Abstract
Campylobacter jejuni (C. jejuni) are able to colonise and infect domestic poultry and also pose a risk for humans. The aim of this study was to determine the extent of genotypic diversity among C. jejuni isolates recovered from avian and human sources in Egypt. Furthermore, the short variable region (SVR) of flagellin A (flaA) gene was analysed for the presence of allelic variants. Our results showed that C. jejuni isolates differ in their capacity to harbour each of the virulence genes alone or when present in various combinations. The flaA gene was detected in all C. jejuni strains and none of the strains had all the studied virulence genes together. When considering C. jejuni strains from the investigated sources, the cdtC gene was the most similar, while the cdtB and iam genes were the most dissimilar. We could identify 13 novel alleles in the analysed strains. The analyses of virulence gene patterns, flaA gene sequences and allelic variants showed that C. jejuni strains from different sources overlapped largely suggesting potential involvement of poultry in transmitting C. jejuni to humans. We also found that the strains isolated from the same host were highly heterogeneous, with chicken strains exhibiting the highest diversity. Moreover, the human strains were clustered closer to chicken ones than to those from pigeon. The results of this study should be taken into consideration when assessing the epidemiology and risk potential of Egyptian C. jejuni not only in poultry, but also in humans.
Keywords: alleles, C. jejuni, flaA typing, humans, poultry, virulence
Introduction
Campylobacter jejuni (C. jejuni) is one of the most frequent bacterial causes of foodborne gastroenteritis worldwide (European Food Safety Authority, 2014). It has been isolated from chickens, turkeys, pigeons and quails (Vazquez et al., 2010; Kovacic et al., 2013; Ramees et al., 2015), where they colonise the intestinal tract and could contaminate the carcasses during processing (e.g., defeathering and evisceration) (Hermans et al., 2011). Human infection occurs commonly through consumption of contaminated poultry meat (Humphrey et al., 2007).
Many studies addressed the virulence characteristics of C. jejuni (Frazão et al., 2017; Maansi et al., 2018). In Egypt, limited studies have focussed on the genotypic diversity (Ahmed et al., 2015) and the antimicrobial resistance of C. jejuni (Abd El-Tawab et al., 2018), yet none of these studies explored the virulence patterns of C. jejuni among different sources, particularly pigeons.
It is well established that C. jejuni exhibits high diversity with regard to the presence of virulence and/or pathogenicity traits including adherence (Coote et al., 2007), invasion (Zheng et al., 2006), toxicity (Abuoun et al., 2005), and molecular mimicry (Datta et al., 2003). Determining the extent of genetic heterogeneity of C. jejuni will tell about the disease burden in certain populations and will aid in predicting the potential source of infection (McCarthy et al., 2007); all are valuable information that can be delivered to surveillance and control programmes with an overarching goal of reducing the disease in poultry and its transmission to humans.
The genomic instability of the flagellin gene, due to frequent occurrence of genomic recombination (Harrington et al., 1997), makes it a good candidate for studying the genetic diversity of C. jejuni. Therefore, molecular typing of flaA gene, and in particular its short variable region (SVR; ∼150 base pairs), has been widely used in studying the genotypic diversity of such bacteria (Meinersmann et al., 1997, 2005; Wardak and Jagielski, 2009). flaA typing represents a convenient and cost-effective typing-scheme, which suits the situation in developing countries, particularly when quick characterisation of the strains is needed. Previous studies have demonstrated that direct sequencing of PCR-amplified flaA-SVRs is useful for Campylobacter genotyping, particularly in short-term and localised epidemiological investigations, allowing similar or higher discriminatory power than multilocus sequence typing (MLST). Furthermore, MLST is unable to distinguish closely related strains in small-scale outbreak investigations and additional methods like flaA typing may be required in order to obtain sufficient resolution (Meinersmann et al., 1997; Sails et al., 2003).
Campylobacter jejuni isolated from humans and poultry are known to be genetically diverse (Ramonaite et al., 2017). Although C. jejuni in Egypt is widely distributed in avian hosts (Rahimi and Ameri, 2011; Abd El-Tawab et al., 2018) and humans (Sainato et al., 2018), there are limited studies that addressed the genetic heterogeneity among Egyptian C. jejuni.
To unravel the extent of genetic diversity of Egyptian C. jejuni, we analysed both virulence gene profiles and flaA allelic variants in C. jejuni isolated from chickens, pigeons and humans. The knowledge gained from this study is highly relevant to the epidemiology and control efforts geared toward reducing C. jejuni infection in Egypt.
Materials and Methods
Samples
The study was conducted during the period from 2015–2018. A total of 270 samples were collected from individual broiler chickens (n = 90) [chicken meat (n = 65) and cloacal swabs (n = 25)] and fresh pigeon droplets (n = 180) of different ages from 10 retail outlets in Zagazig city, Sharkia Governorate, Egypt. Moreover, 270 human stool samples were collected from gastroenteritis patients attending Zagazig University hospital located in the same city, Zagazig. The collected samples were transported in an ice box within 3 h to the laboratory for bacteriological analysis. The animal study was approved by the committee of Animal Welfare and Research Ethics, Faculty of Veterinary Medicine, Zagazig University. Regarding the human study, it was approved by the research ethical committee of Faculty of Medicine, Zagazig University and the work was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for studies involving humans. Written informed consents were obtained from the patients participating in the research study after a full description of the study purpose.
Isolation and Identification of C. jejuni
For the isolation of Campylobacter species, samples were enriched in Preston Campylobacter selective enrichment broth (Oxoid, United Kingdom) at 42°C for 48 h. The enrichment cultures were first streaked onto modified charcoal cefoperazone deoxycholate agar (mCCDA; Oxoid, United Kingdom), then transferred onto Columbia agar (Oxoid, United Kingdom) plates supplemented with 5% sterile defibrinated horse blood. The plates were incubated at 42°C in darkness for 48 h under microaerobic conditions (Vandepitte et al., 2003). The presumptive isolates were confirmed as C. jejuni by biochemical tests including catalase, oxidase, hippurate and indoxyl acetate hydrolyses, and susceptibility to cephalothin and nalidixic acid (30 μg/disc, each) (Quinn et al., 1994). Genomic DNA was extracted from fresh C. jejuni cultures using the QIAamp DNA Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. Polymerase chain reaction (PCR)-based amplification of 23S rRNA and hipO genes was done to confirm the identification of C. jejuni isolates (Wang et al., 2002).
PCR-Based Detection of Virulence Genes
Campylobacter jejuni isolates were screened for the presence of 7 virulence genes namely flaA, virB11, wlaN, iam, cdtA, cdtB and cdtC. All PCR assays were performed in a 25 μL reaction mixture containing 12.5 μL of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 μL of each primer (20 pM; Metabion, Germany), 6 μL of purified C. jejuni DNA and 4.5 μL of nuclease-free water. Primers used for PCR assays are listed in Table 1.
TABLE 1.
Oligonucleotide primers used for the amplification of genus, species and virulence genes of C. jejuni isolates.
| Target gene | Primer name | Oligonucleotide sequence (5′→3′) | Amplicon size (bp) | Reference |
| 23S rRNA | 23SF | TATACCGGTAAGGAGTGCTGGAG | 650 | Wang et al., 2002 |
| 23SR | ATCAATTAACCTTCGAGCACCG | |||
| hipO | CJF | ACTTCTTTATTGCTTGCTGC | 323 | Wang et al., 2002 |
| CJR | GCCACAACAAGTAAAGAAGC | |||
| cdtA | GNW | GGAAATTGGATTTGGGGCTATACT | 165 | Wieczorek et al., 2012 |
| IVH | ATCACAAGGATAATGGACAAT | |||
| cdtB | VAT2 | GTTAAAATCCCCTGCTATCAACCA | 495 | Wieczorek et al., 2012 |
| WMI-R | GTTGGCACTTGGAATTTGCAAGGC | |||
| cdtC | WMI-F | TGGATGATAGCAGGGGATTTTAAC | 555 | Wieczorek et al., 2012 |
| LPF-X | TTGCACATAACCAAAAGGAAG | |||
| flaA | FLA242FU | CTATGGATGAGCAATT(AT)AAAAT | 402 | Meinersmann et al., 1997 |
| FLA625RU | CAAG(AT)CCTGTTCC(AT)ACTGAAG | |||
| virB11 | virB-232 | TCTTGTGAGTTGCCTTACCCCTTTT | 494 | Datta et al., 2003 |
| virB-701 | CCTGCGTGTCCTGTGTTATTTACCC | |||
| wlaN | wlaN DL-39 | TTAAGAGCAAGATATGAAGGTG | 672 | Kordinas et al., 2005 |
| wlaN DL-41 | CCATTTGAATTGATATTTTTG | |||
| iam | IAMF | GCGCAAATATTATCACCC | 518 | Wieczorek et al., 2012 |
| IAMR | TTCACGACTACTACTATGCGG |
FlaA-SVR Sequencing and Allelic Typing
Sequencing of internal 402 bp fragments of the flaA-SVR amplicons was performed using the previously described primer pair Fla242FU and Fla625RU (Meinersmann et al., 1997). The flaA amplicons were purified using the QIAquick PCR purification kit (Qiagen, United States) and sequenced in an ABI 3130 automated DNA Sequencer (Applied Biosystems, United States) using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, United States). flaA-SVR sequences were compared with those previously published at GenBank using Basic Local Alignment Search Tool (BLAST) available at the National Center for Biotechnology Information (NCBI)1. To investigate the genetic relatedness of C. jejuni isolates, a phylogenetic tree was built based on flaA-SVR sequences using neighbour-joining method (Saitou and Nei, 1987) and the Kimura’s two-parameter method (Kimura, 1980) as an estimator for the evolutionary distance among strains. The topology of the tree was evaluated by 1,000 iterations of a bootstrap analysis. This analysis was done using MEGA software (v. 5) (Tamura et al., 2011). A quantitative assessment of the similarity among the analysed sequences was done by running a Clustal W multiple alignment (Thompson et al., 1994) of all sequences. This enabled the generation of percent identity among pairs of flaA-SVR gene sequences without accounting for phylogenetic relationships. This analysis was done using the MegAlign software in the Lasergene (v. 7.1.0) package (DNASTAR, Madison, WI, United States).
