Susceptibility testing of the polymyxins (colistin and polymyxin B) is challenging for clinical laboratories. The Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Subcommittee evaluated two methods to enable accurate testing of these agents. These methods were a colistin broth disk elution (CBDE) and a colistin agar test (CAT), the latter of which was evaluated using two inoculum volumes, 1 μl (CAT-1) and 10 μl (CAT-10).
KEYWORDS: Acinetobacter, colistin, Enterobacterales, Pseudomonas aeruginosa, susceptibility testing, agar dilution, broth microdilution, colistin broth disk elution
ABSTRACT
Susceptibility testing of the polymyxins (colistin and polymyxin B) is challenging for clinical laboratories. The Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Subcommittee evaluated two methods to enable accurate testing of these agents. These methods were a colistin broth disk elution (CBDE) and a colistin agar test (CAT), the latter of which was evaluated using two inoculum volumes, 1 μl (CAT-1) and 10 μl (CAT-10). The methods were evaluated using a collection of 270 isolates of Enterobacterales, 122 Pseudomonas aeruginosa isolates, and 106 Acinetobacter spp. isolates. Overall, 94.4% of CBDE results were in essential agreement and 97.9% in categorical agreement (CA) with reference broth microdilution MICs. Nine very major errors (VME; 3.2%) and 3 major errors (ME; 0.9%) were observed. With the CBDE, 98.6% CA was observed for Enterobacterales (2.5% VME, 0% ME), 99.3% CA was observed for P. aeruginosa (0% VME, 0.7% ME), and 93.1% CA was observed for Acinetobacter spp. (5.6% VME, 3.3% ME). Overall, CA was 94.9% with 6.8% VME using CAT-1 and improved to 98.3% with 3.9% VME using CAT-10. No ME were observed using either CAT-1 or CAT-10. Using the CAT-1/CAT-10, the CA observed was 99.4%/99.7% for Enterobacterales (1%/0.5% VME), 98.7%/100% for P. aeruginosa (8.3%/0% VME), and 88.5%/92.3% for Acinetobacter spp. (21.4%/14.3% VME). Based on these data, the CLSI antimicrobial susceptibility testing (AST) subcommittee endorsed the CBDE and CAT-10 methods for colistin testing of Enterobacterales and P. aeruginosa.
INTRODUCTION
Use of polymyxin drugs (colistin and polymyxin B) has reemerged over the past decade as salvage treatment of multidrug-resistant (MDR) Gram-negative infections (1). While significant data are now available that bring into question the clinical and pharmacokinetic/pharmacodynamic (PK/PD) efficacy of these agents (2–6), their use continues either due to the lack of active alternative antimicrobials (for example, metallo-beta-lactamase producers) or due to the lack of availability and high cost of newer antimicrobial agents with specific activity against MDR infections. High rates of renal and central nervous system toxicity are associated with polymyxin use, even at optimal doses (2). As such, knowledge of an infecting organism’s MIC and categorical interpretation is of significant clinical value when using these toxic agents (2). In 2019, the Clinical and Laboratory Standards Institute (CLSI) approved clinical breakpoints for colistin and polymyxin B for the Enterobacterales and revised the Pseudomonas aeruginosa and Acinetobacter spp. breakpoints (Table 1). CLSI set intermediate and resistant interpretive categories but no susceptible category for these drugs. The rationale behind this decision is summarized elsewhere (CLSI meeting minutes, posted at https://clsi.org/meetings/ast/ast-meeting-files-resources/) and was based primarily on the absence of existing clinical and PK/PD evidence demonstrating that any colistin or polymyxin B MIC predicts clinical efficacy and should not be labeled as “susceptible.”
TABLE 1.
Colistin and polymyxin B breakpoints approved in June 2019 by the CLSI Subcommittee for Antimicrobial Susceptibility Testing for publication in document M100, 30th ed., January 2020a
| Organism | Susceptibleb | Intermediate | Resistant |
|---|---|---|---|
| Enterobacterales | — | ≤2 μg/ml | ≥4 μg/ml |
| P. aeruginosa | — | ≤2 μg/ml | ≥4 μg/ml |
| Acinetobacter spp. | — | ≤2 μg/ml | ≥4 μg/ml |
Colistin and polymyxin B are considered equivalent agents, so MICs obtained from testing colistin predict MICs to polymyxin B, and vice versa.
—, no susceptible category defined.
