Abstract
Arcobacter have been frequently detected in and isolated from bivalves, but there is very little information on the genus Arcobacter in the abalone, an important fishery resource. This study aimed to investigate the genetic diversity and abundance of bacteria from the genus Arcobacter in the Japanese giant abalone, Haliotis gigantea, using molecular methods such as Arcobacter‐specific clone libraries and fluorescence in situ hybridization (FISH). Furthermore, we attempted to isolate the Arcobacter species detected. Twelve genotypes of clones were obtained from Arcobacter‐specific clone libraries. These sequences are not classified with any other known Arcobacter species including pathogenic Arcobacter spp., A. butzleri, A. skirrowii, and A. cryaerophilus, commonly isolated or detected from bivalves. From the FISH analysis, we observed that ARC94F‐positive cells, presumed to be Arcobacter, accounted for 6.96 ± 0.72% of all EUB338‐positive cells. In the culture method, three genotypes of Arcobacter were isolated from abalones. One genotype had a similarity of 99.2%–100.0% to the 16S rRNA gene of Arcobacter marinus, while the others showed only 93.3%–94.3% similarity to other Arcobacter species. These data indicate that abalones carry Arcobacter as a common bacterial genus which includes uncultured species.
Keywords: abalone, Arcobacter, clone libraries, cultivation, fluorescent in situ hybridization
Diversity of marine Arcobacter associated with the giant abalone Haliotis gigantea
1. INTRODUCTION
Arcobacter, formerly classified as Campylobacter, is a member of the class Epsilonproteobacteria, as proposed by Vandamme et al. (1991). Some Arcobacter bacteria have shown pathogenicity to humans, and thus many studies have focused on livestock. Species isolated from pork, broiler carcasses, cattle, ducks, human stool, or porcine abortions include: Arcobacter butzleri, A. skirrowii, A. cibarius, A. cryaerophilus, A. trophiarum, A. defluvii, A. thereius, A. suis, A. cloacae, A. lanthieri, A. faecis, A. lacus, and A. caeni (Collado, Levican, Perez, & Figueras, 2011; De Smet et al., 2011; Houf et al., 2005, 2009; Kiehlbauch et al., 1991; Levican, Collado, & Figueras, 2013; Neill, Campbell, O'Brien, Weatherup, & Ellis, 1985; Pérez‐Cataluña, Salas‐Massó, & Figueras, 2018b; Vandamme et al., 1992; Whiteduck‐Léveillée et al., 2015, 2016). Among them, A. butzleri, A. cryaerophilus, and A. skirrowii are considered to be of clinical interest because they are associated with gastrointestinal disease and bacteremia in humans, and with reproduction disorders, mastitis, and gastric ulcers in farm animals (Ho, Lipman, & Gaastra, 2006). Arcobacter thereius was also isolated from porcine abortions, but the pathological potential of this species is still unknown (Houf et al., 2009). In contrast, other species have not been directly associated with animal or human diseases.
Recently, Arcobacter spp. have also been isolated from marine environments, such as seawater and coastal sediments, and from marine invertebrates. To date, 18 Arcobacter species from a total of 29 have been isolated from marine environments (Table 1), suggesting that this environment may be one of the main habitats for this genus. Arcobacter are found in bivalves (Collado, Guarro, & Figueras, 2009; Laishram, Rathlavath, Lekshmi, Kumar, & Nayak, 2016; Levican, Collado, Yustes, Aguilar, & Figueras, 2014; Salas‐Massó, Andree, Furones, & Figueras, 2016). Romero, García‐Varela, Laclette, and Espejo (2002) reported Arcobacter spp. are widespread in the Chilean oyster in their analysis using 16S rRNA‐RFLP. In addition to bivalves, it has been reported that Arcobacter spp. are found in European lobsters and abalone (Meziti, Mente, & Kormas, 2012; Tanaka, Ootsubo, Sawabe, Ezura, & Tajima, 2004). These results suggest that Arcobacter spp. are widely distributed in marine invertebrates, and potentially indigenous bacteria may play some important role in the host. However, knowledge on the presence and diversity of Arcobacter associated with marine invertebrates including abalone is still lacking compared to pathogenic Arcobacter.
Table 1.
List of Arcobacter spp. isolated from different marine environments
| Source | Species | Reference |
|---|---|---|
| Roots of Spartina alterniflora, sediments from salt marshes | A. nitrofigilis CI | McClung, Patriquin, and Davis (1983) |
| Water from hypersaline lagoon | A. halophilus LA31B | Donachie, Bowman, On, and Alam (2005) |
| Mussels, brackish water | A. mytili F2075 | Collado, Cleenwerck, Trappen, Vos, and Figueras (2009) |
| Seawater, seaweeds, and starfish | A. marinus CL‐S1 | Kim et al. (2010) |
| Sewage | A. defluvii SW28‐11 | Collado et al. (2011) |
| Mussels | A. ellisii F79‐6 | Figueras, Levican, Collado, Inza, and Yustes (2011) |
| Mussels and oysters | A. molluscorum F98‐3 | Figueras, Collado, et al. (2011) |
| Clams | A. venerupis F67‐11 | Levican et al. (2012) |
| Mussels | A. bivalviorum F4 | Levican et al. (2012) |
| Mussels and sewage | A. cloacae SW28‐13 | Levican et al. (2013) |
| Estuarine sediment | A. anaerophilus JC84 | Sasi Jyothsna, Rahul, Ramaprasad, Sasikala, and Ramana (2013) |
| Mussels | A. ebronensis F128‐2 | Levican, Rubio‐Arcos, Martinez‐Murcia, Collado, and Figueras (2015) |
| Seawater | A. aquimarinus W63 | Levican et al. (2015) |
| Seawater | A. acticola AR‐13 | Park, Jung, Kim, and Yoon (2016) |
| Seawater | A. pacificus SW028 | Zhang, Yu, Wang, Yu, and Zhang (2016) |
| Great scallop larvae and tank seawater | A. lekithochrous LFT 1.7 | Diéguez et al. (2017) |
| Abalone |
A. lekithochrous MA5 (syn. A. haliotis MA5) |
Tanaka et al. (2017) |
| Sewage | A. canalis F138‐33 | Pérez‐Cataluña, Salas‐Masso, and Figueras (2018a) |
Therefore, in order to gain knowledge on Arcobacter spp. in marine invertebrates, we tried to explore the diversity and abundance of the genus Arcobacter in the giant abalone Haliotis gigantea, an important fishery resource inhabiting shallow water environments. We used cultivation‐independent methods, such as Arcobacter‐specific clone libraries and fluorescence in situ hybridization (FISH). We also attempted to isolate Arcobacter strains using selective cultivation, and report here the genetic relationships between successfully isolated strains.
2. MATERIALS AND METHODS
2.1. Sample collection and DNA extraction
Nine cultivated giant abalones, H. gigantea, (sample code: CA) and rearing water samples from two tanks (sample code: RW) were collected from the Owase Farming Fishery Center (Owase, Mie, Japan) in February 2012. These samples were supplied for construction of Arcobacter‐specific clone libraries and for FISH. For isolation of Arcobacter spp., three endemic H. gigantea specimens were collected from fish markets in Mie, Japan, in September 2017.