To retrieve the allelic variants of C. jejuni strains, all nucleotide and peptide sequences of flaA-SVRs, using their accession numbers as queries, were used as inputs in Campylobacter pubMLST flaA Oxford database2; these were then compared against reference C. jejuni strains deposited in this database. The allelic variants of flaA-SVR nucleotides and peptides of C. jejuni strains were assigned accordingly.
Bioinformatics and Data Analyses
PC-ORD Software (v. 5, Oregon, United States), the web-based tools MetaboAnalyst (Chong and Xia, 2018), heatmapper (Babicki et al., 2016) and SPSS software (v. 25, IBM, United States) were used for bioinformatics and statistical analyses of the data. To analyse the overall clustering pattern of C. jejuni strains, the frequency of each virulence gene, scored as binary data (present = 1, absent = 0), was used as input in a non-metric multidimensional scaling (nMDS) analysis based on the Euclidean distance. To visualise the association between the studied virulence genes and thus their contribution to strain clustering pattern, we performed a hierarchical clustering using the virulence genes scored as binary data. To determine the relationship between the strain sources, a hierarchical clustering, based on Euclidean distances, was generated using the frequency of the presence of each virulence gene in each source as inputs. The estimated Euclidean distances were used to assess the degree of convergence among strain sources. To visualise the association between the strain sources and the virulence genes, a 2-D correspondence analysis was done, which explained 96% of the variance in the data. A third dimension was not used as its contribution was very low (proportion of inertia = 0.4). To determine the degree of similarity of each virulence gene among strains isolated from various hosts, we applied a random forest non-parametric classification method as described previously (Babicki et al., 2016). The degree of similarity of each gene among C. jejuni strains was determined considering the following isolation sources: human, chicken meat, chicken cloacal swabs and pigeon; chicken (meat and cloacal swabs together), human and pigeon; chicken (meat and cloacal swabs together) and human; human and pigeon; human and birds (chicken and pigeon). In this approach, the frequency of each virulence gene was firstly used to build up random forest classification model (the model prediction is based on the majority vote of an ensemble of 500 tree trial; out of bag error, OOB error = 0.6) in the respective source comparison. The degree of similarity of each gene was then evaluated by measuring the increase of the OOB error when the respective gene is permuted. To quantify the discriminatory power of each gene, a classic univariate receiver operating characteristic (ROC) curve was created using inputs similar to that described for the random forest classification. This enabled estimating the area under the curve (AUC) for each gene, which ranges from 0 to 1 (0 = least discriminatory power; 1 = highest discriminatory power). It was only technically possible to run the ROC analysis on the virulence genes comparing human and bird isolates.
Genotypic Diversity of C. jejuni Strains
Both Simpson’s (D1) (Simpson, 1949) and Shannon’s (H) (Shannon, 1948) diversity indices were used to assess the diversity of C. jejuni strains among various sources and within each source using the virulence genes profile as input data. Simpson’s diversity index was calculated based on the equation: 1-D. In this equation, D = =, where Σ = summation, n = total number of bacterial strains showing a particular gene profile and N = total number of bacterial strains in the respective source (e.g., in human strains). Simpson’s index has a range of 0–1, with 0 indicates identical bacterial strains and 1 indicates the highest diverse bacterial strains. Shannon’s diversity index (H) was calculated using the following equation: , where pi is the proportion of bacterial strains expressing a particular gene profile divided by the total number of bacterial strains in the respective source, In is the natural log of the pi, Σ is the summation, and s is the number of species. The concordance of the two indices was determined by calculating the Pearson correlation (r). This analysis was done using the Past3 software (v. 3.23), Oslo (Hammer et al., 2001).
Statistical Analysis
Statistical analysis of the frequencies of virulence genes within C. jejuni strains isolated from different hosts was assessed by Fisher’s exact test considering P-value < 0.05 as a cut off level. Data was analysed using Stata statistical software version 12 (TX, United States).
Nucleotide Sequence Accession Numbers
The flaA-SVR nucleotide sequences generated in this study have been deposited into the GenBank database with accession numbers KX066127-KX066135, MG677923-MG677934 and MK281494-MK281513.
Results
Prevalence of C. jejuni in Chickens, Pigeons and Humans
The overall prevalence of C. jejuni in avian samples (i.e., chicken and pigeon samples) was 11.11% (30/270). Fourteen out of 90 (15.56%) chicken samples were positive for C. jejuni; 10 from 65 chicken meat (15.38%) and 4 from 25 cloacal swabs (16%), while the isolation rate of C. jejuni in pigeon droplets was 8.89% (16/180). Moreover, 11 C. jejuni were isolated from 270 human stool samples (4.07%). Typical C. jejuni colonies were small, shiny, round, and grey on mCCDA agar medium. No haemolysis on Columbia blood agar and positive reactions for catalase, oxidase and hippurate and indoxyl acetate hydrolyses presumptively identified C. jejuni isolates. Besides, the analysed isolates were typically sensitive to nalidixic acid and resistant to cephalothin as measured by the inhibition zones around the relevant antibiotic discs. The genus and species identification of the isolates were further confirmed by PCR detection of 23S rRNA and hipO genes, respectively.
Frequency of Virulence Genes in C. jejuni Isolates
As depicted in Table 2, the flaA gene was present in all analysed C. jejuni strains (n = 41). The cdtA, cdtB, and cdtC toxin genes were similarly detected in C. jejuni strains (80.49%, each), whereas the three CDT toxin genes were simultaneously found in 58.54% of the analysed strains. Only two subunits of the CDT toxin gene (e.g., cdtBC, cdtAC, and cdtAB) were detected in 3 (7.32%), 3 (7.32%) and 6 (14.63%) of the strains, respectively. Moreover, 3 C. jejuni strains (7.32%) harboured the cdtC gene only and 2 strains (4.88%) had no CDT toxin genes at all (Supplementary Table S1). Besides, iam, wlaN and virB11 genes were found in 34.15, 12.20, and 9.76% of C. jejuni strains, respectively.
TABLE 2.
Occurrence of virulence genes in C. jejuni strains under study.
| Source | No. of C. jejuni strains |
Frequency of virulence genes, no. (%) |
||||||
| cdtA | cdtB | cdtC | flaA | virB11 | wlaN | iam | ||
| Pigeon | 16 | 14 (87.50) | 16 (100.00) | 14 (87.50) | 16 (100.00) | 1 (6.25) | 0 (0.00) | 8 (50.00) |
| Chicken | 14 | 9 (64.29) | 6 (42.86) | 10 (71.43) | 14 (100.00) | 2 (14.29) | 3 (21.43) | 6 (42.86) |
| Human | 11 | 10 (90.91) | 11 (100.00) | 9 (81.82) | 11 (100.00) | 1 (9.09) | 2 (18.18) | 0 (0.00) |
| Total | 41 | 33 (80.49) | 33 (80.49) | 33 (80.49) | 41 (100.00) | 4 (9.76) | 5 (12.20) | 14 (34.15) |
| P-value∗ | 0.173 | 0.000∗ | 0.545 | 1.000 | 0.763 | 0.164 | 0.021∗ | |
∗Statistical significance was calculated using Fisher’s exact test (P-value < 0.05).
The presence of the cdtB gene in human and pigeon strains was significantly (P = 0.0001) higher (100%, each) than in chicken ones (42.86%). Additionally, iam gene was significantly (P = 0.021) more frequently detected in pigeon strains (50%) when compared to chicken (42.86%) and human ones (0%).
