Abstract
Off-tissue effects are persistent issues of modern inhibition-based therapies. By merging the strategies of photopharmacology and small-molecule degraders, we introduce a novel concept for persistent spatiotemporal control of induced protein degradation that potentially prevents off-tissue toxicity. Building on the successful principle of bifunctional all-small-molecule Proteolysis Targeting Chimeras (PROTACs), we designed photoswitchable PROTACs (photoPROTACs) by including ortho-F4-azobenzene linkers between both warhead ligands. This highly bistable yet photoswitchable structural component leads to reversible control over the topological distance between both ligands. The azo-cis-isomer is observed to be inactive because the distance defined by the linker is prohibitively short to permit complex formation between the protein binding partners. By contrast, the azo-trans-isomer is active since it can engage both protein partners to form the necessary and productive ternary complex. Importantly, due to the bistable nature of the ortho-F4-azobenzene moiety employed, the photostationary state of the photoPROTAC is persistent, with no need for continuous irradiation. This technique offers reversible on/off switching of protein degradation that is compatible with an intracellular environment and, therefore, could be useful in experimental exploration of biological signaling pathways—such as those crucial for oncogenic signal transduction. Additionally, this strategy may be suitable for therapeutic intervention to address a variety of diseases. By enabling reversible activation and deactivation of protein degradation, photoPROTACs offer advantages over conventional photocaging strategies that irreversibly release active agents.
Short abstract
Optical control over protein degradation is achieved by introducing an azobenzene linker to the PROTAC scaffold generating novel photoPROTACs that can be reversibly activated with visible light.
Introduction
In recent years, the transition from inhibition of aberrant protein function to specific degradation of undesired proteins with Proteolysis Targeting Chimeras (PROTACs) has resulted in remarkable progress and is currently effecting a paradigm shift in drug discovery and therapy development.1,2 PROTACs effect highly efficient protein degradation by commandeering the endogenous ubiquitin–proteasome system. PROTACs engage proteins of interest and E3 ligases in a ternary complex, leading to specific polyubiquitination and labeling of proteins for degradation via the proteasome. This approach has allowed quick expansion of the “druggable proteome” beyond proteins that bear distinct functional sites responsible for their respective mode of action. By avoiding high drug doses and acting via novel mechanisms, PROTACs show promise as therapeutic candidates for disease phenotypes that display resistance to conventional inhibition-based therapy.3
With the evolution from peptide- to small-molecule-based PROTACs, the therapeutic potential of PROTACs is quickly expanding. Selective degradation of transcriptional regulators (BRD4,4,5 CDK9,6 TRIM247), trans-membrane receptor tyrosine kinases (EGFR,8 c-Met,8 ALK9), hormone receptors (ERRα, AR),10 or proteins linked to neurodegenerative diseases (Tau11) extends the potential of PROTACs for treatment of a variety of diseases, such as cancer of the hematopoietic tissue12 or hormone receptor-mediated solid malignancies.13 Recent additions to the portfolio of chimeric degraders include functional molecules specifically targeting degradation of extracellular proteins.14,15
In spite of the tremendous advancements outlined, current PROTAC approaches may still have undesired effects because systemic application can affect untargeted tissue, a disadvantage shared with traditional inhibitor-based therapeutics. As an example, ARV-771, a highly active BET protein degrader, has been shown to achieve complete regression of prostate cancer in a CRPC mouse xenograft model.13 However, an on-target, general cytotoxic effect was also observed, most notably with the occurrence of skin deterioration at the injection site. This demonstrates the need for a switchable element within the PROTAC scaffold, that would allow reversible degradation in a spatiotemporal manner.
A strategy to potentially circumvent systemic undesired effects of PROTACs involves the use of designed peptides, known as phosphoPROTACs, that can be conditionally activated via phosphorylation by specific growth-factor stimuli.16 Other approaches couple light stimuli to irreversibly induce protein degradation. This includes photocaging17 and the use of inducible degrons.18 However, both of those techniques require the fusion of protein domains. Nonetheless, the use of light stimuli is a highly attractive approach due to the high spatiotemporal precision with which it can be applied.19 Thus, there is a need for light-based strategies complementing optogenetic approaches. In this regard, the growing field of photopharmacology includes many examples enabling optical control of receptor function by employment of photoswitchable ligands.20,21 This prompted us to evaluate possible ways of introducing a photoswitchable handle within the PROTAC scaffold.
Rationale
Despite the published work with a variety of small molecules, PROTAC linkers have been the subject of only nominal variation, which most often has been restricted to alkyl and polyether linkers of varying lengths. This has highlighted the subtle sensitivity of ternary complex formation to linker length and composition.22,23 Analysis of the small-molecule PROTAC literature suggests that a minimum linker length is necessary between the warheads, because shorter linkers are otherwise unable to span the gap needed to simultaneously bind both the recruited proteins in a ternary complex.24
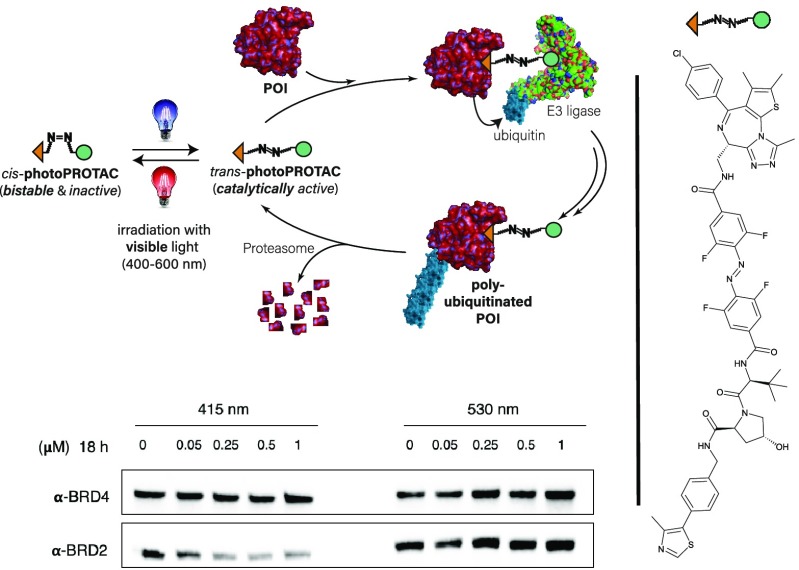
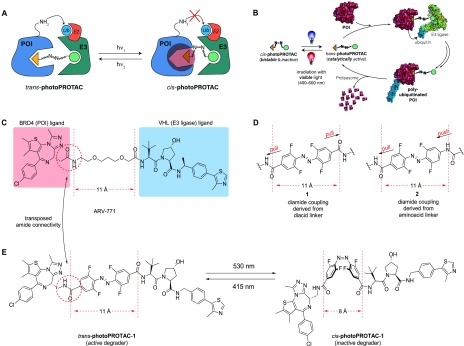
The critical difference in linker length between active and inactive degraders in many of the reported examples is about 3 Å.3,25 Strikingly, the switch between trans- and cis-azobenzenes corresponds to a very similar difference of 3–4 Å in topological distance. Making use of this observation, we envisioned generation of photoswitchable PROTACs (photoPROTACs) in which the typical linear polyether linker is replaced by azobenzenes. This would serve to introduce a novel photoswitchable functionality to the PROTAC linker beyond passive tethering of the two ligands. At the heart of the idea is the design of a trans-photoPROTAC that corresponds to the optimized linker length for efficient induction of ternary complex formation between protein of interest (POI) and E3 ligase (Figure 1A). By contrast, the photoswitched cis-photoPROTAC would span a prohibitively short distance and thus be unable to reach the binding pocket of the second binding partner, precluding ternary complex formation, target ubiquitination, and eventual degradation.
