The Staphylococcus intermedius group (SIG) is a collection of coagulase-positive staphylococci consisting of four distinct species, namely, Staphylococcus cornubiensis, Staphylococcus delphini, Staphylococcus intermedius, and Staphylococcus pseudintermedius. SIG members are animal pathogens and rare causes of human infection. Accurate identification of S. pseudintermedius has important implications for interpretation of antimicrobial susceptibility testing data and may be important for other members of the group.
KEYWORDS: automated biochemical platform, coagulase-positive staphylococci, Staphylococcus delphini, Staphylococcus intermedius group, MALDI-TOF MS
ABSTRACT
The Staphylococcus intermedius group (SIG) is a collection of coagulase-positive staphylococci consisting of four distinct species, namely, Staphylococcus cornubiensis, Staphylococcus delphini, Staphylococcus intermedius, and Staphylococcus pseudintermedius. SIG members are animal pathogens and rare causes of human infection. Accurate identification of S. pseudintermedius has important implications for interpretation of antimicrobial susceptibility testing data and may be important for other members of the group. Therefore, we sought to evaluate the performance of five commercially available identification platforms with 21 S. delphini isolates obtained from a variety of animal and geographic sources. Here, we show that automated biochemical platforms were unable to identify S. delphini to the species level, a function of its omission from their databases, but could identify isolates to the SIG level with various degrees of success. However, all automated systems misidentified at least one isolate as Staphylococcus aureus. One matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) system was able to identify S. delphini to the species level, suggesting that MALDI-TOF MS is the best option for distinguishing members of the SIG. With the exception of S. pseudintermedius, it is unclear if other SIG members should be routinely identified to the species level; however, as our understanding of their role in animal and human diseases increases, it may be necessary and important to do so.
INTRODUCTION
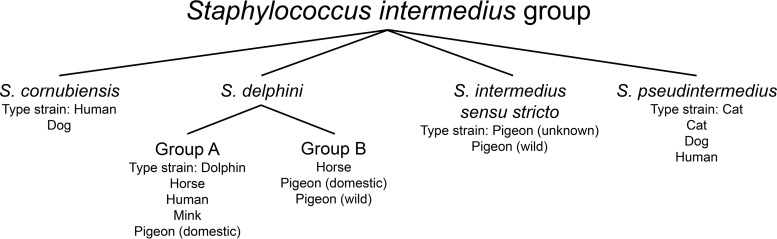
The Staphylococcus intermedius group (SIG) is a collection of coagulase-positive staphylococci composed of four distinct species, namely, Staphylococcus cornubiensis, Staphylococcus intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini (Fig. 1) (1–3). Staphylococcus delphini is further separated into two phylogenetically distinct clades, groups A and B (2). Staphylococcus delphini was first described in 1988 after isolation from two dolphins suffering from suppurative skin lesions (4), but it is typically associated with horses, pigeons, and mustelids, with mustelids serving as the natural hosts of S. delphini group A (1, 2, 5). Staphylococcus pseudintermedius is a zoonotic pathogen for which canines are the presumed source of infection (often as a result of bites), and it has been recovered from a variety of human specimens (6–10). In contrast, there is only a single report each of S. cornubiensis and S. delphini human infection (3, 11). However, it is unclear if the paucity of reports of human infection with non-S. pseudintermedius SIG members is related to the inability of identification platforms to accurately identify them.
FIG 1.
Members of the Staphylococcus intermedius group (SIG) and host associations based upon data presented in references 2, 3, and 11. The animal sources of the type strains are highlighted, and organisms from whom various SIG members have been recovered are also listed. The S. cornubiensis type strain was isolated from a human; however, human isolates associated with all SIG members, including the S. cornubiensis type strain, probably reflect transmission from animals.
