Abstract
Emissions of the strong greenhouse gas methane (CH4) to the atmosphere are mitigated by methanotrophic microorganisms. Methanotrophs found in extremely acidic geothermal systems belong to the phylum Verrucomicrobia. Thermophilic verrucomicrobial methanotrophs from the genus Methylacidiphilum can grow autotrophically on hydrogen gas (H2), but it is unknown whether this also holds for their mesophilic counterparts from the genus Methylacidimicrobium. To determine this, we examined H2 consumption and CO2 fixation by the mesophilic verrucomicrobial methanotroph Methylacidimicrobium tartarophylax 4AC. We found that strain 4AC grows autotrophically on H2 with a maximum growth rate of 0.0048 h–1 and a yield of 2.1 g dry weight⋅mol H2–1, which is about 12 and 41% compared to the growth rate and yield on methane, respectively. The genome of strain 4AC only encodes for an oxygen-sensitive group 1b [NiFe] hydrogenase and H2 is respired only when oxygen concentrations are below 40 μM. Phylogenetic analysis and genomic comparison of methanotrophs revealed diverse [NiFe] hydrogenases, presumably with varying oxygen sensitivity and affinity for H2, which could drive niche differentiation. Our results show that both thermophilic and mesophilic verrucomicrobial methanotrophs can grow as autotrophs on H2 as a sole energy source. Our results suggest that verrucomicrobial methanotrophs are particularly well-equipped to thrive in hostile volcanic ecosystems, since they can consume H2 as additional energy source.
Keywords: Methylacidimicrobium, methanotrophic Verrucomicrobia, acidophilic, H2, [NiFe] hydrogenases, oxygen sensitivity
Introduction
Atmospheric concentrations of the strong greenhouse gas methane (CH4) are increasing due to anthropogenic activities (Kirschke et al., 2013). With a changing climate, it is essential to understand alterations in the sources and sinks of the global methane cycle (Murrell and Jetten, 2009). Methanogenic Archaea constitute the largest methane source, producing this odorless gas in a vast variety of habitats, such as wetlands, rice fields, oceans and the digestive system of termites and ruminants (Conrad, 2009). The main sink can be found in the troposphere, where methane is oxidized photochemically by hydroxyl radicals (OH) (Voulgarakis et al., 2013). In addition, methanotrophs significantly mitigate methane emissions to the atmosphere by oxidizing methane produced by methanogens (Murrell and Jetten, 2009). Methanotrophs either oxidize methane with oxygen (Hanson and Hanson, 1996; Op den Camp et al., 2009; Semrau et al., 2010) or anaerobically, utilizing a variety of alternative electron acceptors (Knittel and Boetius, 2009; Ettwig et al., 2010; Haroon et al., 2013). For many years it was believed that methane oxidation was a feature restricted to the subphyla Alpha- and Gammaproteobacteria (Op den Camp et al., 2009). However, this view was rejected due to the description of novel aerobic methanotrophs belonging to the phylum Verrucomicrobia (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008). All known verrucomicrobial methanotrophs of the genus Methylacidiphilum (strains Kam1, SolV, V4 and sp. RTK17.1) have been isolated from extremely acidic geothermal ecosystems (Pol et al., 2007; Op den Camp et al., 2009; Sharp et al., 2013; Carere et al., 2017). Methanotrophy by thermophilic members of the Verrucomicrobia phylum is an extreme affair, since they can grow on methane below pH 1 and at temperatures up to 65°C (Op den Camp et al., 2009).
All verrucomicrobial methanotrophs of the genus Methylacidiphilum were isolated from hot and acidic environments. Recently, 16S rRNA sequences were retrieved from acidic geothermal areas with a moderate temperature, revealing closely related verrucomicrobial methanotrophs (Sharp et al., 2012, 2014). Strain LP2A was isolated from such a moderate-temperature area and indeed only grows at a moderate temperature (Sharp et al., 2014). This novel verrucomicrobial methanotroph shares only 89.6% 16S rRNA gene sequence identity with representatives of the thermophilic genus Methylacidiphilum (Sharp et al., 2014). Shortly after this finding, van Teeseling et al. (2014) isolated and characterized three new species of acidophilic verrucomicrobial methanotrophs from diverse soil patches at the Solfatara crater, located at the center of the Campi Flegrei caldera, near Naples (Italy). Interestingly, these novel verrucomicrobial methanotrophs are unable to grow at high temperature and they are therefore all mesophiles. The 16S rRNA genes of the new isolates are only about 89% similar to those of the Methylacidiphilum species and 97–98% identical to that of strain LP2A. Therefore, the new genus name Methylacidimicrobium was proposed and the four mesophilic strains were described as Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium fagopyrum 3C, Methylacidimicrobium cyclopophantes 3B and Methylacidimicrobium strain LP2A (van Teeseling et al., 2014). Maximum growth rates (μmax) of 0.013–0.040 h–1 were observed at temperature optima between 35 and 44°C, respectively (van Teeseling et al., 2014). Strain 4AC is the most acid-tolerant methanotroph known to date, growing at pH values as low as 0.5. Similar to the thermophilic verrucomicrobial strains SolV and V4, all isolated mesophilic strains grow autotrophically using the Calvin-Benson-Bassham cycle for CO2 fixation (Khadem et al., 2011; Sharp et al., 2012; van Teeseling et al., 2014). Interestingly, these novel isolates all encode for hydrogen-oxidizing enzymes.
