Clostridioides difficile infection (CDI) is one of the most common health care-associated infections, resulting in significant morbidity, mortality, and economic burden. Diagnosis of CDI relies on the assessment of clinical presentation and laboratory tests. We evaluated the clinical performance of ultrasensitive single-molecule counting technology for detection of C. difficile toxins A and B.
Keywords: C. difficile, ultrasensitivity, single-molecule counting technology
ABSTRACT
Clostridioides difficile infection (CDI) is one of the most common health care-associated infections, resulting in significant morbidity, mortality, and economic burden. Diagnosis of CDI relies on the assessment of clinical presentation and laboratory tests. We evaluated the clinical performance of ultrasensitive single-molecule counting technology for detection of C. difficile toxins A and B. Stool specimens from 298 patients with suspected CDI were tested with the nucleic acid amplification test (NAAT; BD MAX Cdiff assay or Xpert C. difficile assay) and Singulex Clarity C. diff toxins A/B assay. Specimens with discordant results were tested with the cell cytotoxicity neutralization assay (CCNA), and the results were correlated with disease severity and outcome. There were 64 NAAT-positive and 234 NAAT-negative samples. Of the 32 NAAT+/Clarity− and 4 NAAT−/Clarity+ samples, there were 26 CCNA− and 4 CCNA− samples, respectively. CDI relapse was more common in NAAT+/toxin+ patients than in NAAT+/toxin− and NAAT−/toxin− patients. The clinical specificity of Clarity and NAAT was 97.4% and 89.0%, respectively, and overdiagnosis was more than three times more common in NAAT+/toxin− than in NAAT+/toxin+ patients. The Clarity assay was superior to NAATs for the diagnosis of CDI, by reducing overdiagnosis and thereby increasing clinical specificity, and the presence of toxins was associated with negative patient outcomes.
INTRODUCTION
Clostridioides difficile (formerly Clostridium difficile) infection (CDI) is one of the most common health care-associated infections, resulting in significant morbidity, mortality, and economic burden (1). The clinical presentation of CDI ranges in severity from mild diarrhea to fulminant colitis and death, but individuals can also be asymptomatic carriers of C. difficile (2). While 2% to 3% of healthy adults in the general population are colonized with C. difficile, the colonization rate in hospitalized patients can be up to 25% (2). Nosocomial diarrhea is common, and up to one-third of inpatients—or 80% of high-risk groups such as transplant patients—develop diarrhea, mostly due to noninfectious causes (3).
Accurate diagnosis of CDI relies on the assessment of clinical presentation and one or multiple laboratory tests positive for either C. difficile toxins or toxigenic C. difficile. Case definitions of CDI differ, and while the Infectious Diseases Society of America (IDSA) and the Society for Healthcare Epidemiology of America (SHEA) guidelines define CDI by the presence of symptoms (usually diarrhea) and a stool test positive for either toxins or toxigenic C. difficile (4), the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines do not agree on using nucleic acid amplification tests (NAATs) alone for diagnosis and also require the exclusion of non-CDI-related causes of diarrhea (5, 6). Given the high prevalence of both colonization and diarrheal symptoms in an inpatient setting, the detection of toxigenic organisms, either with NAATs or toxigenic culture (TC), has led to overdiagnosis and overtreatment (7, 8). The presence of toxins better correlates with disease than the presence of toxin genes (7, 8), but toxin tests have either poor sensitivity (enzyme immunoassays [EIAs]) or a long turnaround time (cell cytotoxicity neutralization assay [CCNA]) (9, 10).
In this study, we evaluated the clinical performance of an ultrasensitive single-molecule counting technology for the detection of C. difficile toxins and compared it to NAAT, CCNA, clinical outcome, and diagnosis.
MATERIALS AND METHODS
Singulex Clarity C. diff toxins A/B assay.