From a testing perspective, clinical laboratories have struggled to perform polymyxin susceptibility testing (7, 8). No U.S. Food and Drug Administration (FDA)-cleared tests exist for colistin or polymyxin B, as the FDA does not recognize any clinical breakpoints for the polymyxins. The analytical performance of research-use-only disks and gradient strips has been poor (8), and CLSI and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) agree that the only validated test method for the polymyxins is reference broth microdilution (rBMD), which is performed in very few clinical laboratories. The CLSI convened an ad hoc working group (ahWG) to address the testing challenges of the polymyxins. This group evaluated two MIC-based alternative methods for testing colistin, a colistin broth disk elution (CBDE) and a colistin agar test (CAT). The rationale for evaluating these tests was not to develop a more accurate method but, rather, to develop a method as accurate as rBMD that was more amenable to routine clinical laboratory use, i.e., that uses readily available materials and is of low complexity to perform. The CBDE uses commercially available colistin disks, which are added to commercially available prealiquoted cation-adjusted Mueller-Hinton broth (CA-MHB) to achieve a defined concentration of colistin. The CAT is an agar screen plate, which the committee evaluated with the intent that it would be made commercially available by multiple manufacturers. The results from the studies conducted by the ahWG are summarized herein.
MATERIALS AND METHODS
Study design.
CBDE and CAT were evaluated against rBMD in a study designed to meet the CLSI standard M23, 5th edition, criteria for validating alternative reference susceptibility tests (9). Targeted intermethod correlation between CBDE/CAT and rBMD was ≥90% essential agreement (EA) across the clinical isolates evaluated.
Testing was performed across three laboratories using four methods in each laboratory, CBDE, CAT using a 1-μl inoculum (CAT-1), CAT using a 10-μl inoculum (CAT-10), and rBMD, each of which is described in detail below. Given the challenges associated with obtaining precise colistin MICs, rBMD was performed in triplicate for each isolate, using three different brands of cation-adjusted Mueller-Hinton broth (CA-MHB). For each isolate evaluated, a single 0.5 McFarland inoculum was prepared and used to inoculate all four tests in parallel. Each laboratory tested the same panel of challenge isolates to assess interlaboratory agreement, as well as a collection of their own stocked clinical isolates. Challenge isolates were also included in the overall accuracy calculations. For these isolates, the CBDE and CAT results obtained at each site were scored against the reference MIC obtained by that same site.
Reference results were initially defined as the mode of 3 rBMD results. However, as described further in the results section, one of the CA-MHB brands yielded results out of quality control (QC) for Pseudomonas aeruginosa ATCC 27853 and had to be removed from the calculations, yielding two rBMD MICs for each isolate. As a result, the reference MIC definition was modified to take the average of the two rBMD results, which was arbitrarily rounded down to the nearest 2-fold (log2) dilution MIC. If an isolate’s two rBMD results were not in categorical agreement (CA) with each other, testing was repeated by all methods. If results were out of CA a second time, the isolate was excluded from analysis due to the inability to obtain a reference MIC by which to compare results. If the isolate demonstrated skipped wells by rBMD test, testing was repeated with all tests (10). If the skipping phenomenon was observed a second time, the isolate was excluded from the final analysis due to the absence of a reference MIC.
All methods were repeated by the same laboratory to rule out random testing error for each isolate that yielded one or more CBDE or CAT results out of CA with the reference rBMD. Similarly, if skipped dilutions were observed with either CBDE or CAT for an isolate, testing was repeated by all methods. If the repeat tests confirmed initial observations, the results were confirmed as a CA error or an uninterpretable result. If repeat testing did not confirm initial observations, the first set of results was removed from the final analysis.
CA was calculated using the recently approved CLSI colistin breakpoints (Table 1). Given that there is no susceptible category, results that were defined as “intermediate” were treated as “susceptible” for the purpose of performance calculations. In other words, a result of intermediate by rBMD but resistant by CBDE or CAT was considered a major error (ME), and a result of resistant by rBMD but intermediate by CBDE or CAT was considered a very major error (VME). Traditionally, these would be considered minor errors. We elected to evaluate the results in this manner (i) because binary interpretive categories (I/R) exist for the polymyxins, (ii) because the polymyxins are already dosed to achieve maximum safe exposure levels, so no alternative dosing regimens can be used to make up for testing errors, and (iii) because of the narrow margin to achieve an efficacious exposure of polymyxin drug that is predicted by PK/PD if the MIC is 2 μg/ml, even with optimized dosing (2).
CBDE method.
Colistin broth disk elution was performed as described elsewhere with some minor modifications (11). Briefly, 10-μg colistin disks (BD, Sparks, MD) were added to borosilicate tubes containing 25 ml or 10 ml CA-MHB (Remel, Lenexa, KS) to make 0 μg/ml (0 disks in 10 ml), 0.4 μg/ml (1 disk in 25 ml), 1 μg/ml (1 disk in 10 ml), 2 μg/ml (2 disks in 10 ml), and 4 μg/ml (4 disks in 10 ml) final concentrations. The atypical 0.4 μg/ml was used, as 20-ml CA-MHB tubes are not commercially available, and the ahWG was attempting to replicate scenarios that would be typically encountered in clinical practice. Disks were allowed to elute for a minimum of 30 minutes at room temperature, and 125 μl of 0.5 McFarland suspension was added to the 25-ml tubes and 50 μl of inoculum was added to the 10-ml tubes, such that a final concentration of 7.5 × 105 CFU/ml bacteria was achieved. Tubes were vortexed on low speed briefly and incubated at 35°C in ambient air for 16 to 20 h for Enterobacterales and P. aeruginosa and for 20 to 24 h for Acinetobacter spp. MICs were read by visual inspection as the lowest concentration that completely inhibited growth of the test isolates.
