Abstract
Hysteresis is an important feature of enzyme-catalyzed reactions, as it reflects the influence of enzyme regulation in the presence of ligands such as substrates or allosteric molecules. In typical kinetic studies of enzyme activity, hysteretic behavior is observed as a “lag” or “burst” in the time course of the catalyzed reaction. These lags and bursts are due to the relatively slow transition from one state to another state of the enzyme molecule, with different states having different kinetic properties. However, it is difficult to understand the underlying mechanism of hysteresis by observing bulk reactions because the different enzyme molecules in the population behave stochastically. In this work, we studied the hysteretic behavior of mutant β-glucuronidase (GUS) using a high-throughput single-molecule array platform and investigated the effect of thermal treatment on the hysteresis.
Short abstract
We explored the hysteresis of mutant β-glucuronidase at the single-molecule level and found that hysteresis was a substrate-induced activation process, which could be accelerated by a heat-pulse.
Introduction
Hysteresis in enzymology reflects the influence of enzyme regulation in the presence of ligands such as substrates or allosteric molecules.1−3 A hysteretic enzyme will have a slow response to a rapid change in the concentration of substrate, where such a slow response manifests itself as either a “lag” or a “burst” in the kinetic property.4 Hysteresis has physiological significance in enzymology because it functions to control inhibition or activation of a biological pathway. For example, it could modulate the amplitude of inherent oscillations of a metabolic pathway and therefore benefit the cell.5
An enzyme exhibiting hysteresis usually means that it has additional states with different kinetic properties. For example, hysteresis may be a reflection of a ligand-induced slow jump of an enzyme between two forms with different catalytic activities;2,6,7 hysteresis may also result from a ligand-induced slow isomerization reaction of an inactive or partially active enzyme species into a fully active conformation.2,8,9 In addition, different enzyme molecules could remain in different kinetic states at different time points, which makes hysteresis challenging to analyze from bulk measurements.
We and others have developed technologies to measure enzymatic kinetics of populations containing hundreds to thousands of single enzyme molecules and revealed the heterogeneous activity of horseradish peroxidase10 and β-galactosidase.11 In these experiments, instead of observing only one or a few enzyme molecules, the kinetics of many single-molecules can be monitored simultaneously in solution in femtoliter-sized reaction chambers without immobilization, using droplets,12 liposomes,13 or optical fiber bundles.14
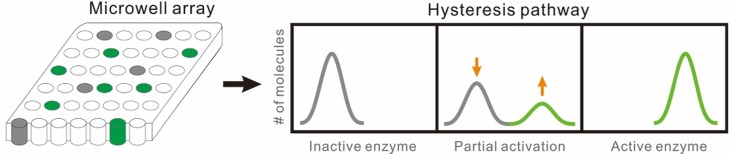
In this paper, we investigated several β-glucuronidase (GUS) variants containing amino acid substitutions at different surface sites. These substitutions caused the enzymes to exhibit hysteresis behavior. We performed a single-molecule enzymology study to understand the hysteresis mechanism. GUS belongs to the glycosidase family of enzymes that catalyzes the breakdown of complex carbohydrates. It has been intensively used as a gene expression reporter and also has been frequently evolved in vitro for a variety of additional functions.15 The single-molecule kinetics of GUS and multiple evolved forms have been studied previously.16 Via protein engineering, we found that a new type of surface modified mutant GUS exhibited hysteresis behavior. To understand how the engineered enzyme transits from one state to another, a microwell array platform, which contains 216 000 microwells, was used to demonstrate the single-molecule kinetic behavior of GUS. In each experiment, single enzyme molecules are trapped in individual 46 fL microwells containing many molecules of the enzyme’s substrate. In the GUS-catalyzed reaction, nonfluorescent resorufin β-d-glucuronide (RDG) substrate is converted to fluorescent resorufin product, which is then read out on a fluorescence microscope. Since the microwells are isolated from each other by oil sealing, kinetic information on individual enzyme molecules can be extracted from the fluorescence images at high throughput (hundreds of enzyme molecules per image). This platform has proven to be a powerful tool for understanding enzyme kinetic properties of populations of molecules with single-molecule resolution.11,17−21 In this work, the activities of hundreds of individual mutant GUS enzyme molecules were monitored simultaneously as a function of time in the microwell array. We observed a gradual population change from inactive to active enzymes, which helps to elucidate the hysteresis pathway (Scheme 1).
Scheme 1. Illustration of the Hysteresis Pathway and Single-Molecule Study.
The microwell array platform can be used to study hundreds of individual enzyme molecules simultaneously and reveal the population change from inactive to active enzymes.
Materials and Methods
Chemicals
Isopropylthio-β-galactoside, pierce protease inhibitor, superior broth, NuPAGE Tris-acetate gels, and spectra multicolor broad range protein ladder were purchased from Thermo Fisher Scientific (Waltham, MA). Benzonase nuclease was bought from EMD Millipore (St. Louis, MO) and used as received. All other chemicals were of analytical grade and were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated.