The Singulex Clarity C. diff toxins A/B assay (Clarity; Singulex, Inc., Alameda, CA, USA) measures C. difficile toxin A (TcdA) and B (TcdB) in stool on the automated Singulex Clarity system, an in vitro diagnostic platform, and was described previously (11). Briefly, the system is based upon a paramagnetic microparticle-based immunoassay powered by single-molecule counting technology that uses single-photon fluorescence detection for analyte quantitation. The quantitative limits of detection for TcdA and TcdB combined are 0.8 and 0.3 pg/ml in buffer, and 2.0 and 0.7 pg/ml in stool, respectively (11), and the cutoff for the assay is set at 12.0 pg/ml in undiluted stool (12). An unformed stool sample volume of 100 μl, or 0.1 g of semisolid stool sample, is diluted 1:20 with 1.9 ml of sample buffer and briefly vortexed. The sample is then centrifuged at 14,000 × g for 10 min, and 300 μl of the supernatant is transferred to a sample tube and loaded onto the Clarity instrument. The instrument automatically performs the immunoassay with a 1:1 mixture of paramagnetic microparticles precoated with anti-TcdA and anti-TcdB monoclonal antibodies (capture reagent) and toxin-specific antibodies labeled with the fluorophore, Alexa Fluor 647 (detection reagent). The Clarity software interpolates the data, using the fluorescent signal, into a combined TcdA/TcdB concentration reported in units of picograms per milliliter stool. The time to the first result after loading is 32 min, and the system can process 1 to 48 samples in an assay run.
Study design.
Unpreserved stool specimens from 298 patients with suspected CDI were collected at MultiCare Health System in Tacoma, WA, USA, from August to December 2018 and tested by the onsite standard of care using NAATs for detection of tcdB, either with the BD MAX Cdiff assay (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) or Xpert C. difficile assay (Cepheid Inc., Sunnyvale, CA, USA), chosen based on workflow considerations. Samples were stored at −80°C and shipped to Singulex (Alameda, CA, USA) for testing with the Clarity assay. Specimens with discordant results were tested with CCNA (C. difficile Tox-B test, TechLab; tested at ARUP Laboratories, Salt Lake City, UT, USA), and results were correlated with clinical outcome parameters, including antibiotic history within 30 days, administration of laxatives 48 h prior to testing, comorbidities, medical chart-confirmed presence of clinically significant diarrhea (≥3 loose stools in 24 h), fever (temperature of >100.4°F or 38.0°C), white blood cell (WBC) count, creatinine, CDI severity classification (4), CDI treatment, admittance to an intensive care unit (ICU), length of stay, resolution of symptoms within 14 days, and 30-day CDI relapse. Non-CDI causes of diarrhea were assessed in NAAT+ patients, including the presence of other gastrointestinal infections, inflammatory bowel disease (IBD) flare-ups, gastrointestinal mechanical or vascular impairment, medication-induced symptoms, and chronic diarrhea of unknown origin.
The study was approved by the MultiCare Health System Institutional Review Board (number 2018/07/3).
Statistical methods.
Patients were classified into mutually exclusive groups based upon their stool C. difficile NAAT and Clarity test results (NAAT+/toxin+, NAAT+/toxin−, NAAT−/toxin−, or NAAT−/toxin+). Categorical CDI outcomes, clinical symptoms, clinical history, and demographics were compared to NAAT/toxin C. difficile test results using the χ2 test for differences between two groups or Fisher’s exact test for differences between two groups with limited expected cell frequencies. Differences in length of stay were compared using the Wilcoxon test. Statistical differences in C. difficile toxin concentrations were assessed by a Mann Whitney U test. All analyses were performed as two-sided tests, and a P value of <0.05 was considered a significant finding. Statistical analysis was performed with SAS v9.4 and GraphPad Prism 8.2.0.
RESULTS
The patients’ mean age was 51.4 years (standard deviation, 22.7 years), and there were 172 (57.7%) women. There were 64 (21.5%) NAAT-positive and 234 (78.5%) NAAT-negative patient samples in the study, of which, 146 (49.0%) were from inpatients. Among the 64 NAAT-positive samples, 32 (50.0%) tested negative with Clarity and 26/32 (81.3%) of the Clarity-negative samples were CCNA negative. Among the NAAT-negative samples, 230 (98.2%) were Clarity-negative and the four Clarity-positive samples were negative by CCNA. The median C. difficile toxin concentrations in NAAT+ and NAAT− samples were 14.2 pg/ml (interquartile range [IQR], 4.3 to 966.1 pg/ml) and 3.7 pg/ml (IQR, 3.0 to 4.6 pg/ml), respectively (Fig. 1).