The internal organs, including the gut and gills, were collected from the abalones followed by the previously described method (Tanaka et al., 2004). To tear off bacterial cells from their host tissue, we used a beads beater on the condition of slower stroke and shorter time (We state that the method will not allow obtaining the whole bacterial diversity present in the host tissue as compared to an approach based on completely grinding the host tissue). Abalone specimens were pooled into each tube and homogenized using a beads beater (4,200 rpm, 30 s; Tietech Co., Nagoya, Japan). Host tissues were removed from CA samples by quick centrifugation (1 s, 8,000 g), and the supernatant was transferred to new tubes and centrifuged for 20 min at 15,000 g to recover bacterial cells. Rearing water samples (RW) were concentrated (50×) using 0.22 µm cellulose membrane filters (Advantec, Tokyo, Japan) and resuspended in sterile phosphate‐buffered saline (PBS: 130 mM NaCl, 10 mM Na2HPO4/NaH2PO4; pH 7.4). Aliquots of bacterial pellets thus obtained from CA and RW samples were subsequently used for FISH analysis and DNA extraction. Bacterial genomic DNA from each sample was extracted using Promega DNA purification system (Promega Corp., Madison, WI, USA) according to the manufacturer's instructions.
2.2. PCR analysis, construction of Arcobacter‐specific clone libraries and sequencing
The Arcobacter genus‐specific primers, ARC94F primer (5′‐TGCGCCACTTAGCTGACA‐3′) and ARC1446R primer (5′‐TAGCATCCCCGCTTCGAATGA‐3′) (Harmon & Wesley, 1996; Snaidr, Amann, Huber, Ludwig, & Schleifer, 1997) were used to amplify the Arcobacter 16S rRNA gene from each sample. PCR reaction mixtures contained 1× PCR reaction buffer, 200 µM dNTP, 5 pmol of each primer, 2.5 units Ex Taq polymerase (TaKaRa Biotechnology Corp., Kyoto, Japan), and 10–100 ng of DNA for a total volume of 50 µl. PCR reactions were performed using an iCycler (Bio‐Rad Lab., Hercules, CA, USA). The amplification conditions were as follows: initial denaturation of 4 min at 95°C followed by 25 cycles of denaturation for 30 s at 95°C, primer annealing for 30 s at 55°C, and primer extension at 72°C for 1.5 min. This was followed by a final extension reaction at 72°C for 7 min. Distilled H2O was used as the template for negative controls; these produced no PCR product, indicating the absence of contaminating DNA in reactions and reagents. The 16S rRNA gene amplicons were purified by the PCR Preps DNA Purification System (Promega Corp.) according to the manufacturer's instructions, and subsequently ligated into the TOPO TA cloning vector (Invitrogen Corp., Carlsbad, CA, USA). Ligation products were transformed into Escherichia coli One Shot TOP10 cells (Invitrogen Corp.) and screened for plasmid insertions by following the manufacturer's instructions. Plasmid DNA containing insertions was sequenced with the ARC94F primer using the Sanger method with an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA). Chromatograms of DNA sequences were examined using Chromas v2.3.3 (Technelysium Pty Ltd., South Brisbane, Australia). All sequences were examined for chimerism using a chimeric sequences detection tool, Bellerophon (Huber, Faulkner, & Hugenholtz, 2004).
2.3. Fluorescent in situ hybridization
The total number of Arcobacter cells in abalone samples was counted using the FISH method (Kepner & Pratt, 1994). Aliquots of bacterial pellets obtained from CA and RW samples were rinsed by 600 µl of sterile PBS and centrifuged at 12,000 g for 5 min, then removed supernatant. These procedures were performed two times. Samples were fixed by adding one volume of PBS to three volumes of 4% paraformaldehyde/PBS, and incubating at 4°C for 3 hr. After centrifugation, the fixative was removed and the bacterial pellet was washed twice with PBS. The washed cells were mixed with 1× PBS and 96% EtOH (1:1) and stored at −20°C (Roller, Wagner, Amann, Ludwig, & Schleifer, 1994). Three microliters of fixed‐cell suspension was spread on the well of an aminopropyl‐silane‐coated 8‐well slide (Matsunami, Japan). The slides were air‐dried and dehydrated by successive immersion in 50, 80, and 99.5%. Hybridization was performed based on a previous study by Ootsubo et al. (2003), with modifications. A 5′‐end TAMRA‐labeled ARC94F probe (5′‐TGCGCCACTTAGCTGACA‐3′; Sigma‐Aldrich Corp, St. Louis, MO; Moreno et al., 2003) was designed to target the 16S rRNA of Arcobacter spp., and a 5′‐end FITC‐labeled EUB338 probe (5′‐GCTGCCTCCCGTAGGAGT‐3′; Sigma‐Aldrich Corp; Amann et al., 1990) to target the 16S rRNA gene of most members of the domain Bacteria. Prior to hybridization, each probe was added to pre‐warmed hybridization buffer (0.9 M NaCl, 20% formamide, 20 mM Tris–HCl [pH 7.4] and 0.1% sodium dodecyl sulfate [SDS]) to a final concentration of 10 p.m. The probe solution was spread on each hybridization well, and the slides incubated at 46°C for 3 hr in an MHS‐2000 hybridization oven (EYELA, Tokyo, Japan). After hybridization, each well was washed twice with washing buffer (20 mM Tris‐HCl [pH 7.4], 180 mM NaCl and 0.01% SDS), rinsed with ddH2O and air‐dried. An epifluorescence light microscope (Eclipse 400; Nikon Corp., Tokyo, Japan), was used for observing the stained cells. Due to a technical error during sampling, we were unable to detect the mean ± SE from RW samples.
2.4. Isolation of Arcobacter spp.
For the isolation of Arcobacter species, the procedure described by Salas‐Massó et al. (2016) was followed, but the media was slightly modified by changing 2.5% NaCl to artificial seawater. The bacterial mixture from three abalones was diluted 10 times using sterile Daigo's Artificial Seawater SP (Nihon Pharmaceutical Co., Tokyo, Japan) and 100 µl of the diluted mixture was inoculated into Arcobacter Broth (Oxoid Ltd., Hampshire, UK, USA) with CAT supplement [cefoperazone at 8 mg/L, amphotericin B at 10 mg/L and teicoplanin at 4 mg/L] (Oxoid Ltd., Atabay & Corry, 1997) suspended in 75% Daigo's Artificial Seawater SP instead of distilled water with 2.5% NaCl. The culture solutions were incubated for 48 or 96 hr at 15°C (sample codes: 15T48H and 15T96H, respectively) or 25°C (sample codes: 25T48H and 25T96H) under aerobic conditions. After cultivation, 200 µl of post‐cultured broth was pipetted onto the surface of polycarbonate membrane filters (pore size, 0.4 µm: Merck Millipore, Burlington, MA) placed on Marine Agar 2216 (Difco, Detroit, MI, USA). The plates were incubated at room temperature for 30 min to allow passive filtration (Atabay & Corry, 1997). Next, the filters were carefully removed and the flow‐through was spread on Marine Agar 2216. The media was incubated at the same temperature and time as the primary culture. After cultivation, presumed Arcobacter colonies (tiny and beige to off‐white in color) were selected and applied to colony PCR in the same way as Arcobacter‐specific clone libraries using ARC94F and ARC1446R primers. Positive PCR products were sequenced using standard Sanger sequencing.