In many volcanic habitats, H2 is available as an additional energy source for methanotrophs (Chiodini et al., 2001; Hanczár et al., 2002; Carere et al., 2017). Hydrogenases catalyze the oxidation of H2 to two protons plus two electrons, and vice versa. Genomic analyses have revealed a plethora of hydrogenases in many different phyla, with either [NiFe], [FeFe] or [Fe] as metals in the active site (Lubitz et al., 2014; Greening et al., 2016). Several methane oxidizers are able to consume H2, as was demonstrated for Methylosinus sp. (de Bont, 1976), Methylocystis sp. and Methylococcus capsulatus Bath (Csáki et al., 2001; Kelly et al., 2005). The presence of genes encoding an uptake hydrogenase and the ribulose-1,5-bisphosphate carboxylase (RuBisCO) in several proteobacterial methanotrophs indicates the possibility of autotrophic growth. However, whereas autotrophic growth of M. capsulatus Bath was observed on solid agar media, physiological studies in liquid media did not support this observation (Dalton and Whittenbury, 1976; Taylor et al., 1981; Stanley and Dalton, 1982; Baxter et al., 2002). The thermophilic verrucomicrobial strains SolV and RTK17.1 were shown to grow autotrophically on H2 (Carere et al., 2017; Mohammadi et al., 2017). Moreover, these extremophiles can utilize CH4 and H2 simultaneously. Reducing equivalents derived from the oxidation of H2 could aid in growth of methanotrophs in times when CH4 availability is low (Hanczár et al., 2002). In geothermal environments with large fluctuations in H2 and CH4 emissions, this mixotrophic lifestyle can provide a major advantage over less metabolically versatile microorganisms (Ward et al., 2017; Power et al., 2018). Consequently, the competence of oxidizing both CH4 and H2 could give an explanation for the dominance of verrucomicrobial methanotrophs in terrestrial volcanic ecosystems (Semrau et al., 2011; Carere et al., 2017).
The closed genome of the thermophilic strain SolV encodes for two uptake hydrogenases that catalyze H2 oxidation: an oxygen-tolerant group 1d and an oxygen-insensitive group 1h H2-uptake [NiFe] hydrogenase (Anvar et al., 2014; Mohammadi et al., 2017). The draft genomes of the mesophilic verrucomicrobial methanotrophs (strains 4AC, 3B, 3C and sp. LP2A) also revealed the presence of an H2-uptake hydrogenase, but it differs significantly from the group 1d and group 1h [NiFe] hydrogenases of the thermophilic strains (Sharp et al., 2014; van Teeseling et al., 2014). Interestingly, the mesophilic strain 4AC was shown to be highly sensitive to O2, whereas the thermophilic strain SolV was shown to consume H2 at ambient air (van Teeseling et al., 2014; Mohammadi et al., 2017). The presence of an uptake hydrogenase and a carbon fixation pathway in all mesophilic strains suggests that they can grow as autotrophs on hydrogen gas. This suggestion implies that hydrogen oxidation by mesophilic verrucomicrobial methanotrophs can be important in the mitigation of greenhouse gas emissions from the natural geothermal environment. We therefore hypothesized that the mesophilic methanotroph Methylacidimicrobium tartarophylax 4AC can grow as an autotroph on H2 as sole energy source under oxygen-limited conditions. To investigate this hypothesis, various physiological experiments were performed in continuous and batch cultures, in the presence or absence of H2, CH4 and CO2 at different oxygen concentrations.
Materials and Methods
Microorganism and Medium Composition
Methylacidimicrobium tartarophylax strain 4AC used in this study was initially isolated and enriched from diverse soil spots at the Solfatara crater, which is at the center of the Campi Flegrei caldera, near Naples (Italy) (Pol et al., 2007; van Teeseling et al., 2014). In this study, the medium was composed of 0.2 mM MgCl2 ⋅ 6 H2O; 0.2 mM CaCl2 ⋅ 2 H2O; 1 mM Na2SO4; 2 mM K2SO4; 2 mM (NH4)2SO4 (10 mM to reach OD600 of 5) and 1 mM NaH2PO4 ⋅ H2O. A trace elements solution was used containing 1 μM NiCl2, CoCl2, Na2MoO4, ZnSO4 and CeCl3, 5 μM MnCl2 and FeSO4, 10 μM CuSO4 and 50 μM nitrilotriacetic acid (NTA). The pH of the medium was adjusted to 3.0 using 1 M H2SO4. To avoid precipitation, CaCl2 ⋅ 2 H2O and the rest of medium were autoclaved separately and mixed after cooling. This medium composition contained all nutrients to obtain an OD600 of 1.0 and was used in batch and continuous cultures, unless stated otherwise.
Chemostat Cultivation on Methane and Hydrogen Gas
The reactor system (Applikon Biotechnology, Delft, NL) was operated at 38°C with a stirring speed of 470 rpm. During growth on methane, the oxygen-limited continuous culture (liquid volume of 950 ml) was supplied with medium at a flow rate of 15.6 ml ⋅ h–1 (D = 0.016 h–1). A gas supply of 6% CH4 (v/v), 5% CO2 (v/v) and 3% O2 was provided by mass flow controllers (MFCs) through a sterile filter and sparged into the medium at a gas flow rate of approximately 13 ml ⋅ min–1. A dissolved oxygen concentration (dO2) of approximately 0–0.02% was obtained when the cells reached a steady state. During growth on hydrogen, the oxygen-limited chemostat (liquid volume of 1.3 L) was supplied with medium at a flow rate of 5 ml ⋅ h–1 (D = 0.004 h–1). A gas supply of 15% H2 (v/v), 5% CO2 (v/v) and 3% O2 was provided by MFCs through a sterile filter and sparged into the medium at a gas flow rate of 13.2 ml ⋅ min–1. A dO2 of about 0–0.01% was obtained when cells reached a steady state. The cell-containing medium was removed automatically from the chemostat by a peristaltic pump when the liquid level reached the sensor in the reactor. During growth on methane or hydrogen gas, the pH was regulated in the steady state at 2.8 and 4.0 using 0.2 M NaOH, respectively.