Staphylococcus aureus and members of the SIG share similar morphologic and biochemical traits, namely, β-hemolysis on blood agar, Gram-positive cocci in clusters, and positive catalase reaction, and many isolates are tube coagulase positive (12), which can confound identification and lead to misidentification of SIG members as S. aureus (6, 13). The misidentification of S. pseudintermedius as S. aureus has important treatment implications as methods and interpretative criteria for detecting mecA-mediated β-lactam resistance differ between the two species (14, 15) and may also be important for S. delphini. Furthermore, as the outcomes of human and animal interactions are better defined, especially in the setting of antimicrobial resistance, it may become increasingly important to accurately differentiate between S. aureus and other coagulase-positive staphylococci to understand the epidemiology and distribution of the various species in these populations.
While there are nucleic acid-based assays for identifying members of the SIG to the species level (16, 17), they are typically not used in routine clinical microbiology laboratories. Therefore, we sought to evaluate the performance of three commercially available automated biochemical identification platforms, MicroScan WalkAway (Beckman Coulter, Inc., Brea, CA), BD Phoenix (Becton, Dickinson and Company [BD], Franklin Lakes, NJ, USA), and Vitek 2 (bioMérieux, Inc., Durham, NC), and two mass spectrometry platforms, MALDI Biotyper (Bruker Daltonics, Inc., Billerica, MA) and Vitek MS (bioMérieux, Inc.) (Table 1), using 21 S. delphini isolates derived from a variety of animals and geographic sources (Table 2) (18).
TABLE 1.
Identification platforms assessed in the study
| Identification platform | Biochemical panel | Software version | MALDI-TOF MS database version | SIG members included in the database |
|---|---|---|---|---|
| MicroScan WalkAway | Pos Combo panel type 33 | 4.42 | NAa | S. intermedius |
| BD Phoenix | PMIC/ID-105 | 6.21A | NA | S. intermedius |
| Vitek 2 | GP ID card | 07.01 | NA | S. intermedius, S. pseudintermedius |
| MALDI Biotyper | NA | NA | IVD: Claim 2 | S. delphini,c S. intermedius,c S. pseudintermedius |
| RUO: 6903/7311b | S. delphini, S. intermedius, S. pseudintermedius | |||
| Vitek MS | NA | NA | IVD: Knowledge Base v2.0 | S. intermedius,d S. pseudintermediusd |
| RUO: Knowledge Base v4.13 | S. delphini, S. delphini/S. intermedius/S. pseudintermedius,e S. intermedius, S. pseudintermedius |
NA, not applicable.
Isolates 8086 and P50 were analyzed using RUO database 7311. The remaining isolates were analyzed using RUO database 6903.
Both S. delphini and S. intermedius were included as non-clinically validated species in the MALDI Biotyper IVD Claim 2 database, which means they were not formally tested in United States Food and Drug Administration clinical trials.
Both S. intermedius and S. pseudintermedius were included as non-clinically validated species in the Vitek MS IVD Knowledge Base v2.0 database.
One possible identification result that can be returned from the Vitek MS RUO Knowledge Base v4.13 is S. delphini/S. intermedius/S. pseudintermedius.
TABLE 2.
Collection of S. delphini isolates analyzed in this study
| Isolate | Group | Host species | Country of Origin |
|---|---|---|---|
| DSM 20771T | A | Dolphin | Italy |
| MI 09-2894 | A | Raccoon | USA |
| MI 09-8445 | A | Sea otter | USA |
| MI 16-1129 | A | Ferret | USA |
| VPP 12 D | A | Horse | Switzerland |
| 19-039 | A | Avian | USA |
| 52-006 | A | Horse | USA |
| 56-021 | A | Llama | USA |
| 62-031 | A | Horse | USA |
| 8086 | B | Horse | UK |
| AV8047 | B | Pigeon | Japan |
| H4A | B | Horse | Japan |
| HT2003-0674 | B | Camel | France |
| P27B | B | Pigeon | Japan |
| P50 | B | Pigeon | Japan |
| 26-037 | B | Horse | USA |
| 33-090 | B | Horse | USA |
| 35-016 | B | Horse | USA |
| 49-046 | B | Horse | USA |
| 53-033 | B | Horse | USA |
| 56-039 | B | Horse | USA |
MATERIALS AND METHODS
Bacterial isolates.