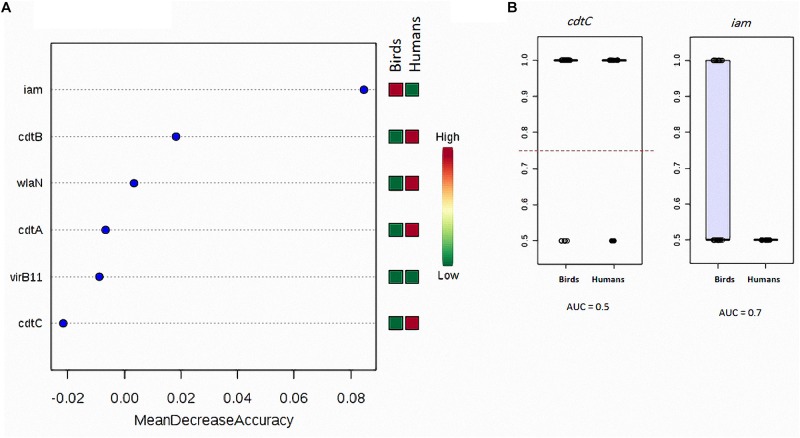
The random forest classification analysis showed that cdtC gene exhibited the highest similarity (the least mean decrease accuracy) when considering C. jejuni isolated from human and birds (chicken meat, chicken cloacal swabs and pigeon together) (Figure 1A). This gene was also the most similar when considering the following strain sources: (i) human, chicken meat, chicken cloacal swabs and pigeon, (ii) human and chicken (meat and cloacal swabs together), (iii) human and pigeon and (iv) human, chicken and pigeon (Supplementary Figure S1). On contrary, iam was the most dissimilar gene when considering C. jejuni strains from (i) human and birds (chicken meat, chicken cloacal swabs and pigeon together) and (ii) human and pigeon. The cdtB gene was the most dissimilar one when considering C. jejuni strains from (i) human, chicken meat, chicken cloacal swabs and pigeon, (ii) human, chicken (meat and cloacal swabs together) and pigeon and (iii) chicken (meat and cloacal swabs together) and humans. ROC-curve analysis (Figure 1B) on the human and bird strains revealed that the cdtC gene had lower AUC value (0.5) than that of the iam gene (0.7).
FIGURE 1.
Degree of similarity and discriminatory power of analysed virulence genes in C. jejuni strains among various sources. (A) Random forest classification showing the predictive accuracy of the studied genes (X-axis). The mini heat map shows the frequency distribution of each gene in the respective strain sources. Each dot refers to one gene with the respective mean decrease accuracy. (B) Box-plot showing the differences in the distribution of cdtC and iam genes between birds and human sources of C. jejuni. The horizontal dotted line refers to the optimal cutoff value. The AUC data were generated by running the ROC curves as described in the “Materials and Methods” section.
The results showed that none of the C. jejuni strains harboured all the seven virulence genes together. More than half of the strains (87.80%) possessed three or more virulence genes (Table 3). Irrespective of the strain source, cdtABC, flaA combination dominated the overall gene profiles, being encoded by 10 strains (24.39%), followed by cdtABC, flaA, iam pattern (7/41, 17.07%) (Supplementary Table S1 and Table 4).
TABLE 3.
Frequency of C. jejuni strains isolated from avian and human sources carrying virulence genes.
| No. of identified virulence genes | Human (n = 11) | Chicken∗ (n = 14) | Pigeon (n = 16) | Human and pigeon (n = 27) | Human and chicken∗ (n = 25) | Chicken∗ and pigeon (n = 30) | Human and chicken∗ and pigeon (n = 41) |
| 1 | 0 (0.00) | 2 (14.29) | 0 (0.00) | 0 (0.00) | 2 (8.00) | 2 (6.67) | 2 (4.88) |
| 2 | 0 (0.00) | 3 (21.43) | 0 (0.00) | 0 (0.00) | 3 (12.00) | 3 (10.00) | 3 (7.32) |
| 3 | 3 (27.27) | 2 (14.29) | 3 (18.75) | 6 (22.22) | 5 (20.00) | 5 (16.67) | 8 (19.51) |
| 4 | 5 (45.45) | 2 (14.29) | 6 (37.50) | 11 (40.74) | 7 (28.00) | 8 (26.67) | 13 (31.71) |
| 5 | 3 (27.27) | 2 (14.29) | 6 (37.50) | 9 (33.33) | 5 (20.00) | 8 (26.67) | 11 (26.83) |
| 6 | 0 (0.00) | 3 (21.43) | 1 (6.25) | 1 (3.70) | 3 (12.00) | 4 (13.33) | 4 (9.76) |
| 7 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Simpson diversity | 0.64 | 0.82 | 0.67 | 0.6722 | 0.80 | 0.82 | 0.77 |
| Shannon diversity | 1.06 | 1.77 | 1.223 | 1.18 | 1.71 | 1.7 | 1.60 |
Numbers of strains and percentages in parentheses indicate the proportion of those carrying the respective virulence genes. ∗Chicken strains refer to those isolated from chicken meat + chicken cloacal swabs.
TABLE 4.
Virulence gene patterns, allelic variants and mutations in flaA-SVR of C. jejuni isolated from avian and human sources.
| Isolate code no. | Source | Virulence genes pattern | flaA-SVR nucleotide allele (No. of variations) | flaA-SVR peptide allele (No. of variations) | ∗∗Translation variation reference → clone | Accession no. |
| 1 | Pigeon droplets | cdtABC, flaA, virB11, iam | 781 (30) | 191 (16) | 49I → 26T, 60N → 37K, 63V → 40E, 64K → 41I, 72S → 49T, 74K → 51R, 76G → 53I, 77L → 54F, 84E → 61R, 85R → 62I, 86I → 63T, 90G → 67E, 93Q → 70P, 94F → 71L, 95T → 72P, 96L → 73F | KX066127.1 |
| 2 | Pigeon droplets | cdtABC, flaA, iam | 731 (32) | 191 (16) | 49I → 27T, 57A → 35E, 60N → 38K, 63V → 41E, 64K → 42I, 66T → 44I, 67I → 45M, 72S → 50T, 74K → 52R, 84E → 62R, 85R → 63I, 86I → 64T, 93Q → 71P, 94F → 72L, 96L → 74F, 101G → 79H | KX066128.1 |
| 3 | Pigeon droplets | cdtABC, flaA, iam | 940∗ | 58 (10) | 49I → 26T, 77L → 54V, 84G → 61A, 86I → 63S, 89S → 66R, 93Q → 70A, 94F → 71V, 96L → 73V, 99Y → 76F, 101G → 78A | KX066129.1 |
| 4 | Pigeon droplets | cdtABC, flaA, iam | 38 (17) | 5 (7) | 49I → 27T, 71Q → 49H, 74K → 52N, 79R → 57C, 89S → 67F, 96L → 74F, 106K → 84P | KX066130.1 |
| 5 | Pigeon droplets | cdtABC, flaA, iam | 1118 (18) | 239 (13) | 56G → 32C, 57A → 33S, 59S → 35L, 62T → 38I, 71Q → 47I, 76G → 52A, 78T → 54K, 79R → 55C, 90G → 66T, 92V → 68E, 99Y → 75S, 101A → 77C, 104D → 80V | KX066131.1 |
| 6 | Pigeon droplets | cdtABC, flaA, iam | 1275 (19) | 209 (13) | 57A → 35S, 62T → 40I, 64K → 42I, 68G → 46A, 70T → 48S, 71Q → 49H, 72S → 50T, 75I → 53M, 94L → 72V, 96L → 74F, 99Y → 77D, 102I → 80K, 104D → 82V | KX066132.1 |
| 7 | Pigeon droplets | cdtABC, flaA | 186 (25) | 356 (17) | 62T → 37A, 63I → 38V, 64K → 39I, 65A → 40V, 72S → 47P, 73S → 48F, 74K → 49Q, 78T → 53S, 85R → 60S, 86I → 61V, 88S → 63T, 93Q → 68S, 96L → 71P, 99Y → 74D, 101G → 76S, 103D → 78H, 104D → 79G | KX066134.1 |
| 8 | Pigeon droplets | cdtABC, flaA | 9 (26) | 60 (15) | 33A → 8T, 49I → 24V, 52E → 27K, 57A → 32E, 66T → 41I, 67I → 42T, 72S → 47T, 77L → 52F, 82T → 57R, 83G → 58V, 90G → 65S, 94F → 69A, 96L → 71H, 102L → 77R, 106Q → 81L | KX066135.1 |
| 9 | Pigeon droplets | cdtAB, flaA | 781 (31) | 191 (14) | 49I → 26T, 57A → 34E, 60N → 37K, 63V → 40E, 64K → 41I, 72S → 49T, 74K → 51R, 76G → 53I, 84E → 61R, 85R → 62I, 86I → 63T, 93Q → 70P, 94F → 71L, 96L → 73F | MK281505 |