Synthesis, Purification, and Characterization of GUS Protein
All types of GUS protein were synthesized in vivo using bacteria containing different plasmids. The plasmids were obtained by cloning the GUS variants’ sequences into the pET28+ vector using the Gibson Assembly method. An N-terminal His6-tag was fused to all enzymes for affinity purification. Briefly, BL21 (DE3) bacteria containing WT (wild-type, pET21-β-glucuronidase), D1, D2, C133, or C262 plasmid (the corresponding mutations are listed in Table S1) were grown overnight in superior broth at 37 °C. Cells were then diluted 1:200 using the same broth, and protein production was induced with 1 mM isopropylthio-β-galactoside during the mid-log phase at 18 °C for 16–18 h. Harvested cells were resuspended and lysed by sonication in lysis buffer (pH 7.4, containing 50 mM sodium phosphate, 300 mM sodium chloride, 20 mM imidazole, pierce protease inhibitor, and Benzonase nuclease). Cell lysate was then centrifuged at 40 000g for 30 min at 4 °C. Cleared cell lysate was then passed through a Ni-NTA agarose column. The column was first washed with five column volumes (CVs) of lysis buffer and then washed for five CVs with washing buffer 1 (pH 7.4, containing 50 mM sodium phosphate, 50 mM imidazole, and 300 mM sodium chloride). Next, the column was eluted with 3 mL of elution buffer (pH 7.4, containing 50 mM sodium phosphate, 500 mM imidazole, and 500 mM sodium chloride). The sample was then dialyzed overnight in dialyzing buffer (pH 7.4, containing 50 mM sodium phosphate, and 1 mM dithiothreitol). The samples were then passed through a size exclusion HiLoad 16/600 Superdex 200 (GE Life Sciences) chromatography column using washing buffer 2 (pH 8.0, containing 20 mM sodium phosphate, 25 mM imidazole, and 500 mM sodium chloride). Fractions containing the target protein were confirmed by SDS-PAGE and then were pooled and dialyzed once again in dialyzing buffer. Samples were then concentrated to 5–10 mg/mL in 4 °C prechilled centrifuges and aliquoted into 50% glycerol for long-term storage at −20 °C. Before use, the enzyme concentration was determined by Nanodrop (Model OneC, Thermo Fisher Scientific, MA).
SDS-PAGE electrophoresis (Figure S1) was carried out with a Mini gel tank (Thermo Fisher) with 1× Novex Tris-acetate SDS running buffer (Thermo Fisher) for 1 h at 150 V (PowerPac power supply, Bio-Rad, Hercules, CA). The gel was stained by Biosafe Coomassie G-250 stain (Bio-Rad, Hercules, CA) and imaged in a Bio-Rad ChemiDoc MP imager. Portions of 2 μg of each GUS protein were loaded into the NuPAGE 7% Tris-acetate protein gel (1.5 mm, 10-well, Invitrogen). A 20 μL portion of the protein ladder solution was loaded in the ladder lane as a reference.
Two mass spectrometry techniques were implemented to determine the quality of the synthesized proteins. The rapifleX MALDI Tissuetyper (Bruker, Billerica, MA) was used to determine the purity and integrity of the protein (Figure S2): each different protein was heated at 70 °C for 10 min and buffer exchanged five times into water containing 0.1% TFA. The matrix contains 1% sinapic acid and 50% acetonitrile. The proteomics results (Table S2) were provided by the Taplin Mass Spectrometry Facility at Harvard Medical School. From Table S2, the protein coverage is in general higher than 90%, and almost all the mutations (except for C253V of D2, due to the loss of the fragment from 251 to 257) were able to be confirmed.
Characterizing the Hysteresis Kinetics by Plate Reader
A plate reader (Tecan Infinite 200 pro, Mannedorf, Switzerland) was used to acquire real-time fluorescence intensities during the catalytic reaction in bulk solution (20 μL). All kinetic measurements for RDG product were carried out at 25 or 37 °C with 550 nm excitation and 590 nm emission, while for 4-methylumbelliferyl-β-d-glucuronide hydrate (4-mug) product, the wavelength was 360 nm excitation and 449 nm emission. Before experiments, the enzyme and substrate solutions were prewarmed, separately, to 25 or 37 °C for 10 min before being mixed and put into the plate reader.