A total of 21 S. delphini isolates were obtained from the following animal sources: avian (n = 1), camel (n = 1), dolphin (n = 1), ferret (n = 1), horse (n = 11), llama (n = 1), pigeon (n = 3), raccoon (n = 1), and sea otter (n = 1) (Table 2). These isolates were recovered from the following six different geographic locations: the United States (n = 13), Japan (n = 4), France (n = 1), Italy (n = 1), Switzerland (n = 1), and the United Kingdom (n = 1). The collection also included the S. delphini type strain DSM 20771. Isolates belonging to group A accounted for 42.9% of isolates (9/21), while the remaining 57.1% (12/21) of isolates were group B. The identity of all isolates was confirmed using a thermonuclease (nuc) gene PCR test that can differentiate between S. aureus, S. delphini groups A and B, Staphylococcus hyicus, S. intermedius, S. pseudintermedius, and Staphylococcus schleiferi (17). The nuc PCR test results were accepted as the reference method for species level identification.
Automated biochemical testing.
The collection was analyzed on the following three commercial automated biochemical platforms: MicroScan WalkAway (identification/antibiotic susceptibility panel, Pos Combo panel type 33; software, LabProV4.42), BD Phoenix (identification/antibiotic susceptibility panel, PMIC/ID-105; software, V6.21A), and Vitek 2 (identification panel, GP ID card; software, Vitek 2 Systems version 07.01) using the manufacturers’ default instructions. Testing on the various platforms was performed at Beckman Coulter, Inc. (Sacramento, CA) for MicroScan WalkAway; Johns Hopkins Hospital (Baltimore, MD) for BD Phoenix; and the University of Tennessee College of Veterinary Medicine (Knoxville, TN) for Vitek 2.
MALDI-TOF MS analysis.
The collection was analyzed on two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) platforms. Isolates were prepared using an organic acid extraction method prior to application to the target (MALDI Biotyper) or applied directly to the target (Vitek MS) according to instructions defined by the manufacturers. Mass spectra were analyzed with the United States Food and Drug Administration-approved in vitro diagnostic (IVD) and research use only (RUO) databases associated with each platform, namely, IVD, Claim 2; RUO, 6903 and 7311 (MALDI Biotyper); IVD, Knowledge Base v2.0; RUO, Saramis Knowledge Base v4.13 (Vitek MS). Testing on the MALDI Biotyper was performed at NewYork-Presbyterian Hospital-Weill Cornell Medical Center (New York, NY), while testing on the Vitek MS was performed at the Emory University School of Medicine (Atlanta, GA). Results were interpreted according to the default instructions provided by the manufacturers.
Data analysis.
All data analysis was performed in Microsoft Excel software.
RESULTS
Performance of the automated biochemical platforms for identifying S. delphini.
The collection of S. delphini isolates was analyzed on the following three automated biochemical testing platforms: MicroScan WalkAway, BD Phoenix, and Vitek 2. For each of these automated biochemical systems, only isolates with high confidence/probability scores (≥90%) were accepted for identification to the species level, while those with low confidence/probability scores (<90%) were considered low discrimination. Data are presented in Table 3. No system identified S. delphini to the species level due to its omission from the associated databases (Table 1). Specifically, the automated systems identified isolates as either S. intermedius or S. pseudintermedius for 52.4% (11/21), 81.0% (17/21), and 57.1% (12/21) of isolates for the MicroScan WalkAway, BD Phoenix, and Vitek 2 platforms, respectively. Interestingly, Vitek 2 identified 88.9% (8/9) of S. delphini group A isolates as S. pseudintermedius, with the remaining isolate being identified as S. aureus. Conversely, only 33.3% (4/12) of the group B isolates were identified as S. pseudintermedius, and 41.7% (5/12) were identified as S. aureus. No other obvious pattern related to the two S. delphini groups was observed for the other platforms. For comparison, S. pseudintermedius LMG 22219T was analyzed on all three platforms, resulting in identification as S. intermedius on the MicroScan WalkAway and BD Phoenix platforms, with high probability and confidence, respectively, and as low-probability S. aureus/S. pseudintermedius on the Vitek 2.