2.5. Cluster analysis of the bacterial community structure
For each sample, sequences were aligned and grouped in Operational Taxonomic Units (OTUs) with >97% sequence identity (Stackebrandt & Goebel, 1994). Homology searches were performed using sequences of approximately 700 bp and the highest homology sequences with each OTU were chosen as the closest relatives. All BLASTn searches were performed with the default parameters available through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Multiple alignments and calculation of distant matrixes were performed by CLUSTAL W (Thompson, Higgins, & Gibson, 1994), using MEGA 7.0 (Kumar, Stecher, & Tamura, 2016). A phylogenetic tree was constructed using the maximum‐likelihood method of MEGA 7.0, with 1,000 replicates in the bootstrap analysis and Kimura's two‐parameter model (Kimura, 1980). Distances were estimated with the Jukes‐Cantor correction.
3. RESULTS
3.1. Arcobacter‐specific clone libraries
To perform a comprehensive search for Arcobacter spp. from abalone and their surrounding seawater, we established Arcobacter‐specific 16S rRNA gene clone libraries. A total of 120 and 30 clones were obtained in our study from abalone (CA) and rearing water (RW) samples, respectively (Table 2). Among these clones, Arcobacter sequences were observed as 12 OTUs from CA and seven OTUs from RW.
Table 2.
16S rRNA gene sequences identified in the clone library and isolation from abalone or seawater
| Methods | Samples | OTUs | No. of clones or isolates | Highest similarity sequence (accession number) |
Identity (%) |
|---|---|---|---|---|---|
| Arcobacter‐specific clone libraries | Abalone (CA) | CA1 | 16 | Uncultured bacterium clone KSTye‐VF1‐B‐003 (JQ611206) | 100 |
| CA2 | 3 | Arcobacter sp. EP1 (LT629996) | 98.6 | ||
| CA3 | 17 | Uncultured bacterium clone SF‐July‐156 (HM591463) | 98.8 | ||
| CA4 | 3 | Uncultured Arcobacter sp. clone DVASD_D318 (KF463610) | 98.9 | ||
| CA5 | 3 | Uncultured Arcobacter sp. clone DVBSW_D345 (KF722009) | 99.5 | ||
| CA6 | 3 | Uncultured bacterium clone AJ‐U‐CD‐41(H) (JX170315) | 98.4 | ||
| CA7 | 20 | Uncultured bacterium clone AJ‐U‐CD‐41(H) (JX170315) | 100 | ||
| CA8 | 1 | Uncultured bacterium clone TopBa31 (EF999357) | 97.2 | ||
| CA9 | 24 | Uncultured epsilon‐proteobacterium clone AT‐pp13 (AY225610) | 95.2 | ||
| CA10 | 4 | Uncultured epsilon‐proteobacterium clone PI_4z10e (AY580424) | 99.6 | ||
| CA11 | 25 | Uncultured bacterium clone SF‐July‐74 (HM591442) | 99.2 | ||
| CA12 | 1 | Epsilon‐proteobacterium Yb‐F (AB496655) | 100 | ||
| Rearing water (RW) | RW1 | 12 | Uncultured bacterium clone HF071 (JX391310) | 98.7 | |
| RW4 | 4 | Uncultured epsilon‐proteobacterium clone PI_4z7d (AY580420) | 98.3 | ||
| RW5 | 4 | Arcobacter pacificus SW028 (JN118552) | 98.5 | ||
| RW6 | 3 | Uncultured bacterium clone SF‐July‐156 (HM591463) | 97.9 | ||
| RW10 | 4 | Arcobacter bivalviorum F4 (FJ573217) | 97.4 | ||
| RW17 | 2 | Uncultured bacterium clone C13W_197 (HM057704) | 98.8 | ||
| RW20 | 1 | Uncultured marine bacterium clone B‐Alg40 (HM437504) | 99.8 | ||
| Isolations |
Abalone 15°C (15T96H) |
15T96H‐1 | 4 | Uncultured bacterium clone HglApr921 (JX016315) | 98.2 |
| 15T96H‐2 | 1 | Uncultured bacterium clone HglApr921 (JX016315) | 98.3 | ||
|
Abalone 25°C (25T96H) |
25T96H | 5 | Arcobacter marinus strain CL‐S1 (EU512920) | 100 |
The 16S rRNA genes identified from abalone (CA) using the Arcobacter‐specific clone library did not show high similarity to known species within the NCBI database. The most abundant genotype in CA was CA11 (25 clones, accession number: LC133145), which had a high similarity score of 99.2% to uncultured bacterium clone SF‐July‐74 (HM591442). CA1 (LC133157) had 100% similarity to bacterium clone KSTye‐VF1‐B‐003 (JQ611206) collected from venting fluid in a yellow vent off Kueishan Island, while CA3 and CA12 (LC133141 and LC133148) each had a similarity of 98.8% and 100% to uncultured bacterium clone SF‐July‐156 (HM591463) and epsilon‐proteobacterium Yb‐F (AB496655) collected from seawater. CA3 did not cluster with any sequences. CA4, CA5, CA8, and CA9 (LC133159, LC133146, LC133160, and LC133142) were assigned to uncultured bacteria collected from marine environments: CA4 and CA5 showed similarity of 98.9% and 99.5% to uncultured Arcobacter sp. clones DVASD_D318 and DVBSW_D345, respectively (KF463610 and KF722009) from a marine coastal ecosystem, CA8 showed 97.2% similarity to uncultured bacterium clone TopBa31 (EF999357) from Pearl River Estuary sediments, and CA9 showed 95.2% similarity to uncultured epsilon‐proteobacterium clone AT‐pp13 (AY225610) from pumice fragments exposed to a Mid‐Atlantic Ridge vent. CA6 and CA7 (LC133158 and LC133140) were assigned to uncultured bacterium clone AJ‐U‐CD‐41(H) (JX170315) isolated from the intestine of a sea cucumber, Apostichopus japonicas, with 98.4% and 100% similarity, respectively. Finally, CA2 and CA10 (LC133161 and LC133156) were closely affiliated with an isolate or a clone collected from protists. CA2 had 98.6% similarity to Arcobacter sp. EP1 (LT629996) isolated from the epibiont of unicellular protists, and CA10 had 99.6% similarity to uncultured epsilon‐proteobacterium clone PI_4z10e from coastal bacterioplankton sampled at Plum Island Sound Estuary.
In the rearing water samples, RW5 (LC133151) showed 98.5% sequence similarity to A. pacificus strain SW028 isolated from seawater (JN118552), and RW10 (LC133153) had a 97.4% high homology to A. bivalviorum strain F4 isolated from bivalves (FJ573217). RW1 (LC133149) showed a sequence similarity of 98.7% to uncultured bacterium clone HF071 (JX391310) detected from marine sediment. RW4 (LC133150) was affiliated with uncultured epsilon‐proteobacterium clone PI_4z7d in a coastal bacterioplankton sample from Plum Island Sound Estuary, at 98.3% similarity. RW17 (LC133154) was closely related to uncultured bacterium clone C13W_197 (HM057704) collected from seawater, at 98.8% similarity. RW6 (LC133152) showed 97.9% similarity to uncultured bacterium clone SF‐July‐156 (HM591463), and RW20 (LC133155) was closely related at 99.8% similarity to uncultured marine bacterium clone B‐Alg40 (HM437504) detected from the surface of algae.