Figure 1.
Concept of photoPROTACs. (A) The functional azobenzene handle allows for photoinduced switching between an active trans- and inactive cis-photoPROTAC isomer which is prohibitively short to engage both proteins in a ternary complex. (B) Features of photoPROTACs: catalytic, bistable, and switchable with visible light. (C) Structure of highly active BET protein degrader ARV-771 displaying a maximal distance of 11 Å between both warhead moieties. (D) “Pull–pull” and “push–pull” modes of modular connectivity for introduction of an o-F4 azobenzene moiety. (E) Devised BRD-degrading photoPROTAC, representatively shown with “pull–pull” diacid linker. The trans-isomer retains the maximal distance displayed by ARV-771 while the cis-isomer is considerably shorter.
In considering the potential applications, the ideal photoswitchable PROTAC-based therapeutic would have long-lived photostationary states (∼days) that are populated following initial light stimuli and persist throughout treatment, thereby avoiding the need for continued or pulsed irradiation exposure. This requires the design of inactive azo-cis-isomers that are configurationally stable in biological settings. In this respect, bistable ortho-tetrafluoroazobenzenes (o-F4-azobenzenes) have been recently introduced,26 and their azo-cis-isomers may display thermal τ1/2’s as high as two years at 25 °C compared to a few hours for the parent cis-azobenzene.27 Within these design boundaries, rationally designed photoPROTACs possess important characteristics summarized in Figure 1B: a highly stable, inactive cis-photoPROTAC is isomerized by means of a visible light stimulus to a catalytically active trans-photoPROTAC, that induces polyubiquitination of a POI by complexation with an E3 ligase. The labeled POI then proceeds to degradation by the proteasome while the trans-photoPROTAC remains active until isomerization via a second light stimulus of a different wavelength regenerates the inactive cis-photoPROTAC.
As a photoPROTAC proof-of-concept, ARV-77113 (Figure 1C) was selected as the lead structure for the generation of a photoswitchable BET protein degrader. To introduce the o-F4-azobenzene fragment within the PROTAC scaffold as part of a highly modular synthesis approach, two different amide linkers (Figure 1D) were envisioned involving either a precursor o-F4-azobenzene diacid, as shown embedded in 1, or, alternatively, an o-F4-azobenzene amino-acid, as shown in 2. The designed replacement of the oligoether linker in ARV-771 with photoswitchable linkers furnishes the isomeric photoPROTAC pair shown in Figure 1E, which maintain an optimal distance of 11 Å between both warheads for the putatively active trans-photoPROTAC-1 and a diminished distance of 8 Å in cis-photoPROTAC-1.
Results
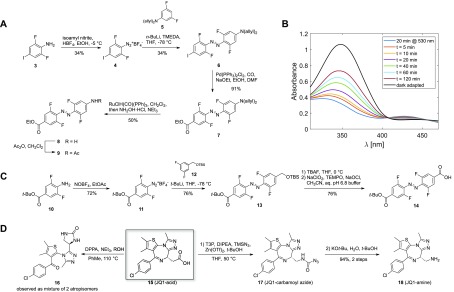
Synthetic efforts commenced with the generation of unsymmetrical amino acid o-F4-azobenzene linker as shown in Figure 1D (right) which had been predicted to possess a cis-τ1/2 of about 80 days.28 The use of Feringa’s method29 gave access to 6 from diazonium salt 4 and the organolithium derivative generated from protected 3,5-difluoroaniline 5 (Figure 2A). Subsequent palladium-catalyzed carbonylative esterification furnished aminoester 7.30 Initial efforts to remove the bisallyl protecting group under classical Pd-mediated conditions31 were not met with success. Instead, ruthenium-catalyzed isomerization and subsequent hydrolysis of the enamine produced gave the targeted aniline 8.32 Acetylation of aniline 8 gave 9, which served as a model to examine bistability. Switching between trans-9 and cis-9 occurred by irradiation at the well-separated n−π* absorption bands at 415 nm (cis–trans) and 530 nm (trans–cis), respectively. Unfortunately, cis-9 generated under 530 nm irradiation quickly isomerized to the thermodynamically more stable trans-9 with a thermal half-life of ∼2 h (Figure 2B). On the basis of our design criteria, this observation rendered azobenzene linker building block 8 unsuitable. It is important to note that this observation is in line with the general concept of diminished bistability of “push–pull” azobenzenes.33,34
Figure 2.