Single-Molecule Analysis Using Microwell Arrays
For single-molecule studies, different GUS samples were loaded into the microwell array (Scheme S1), isolated and sealed with oil, and then monitored individually. First, the microwell array disk (Scheme S1) was prewetted using 100 μL of dimethylsulfoxide, 100 μL of H2O, and 100 μL of 1× PBS buffer (pH 7.4, containing 0.1% bovine serum albumin), sequentially. Then, 50 μL of sample solution (1 pM GUS in 1× PBS buffer containing 100 μM RDG substrate and 0.1% bovine serum albumin) was injected into the microwell array disk at a flow rate of 50 μL/min using a syringe pump. A 40 μL portion of Fluorinert FC-70 oil was then flowed through the fluidic channel to seal the microwells. The microwell array disk was then imaged using a fluorescence microscope (Olympus IX83, 1 s exposure, 20× objective) for kinetic studies. For each experiment, a new microwell array was used. Image analysis was performed using ImageJ software (National Institutes of Health).
Thermal Treatment
The ex situ heat-pulse was conducted by loading 50 μL of target solution in a thermocycler instrument (PCR System 2700, Applied Biosystems). After 1 min of incubation at 37 °C, the solution was air-cooled to room temperature for 2 min before mixing with other components or direct fluorescence measurements either in the plate reader or microwells.
The in situ heating step was carried out by a self-designed custom heating platform (Scheme S3). A 5 mm × 10 mm copper block was fixed on top of the microwell array. The temperature of the heating block was controlled by the temperature controller (Model TC200, Thorlabs). The temperature of the heating block can increase from 25 to 37 °C within 30 s. After sample solution was loaded and sealed in the microwell array, the single-molecule activities of GUS were monitored for a period of time, followed by an increase of temperature to 37 °C while continuous measurements were made.
Results and Discussion
For studies unrelated to the one described in this Article, we originally attempted to generate variants of GUS that would have a single cysteine on their surface, to facilitate surface immobilization.22 Using the Rosetta suite of protein design tools,23,24 the six surface available cysteines on each GUS monomer were mutated to residues predicted to minimally destabilize the protein. As shown in the crystal structure of WT GUS (Scheme S2), the six cysteines were not on or close to the active site (indicated by the substrate structure) so that the changes should not damage or interfere with the active site. The four GUS variants, abbreviated D1, D2, C133, and C262, were generated in which four or five of the six cysteines were replaced (Table S1). D1 and D2 were Rosetta designs, while C133 and C262 were controls in which a rational replacement of cysteine residues with serines was attempted. In the course of characterizing enzyme activities in solution, all four proteins were found to retain substantive activity but showed a lag in activity at 25 °C (Figure S3) that could be repaired in part by incubation at 37 °C (see Figure 3). The appearance of new hysteretic behavior in an engineered enzyme indicated that it might prove possible to learn more about how proteins inhabit energy landscapes, and the variant with the most obvious hysteretic behavior, C133, was chosen for more involved single-molecule studies.
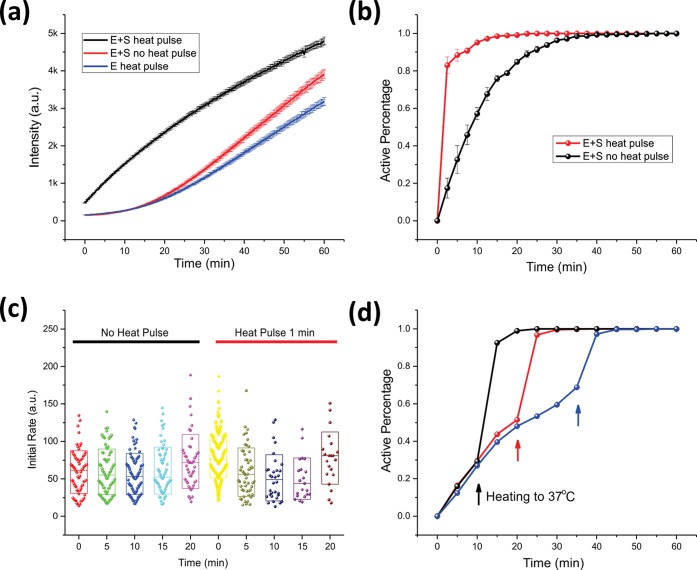
Figure 3.
Effect of thermal treatment on C133. (a) Fluorescence intensity vs time curves of C133 in bulk with or without thermal treatment. The enzyme concentration was 36 pM, and the substrate concentration was 100 μM. (b) Time course of active percentage of C133 in a single-molecule array with and without thermal treatment. The error bars reflect the standard deviation of triplicate measurements. (c) Initial reaction rate of activated C133 molecules in a single-molecule array with and without thermal treatment. The data points were combined from triplicate measurements. The initial reaction rate was defined as Fwell change between the image at t = x + 5 min and t = x min, where x is the time point when a well begins to show obvious fluorescence. (d) In situ heating imposed on C133 in a microwell array. Three microwell arrays were heated to 37 °C at different time points (indicated by color-matching arrows), respectively, and the corresponding active percentage of C133 was recorded as the y axis.