FIG 1.
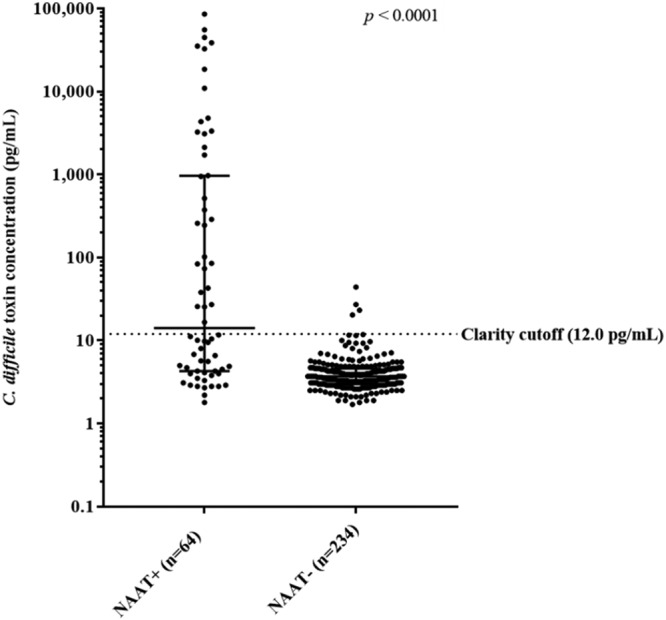
C. difficile toxin concentrations in NAAT+ and NAAT− samples. The median concentrations in NAAT+ and NAAT− samples were 14.2 pg/ml (IQR, 4.3 to 966.1 pg/ml) and 3.7 pg/ml (IQR, 3.0 to 4.6 pg/ml), respectively (P < 0.0001).
Clinical presentation, management, and outcome.
Clinical and laboratory characteristics by different test results are shown in Table 1. There was no difference between the groups in terms of presence of clinically significant diarrhea, elevated WBC, or creatinine, and hence estimated CDI severity. Two patients (0.7%) had received laxatives prior to testing, and 13 (4.4%) presented with fever. The only patient presenting with ileus was toxin positive with a high toxin level (55,526 pg/ml). NAAT+/toxin+ patients had a longer mean length of stay (14.2 days) than NAAT+/toxin− patients (7.6 days), although this difference was not statistically significant (P = 0.7424). Six patients were admitted to the ICU, of which two were NAAT+/toxin+ and four were NAAT−/toxin−. CDI treatment was initiated in 96.9% of NAAT+ patients. Consequently, the groups were similar in terms of symptom resolution within 14 days. Four NAAT+/toxin+ patients but no patients in the other groups experienced CDI relapse (12.5%, P < 0.0389 compared to NAAT+/toxin− patients).
TABLE 1.
Clinical and laboratory characteristics by different test resultsa
| Variable | Value |
P valueb | Value |
P valuec | ||
|---|---|---|---|---|---|---|
| NAAT+/toxin+ (n = 32) | NAAT+/toxin− (n = 32) | NAAT+/toxin− (n = 32) | NAAT−/toxin− (n = 230) | |||
| Antibiotics within 30 days (n [%]) | 14 (43.8) | 12 (37.5) | 0.6107 | 12 (37.5) | 53 (23.0) | 0.0761 |
| History of CDI (n [%]) | 6 (18.8) | 5 (15.6) | 0.7404 | 5 (15.6) | 10 (4.4) | 0.0101 |
| Clinically significant diarrhead (n [%]) | 27/27 (100) | 27/27 (100) | >0.9999 | 27/27 (100) | 167/185 (90.3) | 0.0902 |
| WBC >15,000/mlc (n [%]) | 7/28 (25.0) | 5/24 (20.8) | 0.7222 | 5/24 (20.8) | 23/186 (12.4) | 0.2556 |
| Creatinine >1.5 mg/dlc (n [%]) | 7/28 (25.0) | 6/24 (25.0) | 0.4791 | 6/24 (25.0) | 28/187 (15.0) | 0.6910 |
| Length of stay, inpatients (days) (mean [SD]) | 14.2 (16.9), n = 15 | 7.6 (4.8), n = 14 | 0.7424 | 7.6 (4.8), n = 14 | 7.5 (9.2), n = 123 | 0.1900 |
| Resolution of symptoms within 14 daysc (n [%]) | 21/24 (87.5) | 17/23 (73.9) | 0.3606 | 17/23 (73.9) | 133/160 (83.1) | 0.4990 |
| 30-day CDI relapse (n [%]) | 4 (12.5) | 0 (0) | 0.0389 | 0 (0) | 0 (0) | >0.9999 |
NAAT, nucleic acid amplification test; CDI, C. difficile infection; WBC, white blood cell count.