CAT method.
Colistin agar plates were made by Hardy Diagnostics (Santa Ana, CA). Briefly, these contained 0.25 to 4 μg/ml colistin in Mueller-Hinton Agar (MHA) from BD, Remel, or Hardy. The medium brands were rotated through each testing site, with the exception of one site that tested on all three brands in parallel. Each agar plate in the series (0 to 4 μg/ml) was inoculated with a 1-μl loop and a 10-μl inoculum using either a pipette or a loop of a 1:10 dilution of a 0.5 McFarland suspension of organism. Plates were incubated at 35°C in ambient air for 16 to 20 h for Enterobacterales and P. aeruginosa and for 20 to 24 h for Acinetobacter spp. Any visible growth was read as positive; MICs were determined as the lowest concentration that did not display visible growth.
rBMD method.
Broth microdilution panels were made by Accelerate Diagnostics (Tucson, AZ) and contained 0.25 to 16 μg/ml colistin (Sigma-Aldrich, St. Louis, MO). CA-MHB from Oxoid, BD, and Sigma-Aldrich was used, such that each plate included a control well (0 μg/ml colistin), and colistin test wells spanned the full dilution range with each medium. rBMD panels were frozen, shipped on dry ice, and stored at –80°C at each site prior to use. Prior to shipping, the panels were tested using the quality control (QC) strains P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 in triplicate from panels from the beginning, middle, and end of the rBMD fill run to confirm that each sampling yielded results with these strains that were in QC.
QC testing.
Because E. coli ATCC 25922 yields results off-scale for the tests under evaluation, QC was routinely assessed during the study using the P. aeruginosa ATCC 27853 strain with the expected ranges as published by CLSI (i.e., 0.5 to 4 μg/ml for colistin). In addition, a supplemental QC strain from the CDC and FDA Antibiotic Resistance (AR) Bank was evaluated, Escherichia coli AR Bank no. 0349. This strain was chosen as it was predicted to give on-scale results of 2 to 4 μg/ml by all methods and harbors the mcr-1 gene. Each laboratory generated results for each QC strain, from 13 to 15 times, once per testing day.
Bacterial isolates.
Two sets of isolates were evaluated in this study. The first consisted of a challenge set comprising isolates from each testing laboratory, Accelerate Diagnostics, JMI Laboratories, and the CDC and FDA AR Bank. These isolates were selected based on elevated MICs to colistin, as shown in Fig. S1 in the Supplemental Material. Challenge isolates were collected and shipped to Accelerate Diagnostics to make master stocks which were coded and distributed to each testing laboratory. The exception to this was the CDC and FDA AR Bank isolates, which were requested from the CDC and shipped to each laboratory directly. The second isolate set consisted of stock clinical isolates (referred to herein as stock isolates) from each individual clinical laboratory and tested at that single laboratory for the study. These isolates were collected over time from these institutions and selected as those that either had been previously tested for colistin MICs on request of treating physicians or displayed the MDR phenotypes which might have colistin testing requested. The isolates are summarized in Table 2. Seven isolates, all E. coli, harbored the mcr-1 gene. Prior to testing, each isolate was subcultured from –80°C frozen stocks to a blood agar plate. A colistin disk (BD) was placed in the first quadrant, and a second subculture was performed from growth around the disk to a new blood agar plate.
TABLE 2.
Isolates used in this study
| Species | No. of challenge isolates (no. of results across 3 sites) | No. of stock isolates | No. of resistant results | No. of excluded resultsa |
|---|---|---|---|---|
| Klebsiella aerogenes | 0 (0) | 5 | 0 | 0 |
| Enterobacter cloacae | 6 (18) | 20 | 23 | 0 |
| Escherichia coli | 10 (30) | 62 | 33 | 0 |
| Citrobacter spp. | 1 (3) | 7 | 0 | 0 |
| Klebsiella pneumoniae | 26 (78) | 51 | 90 | 5 |
| Klebsiella spp. | 0 | 72 | 46 | 3 |
| Enterobacter spp. | 0 | 6 | 1 | 0 |
| Providencia stuartii | 0 | 1 | 1 | 0 |
| Serratia spp. | 0 | 1 | 1 | 0 |
| Morganella morganii | 0 | 2 | 2 | 0 |
| Total Enterobacterales | 43 (129) | 227 | 196 | 8 |
| Pseudomonas aeruginosa | 14 (42) | 108 | 13 | 1 |
| Acinetobacter spp. | 21 (63) | 85 | 71 | 17 |
Isolates were excluded if a reproducible rBMD result was not obtained. No mcr-expressing isolates were excluded.