TABLE 3.
Performance of automated biochemical testing for identification of S. delphinia
| Isolate identifier | Group | MicroScan WalkAway | BD Phoenix | Vitek 2 |
|---|---|---|---|---|
| DSM 20771T | A | Low discriminationb | S. saprophyticus | S. pseudintermedius |
| MI 09-2894 | A | Low discriminationc | S. aureus | S. pseudintermedius |
| MI 09-8445 | A | S. aureus | S. intermedius | S. pseudintermedius |
| MI 16-1129 | A | S. intermedius | S. intermedius | S. pseudintermedius |
| VPP 12 D | A | S. intermedius | S. intermedius | S. pseudintermedius |
| 19-039 | A | S. haemolyticus | S. intermedius | S. pseudintermedius |
| 52-006 | A | S. intermedius | S. intermedius | S. pseudintermedius |
| 56-021 | A | S. haemolyticus | S. intermedius | S. pseudintermedius |
| 62-031 | A | S. intermedius | S. intermedius | S. aureus |
| 8086 | B | S. intermedius | S. intermedius | S. pseudintermedius |
| AV8047 | B | Low discriminationd | S. intermedius | S. aureus |
| H4A | B | S. intermedius | S. intermedius | S. aureus |
| HT2003-0674 | B | Low discriminatione | S. intermedius | S. aureus |
| P27B | B | S. intermedius | S. schleiferi | Unidentified organism |
| P50 | B | S. intermedius | S. intermedius | S. pseudintermedius |
| 26-037 | B | S. intermedius | S. intermedius | Low discriminationi |
| 33-090 | B | Low discriminationf | S. aureus | S. pseudintermedius |
| 35-016 | B | Low discriminationg | S. intermedius | S. aureus |
| 49-046 | B | Low discriminationh | S. intermedius | S. aureus |
| 53-033 | B | S. intermedius | S. intermedius | S. pseudintermedius |
| 56-039 | B | S. intermedius | S. intermedius | Low discriminationj |
For each automated biochemical system, only isolates with high confidence/probability scores (≥90%) were accepted as identified to the species level, while those with low confidence/probability scores (<90%) were designated low discrimination.
Low-discrimination identification results: S. aureus (84.52%), S. schleiferi subspecies coagulans (10.61%) and Staphylococcus epidermidis (4.87%).
Low-discrimination identification results: Staphylococcus simulans (51.60%), Staphylococcus haemolyticus (28.19%), S. intermedius (17.02%) and Staphylococcus hyicus (3.19%).
Low-discrimination identification results: S. aureus (79.82%) and S. haemolyticus (20.18%).
Low-discrimination identification results: S. haemolyticus (83.79%), S. hyicus (12.37%), Staphylococcus lugdunensis (2.24%), and Staphylococcus xylosus (1.60%).
Low-discrimination identification results: S. intermedius (43.83%), S. aureus (35.28%), S. simulans (9.44%), Staphylococcus auricularis (7.02%), and S. schleiferi subspecies coagulans (4.43%).
Low-discrimination identification results: S. simulans (76.81%), S. haemolyticus (12.50%), S. auricularis (5.83%), and Staphylococcus warneri (4.86%).
Low-discrimination identification results: S. schleiferi subspecies coagulans (38.63%), S. aureus (30.50%), S. intermedius (20.75%), and S. simulans (10.12%).
Low-discrimination identification results: S. aureus (∼33%), S. hyicus (∼33%), and S. schleiferi (∼33%).
Low-discrimination identification results: S. aureus (∼50%) and S. pseudintermedius (∼50%).
The automated biochemical systems misidentified isolates as S. aureus for 4.8% (1/21), 9.5% (2/21), and 28.6% (6/21) of isolates for the MicroScan WalkAway, BD Phoenix, and Vitek 2 platforms, respectively. Other identification results with high confidence/probability scores included Staphylococcus haemolyticus (9.5% [2/21] of isolates) with the MicroScan WalkAway and Staphylococcus saprophyticus (4.8% [1/21]; for the S. delphini type strain) and Staphylococcus schleiferi (4.8% [1/21] of isolates) with the BD Phoenix. The remaining isolates had low-probability identification scores and included 33.3% (7/21) of the isolates for the MicroScan WalkAway and 9.5% (2/21) of the isolates for the Vitek 2 (Table 3). One isolate, P27B, was not identified by the Vitek 2.