Batch Cultivation
To start a continuous culture on hydrogen gas, we initially tried to grow strain 4AC in batch mode in the bioreactor used for the chemostat cultivation. To obtain the maximum growth rate (μmax) on methane, cells were grown at 38°C at 470 rpm without any limitation in the medium using a gas supply in which the O2 concentration was below 5%. In order to obtain the μmax on hydrogen, cells were grown at 38°C at 470 rpm without any limitation in the medium while the dO2 was kept at 0–0.01%. The batch growth was repeated at least two times for four generations.
Gas Analysis
The consumption of hydrogen gas and carbon dioxide was measured using a HP 5890 gas chromatograph (Agilent, Santa Clara, United States) equipped with a Porapak Q column (1.8 m, ID 2 mm) and a thermal conductivity detector. For hydrogen gas analysis, 30–40 μl gas samples were injected with a glass syringe. The consumption of methane was analyzed using a HP 5890 gas chromatograph (Agilent, Santa Clara, United States) equipped with a Porapak Q column (1.8 m, ID 2 mm) and a flame ionization detector. For methane analysis, 100 μl gas samples were injected with a glass syringe. The consumption of oxygen was measured on an Agilent series 6890 gas chromatograph (GC) equipped with Porapak Q and Molecular Sieve columns and a thermal conductivity detector as described before (Ettwig et al., 2008). For oxygen analysis, 50 μl gas samples were injected with a glass syringe.
Dry Weight Determination and Elemental Analysis
To determine the biomass dry weight concentration, 10 ml of the culture suspension (triplicate) was filtered through pre-weighed 0.45 μm filters and dried to constant weight in a vacuum oven at 60°C. To measure the total content of carbon and nitrogen, 10 ml of the culture suspension (duplicate) was centrifuged at 4,500 × g for 30 min and the clear supernatant was used for the analysis. The nitrogen and carbon content in the supernatant was compared with the corresponding values in the whole cell suspension. The total carbon and nitrogen contents were measured using TOC-L and TNM-1 analyzers (Shimadzu, Kyoto, Japan).
Respiration Experiments
Respiration rates were measured polarographically in a respiration cell with an oxygen microsensor (RC350, Strathkelvin, Motherwell, United Kingdom) using 3 ml of whole cell suspensions of strain 4AC (OD600 = 0.3). Methane-, hydrogen- or oxygen-saturated medium was injected into the respiration chamber to obtain the desired dissolved gas concentrations. The O2 signal was monitored and recorded using SensorTrace Basic software (Unisense, Aarhus, Denmark). The temperature and stirring rate in the respiration chamber were adjusted to 38°C and 1000 rpm, respectively. Rates were expressed as nmol O2 ⋅ min–1 ⋅ mg DW–1 and, when necessary, corrected for endogenous respiration. To avoid high oxygen concentrations at the start of an experiment, samples taken from cultures were immediately transferred into rubber septum sealed bottles under an anoxic atmosphere of nitrogen and carbon dioxide. These bottles contained medium in case dilution was necessary. A subsample was taken from the bottle by a syringe with a long needle and introduced into the respiration chamber at the bottom with the oxygen probe in place while pushing out the air via the inlet channel.
Hydrogenase Classification and Phylogenetic Analysis
The NCBI accession numbers and corresponding microbial species for all [NiFe] hydrogenase large subunit sequences were retrieved from HydDB (Søndergaard et al., 2016). The accession list was initially filtered for methanotrophs by querying NCBI for species that possess both a methane monooxygenase and a methanol dehydrogenase. The resulting list contained a number of methylotrophs (with an annotated ammonia/methane monooxygenase) which were removed by manual inspection. The remaining sequences were retrieved using NCBI Batch Entrez; the sequences from verrucomicrobial methanotrophs were added manually. All sequences were aligned using the default algorithm in T-Coffee v12.00.7fb08c2 (stand-alone). After manual inspection of the multiple sequence alignment, a maximum-likelihood tree was calculated by RAxML v8.2.10. The group 3d [NiFe] hydrogenase was used as outgroup and pruned from the final tree. MEGA 7 was used to visualize the tree and collapse branches that did not contain verrucomicrobial sequences.
