterial meningitis caused by hypervirulent Klebsiella pneumoniae capsular genotype K54 with development of granuloma-like nodal enhancement in the brain during the subacute phase. Intern Med 56:373–376. doi: 10.2169/internalmedicine.56.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR, Backer M, Bajwa R, Catanzaro AT, Crook D, de Almeda K, Fierer J, Greenberg DE, Klevay M, Patel P, Ratner A, Wang J-T, Zola J. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 55:930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 9.Patel PK, Russo TA, Karchmer AW. 2014. Hypervirulent Klebsiella pneumoniae. Open Forum Infect Dis 1:ofu028. doi: 10.1093/ofid/ofu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ. 2007. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 13:986–993. doi: 10.3201/eid1307.070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. 2011. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157:3446–3457. doi: 10.1099/mic.0.050336-0. [DOI] [PubMed] [Google Scholar]
- 14.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 18.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. 2003. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Intensive Care Med 29:530–538. doi: 10.1007/s00134-003-1662-x. [DOI] [PubMed] [Google Scholar]
- 19.Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ. 2002. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137:791–797. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 20.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanda Y. 2013. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, Ip M. 2011. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49:3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada S, Ishii Y, Saga T, Aoki K, Tateda K. 2018. Molecular epidemiology of Klebsiella pneumoniae K1 and K2 isolates in Japan. Diagn Microbiol Infect Dis 91:354–359. doi: 10.1016/j.diagmicrobio.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 25.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 26.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. 2006. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42:1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Sun G, Yu Y, Li N, Chen M, Jin R, Jiao Y, Wu H. 2014. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 58:225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 28.Harada S, Aoki K, Ishii Y, Ohno Y, Nakamura A, Komatsu M, Tateda K. 2019. Emergence of IMP-producing hypervirulent Klebsiella pneumoniae carrying a pLVPK-like virulence plasmid. Int J Antimicrob Agents 53:873–875. doi: 10.1016/j.ijantimicag.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290-17. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maatallah M, Vading M, Kabir MH, Bakhrouf A, Kalin M, Nauclér P, Brisse S, Giske CG. 2014. Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS One 9:e113539. doi: 10.1371/journal.pone.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.