To determine whether the overall stability properties of the engineered protein had changed, or whether the protein was now perhaps trapped in a newly created local free energy minimum, we attempted to measure the enzymatic activities of individual C133 and WT enzyme molecules in microwell arrays. Enzymatic turnover was first measured by monitoring fluorescence intensity changes resulting from the conversion of RDG to resorufin. Following the assessment of background fluorescence intensities at different concentrations of substrate (Figure S4), we chose 100 μM substrate for additional single-molecule studies.
The microarray disk for single-molecule studies contains 24 separate arrays, with each array comprising 216 000 4.25 μm diameter microwells (Scheme S1). Each array is contained within a fluidic channel to enable loading and sealing of the microwells. A 50 μL solution containing substrate and enzyme was flowed through the channel to load the array, and then, 40 μL of fluorinated oil was immediately injected to push out excess enzyme–substrate solution and seal the wells, confining fluorescent products of enzyme activity within each microwell. At sufficiently low concentrations of GUS, each microwell will have either one or zero enzyme molecules.25 As a result, for a reaction that contains 1 pM GUS protein, the kinetics of up to 6000 single enzyme molecules can be monitored simultaneously. While flow cytometry can only monitor end-point signal intensity,26,27 aqueous solutions can be kept inside the microwells for several hours without any leaking,28 allowing the microscope to record signal changes over a long period and thereby derive the kinetic properties of each enzyme molecule.
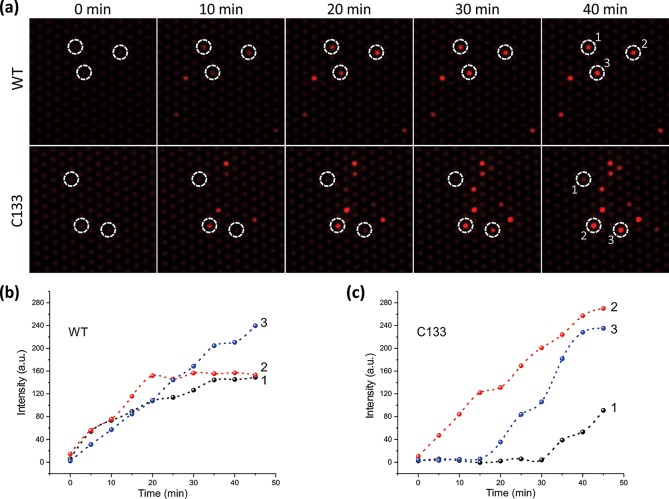
In Figure 1a, the images of both WT and C133 single-molecule assays revealed several bright wells that could be clearly distinguished from the background after 10 min. As expected, the fluorescent intensities of these bright wells increased with time. We chose three representative bright wells from each assay and plotted their average intensities versus time. The fluorescence of all three wells in the WT assay increased continuously from 0 to 40 min (Figure 1b). In contrast, the bright wells of C133 exhibited very different kinetics, with two out of three enzyme molecules showing a period of “dormancy” before converting RDG to resorufin (Figure 1c). Consistent with this result, the overall number of C133 bright wells increased over the observation time.
Figure 1.
Single-molecule study of WT and C133. (a) Fluorescent microscopic images of the single-molecule array at different time points. Bright red microwells indicate the accumulated fluorescent resorufin produced by GUS-catalyzed hydrolysis. White circles and labels highlight several representative GUS molecules. (b, c) Fluorescence intensity vs time curves of the corresponding circled molecules in part a.
For comparison, we conducted fluorescence measurements in a plate reader with a much larger reaction volume (20 μL in the plate reader vs 46 fL in the microwell). The enzyme concentration was 36 pM in bulk solution, which should be equivalent to the concentration for one enzyme molecule present in one femtoliter microwell. As shown in Figure S3, the fluorescence intensity of WT increased linearly from the very beginning of the experiment while the kinetics of C133 had a “lag” phase at the beginning and increased linearly only after 30 min.
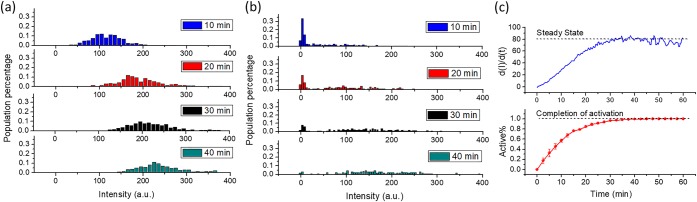
While C133 consistently showed a hysteretic catalytic behavior, the fact that the lag period was clearly different in single-molecule and bulk studies reflected what we had previously seen:11,20 that individual enzyme molecules have divergent activities that can be averaged to recapitulate the bulk kinetic values typically observed. Thus, to better understand the molecular basis for C133’s lag phase, we further analyzed hundreds of single molecules and created activity distribution histograms, as shown in Figure 2a (WT) and Figure 2b (C133). For WT, all molecules were initially active with fluorescence intensities increasing to a mean of 110 a.u. at t = 10 min and then increasing linearly afterward. In contrast, Figure 2b shows that the C133 population exhibited heterogeneous behavior following substrate addition. In one population, Fwell (average background-subtracted fluorescence intensity of each individual microwell) remained close to 0 a.u. during the time course, but the size of this population decreased as time passed. For the other population, Fwell increased linearly with a rate similar to the WT molecules, and as time passed, the size of this population grew. When the initial reaction rates of activated C133 molecules were quantitated, there was no significant difference, irrespective of whether an individual molecule of C133 became active at t = 0, 5, or 10 min (Figure S5). All C133 molecules, once activated, had activities similar to WT. These results strongly suggest a two-state model for activation.