Results
M. tartarophylax Strain 4AC Consumes Hydrogen at Low Oxygen Conditions
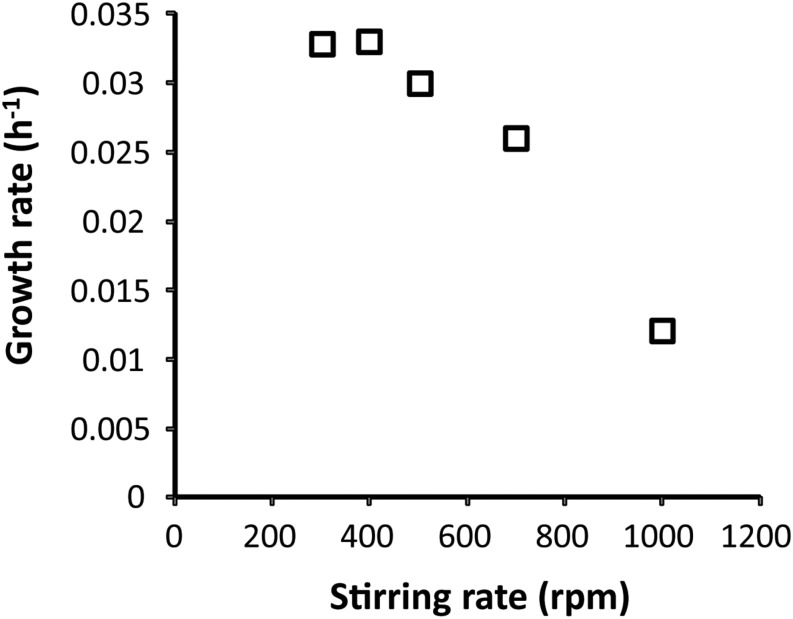
To show H2 consumption under oxygen-limited conditions by strain 4AC, cells were initially cultivated on methane in the bioreactor using batch and chemostat conditions. Considering the high sensitivity to oxygen of strain 4AC (van Teeseling et al., 2014), the maximal stirring rate that could be used during the growth was determined. High stirring rates may cause oxygen stress to strain 4AC by changing the actual oxygen gradient. Lower stirring rates may allow for an oxygen gradient close to the outside of the cell. We observed that increases of the stirring rate from 400 to 1000 rpm resulted in a decrease of the growth rate on methane from 0.033 to 0.012 h–1, respectively (Figure 1). At stirring rates lower than 400 rpm, no further increase in the growth rate was observed compared to the μmax reported previously (0.035 h–1, van Teeseling et al., 2014). Therefore, growth experiments were performed at 400 to 500 rpm. A comparison of batch growth on methane and hydrogen is given in Supplementary Figure S1. Cells from batch cultures of strain 4AC growing on methane at a growth rate of 0.033 h–1 and an oxygen concentration of 1% did not show any hydrogen oxidation when the oxygen concentration in the respiration chamber was 175 μM (Table 1). However, when the O2 concentration became lower than 40 μM, oxidation of 4.1 nmol O2 ⋅ min–1 ⋅ mg DW–1 was measurable, which is approximately 4% of the oxygen consumption with methane (105.9 nmol ⋅ min–1 ⋅ mg DW–1; Table 1). Moreover, cells from a continuous culture grown on methane under O2 limitation showed hydrogen respiration rates below 1 nmol O2 ⋅ min–1 ⋅ mg DW–1 when the oxygen concentration in the respiration chamber was above 100 μM (Table 1) and again elevated rates were measured when O2 concentrations were below 30 μM (4.1 and 2.8 nmol O2 ⋅ min–1 ⋅ mg DW–1; Table 1). Based on our observations, we assume that strain 4AC could oxidize hydrogen only when the oxygen concentration in the respiration chamber was less than 40 μM. To test this hypothesis, we transferred the methane-grown cells directly from the continuous culture under oxygen limitation to a closed serum bottle with a headspace gas of hydrogen and nitrogen. A subsample was taken from this bottle by a syringe with a long needle and introduced at the bottom of the respiration chamber. Using this procedure, we obtained a 10 μM O2 concentration in the respiration chamber at the start of the experiment and this clearly resulted in higher hydrogen respiration rates (8.1 nmol O2 ⋅ min–1 ⋅ mg DW–1; Table 1). These results strongly indicate that the hydrogenase responsible for the hydrogen oxidation activity is only active under oxygen-limited conditions.
FIGURE 1.
Effect of increased stirring on the growth rate of Methylacidimicrobium tartarophylax 4AC with methane as a substrate. Each data point represents the average of two independent experiments at the same stirring speeds.
TABLE 1.
Oxygen respiration rates of Methylacidimicrobium tartarophylax 4AC cells with CH4 or H2.
| Growth condition | e– donor | Limitation | Growth rate (h–1)a | dO2 (%)b | O2 (μM)c | Respiration rated | |
| H2 | CH4 | ||||||
| Batch | CH4 | None | 0.033 ± 0.003 | 1 | 175 <40 |
≈ 0 4.1 |
105.9 |
| Continuouse | CH4 | O2 | 0.016 | <0.02 | 120 <30 |
<1 4.1 |
112.9 |
| Continuousf | CH4 | O2 | 0.016 | <0.02 | 175 <30 |
<1 2.8 |
100.6 |
| Continuousg | CH4 | O2 | 0.016 | <0.02 | 100 <40 |
<1 3.9 |
115.7 |
| Continuoush | CH4 | O2 | 0.016 | <0.02 | <10 | 8.1 | n.d. |
| Batch | H2 | None | 0.0048 ± 0.0006 | 0.05–0.1 | <10 | 11.2 | n.d. |
| Continuous | H2 | O2 | 0.004 | <0.02 | <10 | 10.5 | n.d. |
aIn batch and continuous cultures, growth rate refers to μ and D, respectively. The concentrations of the gasses provided by mass flow controllers in the chemostat cultures were (v/v): 5% CO2 and 6% CH4 or 15% H2. bFor the batch culture on methane the actual oxygen concentration was regulated at 1%. For the hydrogen batch culture regulation was at 0.05–0.1%, while the hydrogen and methane continuous cultures under oxygen limitation showed values below detection limit (0.02%). cThe μM units refer to O2 concentrations in the liquid phase in the respiration chamber. dThe respiration rates measured in the respiration chamber are expressed in nmol O2 ⋅ min–1 ⋅ mg DW–1 and are averages of duplicate measurements that did not deviate more than 10%. eCells were aerated for 10 min prior to the respiration test. fCells were aerated for 1 h prior to the respiration test. gCells without aeration prior to the respiration test. hCells were transferred to a capped serum bottle (60 ml) containing an anoxic atmosphere of hydrogen and nitrogen. n.d. = not determined.