Performance of the MALDI-TOF MS systems for identifying S. delphini.
All S. delphini isolates were analyzed using the MALDI Biotyper and Vitek MS systems. Data are tabulated in Table 4. Staphylococcus delphini was not included as a clinically validated species in either of the IVD database versions used at the time of testing; however, it was included as a non-clinically validated species (i.e., S. delphini was not formally tested in United States Food and Drug Administration clinical trials) in the MALDI Biotyper IVD Claim 2 database (Table 1). Upon analysis with the MALDI Biotyper IVD database, three isolates (14.3%) were not identified (log [score] values, <1.70), namely MI 09-2894, 19-039, and 33-090, and the remainder were identified as S. pseudintermedius (a clinically validated species in the IVD database), although the log (score) values were <2.00 (low-confidence identification) except for that of a single isolate (8086 [group B]; log [score] value, ≥2.00 [high-confidence identification]). For all isolates, including the three not identified, a non-clinically validated identification of S. delphini was returned. All but one isolate (P50 [group B]) had log (score) values ≥2.00. With the Vitek MS IVD database (Knowledge Base v2.0) isolates were identified as either S. pseudintermedius (15/21 [71.4%] of the isolates; confidence values, 99.9%) or low-discrimination identifications of either S. pseudintermedius or S. intermedius (6/21 [28.6%] of the isolates; confidence values, <60%), with all identifications generated from the non-clinically validated portion of the database.
TABLE 4.
Performance of MALDI-TOF MS for identification of S. delphini
| Isolate identifier | Group | MALDI Biotyper full extraction preparative method |
Vitek MS direct application preparative method |
|||
|---|---|---|---|---|---|---|
| IVD database |
RUO database identification (log [score] value)a | IVD database non-clinically validated identificationb (confidence value [%])c | RUO database identification (confidence value [%])c | |||
| Clinically validated identification (log [score] value)a | Non-clinically validated identification (log [score] value)a | |||||
| DSM 20771T | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| MI 09-2894 | A | No identification (<1.70) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| MI 09-8445 | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| MI 16-1129 | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| VPP 12 D | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 19-039 | A | No identification (<1.70) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (48.2%), S. intermedius (51.7%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 52-006 | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 56-021 | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 62-031 | A | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 8086 | B | S. pseudintermedius (≥2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| AV8047 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| H4A | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (50%), S. intermedius (49.9%) | S. delphini/S. intermedius/S. pseudintermedius (92.1%) |
| HT2003-0674 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| P27B | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (50%), S. intermedius (50%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| P50 | B | S. pseudintermedius (<2.00) | S. delphini (<2.00) | S. delphini (≥2.00) | S. pseudintermedius (45%), S. intermedius (54.9%) | S. delphini/S. intermedius/S. pseudintermedius (76.1%) |
| 26-037 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 33-090 | B | No identification (<1.70) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 35-016 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (50%), S. intermedius (50%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 49-046 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (88.1%) |
| 53-033 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (99.9%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
| 56-039 | B | S. pseudintermedius (<2.00) | S. delphini (≥2.00) | S. delphini (≥2.00) | S. pseudintermedius (49.2%), S. intermedius (50.8%) | S. delphini/S. intermedius/S. pseudintermedius (99.9%) |
Log (score) value of 2.00 to 3.00, high-confidence identification; log (score) value of 1.70 to1.99, low-confidence identification; log (score) value of <1.70, no organism identification possible.
No member of the SIG was included in the clinically validated Vitek MS IVD Knowledge Base v2.0 database; however, S. intermedius and S. pseudintermedius were included as non-clinically validated species in the IVD Knowledge Base v2.0 database.