RESULTS
rBMD.
Three brands of medium were used to perform rBMD, one of which (Oxoid) consistently yielded results out of QC range (≤0.25 μg/ml) for P. aeruginosa ATCC 27853. All results were within QC for E. coli ATCC 25922, which was assessed at the time of panel manufacturing. As a result, MICs obtained with this brand of CA-MHB were excluded from the remainder of the study. In contrast, MIC results were within QC for 98.8% of tests using CA-MHB from Sigma and BD.
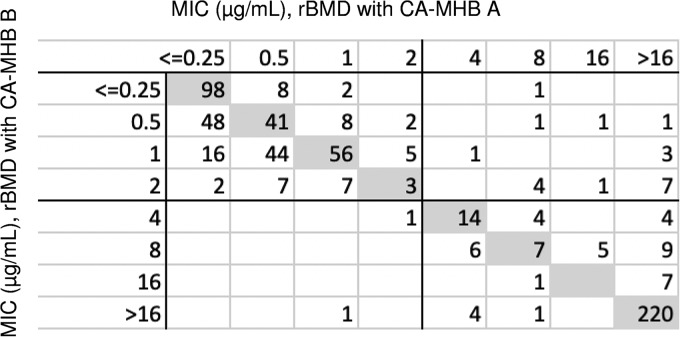
Colistin MICs obtained by Sigma were compared to those obtained by BD CA-MHB. In general, results showed good CA (Fig. 1), with 629 of the 651 results in CA (96.6%) and 67.4% of the results yielding the same MIC (i.e., absolute agreement [AA]). The majority of isolates with results out of CA were Acinetobacter spp. (n = 15, 10.1% of all Acinetobacter spp. results). Only 1.1% and 1.3% of Enterobacterales and P. aeruginosa, respectively, had rBMD results out of CA (not shown). Seventeen (11.5%) A. baumannii isolates were excluded from the study due to either no reference MIC (n = 15) or repeated skipping by rBMD (n = 2). Two (0.6%) P. aeruginosa isolates were excluded for CA errors between the two rBMD results, as were 5 Klebsiella spp. isolates. An additional 3 Klebsiella spp. isolates were excluded due to skipping, for an overall 2.9% of Enterobacterales excluded. Reference MICs calculated from the two rBMD MICs are shown in Fig. S1.
FIG 1.
Scatterplot for reference broth microdilution MICs obtained using two different brands of CA-MHB.
CBDE.
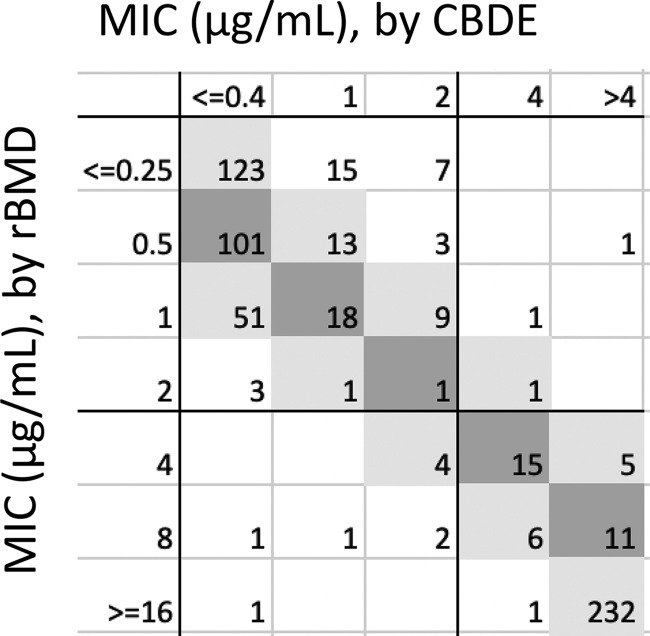
Overall, 94.4% of CBDE results were in EA and 97.9% in CA compared to the reference MIC (Fig. 2, Table 3). Across all isolates, 11 VME and 14 ME were observed on initial testing. Two VME and 11 ME corrected on repeat, yielding overall, 9 VME (3.2%) and 3 ME (0.9%). These are summarized in Table S2. When evaluated individually, 98.6% CA was observed for Enterobacterales (2.5% VME, 0% ME), 99.3% CA was observed for P. aeruginosa (0% VME, 0.7% ME), and 93.1% CA was observed for Acinetobacter spp. (5.6% VME, 3.3% ME). All but one of the VME and ME observed for Acinetobacter spp. were out of EA, whereas the majority of E. coli (4/5) errors were in EA and caused by mcr-1-expressing strains, which are known to have MICs that straddle the breakpoint.
FIG 2.