Figure 2.
Population analysis of active WT and C133. Fwell distribution histograms of (a) WT and (b) C133 at different time points. (c) Comparison of the hysteresis kinetics between a single-molecule array and a bulk assay: the blue curve represents the change of the reaction rate of C133 in the bulk measurement, defined as the derivative of intensity vs time curve (d[I]/d[t]) in Figure S3. The dashed line indicates the steady-state rate, where the d(I)/d(t) is at its maximum value. The red curve represents the time course of the active C133 population percentage in part b. The dashed line represents when all the C133 molecules reached 100% activation. The error bars reflect the standard deviation of triplicate measurements.
To compare the kinetics of enzyme activation between bulk and single-molecule measurements, we calculated: (a) the slope of fluorescence intensity as a function of time (d(I)/d(t)) for the bulk enzyme, and (b) the active population percentage for C133 as a function of time (Figure 2c). These curves were remarkably similar. Overall, we conclude that, as with other kinetic properties, the observed hysteresis in bulk is essentially an average of differential temporal activation of individual C133 molecules.
Previous work has shown that environmental conditions such as pH or temperature can change the hysteresis behavior by affecting the protein’s conformation, via unfolding and folding.29,30 To explore whether single-molecule GUS hysteresis was uniformly or divergently perturbed, a 1 min 37 °C heat-pulse was imposed on either the enzyme alone or the enzyme in the presence of substrate (where both enzymes maintain activity, Figure S6), and then cooling the reactions to 25 °C prior to taking fluorescence measurements. In Figure 3a, the bulk measurement proved that heating C133 in the absence of substrate had no significant effect on the reaction’s hysteresis stage, while heat treatment in the presence of substrate led to the elimination of hysteresis. For single-molecule measurements, the heat-pulse pretreatment in the presence of substrate significantly affected hysteresis, with the time needed for complete activation of C133 being 30 min without the heat-pulse and 10 min with the heat-pulse (Figure 3b). Triplicate measurements indicated that there was a 10–20% variation of the active population in the first 10 min while the variation decreased to less than 5% after more than 75% of the population was activated. When the initial reaction rate distributions of C133 with and without initial added substrate were plotted (Figure 3c), it was clear that the ex situ heat-pulse did not change the distribution of the initial rate. We have measured the fluorescence change of WT GUS in the first 5 min with three parallel experiments and determined the average initial turnover rate (ki,ave) in the first 5 min was about 32 ± 3 s–1, with a CV = 33% (Figures S7 and S8). The ki,ave of C133 molecules was also calculated and is summarized in Table S3 and Figure S9. It was found that ki,ave of C133 molecules that were activated at different time points varied from 26 to 33 s–1, and the ki,ave of heat-pulse activated C133 was 41 s–1, which are close to WT ki,ave, again supporting a two-state model for activation.
To further investigate this thermal effect at the single enzyme molecule level, and to exclude the possibility of activation kinetics being impacted by enzyme:enzyme interactions, we designed and built an in situ heating plate for the microwell array (Scheme S1). This allows the heat treatment to be implemented on single molecules after they are isolated and confined in the microwell. As shown in Figure 3d, we carried out three independent microwell assays and implemented heating at three different time points (indicated by arrows at t = 10, 20, and 40 min). An increase in temperature inevitably resulted in an increase in the active population of single molecules, consistent with activation in bulk.
On the basis of these results, we conclude that the hysteresis exhibited by mutant GUS is a reflection of a slow substrate-induced activation process, which is accelerated by thermal treatment. By comparing the difference between WT and mutant GUS, we hypothesize that the slow activation of mutant GUS is due to a kinetically trapped, inactive form of the newly synthesized enzyme. Binding to substrate can slowly drive the enzyme into its active conformation, and the kinetics of this process are accelerated by thermal energy. The wild-type enzyme either lacks the local free energy minimum exhibited by the mutant or has a substantially lower barrier for activation, allowing it to readily populate the active state even in the absence of a heat-pulse. To determine whether or not the conformational change is reversible, we first preactivated the enzyme molecules with a limited amount of substrate, as shown in Figure S10. After the original substrate was depleted, the solution was diluted and spiked with more substrate to test the enzyme kinetics. As can be seen in the figure, both preactivated enzyme and nonactivated controls have the same kinetic properties, which prove that once the substrate was depleted, the preactivated enzyme returns to its inactive state and exhibits hysteretic behavior.