| 10 | Pigeon droplets | cdtAB, flaA, iam | 1118 (17) | 239 (13) | 56G → 33C, 57A → 34S, 59S → 36L, 62T → 39I, 68G → 45A, 71Q → 48N, 76G → 53A, 78T → 55K, 79R → 56C, 90G → 67T, 92V → 69E, 101A → 78C, 104D → 81V | MK281506 |
| 11 | Pigeon droplets | cdtBC, flaA | 938 (19) | 209 (12) | 57A → 34S, 62T → 39I, 64K → 41I, 68G → 45A, 70T → 47S, 71Q → 48N, 72S → 49T, 75I → 52M, 94L → 71V, 96L → 73F, 102I → 79K, 104D → 81V | MK281507 |
| 12 | Pigeon droplets | cdtABC, flaA | 177 (10) | 74 (5) | 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L, 102I → 108L | MK281508 |
| 13 | Pigeon droplets | cdtABC, flaA, iam | 1486 (11) | 92 (8) | 19Q → 11L, 26M → 18I, 29L → 21I, 63V → 55M, 77V → 69F, 96I → 88V, 97K → 89Q, 99Y → 91F | MK281509 |
| 14 | Pigeon droplets | cdtABC, flaA | 756 (26) | 58 (9) | 49I → 26T, 74K → 51N, 86I → 63S, 89S → 66R, 94F → 71V, 96L → 73V, 99Y → 76F, 101G → 78A, 106K → 83P | MK281510 |
| 15 | Pigeon droplets | cdtBC, flaA | 38 (18) | 239 (5) | 49I → 27T, 74K → 52N, 89S → 67R, 96L → 74F, 106Q → 84P | MK281511 |
| 16 | Pigeon droplets | cdtABC, flaA | 1183∗ | 239 (15) | 57A → 32S, 59S → 34L, 62T → 37I, 64K → 39I, 70T → 45S, 71Q → 46I, 72S → 47T, 75I → 50M, 90G → 65T, 94F → 69V, 96L → 71F, 99Y → 74S, 101A → 76C, 102I → 77K, 104D → 79V | MK281512 |
| 17 | Chicken meat | cdtABC, flaA, wlaN, iam | 177 (8) | 74 (5) | 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L, 102I → 108L | MG677923.1 |
| 18 | Chicken meat | cdtABC, flaA, virB11, iam | 177 (9) | 74 (5) | 10Q → 10E, 23N → 23Y, 39N → 39D, 76G → 76D, 89S → 89L | MG677924.1 |
| 19 | Chicken meat | cdtABC, flaA, iam | 1486 (11) | 92 (7) | 19Q → 13L, 26M → 20I, 63V → 57M, 80F → 74I, 87F → 81L, 97K → 91Q, 104D → 98E | MG677925.1 |
| 20 | Chicken meat | cdtAB, flaA, iam | 177 (12) | 74 (8) | 5A → 11V, 32I → 38T, 36T → 42A, 45S → 51N, 58S → 64G, 71Q → 77L, 81E → 87A, 102I → 108L | MG677926.1 |
| 21 | Chicken cl. swab | cdtAB, flaA | 526∗ | 211 (13) | 62T → 38I, 67I → 43L, 71Q → 47H, 72S → 48T, 74K → 50I, 80F → 56V, 85K → 61S, 86I → 62V, 94F → 70A, 95T → 71A, 99Y → 75A, 102I → 78V, 103D → 79G | KX066133.1 |
| 22 | Chicken cl. swab | cdtABC, flaA, virB11, wlaN | 177 (9) | 74 (6) | 5A → 11V, 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L, 102I 108L | MK281494 |
| 23 | Chicken cl. swab | cdtAC, flaA, wlaN, iam | 177 (12) | 74 (8) | 5A → 11V, 32I → 38T, 36T → 42A, 45S → 51N, 58S → 64G, 71Q → 77L, 81E → 87A, 102I → 108L | MK281513 |
| 24 | Chicken cl. swab | flaA | 288∗ | 92 (9) | 10Q → 8E, 23N → 21Y, 26M → 24I, 39N → 37D, 63V → 61S, 76G → 74D, 80F → 78I, 87F → 85L, 89S → 87L | MK281495 |
| 25 | Chicken meat | cdtC, flaA | 1486 (12) | 92 (7) | 19Q → 15L, 26M → 22I, 29L → 25I, 39N → 35D, 63V → 59M, 97K → 93Q, 104D → 100E | MK281496 |
| 26 | Chicken meat | flaA | 177 (5) | 74 (2) | 39N → 15D, 80F → 56I | MK281497 |
| 27 | Chicken meat | cdtC, flaA | 288 (8) | 92 (4) | 26M → 21I, 63V → 58M, 87F → 82L, 104D → 99E | MK281498 |
| 28 | Chicken meat | cdtAC, flaA, iam | 9 (27) | 211 (15) | 33A → 8T, 62T → 37I, 67I → 42L, 72S → 47T, 74K → 49I, 80F → 55V, 82T → 57R, 85K → 60S, 86I → 61V, 94F → 69A, 95T → 70A, 96L → 71H, 99Y → 74A, 102I → 77V, 103D → 78G | MK281499 |
| 29 | Chicken meat | cdtC, flaA | 526 (23) | 211 (13) | 49I → 24V, 67I → 42L, 71Q → 46H, 72S → 47T, 74K → 49I, 80F → 55V, 83G → 58V, 85K → 60S, 86I → 61V, 94F → 69A, 95T → 70A, 102I → 77V, 103D → 78G | MK281500 |
| 30 | Chicken meat | cdtAC, flaA | 526∗ | 60 (14) | 52E → 28K, 62T → 38I, 66T → 42I, 67I → 43T, 71Q → 47H, 72S → 48T, 74K → 50I, 80F → 56L, 90G → 66S, 94F → 70A, 95T → 71A, 99Y → 75A, 102L → 78R, 103D → 79G | MK281501 |
| 31 | Human stool | cdtABC, flaA, virB11 | 177 (8) | 74 (5) | 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L, 102I → 108L | MG677927.1 |
| 32 | Human stool | cdtABC, flaA | 239 (10) | 9 (8) | 1K → 7R, 12L → 18S, 27E → 33D, 36T → 42S, 42Q → 48H, 73S → 79A, 92V → 98D, 98N → 104S | MG677928.1 |
| 33 | Human stool | cdtABC, flaA, wlaN | 288 (11) | 92 (7) | 8D → 14A, 26M → 32I, 29L → 35V, 63V → 69M, 71Q → 77L, 87F → 93L, 104D → 110E | MG677929.1 |
| 34 | Human stool | cdtABC, flaA | 177 (15) | 74 (7) | 10Q → 10E, 23N → 23Y, 52E → 52Q, 65A → 65G, 76G → 76D, 88T → 88A, 89S → 89L | MG677930.1 |
| 35 | Human stool | cdtABC, flaA, wlaN | 1064 (10) | 267 (10) | 4Q → 10P, 17M → 23L, 31N → 37T, 38F → 44I, 50N → 56Y, 66T → 72I, 79R → 85I, 91V → 97A, 102I → 108L, 107Y → 113L | MG677931.1 |
| 36 | Human stool | cdtABC, flaA | 239 (9) | 9 (8) | 8D → 14E, 10Q → 16H, 19Q → 25K, 39N → 45Y, 46G → 52R, 62T → 68P, 74K → 80Q, 106K → 112Q | MG677932.1 |
| 37 | Human stool | cdtABC, flaA | 177 (14) | 74 (9) | 3T → 3I, 19Q → 19E, 34N → 34Y, 52E → 52D, 70T → 70S, 77V → 77A, 96I → 96V, 101G → 101S, 106K → 106E | MG677933.1 |
| 38 | Human stool | cdtABC, flaA | 288 (7) | 92 (5) | 9G → 2V, 34N → 27S, 58S → 51T, 63V → 56M, 106K → 99R | MG677934.1 |
| 39 | Human stool | cdtAB, flaA | 177 (8) | 74 (5) | 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L, 102I → 108L | MK281502 |
| 40 | Human stool | cdtBC, flaA | 177 (8) | 74 (4) | 32I → 38T, 36T → 42A, 58S → 64G, 71Q → 77L | MK281503 |
| 41 | Human stool | cdtAB, flaA | 177 (13) | 74 (8) | 19Q → 12E, 34N → 27Y, 52E → 45D, 70T → 63S, 77V → 70A, 96I → 89V, 101G → 94S, 106K → 99E | MK281504 |
SVR, short variable region; cl. swab, cloacal swabs; A, alanine; R, arginine; N, asparagine; D, aspartic acid; C, cysteine; E, glutamic acid; Q, glutamine; G, glycine; H, histidine; I, isoleucine; L, leucine; K, lysine; M, methionine; F, phenylalanine; P, proline; S, serine; T, threonine; Y, tyrosine; V, valine. ∗ Nucleotide alleles with asterisks have frameshift rather than point mutations. Novel allelic variants identified in this study are shown in bold. ∗∗The information to the left of the arrow refer to the identity and the position on the reference strains (deposited on http://pubmlst.org/campylobacter/) and the information to the right of the arrow show the corresponding identity and position on the query strains.