Scatterplot of CBDE versus reference MIC, obtained from the MIC mode from two unique rBMD tests.
TABLE 3.
Summary of performance, post-discrepancy resolution, for CBDE and CAT methods compared to reference MICsa
| Test results | CBDE | CAT-1 | CAT-10 |
|---|---|---|---|
| All isolates, n = 627 tests | |||
| CA (%) | 97.9 | 97.0 | 98.3 |
| EA (%) | 94.4 | 94.9 | 96.2 |
| No. of VME | 9 (3.2%) | 19 (6.8%) | 11 (3.9%) |
| No. of ME | 3 (0.9%) | 0 (0%) | 0 (0%) |
| Enterobacterales, n = 348 tests | |||
| CA (%) | 98.6 | 99.4 | 99.7 |
| EA (%) | 94.3 | 98.3 | 99.7 |
| No. of VME | 5 (2.5%) | 2 (1.0%) | 1 (0.5%) |
| No. of ME | 0 (0%) | 0 (0%) | 0 (0%) |
| P. aeruginosa, n = 148 tests | |||
| CA (%) | 99.3 | 98.7 | 100 |
| EA (%) | 96.6 | 99.3 | 99.3 |
| No. of VME | 0 (0%) | 1 (8.3%) | 0 (0%) |
| No. of ME | 1 (0.7%) | 0 (0%) | 0 (0%) |
| Acinetobacter spp., n = 131 tests | |||
| CA (%) | 95.4 | 88.5 | 92.3 |
| EA (%) | 93.1 | 83.1 | 88.5 |
| No. of VME | 4 (5.6%) | 15 (21.4%) | 10 (14.3%) |
| No. of ME | 2 (3.3%) | 0 (0%) | 0 (0%) |
CAT-1 = 1 μl inoculum; CAT-10 = 10 μl inoculum.
Interlaboratory agreement of the CBDE method was evaluated with the challenge strain set, which was tested in all three laboratories. This set comprised 43 Enterobacterales, 21 Acinetobacter spp., and 14 P. aeruginosa isolates. CA was 90.7% (39/43 strains in CA across all three laboratories) for the Enterobacterales and 100% for Acinetobacter and P. aeruginosa isolates. The 4 isolates that demonstrated one or more categorical errors were E. coli isolates, 3 of which harbored mcr-1. All four isolates had MICs of 4 or 8 μg/ml by rBMD and one or more result of 2 μg/ml by CBDE; in other words, these errors were observed for isolates with MICs that were at the breakpoint and were in EA with each other and with the rBMD method.
CAT.
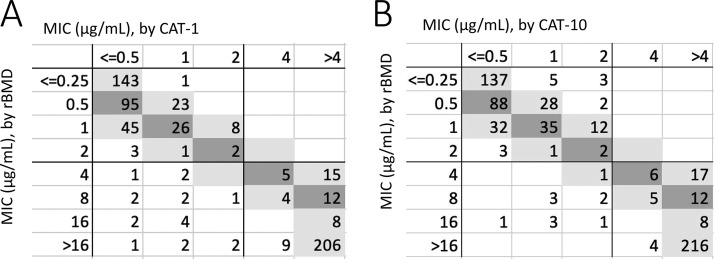
The CAT was evaluated using both a 1-μl (CAT-1) and a 10-μl (CAT-10) inoculum. Overall, performance was better for CAT-10 (Fig. 3, Table 3). CA was 94.9% with 6.8% VME for CAT-1, whereas CA improved to 98.3% with 3.9% VME for CAT-10. On initial testing, 3 ME were observed with CAT-10 and 0 ME with CAT-1. All three ME resolved on repeat testing, yielding no ME with either CAT-1 or CAT-10 (Table 3). Twenty-one initial VME were observed, two of which resolved on repeat testing (both with the CAT-1 method). The remaining VME are detailed in Table S2, 0/19 of which were in EA for CAT-1 and 1/11 of which were in EA for CAT-10. The difference between CAT-1 and CAT-10 was most apparent when evaluating Acinetobacter spp. isolates, where CA was 88.5% with 21.4% VME for CAT-1 versus 92.3% CA with 14.3% VME for CAT-10.
FIG 3.
Scatterplot of CAT-1 (A) and CAT-10 (B) versus reference MIC, obtained from the MIC mode from two unique rBMD tests.
Interlaboratory agreement of the CAT-1 and CAT-10 methods across the challenge strain set tested in all three laboratories demonstrated 100% CA for the Enterobacterales isolates. CA across the P. aeruginosa isolates was 92.9% (13/14) with CAT-1 and 100% with CAT-10. For the A. baumannii isolates, CA was 91.5% (19/21) for CAT-1 and 95.2% (20/21) for CAT-10. None of the category errors were in EA across laboratories (not shown).