(A) Synthetic approach toward unsymmetrical amino acid azobenzene linker 8. (B) Bistability measurement of model substrate 9, starting with enriched cis-9 (t = 0) after a 20 min irradiation at 530 nm (50 μM in CH3CN). (C) Synthetic approach toward monoprotected diacid azobenzene building block 14. (D) Classical Curtius conditions mainly generated urea 16 under reflux conditions, preventing access to 18. Milder Curtius conditions under Lewis-acid catalysis allowed isolation of 17 which could be transformed into JQ-1 amine 18.
The results with 9 prompted us to revise the design of the photoPROTAC conjugate to include a “pull–pull” system by the introduction of diacid linker as shown for 1 (Figure 1D,E), bearing two electron-withdrawing substituents. Notably, this permutation in design necessitated transposition of the amide bond joining BET protein ligand JQ-1 and the azobenzene linker moiety (Figure 1C,E). For ease of handling, generation of building block 14 was targeted, which would allow for facile discrimination of both substitution sites. Aniline 10 was generated in a 3-step sequence starting from 2,6-difluoro-4-iodoaniline (Figure 2C). Treatment of 10 with NOBF4 in EtOAc afforded diazonium tetrafluoroborate 11.35 The diazonium salt was trapped with lithiated TBS-protected 3,5-difluorobenzylalcohol 12 to furnish unsymmetrically substituted o-F4-azobenzene 13.29 After TBS deprotection with TBAF and oxidation36 of the obtained benzylic alcohol desired bifunctional azobenzene linker 14 was generated.
To synthesize required JQ1-amine 18, a sequence was devised to convert JQ1-acid 15 to the corresponding amine via Curtius rearrangement. However, under classical conditions, the isocyanate generated could not be trapped by alcohols such as tert-butanol or benzyl alcohol but instead led to the formation of urea 16 (Figure 2D). We hypothesized that intramolecular reaction of the intermediate isocyanate with the diazepine was leading to complications and examined alternative conditions involving the use of TMSN3 under mild conditions.37,38 Under these conditions JQ1-carbamoyl azide 17 was isolated, which could then be easily deprotected within minutes by employing KOt-Bu in aq. t-BuOH. This facile sequence may be generally applicable with challenging substrates for the Curtius rearrangement.
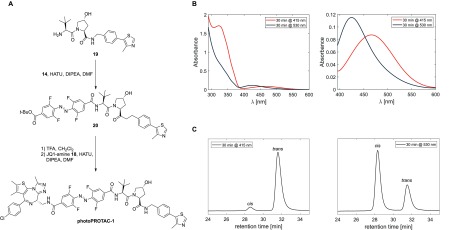
With all necessary building blocks in hand the desired BRD-photoPROTAC-1 was assembled by a series of amide couplings (Figure 3A). After coupling of VHL ligand39,4019 to the azobenzene, the tert-butyl ester was hydrolyzed with TFA, and JQ-1 amine 18 was attached furnishing o-F4-azobenzene linked BRD-photoPROTAC-1. Next, the photochemical properties of the conjugate generated were examined. In DMSO, the cis–trans switch occurred efficiently by irradiation at 415 nm (Figure 3B), producing a photostationary state (PSS) consisting of 95% trans-photoPROTAC-1 as determined by separation of both isomers via HPLC (Figure 3C). Irradiation at 530 nm established a PSS consisting of 68% cis-photoPROTAC-1, which is slightly reduced when compared to examples of o-F4-azobenzenes with higher cis-PSSs27,41 although the n−π* absorption maxima of both isomers are well-separated by 47 nm (Figure S7 for calculated spectra of pure cis- and trans-photoPROTAC-1).
Figure 3.
(A) Modular assembly of photoPROTAC-1. (B) UV–vis spectra of photoenriched trans-photoPROTAC-1 (irradiated at 415 nm, shown in orange) and cis-photoPROTAC-1 (irradiated at 530 nm, shown in dark blue) in DMSO; left panel, UV–vis spectra in the range 290–600 nm; right panel, enlargement of the spectra in the range 390–600 nm. (C) RP-HPLC chromatograms of photoenriched trans-photoPROTAC-1 (415 nm, left panel) and cis-photoPROTAC-1 (530 nm, right panel) in DMSO shown at isosbestic wavelength (275 nm in CH3CN/H2O).
In addition, quantum yields for both isomerization reactions at either 415 or 530 nm irradiation were calculated on the basis of kinetic data (Figures S6–S10; see section S2.9 for technical details). The obtained quantum yields [ϕEZ (530 nm) = 0.28, ϕZE (415 nm) = 0.65] were of similar order as those reported previously for underivatized o-F4-azobenzenes27 indicating efficient isomerization upon photon absorption. Importantly, the reported bistability of the “pull–pull” ortho-F4 azobenzene was also retained in the photoPROTAC-1 derivative, and no thermal back-isomerization of cis-photoPROTAC-1 was observed in DMSO, acetonitrile, or aq. buffer for several days at 37 °C. The suitability for biological applications was further demonstrated by retained stability of a 50 μM solution of photoPROTAC-1 in the presence of 10 mM reduced glutathione over 3 days (Figure S5A). Additionally, stability of photoPROTAC-1 in cells was evaluated. Cells were incubated with or without photoPROTAC-1 at 25 μM, and after different time periods of up to 24 h, cells were harvested and lysed in PBS by multiple freeze–thaw cycles. Then, cell lysates were analyzed by LC-MS for nonreduced and reduced forms of photoPROTAC-1. Consistent with the data presented in Figure S5A, masses matching a potentially reduced form were not detected. However, fully retained isotopic distribution after 24 h matching the nonreduced form of the photoPROTAC indicated the stability of the photoPROTAC under biological conditions (Figure S5B,C).