M. tartarophylax 4AC Grows Autotrophically on Hydrogen
We next asked whether strain 4AC is able to grow as an autotroph on H2. To answer this question, H2 was introduced into a bioreactor batch culture on CH4 with a growth rate of 0.02 h–1. Oxygen was supplied to the reactor so that due to consumption the dissolved oxygen value (dO2) was kept below the detection limit (reading value between 0 and 0.01%). The total gas inflow was kept at 13.2 ml ⋅ min–1. After a period of 4 days in which both H2 and CH4 were simultaneously consumed, methane was removed from the gas mixture while keeping the total gas inflow constant at 13.2 ml ⋅ min–1. In the batch growth of strain 4AC with H2 only, we measured a growth rate of 0.0048 h–1 (±0.0005), indicating a doubling time of 144 h. To obtain higher growth rates, we gradually increased the O2 supply using an MFC until dO2 spikes (maximum 0.02%) were being observed. At this point we stopped increasing the O2 supply, and after a few hours the same procedure was repeated. The optical density (OD600) and the consumption rates of gases were measured daily. The highest hydrogen consumption rate (using a GC) during batch growth was measured at 63 nmol H2 ⋅ min–1 ⋅ mg DW–1. After successful batch growth on H2 only, the system was switched to a continuous culture mode with a growth rate (D) at 0.004 h–1, which is about 83% of the μmax (Table 1). During the steady state, H2, CO2 and O2 consumption rates were measured, resulting in rates of 36.6, 6.4 and 14.1 nmol ⋅ min–1 ⋅ mg DW–1, respectively. In both batch and continuous cultures using H2, the dO2 value was kept at zero. To confirm the high sensitivity of the hydrogenase of strain 4AC to O2 that was observed previously in the respiration experiments, growth in the batch condition was monitored when O2 was in excess. As soon as the cells were exposed to a surplus amount of O2, first a slow increase of the dO2 signal to 0.05–0.1% was observed, followed by a rapid increase showing that growth ceased due to inhibition of the hydrogenase. These results confirm the high sensitivity of the hydrogenase of strain 4AC toward oxygen and the ability of strain 4AC to grow on H2 as an autotroph.
Yield
In order to quantify growth of strain 4AC on hydrogen compared to methane, the growth yields on both gasses were determined. The growth yield on CH4 was obtained by measuring the methane consumption of an oxygen-limited continuous culture (D = 0.016 h–1). Based on dry weight (DW) measurements a yield value of 5.1 ± 0.2 g DW ⋅ mol CH4–1 was calculated. Oxygen consumption measurements (for CH4 versus O2) and gas chromatographic analysis (for CH4 versus CO2) measurements were used to quantify CH4 oxidation, leading to the following stoichiometry:
Organic carbon analysis of centrifuged culture samples revealed the presence of 12% of the total organic matter in the supernatant. In addition, a continuous culture on H2 under O2 limitation (D = 0.004 h–1) was used to assess the growth yield on H2. Based on the consumption of hydrogen and dry weight measurements a yield value of 2.1 ± 0.2 g DW ⋅ mol H2–1 was calculated. Oxygen consumption measurements (for H2 versus O2) and gas chromatographic analysis (for H2 versus CO2) measurements were used to quantify H2 oxidation, leading to the following stoichiometry:
Hydrogenase Classification and Oxygen Sensitivity
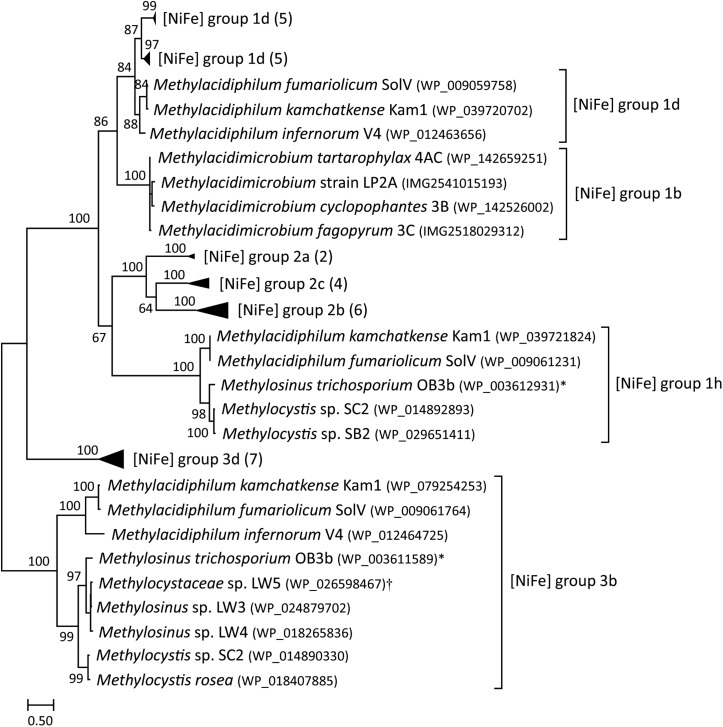
All known Methylacidiphilum isolates encode for a group 1d [NiFe] hydrogenase, whereas, all known Methylacidimicrobium isolates encode for a group 1b [NiFe] hydrogenase (Figure 2) (Hou et al., 2008; Anvar et al., 2014; Sharp et al., 2014; van Teeseling et al., 2014; Erikstad and Birkeland, 2015). Methylacidiphilum strains also encode for a group 3b [NiFe] hydrogenase that could be involved in CO2 fixation (Carere et al., 2017). Moreover, M. fumariolicum SolV and M. kamchatkense encode for a group 1h [NiFe] hydrogenase, proposed to be an oxygen-insensitive enzyme with a high affinity for H2 (Constant et al., 2010). In addition, a few other methanotrophs, such as Methylocystis and Methylosinus, possess multiple different [NiFe] hydrogenases (Figure 2). Genome analysis of the Methylacidimicrobium isolates revealed a membrane-bound b-type cytochrome protein (hupZ) in the operon of group 1b and group 1d [NiFe] hydrogenases that shuttles electrons to the terminal oxidase. The group 1h type is involved in energy conservation, but how electrons are shuttled to the electron transport chain is currently unknown (Greening et al., 2015).