The strains from pigeon and human sources (n = 27) harbored at least 3–6 virulence genes. In contrast, 50% of the chicken strains were positive for up to 3 virulence genes (Table 3). As shown in Supplementary Table S1 and Table 5, cdtABC, flaA dominated the gene profiles in human strains (5/11, 45.45%), was not detected at all in chicken strains and was the second frequent pattern in pigeon strains (5/16, 31.25%). While, cdtABC, flaA, iam pattern was the most frequent in pigeon (6/16, 37.50%), it was not detected in any of human strains and was found in only one chicken strain (1/14, 7.14%). The cdtC, flaA was the dominant pattern in chicken strains (3/14, 21.43%), but none of human or pigeon strains possessed this pattern.
TABLE 5.
Distribution of virulence associated genes and identified allele numbers (at both nucleotide and peptide levels) among the entire collection of C. jejuni strains.
| Isolate source | Virulence gene pattern∗ |
flaA-SVR types (alleles) |
|
| Nucleotide | Peptide | ||
| Human (n = 11) | cdtABC, flaA (5); cdtAB, flaA (2); cdtABC, flaA, wlaN (2); cdtBC, flaA (1); cdtABC, flaA, virB11 (1) | 177 (6); 288 (2); 239 (2); 1064 (1) | 74 (6); 92 (2); 9 (2); 267 (1) |
| Chicken (n = 14) | cdtC, flaA (3); flaA (2); cdtABC, flaA, iam (1); cdtABC, flaA, virB11, iam (1); cdtAB, flaA (1); cdtAB, flaA, iam (1); cdtABC, flaA, wlaN, iam (1); cdtABC, flaA, virB11, wlaN (1); cdtAC, flaA, wlaN, iam (1); cdtAC, flaA, iam (1); cdtAC, flaA (1) | 177 (6); 526 (3); 288 (2); 1486 (2); 9 (1) | 74 (6); 92 (4); 211 (3); 60 (1) |
| Pigeon (n = 16) | cdtABC, flaA, iam (6); cdtABC, flaA (5); cdtBC, flaA (2); cdtABC, flaA, virB11, iam (1); cdtAB, flaA (1); cdtAB, flaA, iam (1) | 781 (2); 38 (2); 1118 (2); 731 (1); 940 (1); 1275 (1); 186 (1); 9 (1); 938 (1); 177 (1); 1486 (1); 756 (1); 1183 (1) | 239 (4); 191 (3); 58 (2); 209 (2); 5 (1); 356 (1); 60 (1); 74 (1); 92 (1) |
SVR, short variable region. ∗Numbers in parentheses indicate the number of strains containing the respective virulence genes combination.
Clustering Pattern of the Virulence Genes and Their Association With C. jejuni Strains
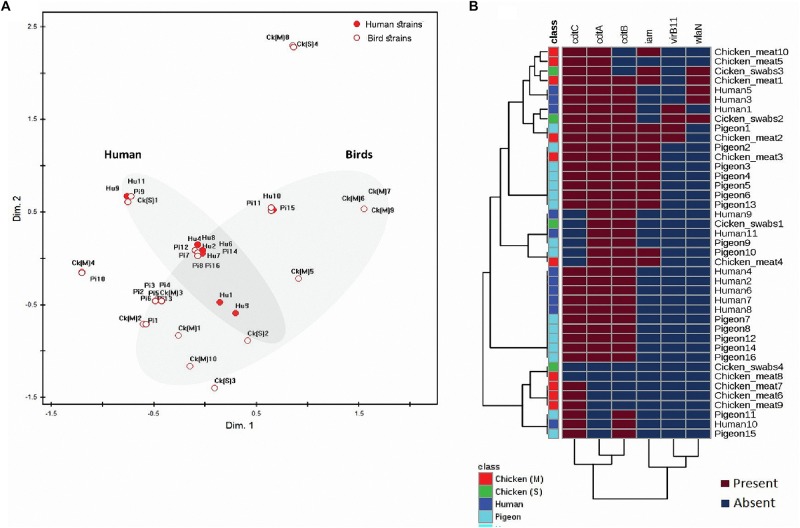
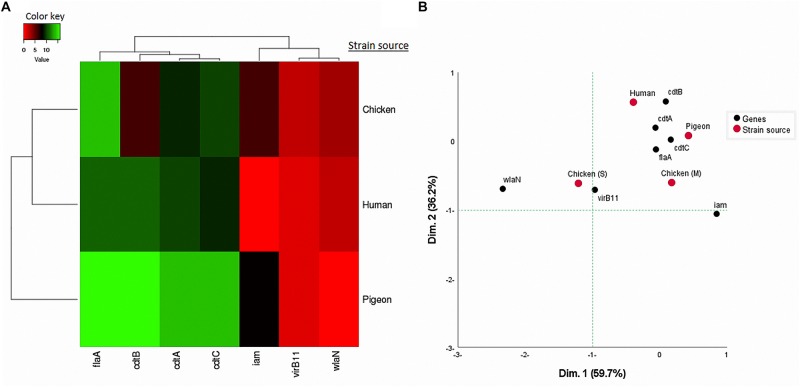
Considering the 41 C. jejuni strains separately (Figure 2B) and when grouped by their sources (Figure 3A), the virulence genes formed two separate clusters. The first cluster contained cdtA, cdtB, and cdtC genes and the second one included iam, virB11 and wlaN genes. The flaA gene was not included in this analysis as it was detected in 100% of the strains. By running a corresponding analysis (Figure 3B), it was evident that cdtA, cdtB, cdtC, and flaA genes were more associated with C. jejuni strains from humans, chicken meat and pigeons. The remaining genes (i.e., iam, virB11 and wlaN) showed a scattered pattern, being more associated with the chicken swab strains.
FIGURE 2.
Distribution and clustering of C. jejuni strains and the encoded virulence genes. (A) Non-metric multidimensional scaling (nMDS) showing the overlap between the bacterial strains (n = 41) represented by their sources. (B) Hierarchical clustering analyses showing the clustering pattern of both bacterial strains (n = 41) and the associated virulence genes (n = 6). FlaA gene was excluded from this analysis as it was expressed in all strains. The nMDS and the heat map were generated based on the expression profile of the studied virulence genes (cdtC, cdtA, cdtB, iam, virB11, and wlaN) in all strains (n = 41). Hu, human; pi, pigeon; ck (M), chicken meat and ck (S), chicken swabs. Bird strains refer to all strains from chicken and pigeon.
FIGURE 3.
(A) Hierarchical clustering of C. jejuni strain sources (human, pigeon, and chicken) and the associated virulence genes based on Euclidian distance. (B) Two-dimensional correspondence analysis (CA) showing the association between the sources of C. jejuni strains [human, pigeon, chicken meat (M) and cloacal swabs (S)] and the studied virulence genes.
Genetic Diversity of C. jejuni Strains Based on Their Virulence Genes Profiles
The nMDS and the dendogram analyses (Figures 2, 3A) of C. jejuni strains (n = 41) showed a non-consistent clustering pattern and high diversity among the analysed strains. The human and bird strains overlapped largely. A similar overlap was observed when analysing the 41 strains grouped by their sources (Figure 3A). The dendogram analysis showed that the human strains were genetically closer to those from chickens (Euclidean distance = 0.19) than to those from pigeons (Euclidean distance = 0.26) (Figure 3A). As shown in Table 3, irrespective of the strain source, the overall Simpson’s and Shannon’s diversity indices of all strains were 0.77 and 1.60, respectively. The highest diversity was observed in C. jejuni strains isolated from chickens, followed by those from pigeons and humans. Overall, the results of both diversity indices were highly positively correlated in a significant manner (Pearson correlation, r = 0.9; P = 0.001).
Genetic Diversity of C. jejuni Based on flaA-SVR Gene Sequences
Using the BLAST tool, it was found that our flaA-SVR sequences and the flanking regions had 92–99% similarity with those of the previously published C. jejuni strains.