Test for comparison between the NAAT+/toxin+ and NAAT+/toxin− groups.
Test for comparison between the NAAT+/toxin− and NAAT−/toxin− groups.
Testing for WBC and creatinine was not performed on 58 and 57 patients, respectively. There was no information available on symptom resolution and medical chart-confirmed presence of clinically significant diarrhea in 87 and 57 patients, respectively. Percentages are reported as proportions of patients in each group with data available.
Antibiotic exposure and CDI history.
Eighty patients had received antibiotics 30 days prior to testing. A trend was observed that a higher proportion of NAAT+/toxin+ patients had received antibiotics (43.8%) than NAAT+/toxin− (37.5%) and NAAT−/toxin− (23.0%) patients, but this was not statistically significant. The NAAT+/toxin+ group had a similar proportion of patients with a history of CDI (18.8%) as the NAAT+/toxin− group (15.6%; P = 0.7404), while more NAAT+/toxin− patients had a CID history than NAAT−/toxin− (4.4%) patients (P = 0.0101).
Clinical specificity.
In the study, 68.8% (22 of 32) of NAAT+/toxin− patients and 21.9% (7 of 32) of NAAT+/toxin+ patients had a non-CDI-related cause of diarrhea (P = 0.0004) (Table 2). Using the guideline CDI case definition (5), 35/64 NAAT+ and 25/32 NAAT+/toxin+ patients were classified as having CDI. If used in clinical practice, Clarity would have achieved 97.4% (95% confidence interval [CI], 94.6 to 98.9%) clinical specificity and 78.1% positive predictive value (PPV; 95% CI, 59.6 to 90.1%), and NAAT had 89.0% (95% CI, 84.4 to 92.4%) clinical specificity and 54.7% PPV (95% CI, 41.8 to 67.0%), assuming all NAAT− patients were CDI negative. If the 4 NAAT−/toxin+ patients were regarded as negative for CDI, the Clarity specificity was 96.0%. The proportion of overdiagnosis was more than three times higher in the NAAT+/toxin− group than in the NAAT+/toxin+ group (Table 2). Among the 29 NAAT+ patients not classified as CDI, 5 (17.2%) had WBC of >15,000/ml, 6 (20.7%) had creatinine of >1.5 mg/dl, and 17 (58.6%) were inpatients. Among the ten NAAT+/toxin− patients where CDI could not be ruled out based on available medical chart data, eight presented as outpatients or at the emergency department and were not admitted, one did not receive treatment, and none had known severe outcomes, indicating that these patients may have had a low likelihood of severe CDI.
TABLE 2.
Presence of non-CDI-related causes of diarrhea by different test resulta
| Variable | No. (%) |
P value | |
|---|---|---|---|
| NAAT+/toxin+ (n = 32) | NAAT+/toxin− (n = 32) | ||
| Gastrointestinal infection other than CDI | 0 (0) | 7 (21.9) | 0.0108 |
| Inflammatory bowel disease flare-up | 5 (15.6) | 6 (18.8) | 0.7424 |
| Gastrointestinal mechanical or vascular impairment | 1 (3.1) | 4 (12.5) | 0.3547 |
| Medication-induced diarrhea | 0 (0) | 3 (9.4) | 0.2381 |
| Chronic diarrhea, unknown origin | 1 (3.1) | 2 (6.3) | 1.0000 |
| Total, non-CDI-related causes of diarrhea | 7 (21.9) | 22 (68.8) | <0.0004 |
NAAT, nucleic acid amplification test; CDI, C. difficile infection.