Confidence value of 60.0 to 99.9%, high-confidence identification (one significant organism or organism group identification returned); confidence value of <60%, low-discrimination identification. Low discrimination identifications are displayed when more than one, but not more than four, organism or organism group identifications are returned.
All isolates were identified as S. delphini using the MALDI Biotyper RUO databases with a log (score) value of ≥2.00, implying accurate identification to the species level. Mass spectra generated for 19 isolates were analyzed using RUO database 6903, while two isolates (8086 and P50) were analyzed using RUO database 7311. Finally, mass spectra analyzed using the Vitek MS RUO database (Saramis Knowledge Base v4.13) resulted in all isolates being identified as S. delphini/S. intermedius/S. pseudintermedius (confidence values ranging from 76.1 to 99.9%). Of particular importance, no isolate was misidentified as S. aureus using either platform. Again, for comparison, S. pseudintermedius LMG 22219T was analyzed. Using the MALDI Biotyper, the S. pseudintermedius type strain was identified as S. pseudintermedius using both the IVD and RUO databases (log [score] values, ≥2.00), while the Vitek MS IVD database generated a non-clinically validated identification of S. pseudintermedius (confidence value, 99.9%) and S. delphini/S. intermedius/S. pseudintermedius (confidence value, 99.9%) with the RUO database.
DISCUSSION
In this study, we evaluated the performance of three automated biochemical platforms and two MALDI-TOF MS systems for the ability to identify S. delphini, a member of the SIG. Although the number of isolates included in the study was limited, the collection contained isolates of both phylogenetic clades recovered from a diverse array of host species from a wide geographic area.
No automated biochemical system identified S. delphini to the species level due to its omission from their associated databases. All systems identified some isolates to the SIG level (range, 52.4 to 81.0% of isolates depending on the platform). However, only six isolates (MI 16-1129 [group A], VPP 12 D [group A], 52-006 [group A], 8086 [group B], P50 [group B], and 53-033 [group B]) were identified as a SIG member with high confidence/probability by all three platforms. Additionally, all three systems misidentified at least one isolate as S. aureus, with Vitek 2 misidentifying the most—six isolates (one group A and five group B isolates), compared to one and two isolates by the MicroScan WalkAway and BD Phoenix platforms, respectively.
We and others have shown that pyrrolidonyl arylamidase (PYR) activity can generally differentiate between S. aureus (PYR negative) and SIG members (PYR positive) (3, 12, 18). In a study by Compton et al., using the same collection of 21 S. delphini isolates described here, all isolates tested positive for PYR activity using PYR broth (reference method), and only one isolate at a single testing site tested negative with one of the four rapid PYR test kits assayed, for an overall interlaboratory agreement of 99.4% (167/168 positive tests obtained for the 21 isolates with four different rapid PYR kits after testing at two different sites) (18). In this study, all isolates tested positive for PYR on the MicroScan WalkAway and Vitek 2 systems, while 71.4% (15/21) of isolates tested PYR positive on the BD Phoenix platform, suggesting that PYR reactivity on these platforms could assist with differentiating between S. aureus and SIG members in instances where there is suspicion for SIG, yet identification is not definite. However, we note that S. aureus can also test PYR positive on an automated biochemical platform (19), and thus PYR may not reliably differentiate between S. aureus and SIG members when extrapolated from such platforms.
In contrast to automated biochemical systems, the MALDI Biotyper was able to accurately identify S. delphini to the species level with log (score) values of ≥2.00 using RUO databases 6903 and 7311, which is in agreement with previous reports probing the performance of MALDI-TOF MS for identifying SIG members to the species level (20, 21). Using the Axima Confidence MALDI-TOF MS (Shimadzu Corp., Kyoto, Japan), Decristophoris et al. showed that 63.6% (14/22) of isolates were identified with confidence values of ≥90% (20). In the study conducted by Murugaiyan and coworkers, 78.9% (15/19) of isolates had log (score) values of ≥2.00 using either the RUO 4613 database or by applying an in-house reference spectra extended version using the Bruker platform (21). Based upon those data, it is clear that a higher proportion of S. delphini isolates were identified to the species level in this study.