The impact of MHA brand was evaluated in one laboratory, which performed CAT using media from three manufacturers in parallel for 190 isolates (112 stock and 78 challenge). Intermedium AA ranged from 90 to 92.1%, EA from 99.0 to 100%, and CA was 100% for CAT-1. Intermedium AA was 92.6 to 93.6%, EA was 99.5 to 100%, and CA was 99.5 to 100% for CAT-10 (not shown).
Quality control results.
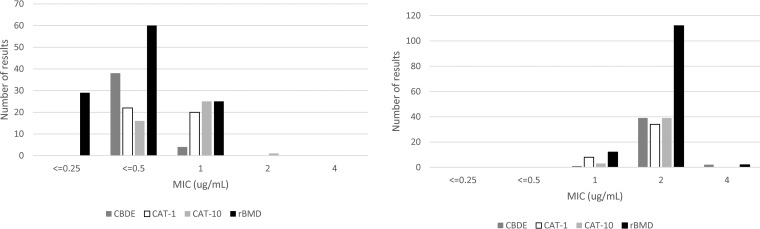
QC testing was in-control for all methods using P. aeruginosa ATCC 27853 (Fig. 4), with the exception of the aforementioned rBMD performed with Oxoid brand CA-MHB, which was excluded from the study. The MIC mode for rBMD using Sigma CA-MHB was 0.5 μg/ml, whereas the mode for BD CA-MHB was 1 μg/ml. With rBMD, bimodal MIC results of 2 to 4 μg/ml were obtained with AR Bank no. 0349 by all three manufacturers of CA-MHB.
FIG 4.
Colistin MIC distribution for quality control indicator strains P. aeruginosa ATCC 27853 (left panel) and E. coli CDC FDA AR no. 0349 (right panel) by the CBDE, CAT-1, CAT-10, and rBMD methods. The lowest concentrations tested were 0.4 μg/ml for CBDE, 0.5 μg/ml for CAT-1 and CAT-10, and 0.25 μg/ml for BMD.
QC results using the CBDE and CAT are shown in Fig. 4A and B. For P. aeruginosa ATCC 27853, 38/42 (90.5%) results were off-scale at an MIC of ≤0.4 μg/ml with the CBDE (Fig. 4A). In contrast, all results were in QC and on-scale for this strain using the CAT, as this method includes a 0.25-μg/ml concentration, which allowed reporting of an MIC of 0.5 μg/ml for 22/42 (52.4%) of the CAT-1 and 16/42 (38.1%) of the CAT-10 tests. For AR Bank no. 0349, all MIC results were on-scale, with a mode at 2 μg/ml for all three alternative methods, CBDE, CAT-1, and CAT-10 (Fig. 4B).
DISCUSSION
In June 2019, the CLSI Antimicrobial Susceptibility Testing (AST) Subcommittee provisionally approved both the CBDE and CAT-10 for testing of Enterobacterales and P. aeruginosa, based on the results presented herein. The methods will not be recommended for testing of Acinetobacter spp. by CLSI at the recommendation of the ahWG, due to high error rates and the large number of isolates excluded from analysis due to the absence of reference MIC. The CAT-1 method was also not approved, as performance was marginally better by CAT-10 (Table 3) and because of anecdotal reports from the participating laboratories that CAT-10 was easier to interpret than the CAT-1, although this claim was not systematically evaluated in this study.
The provisional approval status of the tests stemmed from two concerns raised by the subcommittee, namely, that existing CLSI QC strains for colistin (E. coli ATCC 25922 and P. aeruginosa ATCC 28753) will yield off-scale MICs using both methods and that only one brand of CA-MHB and colistin disk were used during evaluation of the CBDE. For the latter concern, CLSI standard M23 does not stipulate evaluation of multiple brands of media/disks for new test methods, but this concern is nonetheless legitimate, given the challenges associated with colistin testing documented to date.
Colistin and polymyxin B have presented QC challenges at CLSI meetings for several years. In recent history, QC ranges for E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were assessed according to current standards through a tier 2 QC study performed in 2002. These QC ranges were first published in 2003, when the drugs were reinstated into CLSI document M100. The ranges were then reassessed using a second tier 2 study performed in 2012, due to several laboratories reporting issues with E. coli ATCC 25922 falling outside published QC ranges on the lower end (12). At this time, QC ranges were confirmed for the rBMD method and established for BMD with 0.002% polysorbate 80 (surfactant), which yielded lower MICs. A CLSI/EUCAST joint working group then determined that polysorbate 80 should not be used in BMD testing for the polymyxins (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf), and this second set of QC ranges was deleted. At present, colistin QC ranges are 0.25 to 2.0 μg/ml for E. coli ATCC 25922 and 0.5 to 4.0 μg/ml for P. aeruginosa ATCC 27853. The modal MIC for E. coli ATCC 25922 was 0.5 μg/ml (152/246 observations) in studies performed in 2012 and 1.0 μg/ml (129/243 observations, 53.1%) for P. aeruginosa ATCC 27853, with 93 observations (38.3%) of 0.5 μg/ml. Neither strain provides good QC of the CBDE or CAT methods, as results are often off-scale (i.e., no growth in any tube/concentration). It should be noted that this type of off-scale MIC result is common for AST QC strains, as ranges are often below clinical breakpoints and therefore also below the concentrations of antimicrobial tested clinically.