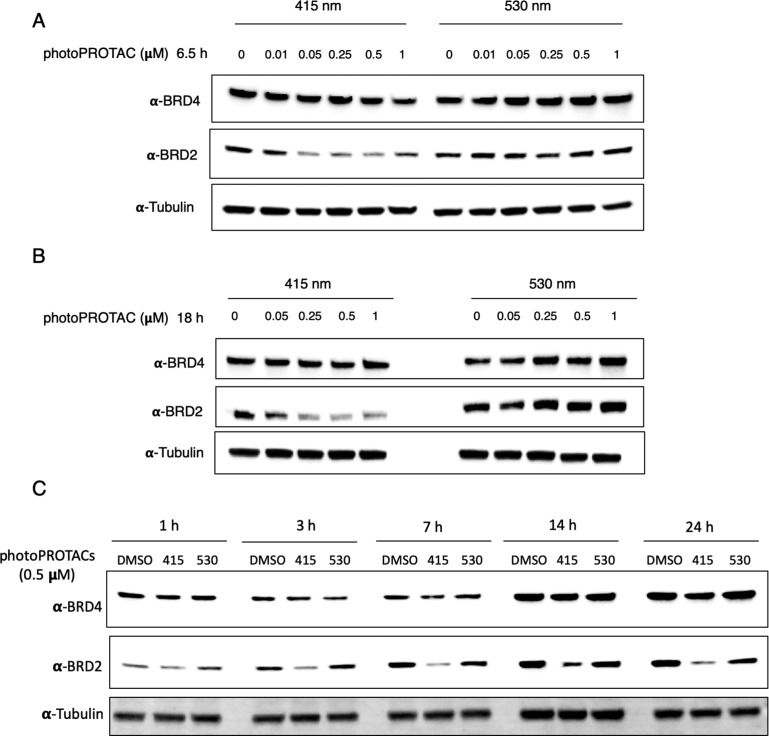
Having confirmed stability under biological conditions and efficient switching between cis- and trans-configurations upon irradiation at 530 and 415 nm, we next carried out biological experiments in Ramos cells. PhotoPROTAC-1 solutions were irradiated using 415 or 530 nm LEDs initially for 20 min to obtain trans- and cis-isomers, respectively; after 1 min of rest and brief vortexing, irradiation was continued for another 10 min. Trans- and cis-photoPROTAC-1 solutions were diluted to the desired concentrations and added to Ramos cells. Incubation of trans-photoPROTAC-1 for 6.5 h significantly induced BRD2 degradation at low nanomolar concentrations (Figure 4A). Conversely, cis-photoPROTAC-1 did not induce BRD2 degradation in the range of concentrations tested. Longer 18 h incubation with either cis- or trans-photoPROTAC-1 did not improve or affect the BRD2 degradation efficiencies (Figure 4B). Interestingly, photoPROTAC-1 did not induce efficient BRD2 degradation in cells without an initial 415 nm irradiation indicating that the presence of a mixture of trans- and cis-photoPROTAC-1 (35% cis-isomer after equilibrating for 5 days under daylight) led to diminished BRD2 degradation (Figure S3). One possible explanation for this observation would be that the inactive cis-isomer has a higher affinity for either BRD2 or VHL than does trans-photoPROTAC-1, which blocks binding of the active isomer and thereby prevents induction of degradation. As a result, ratios well below the PSS of 68% cis at 530 nm evidently suffice to prevent degradation.
Figure 4.
PhotoPROTAC-1 can be converted to active trans- and inactive cis-isomers to induce differential degradation of BRD2. PhotoPROTAC-1 working solutions were irradiated for 30 min using 415 or 530 nm LEDs and added to Ramos cells at varying concentrations. Cells were incubated in the dark for (A) 6.5 h or (B) 18 h prior to cell lysis. Treatment of active trans-photoPROTAC induced significant BRD2 degradation within 6.5 h whereas inactive cis-photoPROTAC-1 did not show a significant effect on BRD2 levels. BRD4 degradation was not observed under indicated conditions. (C) Time-dependent degradation of BRD proteins by photoPROTAC-1. Singly irradiated photoPROTAC-1 (trans and cis) were treated at 500 nM concentrations, and BRD protein levels were analyzed after indicated time points.
To investigate time-dependent degradation profiles of trans- and cis-photoPROTAC-1, Ramos cells were harvested at different time points ranging from 1 to 24 h. Within 1 h, significant BRD2 degradation was not observed. However, after 3 h of incubation, trans-photoPROTAC-1 induced BRD2 degradation, which increased over the next 4 h. After 7 h, BRD2 degradation reached the maximum level and remained unchanged over the following 17 h (Figure 4C). To confirm that trans-photoPROTAC-1 induced BRD2 degradation through the proteasomal pathway, we analyzed the BRD2 degradation profile in the presence or absence of the selective NEDD8 inhibitor MLN-4924. Treatment with MLN-4924 rescued BRD2 degradation indicating that BRD2 degradation took place via the proteasomal pathway (Figure S4).
Curiously, the data did not show a significant degradation of BRD4 in response to cis- or trans-photoPROTAC-1 treatments even though ARV-771 could degrade both BRD4 and BRD2, and with greater potency (Figure S1). Although no structural data are currently available for trans-photoPROTAC-1 bound to BRD4, hypotheses for the observed differential degradation include a newly gained selectivity of the trans-photoPROTAC-1 toward BRD2 over BRD4 due to the reversed amide bond between JQ-1 and o-F4-azobenzene moiety—a structural feature differentiating it from ARV-771 (Figure 1C,E). Also, introduction of the azobenzene to the PROTAC scaffold increased the overall stiffness19 of the conjugate that could further account for a potential loss of important interactions with BRD4. Both structural changes might result in a less stable ternary complex, leading to a more rapid dissociation and consequently to inefficient ubiquitination and proteasomal degradation of BRD4. Overall, these data suggested that the o-F4-azobenzene moiety within photoPROTAC-1 allows interconversion between active trans-photoPROTAC-1 and inactive cis-photoPROTAC-1 that is rapid and stable—the latter made evident by the finding that continuous irradiation does not yield a superior differential BRD2 degradation (Figure S3). Concordantly, the biological data indicate efficient degradation of BRD2 by trans-photoPROTAC-1 and minimum degradation by cis-photoPROTAC-1 obtained from a single, initial irradiation.