It was surprising that we were able to identify hysteretic behavior in an enzyme that had been rationally designed for other purposes, suggesting that kinetically trapped states might be far more common than is generally realized. To test this hypothesis, we analyzed the kinetics of several other mutant GUS enzymes that were designed and generated in parallel with C133 (D1, D2, and C262, see Table S1). Our results confirmed that all of the mutants exhibited hysteresis (Figures S3, S10, and S11) and that the lag phase can be eliminated by heating (Figure S12). After hysteresis, the single-molecule initial rates of D2 proved to be the same as the WT and C133 (Figure S5); however, the variant from Rosetta design has a shorter lag time than the manual design. Figure S11b shows that it took only 10 min for D2 to be completely activated, compared to 30 min for C133 as shown in Figure 3b. We assume that the variants based on Rosetta design were close to the original structure because the best amino acid substitutions (from the calculation) were used to maintain the stability of the new structure while the rational replacement with serines do not have such constraints.
Using a molecular dynamics (MD) simulation31 we are able to observe how mutations in GUS influence the structure of the protein and the transition between conformations (Figure S13). In Figure S13, when starting from homologues32 of the crystal structure, both the WT GUS and the two mutants transition into stable conformations that are each subtly unique. The conformational positions of residues found near the active site and backbone positions located across the entire structure undergo intramolecular rearrangement during the simulation and eventually form stable structures. Using radius of gyration (Rg) as an indicator of intramolecular dynamics,33 we observe that the WT GUS transitions into a conformation with a lower Rg relatively quickly when compared to both mutants. In contrast, D2 and C133 maintain conformations closely related to the crystal structure for longer than the WT, before eventually transitioning into their more compact forms (C133 transitions slower than D2, which may explain why C133’s hysteresis is longer). The compact conformations that exist at the end of the simulation (Figure S13b) also exhibit some variation between WT and mutants. When taken together, the conformations after 20 ns of MD and the Rg seen throughout the MD, we conclude that intramolecular rearrangements and intramolecular dynamics of GUS have been affected by the mutations. We speculate that the interaction between the hysteretic enzyme and substrate is similar to the induced fit model of enzyme catalysis.34,35 According to the induced fit model, the substrate enables the enzyme to change its conformation to achieve optimal binding as a part of the catalytic reaction. In the hysteretic enzyme system in this work, the role of substrate may be similar, which is to guide the enzyme to undergo a conformational change and become activated. It is likely that mutant GUS does not bind well to substrate in its initial conformation, therefore exhibiting no activity. After interacting with substrate, the mutant GUS undergoes a conformational change and achieves tight binding. Once this substrate-induced process is complete, the mutant GUS remains activated as long as substrate is present. This hypothesis explains the requirement for substrate when the temperature was increased to accelerate the activation. Although there is no direct structural data to support the substrate-induced enzyme conformational change, the results described in this work provide a convincing mechanistic explanation for hysteresis in mutant GUS.
During the MD simulation, we observed that the intramolecular transitions from GUS homologues to their compact forms are due to conformational changes in the structure of the entire tetrameric complex, in contrast to independent changes of each monomeric unit. Therefore, any substrate-induced change will likely affect the structure of the complex as a whole. In addition, the active sites of GUS are located at the interfaces of subunits, and substrate binding should affect the adjacent units simultaneously; therefore, it is reasonable to assume that multiple substrate molecules bind to GUS protein cooperatively. The results in Figures 1 and 3 show that the activation of C133 is clearly a two-state process rather than a four-step activation and that the activated enzyme has the same activity as native GUS, which also supports this assumption.
To further explore the hysteresis behavior, we tested the cross activation between two types of substrates, 4-methylumbelliferyl-β-d-glucuronide hydrate (4-mug) and RDG. Figure S14 shows that both D2 and C133 demonstrated hysteresis with 4-mug, and a heat-pulse at 37 °C eliminated the hysteresis, as was the case with RDG. It was also found that both D2 and C133 activated by 4-mug (through heat-pulse) react with RDG with no hysteresis (Figure S14c) and vice versa (Figure S14d). These results show that cross activation occurs between the two substrates. It is worth noting that when WT GUS reacted with 4-mug, there was a short lag at the very beginning; this phenomenon proved our assumption that WT GUS may contain an inactivated state, but the lower energy barrier allows conversion from an inactive to an active state at a rate that is too fast to be observed, when it reacts with RDG.
Safety Statement
No unexpected or unusually high safety hazards were encountered.