The importance of identifying S. pseudintermedius to the species level when recovered from animal and human clinical specimens is now widely recognized. The significance of doing so for other SIG members is not obvious at this time. There is only a single report of human infection due to S. delphini (although this may be a function of the inability of biochemical systems to accurately identify this organism, resulting in underestimation of its prevalence in the human population), and to the best of our knowledge, mecA-mediated β-lactam resistance has not been documented in S. delphini (all 21 isolates included in this study were negative for mecA). However, methicillin resistance in S. pseudintermedius increased in canine isolates from less than 5% in 2001 to near 30% in 2007 (14), and it has subsequently emerged in human-associated isolates (9, 10, 15). In many instances, mecA-mediated β-lactam resistance in S. pseudintermedius is accompanied by resistance to other classes of antimicrobials (e.g., fluoroquinolone, tetracycline, and lincosamide) commonly used in veterinary and human medicine (22). Recently, S. delphini isolates recovered from Danish mink harboring high rates of resistance to erythromycin (20% of isolates), penicillin (47%), and tetracycline (51%) have been described (23). Taken together, these data demonstrate that SIG members have the ability to readily acquire resistance to antimicrobials of central importance to human and animal medicine, and they suggest that identification of SIG members apart from S. pseudintermedius to the species level may be important for therapeutic and epidemiologic purposes.
In conclusion, we demonstrate that MALDI-TOF MS is currently the best identification platform to identify S. delphini to the species level. Subsequent to this study, S. delphini is now included as a clinically validated organism in the MALDI Biotyper Claim 4 IVD database, and is a non-clinically validated organism in the Vitek MS Knowledge Base v3.2 database. In contrast, S. delphini has not been added to the databases of any of the commercial automated biochemical identification platforms tested herein. Thus, it may be possible for manufacturers of these automated biochemical platforms to enhance their identification databases through inclusion of biochemical data acquired from S. delphini isolates obtained from different host species and geographic locations.
ACKNOWLEDGMENTS
We are grateful to Craig Altier, Jean Brinkmeyer, Markus Kostrzewa, David Pincus, and Stefanie Richter for review of the manuscript.
B. L. Zimmer is an employee of Beckman Coulter, Inc.
REFERENCES
- 1.Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, van den Broek AHM, Fitzgerald JR. 2007. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol 189:8685–8692. doi: 10.1128/JB.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. 2007. Reclassification of phenotypically identified Staphylococcus intermedius strains. J Clin Microbiol 45:2770–2778. doi: 10.1128/JCM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray AK, Lee J, Bendall R, Zhang L, Sunde M, Schau Slettemeås J, Gaze W, Page AJ, Vos M. 2018. Staphylococcus cornubiensis sp. nov., a novel member of the Staphylococcus intermedius group (SIG). Int J Syst Evol Microbiol 68:3404–3408. doi: 10.1099/ijsem.0.002992. [DOI] [PubMed] [Google Scholar]
- 4.Varaldo PE, Kilpper-Bälz R, Biavasco F, Satta G, Schleifer KH. 1988. Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int J Syst Bacteriol 38:436–439. doi: 10.1099/00207713-38-4-436. [DOI] [Google Scholar]
- 5.Guardabassi L, Schmidt KR, Petersen TS, Espinosa-Gongora C, Moodley A, Agersø Y, Olsen JE. 2012. Mustelidae are natural hosts of Staphylococcus delphini group A. Vet Microbiol 159:351–353. doi: 10.1016/j.vetmic.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Börjesson S, Gómez-Sanz E, Ekström K, Torres C, Grönlund U. 2015. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur J Clin Microbiol Infect Dis 34:839–844. doi: 10.1007/s10096-014-2300-y. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Murray A, Bendall R, Gaze W, Zhang L, Vos M. 2015. Improved detection of Staphylococcus intermedius group in a routine diagnostic laboratory. J Clin Microbiol 53:961–963. doi: 10.1128/JCM.02474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuan EC, Yoon AJ, Vijayan T, Humphries RM, Suh JD. 2016. Canine Staphylococcus pseudintermedius sinonasal infection in human hosts. Int Forum Allergy Rhinol 6:710–715. doi: 10.1002/alr.21732. [DOI] [PubMed] [Google Scholar]