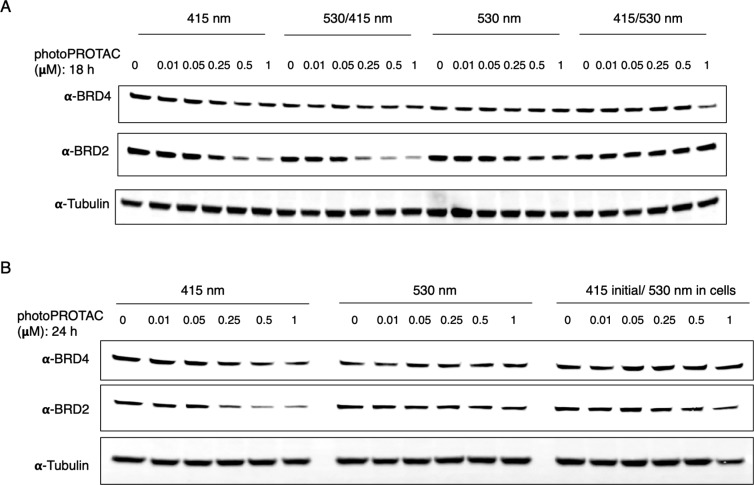
Next, to test the reversible photoswitching between cis-photoPROTAC-1 and trans-photoPROTAC-1, we performed a two-step irradiation process using both 415 and 530 nm LEDs before incubation with cells. Initial 415 and 530 nm irradiations were carried out separately for 30 min to obtain active trans-photoPROTAC-1 and inactive cis-photoPROTAC-1, respectively. Trans-photoPROTAC-1 and cis-photoPROTAC-1 were then subdivided (50:50) into two tubes each. One tube of each photoPROTAC-1 isomer was set aside, while the remaining tube of trans-photoPROTAC-1 was subjected to a second round of irradiation at 530 nm, and the remaining tube of cis-photoPROTAC-1 was exposed to the 415 nm LED. To assess the reversible photoisomerization of photoPROTAC-1, Ramos cells were separately treated with varying concentrations of either singly irradiated (415 nm, trans-photoPROTAC-1; or 530 nm, cis-photoPROTAC-1) or doubly irradiated (530/415 nm, trans-photoPROTAC-1; or 415/530 nm, cis-photoPROTAC-1) photoPROTAC-1. After 18 h of incubation with photoPROTAC-1 obtained from different irradiation combinations, cells were lysed and analyzed for levels of BRD4 and BRD2. In line with the previous experiment (Figure 4), similar BRD4/2 degradation patterns were observed for trans-photoPROTAC-1 and cis-photoPROTAC-1 that were obtained through single irradiation (Figure 5A, 415 or 530 nm). Regarding double-irradiated photoPROTAC-1 (Figure 5A, 530/415 or 415/530 nm), the data demonstrate a complete reversal of their degradation potential following a second irradiation (i.e., 530 nm followed by 415 nm LEDs or vice versa) indicating the dynamic switching of photoPROTAC between active and inactive states.
Figure 5.
PhotoPROTACs are dynamically interchangeable between active trans- and inactive cis-configurations. (A) PhotoPROTAC-1 working solutions were initially irradiated with 415 or 530 nm LEDs followed by second irradiation using 530 or 415 nm (415/530 and 530/415 nm) to reverse the initial configurations. The treatment of singly irradiated trans-photoPROTAC-1 (415 nm) induced a significant BRD2 degradation relative to the cis-photoPROTAC-1 (530 nm) whereas no significant BRD4 degradation was observed in response to both trans- and cis-photoPROTAC-1. Second irradiation with either 530 or 415 nm significantly shifted the biologically active trans-isomer to inactive cis-isomer and vice versa. (B) Temporal control of photoPROTAC-1 activity. Singly irradiated photoPROTAC-1 (trans/415 nm and cis/530 nm) were added to cells. One set of cis- and trans-photoPROTAC-1 treated cells were kept in the dark, and a parallel set of cells treated with trans-photoPROTAC-1 were exposed to irradiation at 530 nm (415 initial/530 nm in cells) to induce the trans to cis isomerization in cells.
To further investigate the reversibility and temporal controllability of photoPROTAC-1, one set of cells treated with trans-photoPROTAC-1 was incubated in the dark, and another, parallel set of cells (trans-photoPROTAC-1 treated) was incubated under 530 nm irradiation. As anticipated, cis-photoPROTAC-1 was recovered by illuminating 530 nm light on cells at the desired time point. Intracellular conversion was confirmed by suspension of BRD2 degradation in contrast to BRD2 levels in cells kept in the dark (Figure 5B). In addition, significant BRD2 degradation was observed when cis-photoPROTAC-1 treated cells were incubated under 415 nm LED (Figure S2) suggesting that the o-F4-azobenzene moiety can be successfully incorporated into PROTAC linkers for light-induced, spatiotemporal control of targeted protein degradation in a reversible manner.
Overall, our data indicate that initial irradiation of photoPROTAC-1 holds the potential for successful spatiotemporal control of photoPROTAC-1 and avoids laborious continuous-irradiation efforts to induce the photoswitching between active and inactive states of PROTACs. Moreover, sustained BRD2 degradation after an 18 h incubation with trans-photoPROTAC-1 as well as sustained suspension of BRD2 degradation after an 18 h incubation with cis-photoPROTAC-1 (Figure 4B) indicate the bistability of o-F4-azobenzene containing photoPROTAC-1. Had the active trans-isomer switched to the inactive cis-isomer over the course of the experiment, persistent BRD2 degradation would have not been prominent considering the significant resynthesis of BRD proteins that occurs as a feedback mechanism in response to inhibition (Figure 4A,B, 530 nm treated cells).42 Vice versa, had the cis-isomer switched to the thermodynamically more stable active trans-isomer within the time scale of the experiment, degradation of BRD2 would have been induced over time. Hence, our approach provides robust control over induced degradation. Taken together, our data provide evidence to support the use of o-F4-azobenzene moiety in PROTAC design to provide a powerful strategy to spatiotemporally control PROTAC activity with a single initial irradiation step.