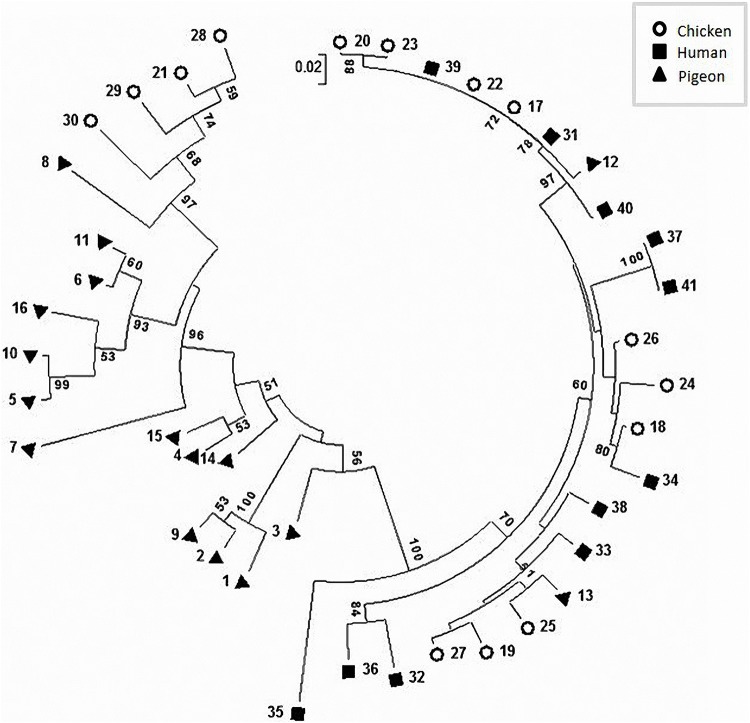
The phylogenetic analysis (Figure 4) and the proximity matrix (Supplementary Table S2) revealed sequence heterogeneity among our C. jejuni strains. While pigeon strains were phylogenetically related, some of them were clustered close to (e.g., strain No. 13) or shared the same lineage with (e.g., strain No. 12) chicken or human strains. The proximity matrix (Supplementary Table S2) indicated that the bacterial population within chickens and pigeons had less similarity on average (average similarity, each = 86.9%) than those within humans (average similarity = 93.4%).
FIGURE 4.
Circular phylogenetic tree of the flaA-SVR sequences generated using the neighbour-joining method. The numbers on each branch indicate the calculated bootstrap values. The scale-bar indicates a branch length equivalent to a 2% difference in the nucleotide sequence.
Allelic Diversity of C. jejuni
We identified 17 and 12 allele variants at nucleotide and peptide levels, respectively (Table 4 and Supplementary Table S1) with 13 novel alleles at the nucleotide level (Bold numbers in Supplementary Table S1 and Table 4). Irrespective of the strain source, allele 177 was the most frequently identified nucleotide variant (13/41, 31.71%), followed by allele 288 (n = 4, 9.76%). The flaA-SVR nucleotide allele 177 was the dominant one in human (6/11, 54.55%) and chicken (6/14, 42.86%) strains, whereas alleles 718, 38 and 1118 were the most frequent in pigeon strains (2/16, 12.5%, each). Certain allele variants were not identified in human and chicken strains, yet were detected in pigeon strains (Supplementary Table S1). At the peptide level, allele 74 was the most frequently identified variant (13/41, 31.71%), followed by allele 92 (7/41, 17.07%) and allele 239 (4/41, 9.76%). The flaA-SVR peptide allele 74 was the most prevalent in human (6/11, 54.55%) and chicken (6/14, 42.86%) strains, whereas allele 239 predominated in pigeon strains (4/16, 25%). The nMDS plots that was built based on all identified alleles (Supplementary Figures S2A,B) indicated that C. jejuni strains isolated from humans and birds were highly diverse and overlapped largely. Considering all identified alleles (at both the nucleotide and peptide levels), human strains were clustered with chicken strains and were positioned apart from pigeon ones (Supplementary Figures S2C,D).
Discussion
Campylobacter jejuni can colonise and infect domestic poultry and pose a risk for humans. In the current study, we analysed the genetic heterogeneity of Egyptian C. jejuni isolates recovered from avian and human sources. Characterisation of C. jejuni isolates from different sources is vital for a better understanding of the impact of multiple sources on the disease burden. The current study revealed low overall prevalence rates of C. jejuni in chicken and human samples (15.56 and 4.07%, respectively). Similar prevalence rates have been reported in a recent study conducted in Egypt (18.12 and 4.1%, respectively) (Ahmed et al., 2015). Moreover, the carriage rate of C. jejuni in pigeon droplets (8.89%) was found to be much lower than that in chickens. A previous Canadian study reported also a low infection rate of C. jejuni in pigeons (9.1%) (Gabriele-Rivet et al., 2016).
The observation that the flaA gene was encoded by all analysed strains suggests the importance of this gene as a virulence marker in our Egyptian C. jejuni strains and possibly other C. jejuni. Our results corporate previous reports (e.g., Datta et al., 2003; Talukder et al., 2008; Rizal et al., 2010; Wieczorek et al., 2012; Shyaka et al., 2015), which also identified flaA gene in 100% of C. jejuni strains. However, a lower prevalence of flaA gene (75.5%) was reported in C. jejuni isolated from chicken meat in Brazil (Melo et al., 2013). This difference may be attributed to the isolate source, sample type and geographical location (Corcoran et al., 2006).
The cdtA, cdtB, and cdtC were detected in the analysed strains by the same percentage (80.49%), and more than half (58.54%) of the strains possessed the three CDT toxin genes together. Previous studies (Datta et al., 2003; Rozynek et al., 2005) reported that the prevalence of each of cdtA, cdtB, or cdtC genes in C. jejuni isolated from poultry and humans exceeded 80%. The high frequency of the co-existence of these subunits and their close genotypic association (Figures 2B, 3A) was previously reported in Poland (70.8%) (Wieczorek and Osek, 2011) indicating that they are highly likely to co-express during an infection event. This is supported by the knowledge that these three subunits are required for full CDT toxin activity (Martinez et al., 2006).
The random forest classification and ROC analyses applied herein showed that the cdtC gene exhibited the highest similarity and thus the least discriminatory power (low AUC values) among C. jejuni strains from all examined sources, whereas iam and cdtB were the highest dissimilar genes among strains from different sources and that iam gene has relatively high discriminatory power (high AUC value) than cdtC gene. It is plausible to assume that the genes (e.g., cdtC), which have similar frequency and low discriminatory power among strains isolated from different hosts could infer about the potential sources of infection if their expression potential was tracked back after an outbreak. However, the genes (e.g., iam and cdtB), which tend to have dissimilar frequencies and have high discriminatory power could differentiate among infection sources. While this result was based on a small data set, it suggests the potential importance of certain genes in tracking and differentiating the sources of C. jejuni infection in human in small outbreaks. An additional wide scale profiling that includes more virulence genes in a large number of strains is needed to generalise these data.
In the current study, we analysed both virulence gene patterns and flaA-SVR sequences and alleles to better understand the extent of genetic diversity of C. jejuni strains among and within each host. Irrespective of the strain sources, we found a clear overlap among human and bird strains. In support of this, the overall Simpson’s and Shannon’s diversity indices were relatively high (Table 3). There was also a random distribution of the virulence gene patterns across the strains. Similar variabilities in the prevalence of virulence genes among C. jejuni have been reported previously (Abu-Madi et al., 2016). Taken together, these data reflect the high genetic diversity of our Egyptian C. jejuni strains.
Sequence-based flaA-SVR typing has been reported as a reliable genotyping method yielding reproducible results (Acke et al., 2010; El-Adawy et al., 2013). In the current study, the distribution of flaA-SVR alleles highlighted the diversity of the studied Egyptian C. jejuni strains when compared to those described previously in the PubMLST database. The variability in flaA-SVR gene sequences and alleles (Supplementary Figure S2) supported the impression from the virulence gene heterogeneity and further indicated the genotypic diversity of C. jejuni. The observed variation in the alleles/peptides might be due to the occurrence of distinct mutations (e.g., transition, transversion, additions or deletions). These genetic alterations could afford a survival advantage of the bacteria, which in turn might lead to evasion of the host immune response (Jerome et al., 2011). A similar heterogeneity was also obtained by Corcoran et al. who studied 41 C. jejuni strains from humans and poultry (Corcoran et al., 2006). Moreover, Ramees et al. (2015) reported a diversity of 65% among 32 analysed C. jejuni strains irrespective of their sources.
The current analysis showed that C. jejuni strains isolated from the same host did not reveal a similar frequency or distribution of both virulence genes and the flaA allelic variants (Supplementary Table S1), suggesting that C. jejuni strains studied here are not host-specific. Similar diversity was also shown previously (Corcoran et al., 2006) after analysing flaA gene of C. jejuni isolated from humans and poultry. Moreover, Vidal et al. (2016) reported a high genotypic diversity of C. jejuni isolated from a chicken broiler flock. The particular distribution of certain strains in certain niche could be attributed to differential response to environmental factors and/or management practices. This, in turn, will modulate the bacterial phenotypic properties (e.g., infectivity, survival and pathogenicity) and ultimately the colonisation in the host (Ahmed et al., 2002; Vidal et al., 2016). Taken together, these results indicate the existence of C. jejuni as quasi-species colonising the same host, which demonstrates the low host adaptability and the weak clonality of these C. jejuni strains as was suggested previously (Corcoran et al., 2006).