3.2. Detection of ARC94F‐positive cells by FISH
The amount of Arcobacter spp. in homogenized cultured abalone and rearing water samples from Minami‐ise, Mie, Japan, was determined by FISH (Table 3). From the abalone samples (n = 9), with a total bacteria count of 1.18 ± 0.71 × 107 cells/g, the number of ARC94F‐positive bacteria was 8.06 ± 0.05 × 105 cells/g, dominating the total bacteria count at 6.96 ± 0.72% (Figure 1). In the abalone rearing water (n = 2), the total bacteria count for each sample was 2.76 × 104 cells/ml and 4.63 × 104 cells/ml, respectively. Among these, the number of ARC94F‐positive bacteria was 1.33 × 103 cells/ml and 1.08 × 103 cells/ml, dominating 4.8% or 2.3% of the total bacteria, respectively.
Table 3.
Total and Arcobacter bacterial counts from abalones or in rearing water by direct microscopy
| Abalone (n = 9, mean ± SE) | Rearing water (n = 2, mean) | |
|---|---|---|
| EUB338 (cells/g or ml) | 1.18 ± 0.71 × 107 | 3.70 × 104 |
| ARC94 (cells/g or ml) | 8.06 ± 0.05 × 105 | 1.21 × 103 |
|
Rate of Arcobacter (% of total bacterial count) |
6.96 ± 0.72 | 3.55 |
Figure 1.
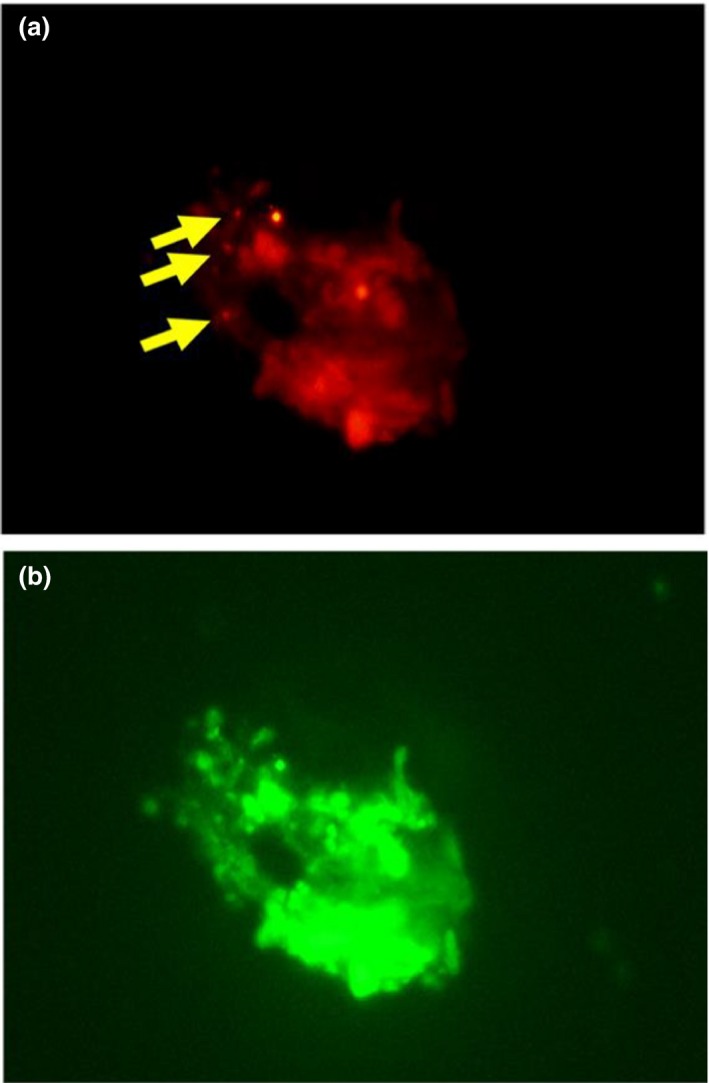
FISH photograph using (a) probe ARC94F and (b) probe EUB338, showing positive cells attached to abalone tissues. Yellow arrows indicate Arcobacter‐stained cells. FISH, fluorescence in situ hybridization
3.3. Isolation of Arcobacter Spp.
When incubation of samples from natural abalones collected in Mie, Japan, was performed for 96 hr at 15°C or 25°C (sample codes: 15T96H or 25T96H), the total numbers of colonies were 871 and 338, respectively. Upon further selection, 12 and eight colonies from the 15T96H and 25T96H samples were presumed to be Arcobacter colonies (tiny and beige to off‐white in color). Colony PCR subsequently confirmed that five of the selected Arcobacter‐like colonies for each sample were affiliated to Arcobacter spp. Finally, colonies with sequence similarity of >97% were grouped and given a similar designation. We obtained four strains designated as 15T96H‐1, one strain designated as 15T96H‐2 from the 15T96H sample, and five strains designated as 25T96H‐1 from the 25T96H sample. The 16S rRNA gene sequences of 15T96H‐1 and 15T96H‐2 showed high similarity (98.2% and 98.3%, respectively) with uncultured bacterium clone HglApr921 (JX016315). Since both these strains have low homology with other isolated Arcobacter species (93.3%–94.3% similarity), they suggest potential new species. On the other hand, all strains of 25T96H‐1 showed similarity ranging from 99.2% to 100% with A. marinus. For the samples incubated at 15°C or 25°C for 48 hr, a few colonies were isolated but they were not identified as Arcobacter spp.
4. DISCUSSION
Thus far, research on Arcobacter has mainly been focused on detection since several Arcobacter species are pathogenic to humans (Collado et al., 2011; Ho et al., 2006; Houf, Tutenel, Zutter, Hoof, & Vandamme, 2000). Meat and seafood are the most common sources of Arcobacter reported (Atabay & Corry, 1997; Houf, Zutter, Hoof, & Vandamme, 2002; Hume et al., 2001; Romero et al., 2002; Collado, Guarro, et al., 2009; Levican et al., 2014; Salas‐Massó et al., 2016; Mottola et al., 2016; Rathlavath, Kohli, et al., 2017; Rathlavath, Kumar, & Nayak, 2017; Vicente‐Martins, Oleastro, Domingues, & Ferreira, 2018). Arcobacter is also detected or isolated from seawater, sewage, and drinking water, and these environments are considered important as they could be one of the possible routes of transmission of Arcobacter to human and animal intestinal tracts (Collado et al., 2011). Various methods including enterobacterial repetitive intergenic consensus PCR (ERIC‐PCR), randomly amplified polymorphic DNA‐PCR (Houf et al., 2002), and amplified fragment length polymorphism (On, Harrington, & Atabay, 2003,2004) have been used to detect and elucidate the transmission routes or to trace the sources of Arcobacter outbreaks (Collado & Figueras, 2011). Multiplex‐PCR that can detect multiple species simultaneously has also been used (Brightwell et al., 2007; Houf et al., 2000; Khan et al., 2017). ERIC‐PCR in particular has been successfully applied to outbreak investigations (Vandamme et al., 1993) in food. Although these methods have advantages due to its simplicity and cost, they detect only a specific species from isolated strains or from mixed cultures based on specific culture conditions (González, Bayas Morejón, & Ferrús, 2017).