Outlook
Bistable heterodiazocine motifs, displaying >99% cis-content in the dark with stable trans-isomers bearing thermal half-lives of several days, are promising candidates that may be amalgamated with our overarching strategy to generate cis-inactive photoPROTACs.43 Future approaches could include azobenzenes sufficiently red-shifted to allow irradiation in the NIR/IR window for optimal tissue penetration.44,45 Additionally, taking advantage of two-photon excitation processes may further enable switching in a more favorable window.28,41
Applications of our photoPROTAC method to target proteins beyond BET domains are currently under investigation and will be reported in due course. This also includes engagement of other E3 ligases such as cereblon or MDM2. Moreover, photoPROTACs may serve as a reversibly controllable platform for aggressively targeting central parts of the cell machinery with high resolution, for example, for the degradation of antiapoptotic proteins. Dilution via diffusion of the on-site activated trans-photoPROTAC may suffice to prevent off-tissue effects; however, for even higher resolution, back-switching of the activated trans-photoPROTAC to the inactive azo-cis-isomer by a second optical fiber light source at a defined distance from the application site can be envisioned. As part of our studies it was demonstrated that isomer compositions in ratios well below the cis-rich PSS at 530 nm suffice to halt degradation, which indicates that very short irradiation periods in the range of minutes (for kinetic data cf. Figures S8 and S10) could be already satisfactory to persistently turn off degradation activity. This is an advantage over conventional photocaging strategies which release active drugs only irreversibly.
While in the process of preparing this manuscript, we became aware of a preprint demonstrating an alternative approach by generating cis-active photoswitchable PROTACs.46 In their complementary approach, Trauner et al. introduced azobenzene moieties to ligands of E3 ligase cereblon. In contrast to our bistable system, their strategy necessitates continued light pulses at 390 nm to induce prolonged degradation otherwise their degrader remains inactive in the dark.
Conclusion
The combination of two recently emerging areas in drug discovery, namely, photopharmacology and small-molecule degraders, has led us to develop the concept of photoswitchable, bistable photoPROTACs with potentially far reaching implications for manifold applications. In combination with modern methods of proteomics, photoPROTACs offer further opportunity for studying downstream effects of signaling pathways which are yet insufficiently understood.47 More broadly, it should be noted that spatiotemporal activation/deactivation of photoswitchable PROTACs by a single irradiation event may find use in novel therapeutics.
Acknowledgments
C.M.C. gratefully acknowledges support from the NIH (R35 CA197589). E.M.C. gratefully acknowledges ETH Zurich for financial support. P.P. acknowledges a fellowship of the Stipendienfonds Schweizerische Chemische Industrie (SSCI). Dr. John Hines is acknowledged for assistance in editing the manuscript. We are also thankful to Dr. Saul Jaime-Figueroa and Dr. Fabian Menges for their valuable inputs.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00713.
Additional figures and data, experimental procedures, and photochemical and analytical characterization of synthetic compounds (PDF)
Author Contributions
⊥ P.P. and K.T.G.S. contributed equally to this work. P.P. and E.M.C. conceived the project with feedback from C.M.C. P.P. synthesized the compounds. K.T.G.S. performed biological experiments.
The authors declare the following competing financial interest(s): C.M.C. is a shareholder in and consultant to Arvinas, Inc., which partially supported this work.
Supplementary Material
References
- Lai A. C.; Crews C. M. Induced Protein Degradation: An Emerging Drug Discovery Paradigm. Nat. Rev. Drug Discovery 2017, 16 (2), 101–114. 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G. M.; Crews C. M. Small-Molecule Modulation of Protein Homeostasis. Chem. Rev. 2017, 117 (17), 11269–11301. 10.1021/acs.chemrev.7b00077. [DOI] [PubMed] [Google Scholar]
- Buhimschi A. D.; Armstrong H. A.; Toure M.; Jaime-Figueroa S.; Chen T. L.; Lehman A. M.; Woyach J. A.; Johnson A. J.; Byrd J. C.; Crews C. M. Targeting the C481S Ibrutinib-Resistance Mutation in Bruton’s Tyrosine Kinase Using PROTAC-Mediated Degradation. Biochemistry 2018, 57 (26), 3564–3575. 10.1021/acs.biochem.8b00391. [DOI] [PubMed] [Google Scholar]
- Winter G. E.; Buckley D. L.; Paulk J.; Roberts J. M.; Souza A.; Dhe-Paganon S.; Bradner J. E. Phthalimide Conjugation as a Strategy for in Vivo Target Protein Degradation. Science (Washington, DC, U. S.) 2015, 348 (6241), 1376–1381. 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Qian Y.; Altieri M.; Dong H.; Wang J.; Raina K.; Hines J.; Winkler J. D.; Crew A. P.; Coleman K.; et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22 (6), 755–763. 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson C. M.; Jiang B.; Erb M. A.; Liang Y.; Doctor Z. M.; Zhang Z.; Zhang T.; Kwiatkowski N.; Boukhali M.; Green J. L.; et al. Pharmacological Perturbation of CDK9 Using Selective CDK9 Inhibition or Degradation. Nat. Chem. Biol. 2018, 14 (2), 163–170. 