Conclusion
In conclusion, we observed multiple GUS mutants that exhibited hysteresis behavior after several surface cysteines were replaced with other amino acids. The single-molecule kinetic analysis of mutant GUS indicates the existence of an inactive intermediate before the enzyme folds into its active form. The hysteresis is a result of the slow conversion from an inactive population to an active population. The single-molecule initial rate results indicate that there is no significant difference in the activity of WT and activated mutant GUS. The hysteresis can be eliminated when the reaction temperature is increased in the presence of substrates. Furthermore, we observed a return to the inactive state when substrate was removed. On the basis of these observations, we propose a possible hysteresis mechanism based on an induced fit model, in which the interaction of substrate with the hysteretic enzyme leads to a conformational change and activates the enzyme. The finding of thermal activation could be potentially useful in various applications. For example, a preheating step could be used for the enhancement of substrate or antibody binding in immunoassays. The mutant GUS with thermal switch-on properties can be used as a potential protein switch to sense and actuate molecular functions, which can be applied in diagnostics, drug metabolism, and other biosignaling transduction networks.36
Acknowledgments
This work is funded by Brigham and Women’s Hospital and the Wyss Institute.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00718.
Additional data and figures including microwell array configuration, crystal structures, heating platform schematic, SDS-PAGE gel image, MALDI-TOF results, hysteresis behavior in bulk, steady-state reaction rate vs substrate concentration, rate distributions, fluorescence intensity calibration, molecular dynamics simulation, and cross activation (PDF)
Author Contributions
∥ Y.J. and X.L. contributed equally to the work.
The authors declare no competing financial interest.
Supplementary Material
References
- Rabin B. Co-operative effects in enzyme catalysis: a possible kinetic model based on substrate-induced conformation isomerization. Biochem. J. 1967, 102, 22C. 10.1042/bj1020022C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J. Biol. Chem. 1970, 245, 5788. [PubMed] [Google Scholar]
- Hanozet G.; Pircher H. P.; Vanni P.; Oesch B.; Semenza G. An example of enzyme hysteresis - the slow and tight interaction of some fully competitive inhibitors with small intestinal sucrase. J. Biol. Chem. 1981, 256, 3703. [PubMed] [Google Scholar]
- Neet K. E.; Ainslie G. R. Jr In Methods in enzymology; Elsevier, 1980; Vol. 64. [DOI] [PubMed] [Google Scholar]
- Lalanne M.; Henderson J. F. Effects of hormones and drugs on phosphoribosyl pyrophosphate concentrations in mouse liver. Can. J. Biochem. 1975, 53, 394. 10.1139/o75-055. [DOI] [PubMed] [Google Scholar]
- Singer S. C.; Holmes E. W. Human glutamine phosphoribosylpyrophosphate amidotransferase. Hysteretic properties. J. Biol. Chem. 1977, 252, 7959. [PubMed] [Google Scholar]
- Frieden C. Slow transitions and hysteretic behavior in enzymes. Annu. Rev. Biochem. 1979, 48, 471. 10.1146/annurev.bi.48.070179.002351. [DOI] [PubMed] [Google Scholar]
- Kurganov B. I.; Dorozhko A. I.; Kagan Z. S.; Yakovlev V. A. The theoretical analysis of kinetic behaviour of “hysteretic” allosteric enzymes III. Dissociating and associating enzyme systems in which the rate of installation of equilibrium between the oligomeric forms is comparable to that of enzymatic reaction. J. Theor. Biol. 1976, 60, 287. 10.1016/0022-5193(76)90061-8. [DOI] [PubMed] [Google Scholar]
- Kaiser W. M.; Huber S. C. Correlation between apparent activation state of nitrate reductase (NR), NR hysteresis and degradation of NR protein. J. Exp. Bot. 1997, 48, 1367. 10.1093/jxb/48.7.1367. [DOI] [Google Scholar]
- Gorris H. H.; Walt D. R. Mechanistic aspects of horseradish peroxidase elucidated through single-molecule studies. J. Am. Chem. Soc. 2009, 131, 6277. 10.1021/ja9008858. [DOI] [PubMed] [Google Scholar]
- Rissin D. M.; Gorris H. H.; Walt D. R. Distinct and long-lived activity states of single enzyme molecules. J. Am. Chem. Soc. 2008, 130 (15), 5349. 10.1021/ja711414f. [DOI] [PubMed] [Google Scholar]
- Rotman B. Measurement of activity of single molecules of β-D-galactosidase. Proc. Natl. Acad. Sci. U. S. A. 1961, 47, 1981. 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin T. M.; Yeung E. S. Single-molecule reactions in liposomes. Angew. Chem., Int. Ed. 2007, 46, 8032. 10.1002/anie.200702348. [DOI] [PubMed] [Google Scholar]
- Gorris H. H.; Blicharz T. M.; Walt D. R. Optical-fiber bundles. FEBS J. 2007, 274, 5462. 10.1111/j.1742-4658.2007.06078.x. [DOI] [PubMed] [Google Scholar]