To prevent bias resulting from culture‐dependent methods in this study, we used Arcobacter‐specific clone libraries to directly identify 16S rRNA gene sequences. Twelve OTUs relating to Arcobacter were detected from abalones using Arcobacter‐specific clone libraries (Table 2), all clustered with previously reported Arcobacter sequences (Figure 2). All the OTUs showed similarity to 16S rRNA genes detected from marine environments such as marine invertebrates or seawater, but not from those identified from terrestrial sources such as poultry. Furthermore, these sequences are not classified with any other known Arcobacter species. Interestingly, using our analytical approach, the samples from abalones also did not show the presence of pathogenic Arcobacter spp., A. butzleri, A. skirrowii, and A. cryaerophilus, commonly isolated or detected from bivalves. Bivalves such as mussels and clams are filter feeders that feed plankton using gills, while abalones feed on brown algae. Thus, they have a more developed digestive system compared to bivalves. The result suggests that gastropods such as abalone may not be host or harbor pathogenic Arcobacter species, perhaps due to their different feeding habits and digestive system.
Figure 2.
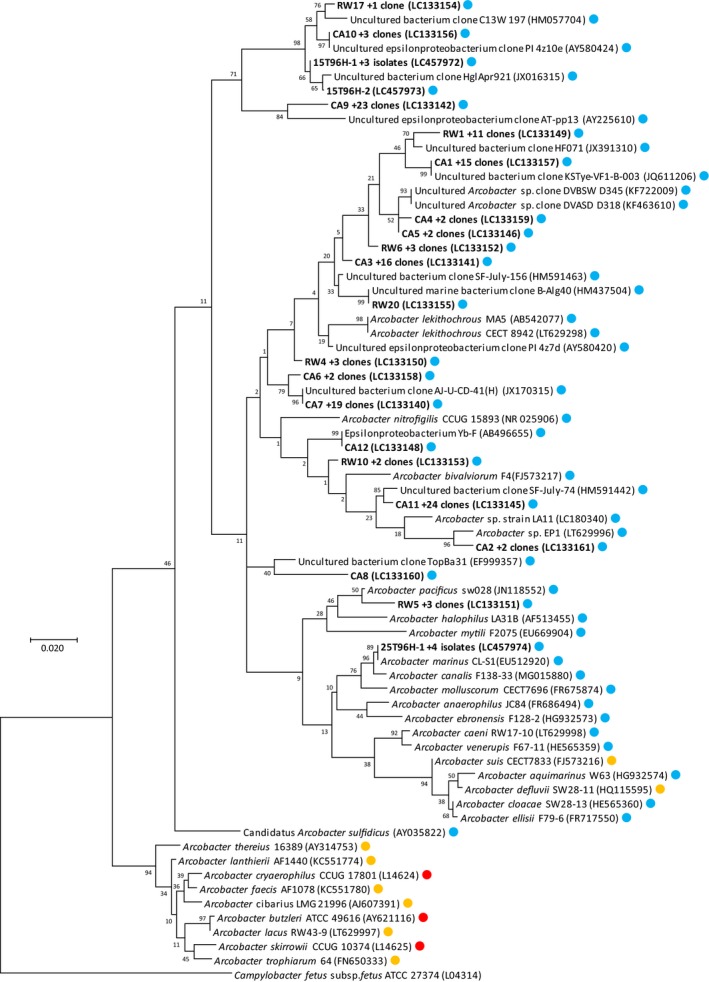
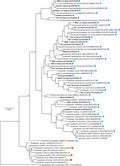
16S rRNA gene‐based phylogenetic tree of Arcobacter spp. from abalone and environmental samples. Circles colors indicate origins or pathogenicity of Arcobacter spp. as follows: blue, marine habitats, orange, terrestrial environments and red, pathogenic species. The tree was generated using the maximum likelihood (ML) method with 1,000 replicates in the bootstrap analysis. The distances were estimated with the Jukes‐Cantor correction. The tree was rooted with Campylobacter fetus subsp. fetus ATCC 27374, and gene sequences are followed by GenBank accession numbers in parentheses. Scale bar represents 2% sequence divergence
In this study, we employed the ARC94F Arcobacter‐specific probe for FISH against cells isolated from abalone for detection and quantification. This ARC94F probe has been used for specific counting of genus Arcobacter in seawater (Fera et al., 2008, 2010; Moreno et al., 2003). The ratio of ARC94F‐positive cells suggested that Arcobacter might be a common bacterial genus in abalones (6.96%, Table 3). Haliotis gigantea appears to be a habitat for Arcobacter species, but the role and effect on their hosts are still unclear.
Regarding the cultivation of Arcobacter spp., several conditions have been introduced, such as altering the NaCl concentration (Salas‐Massó et al., 2016) or the requirement of sea salts (Diéguez, Balboa, Magnesen, & Romalde, 2017). Hence, we used artificial sea salt instead of NaCl and added more than 2.5% sea salt during isolation. In addition, the incubation temperature used for Arcobacter isolation was set to 15 or 25°C, which are closer to the seawater temperatures of the natural habitat of the abalones at Ise Bay, Mie prefecture, against common methods (30 to 37°C: Vandamme et al., 1991; Houf et al., 2000; Collado, Guarro, et al., 2009; Merga et al., 2011; Salas‐Massó et al., 2016; Salas‐Massó, Figueras, Andree, & Furones, 2018; Laishram et al., 2016; González et al., 2017). As a result, 10 Arcobacter isolates (five known and five novels) were recovered from samples 15T96H and 25T96H. These were classified into three genotypes based on 97% sequence similarity (15T96H‐1, 15T96H‐2, and 25T96H‐1). The 16S rRNA gene of 25T96H‐1 had a high similarity of 99.2%–100% to A. marinas, which has been isolated from a mixture of seawater and starfish (Kim, Hwang, & Cho, 2010). In contrast, the isolates 15T96H‐1 and 15T96H‐2 had no closely related sequences in all other known Arcobacter isolates. Including comparisons with uncultured clones, the 16S rRNA genes of 15T96H‐1 or 1596H‐2 have related to an uncultured clone detected from the Pacific oyster Crassostrea gigas, an invertebrate living in shallow water (Madigan et al., 2014) and marine bulk water (Teeling et al., 2012). Both 15T96H‐1 and 15T96H‐2 were isolated only at the 15°C incubation temperature. From these observations, we believe that 15T96H‐1 and 15T96H‐2 will be able to be isolated from various marine invertebrates with sea salt medium at lower temperatures. In terms of incubation time, no Arcobacter species were isolated within 2 days of incubation. This implies that when incubation temperature is lower than 37°C, Arcobacter requires more than 96 hr to grow before colonies can be detected. There are still many species of Arcobacter detectable by molecular methods in abalones that are not cultivable. We feel that isolation methods should be improved to obtain these uncultured Arcobacter species.