As evidenced by the measurement of diversity indices, virulence gene patterns and the average similarity of flaA-SVR sequences, chicken strains were found to be highly diverse relative to those from human or pigeon. The rationale behind the high diversity of C. jejuni in this particular host is not yet clear. However, the uniqueness of the chicken as a host for such bacteria (Awad et al., 2018) and the high load of C. jejuni (106 – 108 colony forming units/g) (Beery et al., 1988) in chicken gut might account for this heterogeneity, which usually stems from frequent recombination and plasmid transfer among strains in the same niche. These data demonstrate the importance of chickens in the epidemiology and infection dynamics of C. jejuni in Egypt.
Applying a hierarchical clustering on the frequency of the presence of each virulence gene (Figure 3A) and allelic/peptide variants (Supplementary Figures S2C,D) in C. jejuni strains revealed that human strains were genotypically clustered close to chicken strains rather than to pigeon ones. While this needs to be interpreted using large studies, it suggests the importance of chicken as a source of human infection with C. jejuni as was stated previously (Oh et al., 2017).
A shortcoming of this study is due to the limited funding resources. Therefore, we described the diversity of C. jejuni depending on sequence-based typing of flaA-SVR gene, which is less costly and thus fits more the situation in developing countries, notably Egypt. While this approach is convenient for small-scale investigations, using advanced techniques such as pulsed-field gel electrophoresis (PFGE) and MLST would have added more sensitivity and accuracy to the results and would allow comparing different typing methods at once. Future studies are warranted in this direction. In conclusion, this is the first report in Egypt that describes the extent of genetic diversity of C. jejuni isolated from chickens, pigeons and humans. It suggests the possible role of poultry, particularly chickens, in the transmission of C. jejuni to humans. It is recommended to consider these observations in the future epidemiological evaluation and risk assessment of C. jejuni not only in poultry, but also in humans.
Data Availability Statement
The datasets generated for this study can be found in the GenBank database with accession numbers KX066127–KX066135, MG677923–MG677934, and MK281494–MK281513.
Ethics Statement
The animal study was approved by the committee of Animal Welfare and Research Ethics of Faculty of Veterinary Medicine, Zagazig University. The study involving human participants was reviewed and approved by the research ethical committee of Faculty of Medicine, Zagazig University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MA, NA, and EA designed the study. MA and NA carried out the molecular analyses and participated in the data analysis. MS performed the bioinformatics and statistical analyses of the data. EE, MB, and RM conceived the study and participated in the design. MA, NA, and MS wrote the initial draft of the manuscript. All authors approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors express their deep gratitude to Dr. Ahmed Hefny (Veterinary Hospital, Faculty of Veterinary Medicine, Zagazig University, Egypt) for his assistance during the collection of chicken samples and isolation of the bacteria. The authors are thankful to Dr. Hazem Ramadan (Hygiene and Zoonosis Department, Faculty of Veterinary Medicine, Mansoura University, Egypt) for his critical reading of the manuscript.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02353/full#supplementary-material
Degree of similarity of analysed virulence genes across the sources of C. jejuni strains. The figures show random forest classification with the predictive accuracy of each of the studied genes plotted on the X-axis against the respective gene name on the Y-axis. The mini heat maps show the frequency distribution of each gene in the respective strain sources.
Clustering patterns and diversity of C. jejuni strains based on frequency of flaA-SVR alleles. (A,B) Show a non-metric multidimensional scaling of the 41 analysed strains based on the presence and absence of all alleles of the flaA-SVR nucleotides (A) and proteins (B). (C,D) Show clustering pattern of C. jejuni strain sources (based on Euclidean distance) using the frequency of alleles at the nucleotide (C) and protein (D) levels as input data.
Frequency of virulence gene combinations, and nucleotide and peptide alleles among 41C. jejuni strains from different sources.
Distance matrix showing the percentage of similarity between each pair of the strains based on the alignment of flaA-SVR sequences. HU: humans, PI: pigeon, CK: chicken. ∗∗∗ indicates 100% similarity.
References
- Abd El-Tawab A. A., Ammar A. M., Ahmed H. A., Ei Hofy F. I., Hefny A. A. (2018). Fluoroquinolone resistance and gyrA mutations in Campylobacter jejuni and Campylobacter coli isolated from chicken in Egypt. J. Glob. Antimicrob. Resist. 13 22–23. 10.1016/j.jgar.2018.02.019 [DOI] [PubMed] [Google Scholar]
- Abu-Madi M., Behnke J. M., Sharma A., Bearden R., Al-Banna N. (2016). Prevalence of virulence/stress genes in Campylobacter jejuni from chicken meat sold in Qatari retail outlets. PLoS One 11:e0156938. 10.1371/journal.pone.0156938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuoun M., Manning G., Cawthraw S. A., Ridley A., Ahmed I. H., Wassenaar T. M., et al. (2005). Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect. Immun. 73 3053–3062. 10.1128/IAI.73.5.3053-3062.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acke E., McGill K., Lawlor A., Jones B. R., Fanning S., Whyte P. (2010). Genetic diversity among Campylobacter jejuni isolates from pets in Ireland. Vet. Rec. 166 102–106. 10.1136/vr.c357 [DOI] [PubMed] [Google Scholar]
- Ahmed H. A., El Hofy F. I., Ammar A. M., Abd El Tawab A. A., Hefny A. A. (2015). ERIC-PCR genotyping of some Campylobacter jejuni isolates of chicken and human origin in Egypt. Vector Borne Zoonotic Dis. 15 713–717. 10.1089/vbz.2015.1836 [DOI] [PubMed] [Google Scholar]
- Ahmed I. H., Manning G., Wassenaar T. M., Cawthraw S., Newell D. G. (2002). Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiol. 148 1203–1212. 10.1099/00221287-148-4-1203 [DOI] [PubMed] [Google Scholar]
- Awad W. A., Hess C., Hess M. (2018). Re-thinking the chicken-Campylobacter jejuni interaction: a review. Avian. Pathol. 47 352–363. 10.1080/03079457.2018.1475724 [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J. R., Maciejewski A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44 W147–W153. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beery J. T., Hugdahl M. B., Doyle M. P. (1988). Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Xia J. (2018). MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34 4313–4314. 10.1093/bioinformatics/bty528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G., Stewart-Tull D. E., Owen R. J., Bolton F. J., Siemer B. L., Candlish D., et al. (2007). Comparison of virulence-associated in vitro properties of typed strains of Campylobacter jejuni from different sources. J. Med. Microbiol. 56 722–732. 10.1099/jmm.0.47130-0 [DOI] [PubMed] [Google Scholar]
- Corcoran D., Quinn T., Cotter L., Whyte P., Fanning S. (2006). Antimicrobial resistance profiling and fla-typing of Irish thermophillic Campylobacter spp. of human and poultry origin. Lett. Appl. Microbiol. 43 560–565. 10.1111/j.1472-765X.2006.01987.x [DOI] [PubMed] [Google Scholar]
- Datta S., Niwa H., Itoh K. (2003). Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 52 345–348. 10.1099/jmm.0.05056-0 [DOI] [PubMed] [Google Scholar]
- El-Adawy H., Hotzel H., Tomaso H., Neubauer H., Taboada E. N., Ehricht R., et al. (2013). Detection of genetic diversity in Campylobacter jejuni isolated from a commercial turkey flock using flaA typing, MLST analysis and microarray assay. PLoS One 8:e51582. 10.1371/journal.pone.0051582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority. (2014). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 12 99–107. 10.2903/j.efsa.2014.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão M. R., Medeiros M. I., Duque S. D., Falcão J. P. (2017). Pathogenic potential and genotypic diversity of Campylobacter jejuni: a neglected food-borne pathogen in Brazil. J. Med. Microbiol. 66 350–359. 10.1099/jmm.0.000424 [DOI] [PubMed] [Google Scholar]
- Gabriele-Rivet V., Fairbrother J. H., Tremblay D., Harel J., Côté N., Arsenault J. (2016). Prevalence and risk factors for Campylobacter spp., Salmonella spp., Coxiella burnetii, and Newcastle disease virus in feral pigeons (Columba livia) in public areas of Montreal, Canada. Can. J. Vet. Res. 80 81–85. [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper D. A. T., Ryan P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4 4–9. [Google Scholar]