FIGURE 2.
A maximum-likelihood phylogenetic tree based on the protein sequences of [NiFe] hydrogenases (large subunit) found in methanotrophs. Hydrogenase group labels are based on HydDB. The number of sequences in collapsed branches is shown in parentheses. Bootstrap scores are based on 500 replicates. ∗Multispecies record that also includes Methylosinus sp. 3S-1; †Multispecies record that also includes Methylocystis sp. sav-2.
Oxygen tolerance during hydrogen consumption heavily depends on the type of hydrogenase encoded (Table 2). The group 1b type studied here is active only below 40 μM O2, indicating that the enzyme is sensitive to oxygen or expressed only under low oxygen conditions. In contrast, the group 1h [NiFe] hydrogenase of M. fumariolicum SolV is active up to at least ambient oxygen. The group 1d type is classified as an oxygen-tolerant [NiFe] hydrogenase (Greening et al., 2016), but it is only expressed under oxygen-limiting conditions (Khadem et al., 2012; Mohammadi et al., 2017). Group 1d [NiFe] hydrogenases can be inactivated by O2, but have mechanisms to rapidly reactivate. In contrast, oxygen-insensitive hydrogenases do not get inhibited even at high concentrations (Kalms et al., 2018). The group 1b and group 1d type share clear similarities in gene arrangements and maturation proteins (Mohammadi et al., 2017). However, the large subunit of the group 1b [NiFe] hydrogenase in mesophilic strains 3B, 3C, 4AC and LP2A is only 46–48% similar to the large subunit of group 1d [NiFe] hydrogenase present in thermophilic verrucomicrobial methanotrophs. Likewise, the small subunit of the hydrogenase in strains 3B, 3C, 4AC and LP2A is only 41–44% similar to the small subunit of group 1d [NiFe] hydrogenase of Methylacidiphilum species. Considering the phylogenetic position, motif analysis, the lack of supernumerary iron-sulfur clusters involved in oxygen tolerance and the physiological results showing hydrogen consumption under oxygen limitation, we therefore conclude that the hydrogenases present in the Methylacidimicrobium strains are part of the oxygen-sensitive group 1b [NiFe] hydrogenases (Figure 2).
TABLE 2.
The [NiFe] H2-uptake hydrogenases of acidophilic verrucomicrobial methanotrophs.
| Organism | Oxygen tolerance | Uptake hydrogenase | Gene namea | Gene IDb |
| groupa | ||||
| Methylacidimicrobium cyclopophantes 3Bc | Sensitive | Group 1b | hynBd hynAe |
60379.peg.2444 60379.peg.2445 |
| Methylacidimicrobium fagopyrum 3C | Sensitive | Group 1b | hynBd hynAe |
VER3v2_90073-4f VER3v2_90072 |
| Methylacidimicrobium strain LP2A | Sensitive | Group 1b | hynBd hynAe |
MAMLP_v1_11153 MAMLP_v1_11152 |
| Methylacidimicrobium tartarophylax 4AC c | Sensitive | Group 1b | hynBd hynAe |
60380.peg.187 60380.peg.186 |
| Methylacidimicrobium thermophilum A8 | Sensitive | Group 1b | hynBd hynAe |
MTHERMO_v1_1379 MTHERMO_v1_1378 |
| Methylacidiphilum fumariolicum SolV | Tolerantg | Group 1d Group 1h |
hyaBd hyaAe hhyLd hhySe |
Mfumv2_1564 Mfumv2_1565 Mfumv2_0979 Mfumv2_0978 |
| Methylacidiphilum infernorum V4 | Tolerant | Group 1d | hyaBd hyaAe |
Minf_1320 Minf_1321 |
| Methylacidiphilum kamchatkensis Kam1 | Tolerant | Group 1d Group 1h |
hyaBd hyaAe hhyLd hhySe |
JQNX01_v1_10368 JQNX01_v1_10367 JQNX01_v1_60118 JQNX01_v1_60119 |
| Methylacidiphilum sp. RTK17.1 | Tolerantg | Group 1d | hyaBd hyaAe |
ANC58185.1 ANC58184.1 |
aAccording to Søndergaard et al. (2016). bAvailable at the MicroScope annotation platform. cThe draft genomes of strains 4AC and 3B are fragmented. dLarge subunit. eSmall subunit. fThe large subunit of the hydrogenase of strain 3C is present in two pieces. gConflicting data have emerged on the oxygen tolerance of the group 1d type of these strains.
Discussion
In this study, we have shown that the mesophilic Methylacidimicrobium tartarophylax strain 4AC is able to grow as an autotroph on hydrogen gas as sole energy source under oxygen-limited conditions. Hydrogen consumption by microorganisms is an ancient trait: H2 is thought to be the first energy source utilized by microorganisms on Earth (Lane et al., 2010). All known methanotrophs of the Verrucomicrobia phylum encode for one or more [NiFe] hydrogenases, but with distinct properties. These hydrogenases are very different in terms of oxygen tolerance, which could lead to niche differentiation of thermophilic and mesophilic methanotrophs in the natural geothermal environment.