In conclusion, we succeeded in detecting several new Arcobacter genotypes from abalone using Arcobacter‐specific 16S rRNA gene libraries. Furthermore, since most of the clones showed low similarity with other known Arcobacter spp. and no pathogenic Arcobacter were detected or isolated from abalone, we need further investigations for uncultured Arcobacter spp. which remains to be determined in H. gigantea.
CONFLICT OF INTERESTS
All authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
YM, SI, and RO performed the experiments; RT, TM, and SF designed and supervised the study; YM wrote the paper. All authors read and approved the final manuscript.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
This work was supported by a JSPS Research Fellowship (no. 18J14216) for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Mizutani Y, Iehata S, Mori T, Oh R, Fukuzaki S, Tanaka R. Diversity, enumeration, and isolation of Arcobacter spp. in the giant abalone, Haliotis gigantea . MicrobiologyOpen. 2019;8:e890 10.1002/mbo3.890
DATA ACCESSIBILITY
Raw sequencing data are available at the NCBI website (http://www.ncbi.nlm.nih.gov/). Accession numbers of 16S rRNA gene sequence data from Arcobacter‐specific clone libraries are LC133140–LC133161, and from bacteria isolations, LC457972–LC457974.
REFERENCES
- Amann, R. I. , Binder, B. J. , Olson, R. J. , Chisholm, S. W. , Devereux, R. , & Stahl, D. A. (1990). Combination of 16S rRNA‐targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and Environment Microbiology, 56, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabay, H. I. , & Corry, J. E. (1997). The prevalence of campylobacters and arcobacters in broiler chickens. Journal of Applied Microbiology, 83, 619–626. 10.1046/j.1365-2672.1997.00277.x [DOI] [PubMed] [Google Scholar]
- Brightwell, G. , Mowat, E. , Clemens, R. , Boerema, J. , Pulford, D. J. , & On, S. L. (2007). Development of a multiplex and real time PCR assay for the specific detection of Arcobacter butzleri and Arcobacter cryaerophilus . Journal of Microbiol Methods, 68, 318–325. 10.1016/j.mimet.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Collado, L. , Cleenwerck, I. , Van Trappen, S. , De Vos, P. , & Figueras, M. J. (2009). Arcobacter mytili sp. nov., an indoxyl acetate‐hydrolysis‐negative bacterium isolated from mussels. International Journal of Systematic and Evolutionary Microbiology, 59, 1391–1396. 10.1099/ijs.0.003749-0 [DOI] [PubMed] [Google Scholar]
- Collado, L. , & Figueras, M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter . Clinical Microbiology Reviews, 24, 174–192. 10.1128/cmr.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado, L. , Guarro, J. , & Figueras, M. J. (2009). Prevalence of Arcobacter in meat and shellfish. Journal of Food Protection, 72, 1102–1106. 10.4315/0362-028X-72.5.1102 [DOI] [PubMed] [Google Scholar]
- Collado, L. , Levican, A. , Perez, J. , & Figueras, M. J. (2011). Arcobacter defluvii sp. nov., isolated from sewage samples. International Journal of Systematic and Evolutionary Microbiology, 61, 2155–2161. 10.1099/ijs.0.025668-0 [DOI] [PubMed] [Google Scholar]
- De Smet, S. , Vandamme, P. , De Zutter, L. , On, S. L. , Douidah, L. , & Houf, K. (2011). Arcobacter trophiarum sp. nov., isolated from fattening pigs. International Journal of Systematic and Evolutionary Microbiology, 61, 356–361. 10.1099/ijs.0.022665-0 [DOI] [PubMed] [Google Scholar]
- Diéguez, A. L. , Balboa, S. , Magnesen, T. , & Romalde, J. L. (2017). Arcobacter lekithochrous sp. nov., isolated from a molluscan hatchery. International Journal of Systematic and Evolutionary Microbiology, 67, 1327–1332. 10.1099/ijsem.0.001809 [DOI] [PubMed] [Google Scholar]
- Donachie, S. P. , Bowman, J. P. , On, S. L. , & Alam, M. (2005). Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter . International Journal of Systematic and Evolutionary Microbiology, 55, 1271–1277. 10.1099/ijs.0.63581-0 [DOI] [PubMed] [Google Scholar]
- Fera, M. T. , Gugliandolo, C. , Lentini, V. , Favaloro, A. , Bonanno, D. , La Camera, E. , & Maugeri, T. L. (2010). Specific detection of Arcobacter spp. in estuarine waters of Southern Italy by PCR and fluorescent in situ hybridization. Letters in Applied Microbiology, 50, 65–70. 10.1111/j.1472-765x.2009.02767.x [DOI] [PubMed] [Google Scholar]
- Fera, M. T. , Maugeri, T. L. , Gugliandolo, C. , La Camera, E. , Lentini, V. , Favaloro, A. , … Carbone, M. (2008). Induction and resuscitation of viable nonculturable Arcobacter butzleri cells. Applied and Environment Microbiology, 74, 3266–3268. 10.1128/aem.00059-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras, M. J. , Collado, L. , Levican, A. , Perez, J. , Solsona, M. J. , & Yustes, C. (2011). Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Systematic and Applied Microbiology, 34, 105–109. 10.1016/j.syapm.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Figueras, M. J. , Levican, A. , Collado, L. , Inza, M. I. , & Yustes, C. (2011). Arcobacter ellisii sp. nov., isolated from mussels. Systematic and Applied Microbiology, 34, 414–418. 10.1016/j.syapm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- González, A. , Bayas Morejón, I. F. , & Ferrús, M. A. (2017). Isolation, molecular identification and quinolone‐susceptibility testing of Arcobacter spp. isolated from fresh vegetables in Spain. Food Microbiology, 65, 279–283. 10.1016/j.fm.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Harmon, K. M. , & Wesley, I. V. (1996). Identification of Arcobacter isolates by PCR. Letters in Applied Microbiology, 23, 241–244. 10.1111/j.1472-765x.1996.tb00074.x [DOI] [PubMed] [Google Scholar]
- Ho, H. T. , Lipman, L. J. , & Gaastra, W. (2006). Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Veterinary Microbiology, 115, 1–13. 10.1016/j.vetmic.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Houf, K. , De Zutter, L. , Van Hoof, J. , & Vandamme, P. (2002). Assessment of the genetic diversity among Arcobacters isolated from poultry products by using two PCR‐based typing methods. Applied and Environment Microbiology, 68, 2172–2178. 10.1128/aem.68.5.2172-2178.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houf, K. , On, S. L. , Coenye, T. , Debruyne, L. , De Smet, S. , & Vandamme, P. (2009). Arcobacter thereius sp. nov., isolated from pigs and ducks. International Journal of Systematic and Evolutionary Microbiology, 59, 2599–2604. 10.1099/ijs.0.006650-0 [DOI] [PubMed] [Google Scholar]