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechijian L. N.; Buckley D. L.; Lawlor M. A.; Reyes J. M.; Paulk J.; Ott C. J.; Winter G. E.; Erb M. A.; Scott T. G.; Xu M.; et al. Functional TRIM24 Degrader via Conjugation of Ineffectual Bromodomain and VHL Ligands. Nat. Chem. Biol. 2018, 14 (4), 405–412. 10.1038/s41589-018-0010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G. M.; Smith B. E.; Lai A. C.; Jaime-Figueroa S.; McQuaid D. C.; Bondeson D. P.; Toure M.; Dong H.; Qian Y.; Wang J.; et al. The Advantages of Targeted Protein Degradation Over Inhibition: An RTK Case Study. Cell Chem. Biol. 2018, 25 (1), 67–77. 10.1016/j.chembiol.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C. E.; Gao Y.; Tan L.; Donovan K. A.; Nowak R. P.; Loehr A.; Bahcall M.; Fischer E. S.; Jänne P. A.; George R. E.; et al. Chemically Induced Degradation of Anaplastic Lymphoma Kinase (ALK). J. Med. Chem. 2018, 61 (9), 4249–4255. 10.1021/acs.jmedchem.7b01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson D. P.; Mares A.; Smith I. E. D.; Ko E.; Campos S.; Miah A. H.; Mulholland K. E.; Routly N.; Buckley D. L.; Gustafson J. L.; et al. Catalytic in Vivo Protein Knockdown by Small-Molecule PROTACs. Nat. Chem. Biol. 2015, 11 (8), 611–617. 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. C.; Ferguson F. M.; Cai Q.; Donovan K. A.; Nandi G.; Patnaik D.; Zhang T.; Huang H. T.; Lucente D. E.; Dickerson B. C.; et al. Targeted Degradation of Aberrant Tau in Frontotemporal Dementia Patient-Derived Neuronal Cell Models. eLife 2019, 8 (8), e45457 10.7554/eLife.45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz D. T.; Fiskus W.; Qian Y.; Manshouri T.; Rajapakshe K.; Raina K.; Coleman K. G.; Crew A. P.; Shen A.; Mill C. P.; et al. Novel BET Protein Proteolysis-Targeting Chimera Exerts Superior Lethal Activity than Bromodomain Inhibitor (BETi) against Post-Myeloproliferative Neoplasm Secondary (s) AML Cells. Leukemia 2017, 31 (9), 1951–1961. 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K.; Lu J.; Qian Y.; Altieri M.; Gordon D.; Rossi A. M. K.; Wang J.; Chen X.; Dong H.; Siu K.; et al. PROTAC-Induced BET Protein Degradation as a Therapy for Castration-Resistant Prostate Cancer. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (26), 7124–7129. 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalawansha D. A.; Paiva S. L.; Rafizadeh D. N.; Pettersson M.; Qin L.; Crews C. M. Targeted Protein Internalization and Degradation by ENDosome TArgeting Chimeras (ENDTACs). ACS Cent. Sci. 2019, 5 (6), 1079–1084. 10.1021/acscentsci.9b00224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Banik S.; Pedram K.; Wisnovsky S.; Riley N.; Bertozzi C.. Lysosome Targeting Chimeras (LYTACs) for the Degradation of Secreted and Membrane Proteins. ChemRexiv 2019, preprint, 10.26434/chemrxiv.7927061. [DOI] [Google Scholar]
- Hines J.; Gough J. D.; Corson T. W.; Crews C. M. Posttranslational Protein Knockdown Coupled to Receptor Tyrosine Kinase Activation with PhosphoPROTACs. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (22), 8942–8947. 10.1073/pnas.1217206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacour Q.; Li C.; Plamont M. A.; Billon-Denis E.; Aujard I.; Le Saux T.; Jullien L.; Gautier A. Light-Activated Proteolysis for the Spatiotemporal Control of Proteins. ACS Chem. Biol. 2015, 10 (7), 1643–1647. 10.1021/acschembio.5b00069. [DOI] [PubMed] [Google Scholar]
- Bonger K. M.; Rakhit R.; Payumo A. Y.; Chen J. K.; Wandless T. J. General Method for Regulating Protein Stability with Light. ACS Chem. Biol. 2014, 9 (1), 111–115. 10.1021/cb400755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velema W. A.; Szymanski W.; Feringa B. L. Photopharmacology: Beyond Proof of Principle. J. Am. Chem. Soc. 2014, 136 (6), 2178–2191. 10.1021/ja413063e. [DOI] [PubMed] [Google Scholar]
- Westphal M. V.; Schafroth M. A.; Sarott R. C.; Imhof M. A.; Bold C. P.; Leippe P.; Dhopeshwarkar A.; Grandner J. M.; Katritch V.; Mackie K.; et al. Synthesis of Photoswitchable9-Tetrahydrocannabinol Derivatives Enables Optical Control of Cannabinoid Receptor 1 Signaling. J. Am. Chem. Soc. 2017, 139 (50), 18206–18212. 10.1021/jacs.7b06456. [DOI] [PubMed] [Google Scholar]
- Hüll K.; Morstein J.; Trauner D. In Vivo Photopharmacology. Chem. Rev. 2018, 118 (21), 10710–10747. 10.1021/acs.chemrev.8b00037. [DOI] [PubMed] [Google Scholar]
- Bondeson D. P.; Smith B. E.; Burslem G. M.; Buhimschi A. D.; Hines J.; Jaime-Figueroa S.; Wang J.; Hamman B. D.; Ishchenko A.; Crews C. M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25 (1), 78–87. 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. E.; Wang S. L.; Jaime-Figueroa S.; Harbin A.; Wang J.; Hamman B. D.; Crews C. M. Differential PROTAC Substrate Specificity Dictated by Orientation of Recruited E3 Ligase. Nat. Commun. 2019, 10 (1), 131. 10.1038/s41467-018-08027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew A. P.; Raina K.; Dong H.; Qian Y.; Wang J.; Vigil D.; Serebrenik Y. V.; Hamman B. D.; Morgan A.; Ferraro C.; et al. Identification and Characterization of von Hippel-Lindau-Recruiting Proteolysis Targeting Chimeras (PROTACs) of TANK-Binding Kinase 1. J. Med. Chem. 2018, 61 (2), 583–598. 10.1021/acs.jmedchem.7b00635. [DOI] [PubMed] [Google Scholar]