The oxygen tolerance of strain 4AC for hydrogen consumption is low. Strain 4AC is able to grow autotrophically on hydrogen gas when the flux of oxygen is regulated to obtain a dO2 level in the cultivation system below the detection limit. The highest activity was observed when the oxygen concentration was limited to below 10 μM (8.1 O2 nmol ⋅ min–1 ⋅ mg DW–1). At oxygen-limited conditions, the maximum growth rate (μmax) on H2 (0.0048 h–1) is approximately 12% compared to the growth rate on methane (van Teeseling et al., 2014). The measured yield on H2 (2.1 ± 0.2 g DW ⋅ mol H2–1) is about 41% compared to that on CH4 (5.1 ± 0.2 g DW ⋅ mol CH4–1) and lower than the yield reported for M. fumariolicum strain SolV (3.4 g DW ⋅ mol H2–1), for ‘Knallgas’ bacteria like Ralstonia eutropha (4.6 g DW ⋅ mol H2–1; Morinaga et al., 1978) and for Hydrogenomonas eutropha (5 g DW ⋅ mol H2–1; Bongers, 1970). Considering the number of electrons available from CH4 (8e–) and H2 (2e–) assuming complete oxidation, the biomass increase is 0.043 and 0.085 mole CH2O (biomass) per electron for CH4 and H2, respectively, indicating that hydrogen might be a better electron source. The group 1b [NiFe] hydrogenases were thought to be restricted to strict anaerobes for the reduction of alternative electron acceptors (Greening et al., 2016). However, here we show that strain 4AC couples hydrogen oxidation to the reduction of molecular oxygen when the oxygen concentration is below 40 μM. Group 1b and 1d [NiFe] hydrogenases are more oxygen-sensitive and, theoretically, this could result in the net translocation of more protons per molecule of hydrogen oxidized compared to group 1h [NiFe] hydrogenases, which reflects their periplasmic localization (Cordero et al., 2019). We therefore provide strong evidence that group 1b [NiFe] hydrogenases are also involved in aerobic respiration.
The ability of strain 4AC to consume H2 is likely to be a universal trait shared among methanotrophic Verrucomicrobia. The H2-uptake [NiFe] hydrogenases of this guild can be divided over three distinct groups (Op den Camp et al., 2009; Sharp et al., 2014; van Teeseling et al., 2014). Genomic analyses revealed that all known mesophilic methanotrophic Verrucomicrobia encode a membrane-bound group 1b [NiFe] H2-uptake hydrogenase. In contrast, all their thermophilic counterparts encode a membrane-bound group 1d [NiFe] H2-uptake hydrogenase. In addition, the thermophilic strains, SolV and Kam1, possess a group 1h [NiFe] H2-uptake hydrogenase. In the betaproteobacterium Ralstonia eutropha H16, the group 1h type was found to be insensitive to oxygen, which is likely due to an unusual coordination of the proximal iron-sulfur cluster where a cysteine residue is replaced by an aspartic acid residue (Fritsch et al., 2011; Schäfer et al., 2016). In strain SolV, the group 1h [NiFe] hydrogenase oxidizes hydrogen under at least ambient oxygen, whereas autotrophic growth only occurs below an oxygen concentration of 1.5%. This difference could be explained by oxygen sensitivity of the group 3b [NiFe] hydrogenase, which likely couples H2 oxidation to CO2 fixation in Methylacidiphilum (Carere et al., 2017). However, this NADH-producing group 3b type is not necessarily needed for autotrophic growth on hydrogen gas, since it is absent in all mesophilic strains. As for the group 1d [NiFe] hydrogenase, this enzyme is only expressed and active under oxygen-limiting conditions in strain SolV (Mohammadi et al., 2017). However, inconsistent results have emerged from studies on the isolate Methylacidiphilum sp. RTK17.1, where the group 1d type appeared more tolerant toward oxygen (Carere et al., 2017). The answer for this oxygen tolerance could arise from the unusual coordination of the proximal iron-sulfur cluster, in which six instead of four cysteine residues are involved (Shomura et al., 2011). Apparently, other factors are at play that determine oxygen tolerance in the Methylacidiphilum strains. Altogether, the presence of only the group 1b [NiFe] hydrogenase in strain 4AC explains why growth only occurs under strong oxygen limitation, which likely applies to the other mesophilic strains as well.
Hydrogen consumption is a ubiquitous trait among phyla and indeed oxygen limitation largely determines the distribution of different hydrogenase types (Greening et al., 2016). Different types are found in a wide range of habitats, varying from the hypoxic hydrogen-rich animal guts to soils and waters with low H2 and high O2 availability. Group 1b [NiFe] hydrogenases are mostly found in hypoxic environments such as peat bogs (Greening et al., 2016). However, this hydrogenase type is also encoded by the human pathogen Helicobacter pylori, supporting aerobic H2 oxidation in microoxic environments (Olson and Maier, 2002; Greening et al., 2016). Indeed, group 1b [NiFe] hydrogenases were also detected in geothermal environments. The group 1b [NiFe] hydrogenases in the metagenomics survey of these geothermal environments, however, were encoded by members of the Aquificae phylum, and did not include Methylacidimicrobium members. We therefore propose that group 1b [NiFe] hydrogenases are more abundant in volcanic ecosystems than previously thought. Indeed, the mesophilic and thermophilic verrucomicrobial methanotrophs experience various oxygen concentrations in their natural geothermal habitat (Chiodini et al., 2001; Pol et al., 2007). The strong reduction potential of hydrogen may not only decrease the threshold for oxidizing methane (Hanczár et al., 2002), but H2 also sustains growth when methane is absent as was shown in this study and before in Methylacidiphilum (Carere et al., 2017; Mohammadi et al., 2017). In the natural ecosystem of strain 4AC, hydrogen levels in volcanic gasses are much higher than those of methane (Chiodini et al., 2001). Hydrogen gas might allow for the uptake of methane even at atmospheric concentrations, which was observed in the Solfatara ecosystem (Castaldi and Tedesco, 2005). The [NiFe] H2-uptake hydrogenases have been shown to be more widespread in the phylum Verrucomicrobia than previously thought (Greening et al., 2016), suggesting that Verrucomicrobia may play a role in the hydrogen cycle in different ecosystems.