- Houf, K. , On, S. L. , Coenye, T. , Mast, J. , Van Hoof, J. , & Vandamme, P. (2005). Arcobacter cibarius sp. nov., isolated from broiler carcasses. International Journal of Systematic and Evolutionary Microbiology, 55, 713–717. 10.1099/ijs.0.63103-0 [DOI] [PubMed] [Google Scholar]
- Houf, K. , Tutenel, A. , De Zutter, L. , Van Hoof, J. , & Vandamme, P. (2000). Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii . FEMS Microbiology Letters, 193, 89–94. 10.1016/s0378-1097(00)00461-4 [DOI] [PubMed] [Google Scholar]
- Huber, T. , Faulkner, G. , & Hugenholtz, P. (2004). Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics, 20, 2317–2319. 10.1093/bioinformatics/bth226 [DOI] [PubMed] [Google Scholar]
- Hume, M. E. , Harvey, R. B. , Stanker, L. H. , Droleskey, R. E. , Poole, T. L. , & Zhang, H. B. (2001). Genotypic variation among Arcobacter isolates from a farrow‐to‐finish swine facility. Journal of Food Protection, 64, 645–651. 10.4315/0362-028X-64.5.645 [DOI] [PubMed] [Google Scholar]
- Kepner, R. L. Jr. , & Pratt, J. R. (1994). Use of fluorochromes for direct enumeration of total bacteria in environmental samples: Past and present. Microbiological Reviews, 58, 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, I. U. H. , Cloutier, M. , Libby, M. , Lapen, D. R. , Wilkes, G. , & Topp, E. (2017). Enhanced single‐tube multiplex PCR assay for detection and identification of six Arcobacter species. Journal of Applied Microbiology, 123, 1522–1532. 10.1111/jam.13597 [DOI] [PubMed] [Google Scholar]
- Kiehlbauch, J. A. , Brenner, D. J. , Nicholson, M. A. , Baker, C. N. , Patton, C. M. , Steigerwalt, A. G. , & Wachsmuth, I. K. (1991). Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. Journal of Clinical Microbiology, 29, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. M. , Hwang, C. Y. , & Cho, B. C. (2010). Arcobacter marinus sp. nov. International Journal of Systematic and Evolutionary Microbiology, 60, 531–536. 10.1099/ijs.0.007740-0 [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111–120. 10.1007/bf01731581 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram, M. , Rathlavath, S. , Lekshmi, M. , Kumar, S. , & Nayak, B. B. (2016). Isolation and characterization of Arcobacter spp. from fresh seafood and the aquatic environment. International Journal of Food Microbiology, 232, 87–89. 10.1016/j.ijfoodmicro.2016.05.018 [DOI] [PubMed] [Google Scholar]
- Levican, A. , Collado, L. , Aguilar, C. , Yustes, C. , Diéguez, A. L. , Romalde, J. L. , & Figueras, M. J. (2012). Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Systematic and Applied Microbiology, 35, 133–138. 10.1016/j.syapm.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Levican, A. , Collado, L. , & Figueras, M. J. (2013). Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Systematic and Applied Microbiology, 36, 22–27. 10.1016/j.syapm.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Levican, A. , Collado, L. , Yustes, C. , Aguilar, C. , & Figueras, M. J. (2014). Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Applied and Environment Microbiology, 80, 385–391. 10.1128/AEM.03014-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican, A. , Rubio‐Arcos, S. , Martinez‐Murcia, A. , Collado, L. , & Figueras, M. J. (2015). Arcobacter ebronensis sp. nov. and Arcobacter aquimarinus sp. nov., two new species isolated from marine environment. Systematic and Applied Microbiology, 38, 30–35. 10.1016/j.syapm.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Madigan, T. L. , Bott, N. J. , Torok, V. A. , Percy, N. J. , Carragher, J. F. , de Barros Lopes, M. A. , & Kiermeier, A. (2014). A microbial spoilage profile of half shell Pacific oysters (Crassostrea gigas) and Sydney rock oysters (Saccostrea glomerata). Food Microbiology, 38, 219–227. 10.1016/j.fm.2013.09.005 [DOI] [PubMed] [Google Scholar]
- McClung, C. R. , Patriquin, D. G. , & Davis, R. E. (1983). Campylobacter nitrofigilis sp. nov., a nitrogen‐fixing bacterium associated with roots of Spartina alterniflora Loisel . International Journal of Systematic Bacteriology, 33, 605–612. 10.1099/00207713-33-3-605 [DOI] [Google Scholar]
- Merga, J. Y. , Leatherbarrow, A. J. , Winstanley, C. , Bennett, M. , Hart, C. A. , Miller, W. G. , & Williams, N. J. (2011). Comparison of Arcobacter isolation methods, and diversity of Arcobacter spp. in Cheshire, United Kingdom. Applied and Environmental Microbiology, 77, 1646–1650. 10.1128/AEM.01964-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziti, A. , Mente, E. , & Kormas, K. A. (2012). Gut bacteria associated with different diets in reared Nephrops norvegicus . Systematic and Applied Microbiology, 35, 473–482. 10.1016/j.syapm.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Moreno, Y. , Botella, S. , Alonso, J. L. , Ferrús, M. A. , Hernández, M. , & Hernández, J. (2003). Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Applied and Environment Microbiology, 69, 1181–1186. 10.1128/aem.69.2.1181-1186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola, A. , Bonerba, E. , Figueras, M. J. , Pérez‐Cataluña, A. , Marchetti, P. , Serraino, A. , … Di Pinto, A. (2016). Occurrence of potentially pathogenic arcobacters in shellfish. Food Microbiology, 57, 23–27. 10.1016/j.fm.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Neill, S. D. , Campbell, J. N. , O'Brien, J. J. , Weatherup, S. T. C. , & Ellis, W. A. (1985). Taxonomic position of Campylobacter cryaerophila sp. nov. International Journal of Systematic Bacteriology, 35, 342–356. 10.1099/00207713-35-3-342 [DOI] [Google Scholar]
- On, S. L. , Atabay, H. I. , Amisu, K. O. , Coker, A. O. , & Harrington, C. S. (2004). Genotyping and genetic diversity of Arcobacter butzleri by amplified fragment length polymorphism (AFLP) analysis. Letters in Applied Microbiology, 39, 347–352. 10.1111/j.1472-765x.2004.01584.x [DOI] [PubMed] [Google Scholar]
- On, S. L. , Harrington, C. S. , & Atabay, H. I. (2003). Differentiation of Arcobacter species by numerical analysis of AFLP profiles and description of a novel Arcobacter from pig abortions and turkey faeces. Journal of Applied Microbiology, 95, 1096–1105. 10.1046/j.1365-2672.2003.02100.x [DOI] [PubMed] [Google Scholar]
- Ootsubo, M. , Shimizu, T. , Tanaka, R. , Sawabe, T. , Tajima, K. , & Ezura, Y. (2003). Seven‐hour fluorescence in situ hybridization technique for enumeration of Enterobacteriaceae in food and environmental water sample. Journal of Applied Microbiology, 95, 1182–1190. 10.1046/j.1365-2672.2003.02051.x [DOI] [PubMed] [Google Scholar]