- Zhou B.; Hu J.; Xu F.; Chen Z.; Bai L.; Fernandez-Salas E.; Lin M.; Liu L.; Yang C. Y.; Zhao Y.; et al. Discovery of a Small-Molecule Degrader of Bromodomain and Extra-Terminal (BET) Proteins with Picomolar Cellular Potencies and Capable of Achieving Tumor Regression. J. Med. Chem. 2018, 61 (2), 462–481. 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bléger D.; Schwarz J.; Brouwer A. M.; Hecht S. O -Fluoroazobenzenes as Readily Synthesized Photoswitches Offering Nearly Quantitative Two-Way Isomerization with Visible Light. J. Am. Chem. Soc. 2012, 134 (51), 20597–20600. 10.1021/ja310323y. [DOI] [PubMed] [Google Scholar]
- Knie C.; Utecht M.; Zhao F.; Kulla H.; Kovalenko S.; Brouwer A. M.; Saalfrank P.; Hecht S.; Bléger D. Ortho-Fluoroazobenzenes: Visible Light Switches with Very Long-Lived Z Isomers. Chem. - Eur. J. 2014, 20 (50), 16492–16501. 10.1002/chem.201404649. [DOI] [PubMed] [Google Scholar]
- Cabré G.; Garrido-Charles A.; Moreno M.; Bosch M.; Porta-de-la-Riva M.; Krieg M.; Gascón-Moya M.; Camarero N.; Gelabert R.; Lluch J. M.; et al. Rationally Designed Azobenzene Photoswitches for Efficient Two-Photon Neuronal Excitation. Nat. Commun. 2019, 10 (1), 907. 10.1038/s41467-019-08796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. J.; Lerch M. M.; Szymanski W.; Feringa B. L. Direct and Versatile Synthesis of Red-Shifted Azobenzenes. Angew. Chem., Int. Ed. 2016, 55 (43), 13514–13518. 10.1002/anie.201607529. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Liu J.; Lu Z.; Luo X.; Zhang H.; Lan Y.; Lei A. Base-Induced Mechanistic Variation in Palladium-Catalyzed Carbonylation of Aryl Iodides. J. Am. Chem. Soc. 2010, 132 (9), 3153–3158. 10.1021/ja909962f. [DOI] [PubMed] [Google Scholar]
- Garro-Helion F.; Merzouk A.; Guibé F. Mild and Selective Palladium(0)-Catalyzed Deallylation of Allylic Amines. Allylamine and Diallylamine as Very Convenient Ammonia Equivalents for the Synthesis of Primary Amines. J. Org. Chem. 1993, 58, 6109–6113. 10.1021/jo00074a044. [DOI] [Google Scholar]
- Lutz T. A.; Spanner P.; Wanner K. T. A General Approach to Substituted Diphenyldiazenes. Tetrahedron 2016, 72 (12), 1579–1589. 10.1016/j.tet.2016.02.011. [DOI] [Google Scholar]
- Dong M.; Babalhavaeji A.; Samanta S.; Beharry A. A.; Woolley G. A. Red-Shifting Azobenzene Photoswitches for in Vivo Use. Acc. Chem. Res. 2015, 48 (10), 2662–2670. 10.1021/acs.accounts.5b00270. [DOI] [PubMed] [Google Scholar]
- Garcia-Amorós J.; Díaz-Lobo M.; Nonell S.; Velasco D. Fastest Thermal Isomerization of an Azobenzene for Nanosecond Photoswitching Applications under Physiological Conditions. Angew. Chem., Int. Ed. 2012, 51 (51), 12820–12823. 10.1002/anie.201207602. [DOI] [PubMed] [Google Scholar]
- Wannagat U.; Hohlstein G. Zur Fluorierung Mit Nitrosyl-tetrafluoro-borat. Chem. Ber. 1955, 88 (12), 1839–1846. 10.1002/cber.19550881203. [DOI] [Google Scholar]
- Zhao M.; Li J.; Mano E.; Song Z.; Tschaen D. M.; Grabowski E. J. J.; Reider P. J. Oxidation of Primary Alcohols to Carboxylic Acids with Sodium Chlorite Catalyzed by TEMPO and Bleach. J. Org. Chem. 1999, 64 (7), 2564–2566. 10.1021/jo982143y. [DOI] [Google Scholar]
- Lebel H.; Leogane O. Boc-Protected Amines via a Mild and Efficient One-Pot Curtius Rearrangement. Org. Lett. 2005, 7 (19), 4107–4110. 10.1021/ol051428b. [DOI] [PubMed] [Google Scholar]
- Augustine J. K.; Bombrun A.; Mandal A. B.; Alagarsamy P.; Atta R. N.; Selvam P. Propylphosphonic Anhydride (T3P ®)-Mediated One-Pot Rearrangement of Carboxylic Acids to Carbamates. Synthesis 2011, 2011 (9), 1477–1483. 10.1055/s-0030-1259964. [DOI] [Google Scholar]
- Galdeano C.; Gadd M. S.; Soares P.; Scaffidi S.; Van Molle I.; Birced I.; Hewitt S.; Dias D. M.; Ciulli A. Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities. J. Med. Chem. 2014, 57 (20), 8657–8663. 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerle M.; Chan K. H.; Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015, 10 (8), 1770–1777. 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passlick S.; Richers M. T.; Ellis-Davies G. C. R. Thermodynamically Stable, Photoreversible Pharmacology in Neurons with One- and Two-Photon Excitation. Angew. Chem., Int. Ed. 2018, 57 (38), 12554–12557. 10.1002/anie.201807880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riching K. M.; Mahan S.; Corona C. R.; McDougall M.; Vasta J. D.; Robers M. B.; Urh M.; Daniels D. L. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Biol. 2018, 13 (9), 2758–2770. 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- Hammerich M.; Schütt C.; Stähler C.; Lentes P.; Röhricht F.; Höppner R.; Herges R. Heterodiazocines: Synthesis and Photochromic Properties, Trans to Cis Switching within the Bio-Optical Window. J. Am. Chem. Soc. 2016, 138 (40), 13111–13114. 10.1021/jacs.6b05846. [DOI] [PubMed] [Google Scholar]
- Profio A. E. Light Transport in Tissue. Appl. Opt. 1989, 28 (12), 2216–2222. 10.1364/AO.28.002216. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Salo D.; Kim D. M.; Komarov S.; Tai Y.-C.; Berezin M. Y. Penetration Depth of Photons in Biological Tissues from Hyperspectral Imaging in Shortwave Infrared in Transmission and Reflection Geometries. J. Biomed. Opt. 2016, 21 (12), 126006. 10.1117/1.JBO.21.12.126006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders M.; Matsuura B.; Bérouti M.; Simoneschi D.; Marzio A.; Pagano M.; Trauner D. PHOTACs Enable Optical Control of Protein Degradation. ChemRxiv 2019, preprint, 10.26434/chemrxiv.8206688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F.; Mina M.; Armenia J.; Chatila W. K.; Luna A.; La K. C.; Dimitriadoy S.; Liu D. L.; Kantheti H. S.; Saghafinia S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173 (2), 321–337. 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.