DISCUSSION
This study evaluates the diagnostic accuracy of ultrasensitive C. difficile toxin detection using single-molecule counting technology for the diagnosis of CDI compared to the detection of C. difficile toxin genes using NAAT. We show that the ultrasensitive toxin assay has the potential to significantly reduce the number of colonized patients being diagnosed as and treated for CDI, without delaying or withholding treatment in severe CDI cases.
CDI represents a significant burden to health care providers and organizations, and colonized patients may suffer from overtreatment when NAAT is utilized for diagnosis (1, 13). Colonization with C. difficile is common and treatment of carriers is not recommended (4, 5). It has been shown that the detection of toxigenic organisms, with NAAT or TC, leads to overdiagnosis (7, 8), but free-toxin tests are insensitive or take days to perform, leading to missed cases or delayed diagnosis (9). ESCMID guidelines recommend the exclusion of non-CDI-related causes of diarrhea for CDI diagnosis, although the conditions are not mutually exclusive (5). In this study, more than two-thirds of NAAT+/toxin− patients had a non-CDI-related cause of diarrhea, indicating that NAAT utilization leads to significant overdiagnosis. Clarity and NAAT had 97.4% and 89.0% clinical specificity, respectively, but among the NAAT+/toxin− patients where CDI could not be ruled out based on available medical charts, none presented with signs of severe disease, indicating these might not have been CDI cases and the specificity could potentially be revised. Non-CDI-related causes of diarrhea were also present in NAAT+/toxin+ patients, but the proportion was more than three times higher in the NAAT+/toxin− group. CDI is a clinical diagnosis and toxins are essential but not sufficient for disease, and while carriers can have toxins present (14–16), factors related to host immunity are known to impact disease progression (17, 18). Colonized patients with low levels of toxin A IgG have a much greater risk of CDI than carriers with high antibody levels (17), and an antibody response to toxin A, during an initial episode of CDI is associated with protection against recurrence (18).
In this study, only toxin-positive patients experienced CDI relapse. NAAT+/toxin+ patients may have had a longer length of stay than NAAT+/toxin− patients, although this difference was not statistically significant at this sample size. This is in line with previous findings showing that the presence of toxin better correlates with severity and outcome than presence of toxigenic organism only (7, 8). CDI is a toxin-mediated disease, and only 50% of NAAT+ patients in this study had detectable toxins, which is in line with other studies using an ultrasensitive toxin assay (11, 19, 20). Toxin gene expression is regulated by multiple different factors, including quorum signaling, temperature, and metabolism changes (21, 22), and the presence of toxin gene(s) does not equal presence of toxins (23).
There were four patients with NAAT−/Clarity+/CCNA− samples. The NAATs used in this study detect the presence of tcdB and CCNA primarily detects the function of TcdB (9), while Clarity detects both TcdA and TcdB. It has been shown, using ultrasensitive toxin assays, that some C. difficile strains primarily produce TcdA (24, 25).
Studies have shown that testing patients with high clinical probability of CDI prevents the false-positive results obtained with NAAT (26, 27), and according to the recent IDSA/SHEA guidelines, NAAT alone can be used for CDI diagnosis when there are preagreed institutional criteria for patient stool submission (4). We recently described the implementation of a computerized support tool coupled with education to enforce preanalytical screening for C. difficile testing (28). As a direct result, in this study, only 0.7% of the patients had prior laxative use and 92.5% had clinically significant diarrhea, although the later proportion most likely is higher, given that this parameter is not always recorded in the medical chart but is a requirement for ordering a C. difficile test in the electronic system. However, although rigorous stool submission criteria were in place, more than two-thirds of NAAT+/toxin− patients had other causes of diarrhea, thereby not fulfilling the ESCMID definition of a CDI case. In this study, 96.9% of NAAT+ patients were treated, indicating that almost all patients are treated based on a NAAT result, which is in line with previous reports (16, 28–30). Clinical and laboratory parameters related to disease severity had limited value in distinguishing carriers from CDI patients, and there was no difference in WBC or creatinine levels. Factors related to immune and renal status impact levels of WBC and creatinine, and although difference in CDI severity has been assessed using these parameters (7, 8), their role in treatment guidance may be questioned.