- Park, S. , Jung, Y. T. , Kim, S. , & Yoon, J. H. (2016). Arcobacter acticola sp. nov., isolated from seawater on the East Sea in South Korea. Journal of Microbiology, 54, 655–659. 10.1007/s12275-016-6268-4 [DOI] [PubMed] [Google Scholar]
- Perez‐Cataluña, A. , Salas‐Masso, N. , & Figueras, M. J. (2018a). Arcobacter canalis sp. nov., isolated from a water canal contaminated with urban sewage. International Journal of Systematic and Evolutionary Microbiology, 68, 1258–1264. 10.1099/ijsem.0.002662 [DOI] [PubMed] [Google Scholar]
- Pérez‐Cataluña, A. , Salas‐Massó, N. , & Figueras, M. J. (2018b). Arcobacter lacus sp. nov. and Arcobacter caeni sp. nov., two novel species isolated from reclaimed water. Systematic and Applied Microbiology. 10.1099/ijsem.0.003101 [DOI] [PubMed] [Google Scholar]
- Rathlavath, S. , Kohli, V. , Singh, A. S. , Lekshmi, M. , Tripathi, G. , Kumar, S. , & Nayak, B. B. (2017). Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. International Journal of Food Microbiology, 263, 32–37. 10.1016/j.ijfoodmicro.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Rathlavath, S. , Kumar, S. , & Nayak, B. B. (2017). Comparative isolation and genetic diversity of Arcobacter sp. from fish and the coastal environment. Letters in Applied Microbiology, 65, 42–49. 10.1111/lam.12743 [DOI] [PubMed] [Google Scholar]
- Roller, C. , Wagner, M. , Amann, R. , Ludwig, W. , & Schleifer, K. H. (1994). In situ probing of Gram‐positive bacteria with high DNA G + C content using 23S rRNA‐targeted oligonucleotides. Microbiology, 140, 2849–2858. 10.1099/00221287-140-10-2849 [DOI] [PubMed] [Google Scholar]
- Romero, J. , García‐Varela, M. , Laclette, J. P. , & Espejo, R. T. (2002). Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis). Microbial Ecology, 44, 365–371. 10.1007/s00248-002-1063-7 [DOI] [PubMed] [Google Scholar]
- Salas‐Massó, N. , Andree, K. B. , Furones, M. D. , & Figueras, M. J. (2016). Enhanced recovery of Arcobacter spp. using NaCl in culture media and re‐assessment of the traits of Arcobacter marinus and Arcobacter halophilus isolated from marine water and shellfish. Science of the Total Environment, 566–567, 1355–1361. 10.1016/j.scitotenv.2016.05.197 [DOI] [PubMed] [Google Scholar]
- Salas‐Massó, N. , Figueras, M. J. , Andree, K. B. , & Furones, M. D. (2018). Do the Escherichia coli European Union shellfish safety standards predict the presence of Arcobacter spp., a potential zoonotic pathogen? Science of the Total Environment, 624, 1171–1179. 10.1016/j.scitotenv.2017.12.178 [DOI] [PubMed] [Google Scholar]
- Sasi Jyothsna, T. S. , Rahul, K. , Ramaprasad, E. V. , Sasikala, C. H. , & Ramana, C. V. (2013). Arcobacter anaerophilus sp. nov., isolated from an estuarine sediment and emended description of the genus Arcobacter . International Journal of Systematic and Evolutionary Microbiology, 63, 4619–4625. 10.1099/ijs.0.054155-0 [DOI] [PubMed] [Google Scholar]
- Snaidr, J. , Amann, R. , Huber, I. , Ludwig, W. , & Schleifer, K. H. (1997). Phylogenetic analysis and in situ identification of bacteria in activated sludge. Applied and Environment Microbiology, 63, 2884–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt, E. , & Goebel, B. M. (1994). Taxonomic note: A place for DNA‐DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic Bacteriology, 44, 846–849. 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- Tanaka, R. , Cleenwerck, I. , Mizutani, Y. , Iehata, S. , Bossier, P. , & Vandamme, P. (2017). Arcobacter haliotis sp. nov., isolated from abalone species Haliotis gigantea . International Journal of Systematic and Evolutionary Microbiology, 67, 3050–3056. 10.1099/ijsem.0.002080 [DOI] [PubMed] [Google Scholar]
- Tanaka, R. , Ootsubo, M. , Sawabe, T. , Ezura, Y. , & Tajima, K. (2004). Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture‐independent techniques. Aquaculture, 241, 453–463. 10.1016/j.aquaculture.2004.08.032 [DOI] [Google Scholar]
- Teeling, H. , Fuchs, B. M. , Becher, D. , Klockow, C. , Gardebrecht, A. , Bennke, C. M. , … Amann, R. (2012). Substrate‐controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science, 336, 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme, P. , Falsen, E. , Rossau, R. , Hoste, B. , Segers, P. , Tytgat, R. , & De Ley, J. (1991). Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: Emendation of generic descriptions and proposal of Arcobacter gen. nov. International Journal of Systematic Bacteriology, 41, 88–103. 10.1099/00207713-41-1-88 [DOI] [PubMed] [Google Scholar]
- Vandamme, P. , Giesendorf, B. A. , van Belkum, A. , Pierard, D. , Lauwers, S. , Kersters, K. , … Quint, W. G. (1993). Discrimination of epidemic and sporadic isolates of Arcobacter butzleri by polymerase chainreaction‐mediated DNA fingerprinting. Journal of Clinical Microbiology, 31, 3317–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme, P. , Vancanneyt, M. , Pot, B. , Mels, L. , Hoste, B. , Dewettinck, D. , … Goossens, H. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. International Journal of Systematic Bacteriology, 42, 344–356. 10.1099/00207713-42-3-344 [DOI] [PubMed] [Google Scholar]
- Vicente‐Martins, S. , Oleastro, M. , Domingues, F. C. , & Ferreira, S. (2018). Arcobacter spp. at retail food from Portugal: Prevalence, genotyping and antibiotics resistance. Food Control, 85, 107–112. 10.1016/j.foodcont.2017.09.024 [DOI] [Google Scholar]
- Whiteduck‐Léveillée, K. , Whiteduck‐Léveillée, J. , Cloutier, M. , Tambong, J. T. , Xu, R. , Topp, E. , … Khan, I. U. (2015). Arcobacter lanthieri sp. nov., isolated from pig and dairy cattle manure. International Journal of Systematic and Evolutionary Microbiology, 65, 2709–2716. 10.1099/ijs.0.000318 [DOI] [PubMed] [Google Scholar]
- Whiteduck‐Léveillée, K. , Whiteduck‐Léveillée, J. , Cloutier, M. , Tambong, J. T. , Xu, R. , Topp, E. , … Khan, I. U. (2016). Identification, characterization and description of Arcobacter faecis sp. nov., isolated from a human waste septic tank. Systematic and Applied Microbiology, 39, 93–99. 10.1016/j.syapm.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Yu, C. , Wang, X. , Yu, S. , & Zhang, X. H. (2016). Arcobacter pacificus sp. nov., isolated from seawater of the South Pacific Gyre. International Journal of Systematic and Evolutionary Microbiology, 66, 542–547. 10.1099/ijsem.0.000751 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing data are available at the NCBI website (http://www.ncbi.nlm.nih.gov/). Accession numbers of 16S rRNA gene sequence data from Arcobacter‐specific clone libraries are LC133140–LC133161, and from bacteria isolations, LC457972–LC457974.