- Harrington C. S., Thomson-Carter F. M., Carter P. E. (1997). Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35 2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans D., Van Deun K., Martel A., Van Immerseel F., Messens W., Heyndrickx M., et al. (2011). Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42:82. 10.1186/1297-9716-42-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., O’Brien S., Madsen M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117 237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Jerome J. P., Bell J. A., Plovanich-Jones A. E., Barrick J. E., Brown C. T., Mansfield L. S. (2011). Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One 6:e16399. 10.1371/journal.pone.0016399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. 10.1007/bf01731581 [DOI] [PubMed] [Google Scholar]
- Kordinas V., Nicolaou C., Ioannidis A., Papavasileiou E., John Legakis N., Chatzipanagiotou S. (2005). Prevalence of four virulence genes in Campylobacter jejuni determined by PCR and sequence analysis. Mol. Diagn. 9 211–215. 10.1007/bf03260094 [DOI] [PubMed] [Google Scholar]
- Kovacic A., Listes I., Vucica C., Kozacinski L., Tripkovic I., Sisko-Kraljevic K. (2013). Distribution and genotypic characterization of Campylobacter jejuni isolated from poultry in Split and Dalmatia County. Croatia. Zoonoses Public Health 60 269–276. 10.1111/j.1863-2378.2012.01519.x [DOI] [PubMed] [Google Scholar]
- Maansi N. R., Kumar D., Upadhyay A. K. (2018). Virulence typing and antibiotic susceptibility profiling of thermophilic campylobacters isolated from poultry, animal, and human species. Vet. World 11 1698–1705. 10.14202/vetworld.2018.1698-1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Mateo E., Churruca E., Girbau C., Alonso R., Fernandez-Astorga A. (2006). Detection of cdtA, cdtB, and cdtC genes in Campylobacter jejuni by multiplex PCR. Int. J. Med. Microbiol. 296 45–48. 10.1016/j.ijmm.2005.08.003 [DOI] [PubMed] [Google Scholar]
- McCarthy N. D., Colles F. M., Dingle K. E., Bagnall M. C., Manning G., Maiden M. C., et al. (2007). Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13 267–272. 10.3201/eid1302.060620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann R. J., Helsel L. O., Fields P. I., Hiett K. L. (1997). Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35 2810–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann R. J., Phillips R. W., Hiett K. L., Fedorka-Cray P. (2005). Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 71 6368–6374. 10.1128/AEM.71.10.6368-6374.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo R. T., Nalevaiko P. C., Mendonça E. P., Borges L. W., Fonseca B. B., Beletti M. E., et al. (2013). Campylobacter jejuni strains isolated from chicken meat harbor several virulence factors and represent a potential risk to humans. Food Control 33 227–231. 10.1016/j.foodcont.2013.02.032 [DOI] [Google Scholar]
- Oh J. Y., Kwon Y. K., Wei B., Jang H. K., Lim S. K., Kim C. H., et al. (2017). Epidemiological relationships of Campylobacter jejuni strains isolated from humans and chickens in South Korea. J. Microbiol. 55 13–20. 10.1007/s12275-017-6308-8 [DOI] [PubMed] [Google Scholar]
- Quinn P. J., Carter M. E., Markey B. K., Carter G. R. (eds) (1994). “Campylobacter species,” in Clinical Veterinary Microbiology, (London: Mosby-Year Book Europe Limited; ), 268–272. [Google Scholar]
- Rahimi E., Ameri M. (2011). Antimicrobial resistance patterns of Campylobacter spp. isolated from raw chicken, turkey, quail, partridge, and ostrich meat in Iran. Food Control 22 1165–1170. 10.1016/j.foodcont.2011.01.010 [DOI] [Google Scholar]
- Ramees T. P., Suman Kumar M., Dubal Z. B., Sivakumar A. M., Gupta S., Dhama K., et al. (2015). Genotyping of Campylobacter jejuni and C. coli from different sources and human by ERIC-PCR. J. Vet. Public Health 13 93–98. [Google Scholar]
- Ramonaite S., Tamuleviciene E., Alter T., Kasnauskyte N., Malakauskas M. (2017). MLST genotypes of Campylobacter jejuni isolated from broiler products, dairy cattle and human campylobacteriosis cases in Lithuania. BMC Infect. Dis. 17:430. 10.1186/s12879-017-2535-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizal A., Kumar A., Vidyarthi A. S. (2010). Prevalence of pathogenic genes in Campylobacter jejuni isolated from poultry and human. Internet J. Food Saf. 12 29–34. [Google Scholar]
- Rozynek E., Dzierzanowska-Fangrat K., Jozwiak P., Popowski J., Korsak D., Dzierzanowska D. (2005). Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J. Med. Microbiol. 54 615–619. 10.1099/jmm.0.45988-0 [DOI] [PubMed] [Google Scholar]
- Sails A. D., Swaminathan B., Fields P. I. (2003). Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing correlate with strain associations identified by multilocus enzyme electrophoresis. J. Clin. Microbiol. 41 4058–4067. 10.1128/jcm.41.9.4058-4067.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainato R., ElGendy A., Poly F., Kuroiwa J., Guerry P., Riddle M. S., et al. (2018). Epidemiology of Campylobacter infections among children in Egypt. Am. J. Trop. Med. Hyg. 98 581–585. 10.4269/ajtmh.17-0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Shannon C. A. (1948). Mathematical theory of communication. Bell Syst. Tech. J. 27 379–423. 10.1002/j.1538-7305.1948.tb01338.x 30854411 [DOI] [Google Scholar]
- Shyaka A., Kusumoto A., Chaisowwong W., Okouchi Y., Fukumoto S., Yoshimura A., et al. (2015). Virulence characterization of Campylobacter jejuni isolated from resident wild birds in Tokachi area, Japan. J. Vet. Med. Sci. 77 967–972. 10.1292/jvms.15-0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E. H. (1949). Measurement of diversity. Nature 163:688 10.1038/163688a0 [DOI] [Google Scholar]
- Talukder K. A., Aslam M., Islam Z., Azmi I. J., Dutta D. K., Hossain S., et al. (2008). Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 46 1485–1488. 10.1128/JCM.01912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepitte J., Verhaegen J., Engbaek K., Rohner P., Piot P., Heuck C. (2003). Basic Laboratory Procedures in Clinical Bacteriology, 2nd Edn Geneva: World Health Organization. [Google Scholar]
- Vazquez B., Esperon F., Neves E., Lopez J., Ballesteros C., Munoz M. J. (2010). Screening for several potential pathogens in feral pigeons (Columba livia) in Madrid. Acta Vet. Scand. 52:45. 10.1186/1751-0147-52-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A. B., Colles F. M., Rodgers J. D., McCarthy N. D., Davies R. H., Maiden M. C. J., et al. (2016). Genetic diversity of Campylobacter jejuni and Campylobacter coli isolates from conventional broiler flocks and the impacts of sampling strategy and laboratory method. Appl. Environ. Microbiol. 82 2347–2355. 10.1128/AEM.03693-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Clark C. G., Taylor T. M., Pucknell C., Barton C., Price L., et al. (2002). Colony multiplex PCR assay for identification and differentiation of C. jejuni, C. coli, C. lari, C. upsaliensis and C. fetus subsp. fetus. J. Clin. Microbiol. 40 4744–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardak S., Jagielski M. (2009). Evaluation of genotypic and phenotypic methods for the differentiation of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. II. PFGE, ERIC-PCR, PCR-flaA-RFLP and MLST. Med. Dośw. Mikrobiol. 61 63–77. [PubMed] [Google Scholar]
- Wieczorek K., Osek J. (2011). Molecular characterization of Campylobacter spp. isolated from poultry faeces and carcasses in Poland. Acta Vet. Brno 80 19–27. 10.2754/avb201180010019 [DOI] [Google Scholar]
- Wieczorek K., Szewczyk R., Osek J. (2012). Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter jejuni and C. coli isolated from retail raw meat in Poland. Vet. Med. 57 293–299. 10.17221/6016-vetmed [DOI] [Google Scholar]
- Zheng J., Meng J., Zhao S., Singh R., Song W. (2006). Adherence to and invasion of human intestinal epithelial cells by Campylobacter jejuni and Campylobacter coli isolates from retail meat products. J. Food Prot. 69 768–774. 10.4315/0362-028x-69.4.768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Degree of similarity of analysed virulence genes across the sources of C. jejuni strains. The figures show random forest classification with the predictive accuracy of each of the studied genes plotted on the X-axis against the respective gene name on the Y-axis. The mini heat maps show the frequency distribution of each gene in the respective strain sources.
Clustering patterns and diversity of C. jejuni strains based on frequency of flaA-SVR alleles. (A,B) Show a non-metric multidimensional scaling of the 41 analysed strains based on the presence and absence of all alleles of the flaA-SVR nucleotides (A) and proteins (B). (C,D) Show clustering pattern of C. jejuni strain sources (based on Euclidean distance) using the frequency of alleles at the nucleotide (C) and protein (D) levels as input data.
Frequency of virulence gene combinations, and nucleotide and peptide alleles among 41C. jejuni strains from different sources.
Distance matrix showing the percentage of similarity between each pair of the strains based on the alignment of flaA-SVR sequences. HU: humans, PI: pigeon, CK: chicken. ∗∗∗ indicates 100% similarity.
Data Availability Statement
The datasets generated for this study can be found in the GenBank database with accession numbers KX066127–KX066135, MG677923–MG677934, and MK281494–MK281513.