This study has limitations. First, testing with fresh samples might have been preferred over frozen samples to avoid the risk of potential toxin degradation. Studies have shown, however, that toxins are stable in refrigerated or frozen conditions (11, 31–33). Second, more than 50% of the patients were outpatients, which reduced the ability to assess differences in some clinical outcome parameters, such as length of stay. Lastly, this was a single-center study, and the results need to be confirmed in a larger patient cohort and in different geographical regions.
In summary, presence of toxins, as measured with single-molecule counting technology, correlated with CDI relapse. The use of ultrasensitive toxin detection significantly reduced CDI overdiagnosis and thereby improved clinical specificity compared with that of NAAT.
ACKNOWLEDGMENTS
We acknowledge the work of the Microbiology Laboratory at MultiCare Health System.
J.S., J.E., P.K., and N.N. are employees of Singulex, Inc.
This study was supported by Singulex.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett JG, Gerding DN. 2008. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis 46 Suppl 1:S12–S18. doi: 10.1086/521863. [DOI] [PubMed] [Google Scholar]
- 3.Polage CR, Solnick JV, Cohen SH. 2012. Nosocomial diarrhea: evaluation and treatment of causes other than Clostridium difficile. Clin Infect Dis 55:982–989. doi: 10.1093/cid/cis551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debast SB, Bauer MP, Kuijper EJ, European Society of Clinical Microbiology and Infectious Diseases . 2014. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 20 Suppl 2:1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 6.Crobach MJT, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. 2016. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22 Suppl 4:S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang Y-W, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile Infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O'Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham C-A, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 26:604–630. doi: 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang FC, Polage CR, Wilcox MH. 2017. Point-counterpoint: what is the optimal approach for detection of Clostridium difficile infection? J Clin Microbiol 55:670–680. doi: 10.1128/JCM.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandlund J, Bartolome A, Almazan A, Tam S, Biscocho S, Abusali S, Bishop JJ, Nolan N, Estis J, Todd J, Young S, Senchyna F, Banaei N. 2018. Ultrasensitive detection of Clostridioides difficile toxins A and B by use of automated single-molecule counting technology. J Clin Microbiol 56:e00908-18. doi: 10.1128/JCM.00908-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singulex. 2019. Singulex Clarity C. diff toxins A/B assay: instructions for use. LBL-0285 Rev. 03 Singulex, Inc., Alameda, CA. [Google Scholar]
- 13.Gould CV, Edwards JR, Cohen J, Bamberg WM, Clark LA, Farley MM, Johnston H, Nadle J, Winston L, Gerding DN, McDonald LC, Lessa FC, Clostridium difficile Infection Surveillance Investigators, Centers for Disease Control and Prevention . 2013. Effect of nucleic acid amplification testing on population-based incidence rates of Clostridium difficile infection. Clin Infect Dis 57:1304–1307. doi: 10.1093/cid/cit492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollock NR, Banz A, Chen X, Williams D, Xu H, Cuddemi CA, Cui AX, Perrotta M, Alhassan E, Riou B, Lantz A, Miller MA, Kelly CP. 2019. Comparison of Clostridioides difficile stool toxin concentrations in adults with symptomatic infection and asymptomatic carriage using an ultrasensitive quantitative immunoassay. Clin Infect Dis 68:78–86. doi: 10.1093/cid/ciy415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong YKN, Gonzalez-Orta M, Saldana C, Cadnum JL, Jencson AL, Donskey CJ. 2019. Frequency of positive enzyme immunoassay for toxin in stool of asymptomatic carriers of Clostridium difficile. Clin Infect Dis 68:711–711. doi: 10.1093/cid/ciy701. [DOI] [PubMed] [Google Scholar]