- Matsumura I.; Ellington A. D. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J. Mol. Biol. 2001, 305, 331. 10.1006/jmbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- Liebherr R. B.; Renner M.; Gorris H. H. A single molecule perspective on the functional diversity of in vitro evolved beta-glucuronidase. J. Am. Chem. Soc. 2014, 136, 5949. 10.1021/ja412379p. [DOI] [PubMed] [Google Scholar]
- Rondelez Y.; Tresset G.; Tabata K. V.; Arata H.; Fujita H.; Takeuchi S.; Noji H. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat. Biotechnol. 2005, 23, 361. 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- Gorris H. H.; Rissin D. M.; Walt D. R. Stochastic inhibitor release and binding from single-enzyme molecules. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 17680. 10.1073/pnas.0705411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan C. W.; Rivnak A. J.; Campbell T. G.; Piech T.; Rissin D. M.; Mosl M.; Peterca A.; Niederberger H. P.; Minnehan K. A.; Patel P. P.; et al. Isolation and detection of single molecules on paramagnetic beads using sequential fluid flows in microfabricated polymer array assemblies. Lab Chip 2012, 12, 977. 10.1039/C2LC20744C. [DOI] [PubMed] [Google Scholar]
- Li X.; Jiang Y.; Chong S. R.; Walt D. R. Bottom-up single-molecule strategy for understanding subunit function of tetrameric beta-galactosidase. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 8346. 10.1073/pnas.1805690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Hayman R. B.; Walt D. R. Detection of single-molecule DNA hybridization using enzymatic amplification in an array of femtoliter-sized reaction vessels. J. Am. Chem. Soc. 2008, 130, 12622. 10.1021/ja8053018. [DOI] [PubMed] [Google Scholar]
- Robson T.; Shah D. S. H.; Solovyova A. S.; Lakey J. H. Modular protein engineering approach to the functionalization of gold nanoparticles for use in clinical diagnostics. ACS Appl. Nano Mater. 2018, 1, 3590. 10.1021/acsanm.8b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap M. G.; Leij F. J.; van Genuchten M. T. ROSETTA: a computer program for estimating soil hydraulic parameters with hierarchical pedotransfer functions. J. Hydrol. 2001, 251, 163. 10.1016/S0022-1694(01)00466-8. [DOI] [Google Scholar]
- DiMaio F. In Protein Crystallography; Springer, 2017. [Google Scholar]
- Zhang H. B.; Nie S.; Etson C. M.; Wang R. M.; Walt D. R. Oil-sealed femtoliter fiber-optic arrays for single molecule analysis. Lab Chip 2012, 12, 2229. 10.1039/c2lc21113k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.; Allen P. B.; Ellington A. D. A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 2016, 11, 157. 10.1038/nnano.2015.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazutis L.; Gilbert J.; Ung W. L.; Weitz D. A.; Griffiths A. D.; Heyman J. A. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2013, 8, 870. 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B. K.; Cavanagh P. E.; Li X.; Walt D. R. Ultrasensitive single-molecule enzyme detection and analysis using a polymer microarray. Anal. Chem. 2018, 90, 3091. 10.1021/acs.analchem.7b03980. [DOI] [PubMed] [Google Scholar]
- Moon C. P.; Kwon S.; Fleming K. G. Overcoming hysteresis to attain reversible equilibrium folding for outer membrane phospholipase A in phospholipid bilayers. J. Mol. Biol. 2011, 413, 484. 10.1016/j.jmb.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews B. T.; Capraro D. T.; Sukowska J. I.; Onuchic J. N.; Jennings P. A. Hysteresis as a marker for complex, overlapping landscapes in proteins. J. Phys. Chem. Lett. 2013, 4, 180. 10.1021/jz301893w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman P.; Swails J.; Chodera J. D.; McGibbon R. T.; Zhao Y. T.; Beauchamp K. A.; Wang L. P.; Simmonett A. C.; Harrigan M. P.; Stern C. D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N.; Webb B.; Marti-Renom M. A.; Madhusudhan M. S.; Eramian D.; Shen M.-Y.; Pieper U.; Sali A. Comparative protein structure modeling using Modeller. Current protocols in bioinformatics 2006, 15, 5-6-1. 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A.; Meireles L. M.; Bahar I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575. 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester M. R.; Johnson E. F.; Marques-Soares C.; Dijols S.; Dansette P. M.; Mansuy D.; Stout C. D. Structure of mammalian cytochrome P4502C5 complexed with diclofenac at 2.1 angstrom resolution: Evidence for an induced fit model of substrate binding. Biochemistry 2003, 42, 9335. 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- Koshland D. E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 1958, 44, 98. 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein V.; Alexandrov K. Synthetic protein switches: design principles and applications. Trends Biotechnol. 2015, 33, 101. 10.1016/j.tibtech.2014.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.