Given this complex QC history, the ahWG anticipated QC challenges for the CBDE and CAT tests and attempted to establish a new QC strain, E. coli AR Bank no. 0349. This isolate was chosen as it consistently yields MICs of 2 to 4 μg/ml by a variety of methods and should yield on-scale results for both the CBDE and CAT methods. CLSI has rigorous standards for establishing QC, which are outlined in the M23 guideline (9). While these studies were attempted, they were not fully met, largely because one of the three medium brands of CA-MHB yielded results that were consistently out of range for P. aeruginosa ATCC 27853. While data from this brand of CA-MHB were eliminated when evaluating the performance of the two tests, M23 stipulates that three brands of medium must be used when establishing QC ranges, and therefore no official range could be established. Additional studies are under way to resolve this limitation. However, given the global clinical need, the AST Subcommittee did not want to postpone publication of colistin test methods and approved both CBDE and CAT with a provisional QC range of 1 to 4 μg/ml for E. coli AR bank no. 0349. Laboratories should expect to obtain results of 2 μg/ml the majority of the time, and repeated MICs of 1 μg/ml or 4 μg/ml should be investigated. Of note, EUCAST has also established a specialized QC strain for colistin testing, NCTC 13846, which also harbors mcr-1. EUCAST indicates that this strain should have a target MIC of 4 μg/ml, and only occasional MICs of 2 or 8 μg/ml should be observed (13).
The second concern raised by the subcommittee was the use of only one brand of medium for the CBDE. Prealiquoted tubes of CA-MHB in 10-ml and 25-ml volumes are commercially available but only from one manufacturer in the United States. The ahWG attempted to develop a method that had low complexity for laboratories and felt that aliquoting medium was not feasible in many laboratory settings. Several lots of medium were, however, used throughout the study, across the three laboratories, and no trends in performance were noted. It should be noted the CBDE is a low-throughput test, requiring use of multiple disks and tubes for each isolate tested and ample incubator space. As such, laboratories that perform a higher volume of testing may find the CAT-10 more suitable to their needs. A commercial source of colistin agar plates is not yet available, but evaluation of three brands of MHA plates for this test demonstrated no differences in performance. Once prepared, multiple isolates can be tested per plate with the CAT-10 method. While not officially studied, laboratories in this study used CAT plates up to 3 weeks after receipt, suggesting a shelf life of at least 1 month when shipping time is incorporated.
Lastly, the CBDE method to be endorsed by CLSI will not include the 0.4-μg/ml tube (one 10-μg colistin disk into 25 ml CA-MHB) and will only include the 0-μg/ml, 1-μg/ml, 2-μg/ml, and 4-μg/ml tubes as described in the initial publication of the method (11). With the newly approved breakpoints (Table 1), there is no need for the additional dilution, which helps reduce the cost and decrease the complexity of testing (the need for a different volume CA-MHB tube and a different inoculum for the tube).
This study has the limitation of not evaluating polymyxin B tests. Some institutions preferentially use polymyxin B over colistin, and several countries only have access to polymyxin B. However, CLSI recently confirmed that colistin results predict those of polymyxin B, meaning both the CBDE and CAT-10 results could be used to predict polymyxin B results. We did not evaluate a polymyxin B broth disk elution or agar test specifically. It should be noted that commercially available polymyxin B disks contain 300 U of antimicrobial, which is equivalent to 30 μg of polymyxin. As such, laboratories that desire to adapt these methods to polymyxin B must be cognizant that three times as much CA-MHB must be used to achieve the same concentrations of polymyxin B as was done herein for colistin. In contrast, several reports have demonstrated good performance of polymyxin B agar dilution tests, and so we anticipate that a polymyxin B agar test would perform well for the Enterobacterales and P. aeruginosa.
A second limitation to our study is the unconventional definition of a reference MIC. Polymyxin susceptibility testing is fraught with technical challenges, including the cationic nature of the polymyxins, their propensity to adsorb to plastic surfaces, variation in the concentration of each active polymyxin in a given stock powder (7), and the poorly defined heteroresistance phenotype that has been observed by several laboratories (14–18). As such, we chose to only evaluate the CBDE and CAT methods for isolates with a reproducible rBMD result (i.e., CA between the two rBMD tests and no skipped wells). Outside of Acinetobacter spp., for which 10% of rBMD were out of CA, this occurred only rarely, with <1% of P. aeruginosa isolates and 3.3% of Klebsiella spp. tested generating rBMD results out of CA. Only 5 isolates in this study were excluded from analysis due to repeat skipped wells suggestive of a heteroresistance phenotype, 2 Acinetobacter spp. and 3 Klebsiella spp. Some have noted that colistin MICs change over subculture and frozen storage (19), and as such it is probable that clinical laboratories will encounter isolates that demonstrate heteroresistance more frequently than was seen in this study. Laboratories should carefully review results for skipped dilutions by both CBDE and CAT-10 methods and repeat testing for those isolates that demonstrated this phenotype. If skipping occurs a second time, it may be advisable to indicate that the MIC is indeterminate, as the clinical significance of heteroresistance is not known.