- 9.Somayaji R, Priyantha MA, Rubin JE, Church D. 2016. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis 85:471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Yarbrough ML, Lainhart W, Burnham CA. 2018. Epidemiology, clinical characteristics, and antimicrobial susceptibility of Staphylococcus intermedius group. J Clin Microbiol 56:e01788-17. doi: 10.1128/JCM.01788-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magleby R, Bemis DA, Kim D, Carroll KC, Castanheira M, Kania SA, Jenkins SG, Westblade LF. 2019. First reported human isolation of Staphylococcus delphini. Diagn Microbiol Infect Dis 94:274–276. doi: 10.1016/j.diagmicrobio.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Becker K, Skov RL, von Eiff C. 2015. Staphylococcus, Micrococcus, and other catalase-positive cocci, p 354–382. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW, (ed). Manual of clinical microbiology, 11th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 13.Pottumarthy S, Schapiro JM, Prentice JL, Houze YB, Swanzy SR, Fang FC, Cookson BT. 2004. Clinical isolates of Staphylococcus intermedius masquerading as methicillin-resistant Staphylococcus aureus. J Clin Microbiol 42:5881–5884. doi: 10.1128/JCM.42.12.5881-5884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bemis DA, Jones RD, Frank LA, Kania SA. 2009. Evaluation of susceptibility test breakpoints used to predict mecA-mediated resistance in Staphylococcus pseudintermedius isolated from dogs. J Vet Diagn Invest 21:53–58. doi: 10.1177/104063870902100108. [DOI] [PubMed] [Google Scholar]
- 15.Wu MT, Burnham CA, Westblade LF, Dien Bard J, Lawhon SD, Wallace MA, Stanley T, Burd E, Hindler J, Humphries RM. 2016. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J Clin Microbiol 54:535–542. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaiotta G, Fusco V, Ercolini D, Pepe O, Coppola S. 2010. Diversity of Staphylococcus species strains based on partial kat (catalase) gene sequences and design of a PCR-restriction fragment length polymorphism assay for identification and differentiation of coagulase-positive species (S. aureus, S. delphini, S. hyicus, S. intermedius, S. pseudintermedius, and S. schleiferi subsp. coagulans). J Clin Microbiol 48:192–201. doi: 10.1128/JCM.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. 2010. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol 48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton ST, Kania SA, Robertson AE, Lawhon SD, Jenkins SG, Westblade LF, Bemis DA. 2017. Evaluation of pyrrolidonyl arylamidase activity in Staphylococcus delphini. J Clin Microbiol 55:859–864. doi: 10.1128/JCM.02076-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong SY, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. 2011. Identification of Staphylococcus intermedius group by MALDI-TOF MS. Syst Appl Microbiol 34:45–51. doi: 10.1016/j.syapm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Murugaiyan J, Walther B, Stamm I, Abou-Elnaga Y, Brueggemann-Schwarze S, Vincze S, Wieler LH, Lübke-Becker A, Semmler T, Roesler U. 2014. Species differentiation within the Staphylococcus intermedius group using a refined MALDI-TOF MS database. Clin Microbiol Infect 20:1007–1015. doi: 10.1111/1469-0691.12662. [DOI] [PubMed] [Google Scholar]
- 22.Humphries RM, Wu MT, Westblade LF, Robertson AE, Burnham CA, Wallace MA, Burd EM, Lawhon S, Hindler JA. 2016. In vitro antimicrobial susceptibility of Staphylococcus pseudintermedius isolates of human and animal origin. J Clin Microbiol 54:1391–1394. doi: 10.1128/JCM.00270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaisen NK, Lassen DCK, Chriél M, Larsen G, Jensen VF, Pedersen K. 2017. Antimicrobial resistance among pathogenic bacteria from mink (Neovison vison) in Denmark. Acta Vet Scand 59:60. doi: 10.1186/s13028-017-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]