- 16.Anikst VE, Gaur RL, Schroeder LF, Banaei N. 2016. Organism burden, toxin concentration, and lactoferrin concentration do not distinguish between clinically significant and nonsignificant diarrhea in patients with Clostridium difficile. Diagn Microbiol Infect Dis 84:343–346. doi: 10.1016/j.diagmicrobio.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Kyne L, Warny M, Qamar A, Kelly CP. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 18.Kyne L, Warny M, Qamar A, Kelly CP. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357:189–193. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 19.Hansen G, Young S, Wu AHB, Herding E, Nordberg V, Mills R, Griego-Fullbright C, Wagner A, Ong CM, Lewis S, Yoon J, Estis J, Sandlund J, Friedland E, Carroll KC. 2019. Ultrasensitive detection of Clostridioides difficile toxins in stool using single molecule counting technology: comparison with CCCNA free toxin detection. ECCMID, Amsterdam, The Netherlands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandlund J, Mills R, Griego-Fullbright C, Wagner A, Estis J, Bartolome A, Almazan A, Tam S, Biscocho S, Abusali S, Nolan N, Bishop JJ, Todd J, Young S. 2019. Laboratory comparison between cell cytotoxicity neutralization assay and ultrasensitive single molecule counting technology for detection of Clostridioides difficile toxins A and B, PCR, enzyme immunoassays, and multistep algorithms. Diagn Microbiol Infect Dis 95:20–24. doi: 10.1016/j.diagmicrobio.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6:e02569. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. doi: 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banz A, Lantz A, Riou B, Foussadier A, Miller M, Davies K, Wilcox M. 2018. Sensitivity of single-molecule array assays to detect Clostridium difficile toxins in comparison to conventional laboratory testing algorithms. J Clin Microbiol 56:e00452-18. doi: 10.1128/JCM.00452-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katzenbach P, Dave G, Murkherjee A, Todd J, Bishop J, Estis J. 2018. Single molecule counting technology for ultrasensitive quantification of Clostridium difficile toxins A and B. IDWeek, San Francisco, CA. [Google Scholar]
- 26.Madden GR, German Mesner I, Cox HL, Mathers AJ, Lyman JA, Sifri CD, Enfield KB. 2018. Reduced Clostridium difficile tests and laboratory-identified events with a computerized clinical decision support tool and financial incentive. Infect Control Hosp Epidemiol 39:737–740. doi: 10.1017/ice.2018.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong CY, Gombar S, Wilson R, Sundararajan G, Tekic N, Holubar M, Shepard J, Madison A, Tompkins L, Shah N, Deresinski S, Schroeder LF, Banaei N. 2017. Real-time electronic tracking of diarrheal episodes and laxative therapy enables verification of Clostridium difficile clinical testing criteria and reduction of Clostridium difficile infection rates. J Clin Microbiol 55:1276–1284. doi: 10.1128/JCM.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow S-K, Naderpour A, Van Enk J. 2018. It is not about the assay: preanalytical screening is the key to reducing Clostridioides difficile infection. J Clin Microbiol 57: e01553-18. doi: 10.1128/JCM.01553-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckel WR, Avdic E, Carroll KC, Gunaseelan V, Hadhazy E, Cosgrove SE. 2015. Gut check: Clostridium difficile testing and treatment in the molecular testing era. Infect Control Hosp Epidemiol 36:217–221. doi: 10.1017/ice.2014.19. [DOI] [PubMed] [Google Scholar]
- 30.Rock C, Pana Z, Leekha S, Trexler P, Andonian J, Gadala A, Carroll KC, Maragakis LL, CDC Prevention Epicenters Program . 2018. National Healthcare Safety Network laboratory-identified Clostridium difficile event reporting: a need for diagnostic stewardship. Am J Infect Control 46:456–458. doi: 10.1016/j.ajic.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfa MJ, Olson N, Murray B-L. 2014. Fecal specimens for Clostridium difficile diagnostic testing are stable for up to 72 hours at 4°C. J Med Microbiol Diagn 3:140. doi: 10.4172/2161-0703.1000140. [DOI] [Google Scholar]
- 32.Modi C, DePasquale JR, Nguyen NQ, Malinowski JE, Perez G. 2010. Does the handling time of unrefrigerated human fecal specimens impact the detection of Clostridium difficile toxins in a hospital setting? Indian J Gastroenterol 29:157–161. doi: 10.1007/s12664-010-0040-1. [DOI] [PubMed] [Google Scholar]
- 33.Schora DM, Peterson LR, Usacheva EA. 2018. Immunological stability of Clostridium difficile toxins in clinical specimens. Infect Control Hosp Epidemiol 39:434–438. doi: 10.1017/ice.2018.20. [DOI] [PubMed] [Google Scholar]