In summary, we present data that led to CLSI endorsement of the CBDE and CAT-10 as colistin susceptibility testing methods. These methods will be published in the 30th edition of CLSI document M100 in January 2020. The AST Subcommittee will continue work on an alternative QC strain, namely, E. coli AR Bank no. 0349, so that laboratories can appropriately control these tests. In the interim, laboratories can use the E. coli AR Bank no. 0349 strain and P. aeruginosa ATCC 27853 to QC these tests. While CLSI has actively developed several tools to aid with the use of colistin and polymyxin B, including breakpoints and test methods, the limitations of these agents should be carefully weighed prior to their use, in particular, when other antimicrobial agents with proven antimicrobial activity are available.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andre Hsiung, Alani Barajas, and Susie Cuna at Hardy Diagnostics for their help in making colistin agar plates for the CAT-1 and CAT-10 methods. We thank BD for providing colistin disks, Thermo Fisher for providing discounted CA-MHB tubes, and Accelerate Diagnostics for making rBMD panels for this study.
R.M.H. is an employee and shareholder of Accelerate Diagnostics.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01269-19.
REFERENCES
- 1.Lenhard JR, Bulman ZP, Tsuji BT, Kaye KS. 2019. Shifting gears: the future of polymyxin antibiotics. Antibiotics (Basel) 8:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, Giacobbe DR, Viscoli C, Giamarellou H, Karaiskos I, Kaye D, Mouton JW, Tam VH, Thamlikitkul V, Wunderink RG, Li J, Nation RL, Kaye KS. 2019. International consensus guidelines for the optimal use of the polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Fowler VG Jr, Paterson DL, Bonomo RA, Evans S, Antibacterial Resistance Leadership Group . 2018. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 66:163–171. doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wunderink RG, Giamarellos-Bourboulis EJ, Rahav G, Mathers AJ, Bassetti M, Vazquez J, Cornely OA, Solomkin J, Bhowmick T, Bishara J, Daikos GL, Felton T, Furst MJL, Kwak EJ, Menichetti F, Oren I, Alexander EL, Griffith D, Lomovskaya O, Loutit J, Zhang S, Dudley MN, Kaye KS. 2018. Effect and safety of meropenem-vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II randomized clinical trial. Infect Dis Ther 7:439–455. doi: 10.1007/s40121-018-0214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medeiros GS, Rigatto MH, Falci DR, Zavascki AP. 2019. Combination therapy with polymyxin B for carbapenemase-producing Klebsiella pneumoniae bloodstream infection. Int J Antimicrob Agents 53:152–157. doi: 10.1016/j.ijantimicag.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Vardakas KZ, Falagas ME. 2017. Colistin versus polymyxin B for the treatment of patients with multidrug-resistant Gram-negative infections: a systematic review and meta-analysis. Int J Antimicrob Agents 49:233–238. doi: 10.1016/j.ijantimicag.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Humphries RM. 2015. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 35:22–27. doi: 10.1002/phar.1505. [DOI] [PubMed] [Google Scholar]
- 8.Matuschek E, Ahman J, Webster C, Kahlmeter G. 2018. Antimicrobial susceptibility testing of colistin: evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 24:865–870. doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 9.CLSI. 2018. Development of in vitro susceptibility testing criteria and quality control parameters, CLSI M23, 5th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.CLSI. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; M07 standard, 11th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Simner PJ, Bergman Y, Trejo M, Roberts AA, Marayan R, Tekle T, Campeau S, Kazmi AQ, Bell DT, Lewis S, Tamma PD, Humphries R, Hindler JA. 2019. Two-site evaluation of the colistin broth disk elution test to determine colistin in vitro activity against Gram-negative bacilli. J Clin Microbiol 57:e01163-18. doi: 10.1128/JCM.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. 2013. CLSI AST Subcommittee 2013 January agenda book, presented by CMI. https://clsi.org/meetings/ast-file-resources/. Accessed 25 July 2019.
- 13.EUCAST. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0, 2019. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 14.Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, Goossens H, Malhotra-Kumar S. 2018. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis 37:345–353. doi: 10.1007/s10096-017-3140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landman D, Salamera J, Quale J. 2013. Irreproducible and uninterpretable polymyxin B MICs for Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 51:4106–4111. doi: 10.1128/JCM.02129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.