Conclusion
In conclusion, we show that strain 4AC can grow autotrophically on hydrogen gas but only under oxygen limited conditions using an oxygen-sensitive hydrogenase. This is the first study to show hydrogen oxidation by a mesophilic verrucomicrobial methanotroph. Apparently, the group 1b [NiFe] hydrogenase is not only functional in anaerobic respiration, but also in aerobic respiration at low oxygen. We propose that distribution of Methylacidiphilum and Methylacidimicrobium within acidic geothermal environments is influenced by the oxygen concentration, due to major differences in oxygen tolerance of the encoded hydrogenases. Therefore, we postulate that Methylacidimicrobium utilizes hydrogen gas and methane in acidic volcanic systems at moderate temperatures and low oxygen, similarly to the metabolism of its relatives of the Methylacidiphilum genus at higher temperatures and at various oxygen concentrations. This extends the evidence that verrucomicrobial methanotrophs are key players in consuming hydrogen and, therefore, these hydrogenases could aid in mitigation of greenhouse gasses.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
SM, AP, MJ, and HC designed the project and experiments. SM and AP performed the experimental work. SM and AP maintained the chemostat cultures. TB performed the phylogenetic analysis. SM, RS, AP, and HC performed data analysis and data interpretation. RS, SM, and HC wrote the manuscript with feedback from the other authors. HC and MJ supervised the research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. SM was supported by the Spinoza grant of MJ (Netherlands Organization for Scientific Research) and the European Research Council (ERC Advanced Grant project VOLCANO 669371), MJ by the European Research Council (ERC Advanced Grant Eco_MoM 339880), and HC, RS, and TB by the European Research Council (ERC Advanced Grant project VOLCANO 669371).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02352/full#supplementary-material
References
- Anvar S. Y., Frank J., Pol A., Schmitz A., Kraaijeveld K., den Dunnen J. T., et al. (2014). The genomic landscape of the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. BMC Genomics 15:914. 10.1186/1471-2164-15-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter N. J., Hirt R. P., Bodrossy L., Kovacs K. L., Embley M. T., Prosser J. I., et al. (2002). The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch. Microbiol. 177 279–289. 10.1007/s00203-001-0387-x [DOI] [PubMed] [Google Scholar]
- Bongers L. (1970). Yields of Hydrogenomonas eutropha from growth on succinate and fumarate. J. Bacteriol. 102 598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe H., Schmitz O., Yates M. G., Newton W. E. (2010). Nitrogen fixation and hydrogen metabolism in Cyanobacteria. Microbiol. Mol. Biol. R. 74 529–555. 10.1128/MMBR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carere C. R., Hards K., Houghton K. M., Power J. F., McDonald B., Collet C., et al. (2017). Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J. 11 2599–2610. 10.1038/ismej.2017.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldi S., Tedesco D. (2005). Methane production and consumption in an active volcanic environment of Southern Italy. Chemosphere 58 131–139. 10.1016/j.chemosphere.2004.08.023 [DOI] [PubMed] [Google Scholar]
- Chiodini G., Frondini F., Cardellini C., Granieri D., Marini L., Ventura G. (2001). CO2 degassing and energy release at Solfatara volcano, Campi Flegrei, Italy. J. Geophys. Res. 106 16213–16221. 10.1029/2001JB000246 [DOI] [Google Scholar]
- Conrad R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1 285–292. 10.1111/j.1758-2229.2009.00038.x [DOI] [PubMed] [Google Scholar]
- Constant P., Chowdhury S. P., Pratscher J., Conrad R. (2010). Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ. Microbiol. 12 821–829. 10.1111/j.1462-2920.2009.02130.x [DOI] [PubMed] [Google Scholar]
- Coppi M. V., O’Neil R. A., Lovley D. R. (2004). Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe(III) and other electron acceptors by Geobacter sulfurreducens. J. Bacteriol. 186 3022–3028. 10.1128/JB.186.10.3022-3028.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero P. R. F., Grinter R., Hards K., Cryle M. J., Warr C. G., Cook G. M., et al. (2019). Two uptake hydrogenases differentially interact with the aerobic respiratory chain during mycobacterial growth and persistence. bioRxiv [Preprint] 10.1101/769216, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csáki R., Hanczár T., Bodrossy L., Murrell J. C., Kovács K. L. (2001). Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 205 203–207. 10.1111/j.1574-6968.2001.tb10948.x [DOI] [PubMed] [Google Scholar]
- Dalton H., Whittenbury R. (1976). Acetylene reduction technique as an assay for nitrogenase activity in the methane oxidizing bacterium Methylococcus capsulatus strain bath. Arch. Microbiol. 109 147–151. 10.1007/bf00425127 [DOI] [Google Scholar]
- de Bont J. A. M. (1976). Hydrogenase activity in nitrogen-fixing methane-oxidizing bacteria. Antonie Van Leeuwenhoek 42 255–259. 10.1007/bf00394122 [DOI] [PubMed] [Google Scholar]
- Dunfield P. F., Yuryev A., Senin P., Smirnova A. V., Stott M. B., Hou S., et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum verrucomicrobia. Nature 450 879–883. 10.1038/nature06411 [DOI] [PubMed] [Google Scholar]
- Erikstad H.-A., Birkeland N.-K. (2015). Draft genome sequence of “candidatus Methylacidiphilum kamchatkense” strain Kam1, a thermoacidophilic methanotrophic verrucomicrobium. Genome Announc. 3:e00065-15. 10.1128/genomeA.00065-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig K. F., Butler M. K., Le Paslier D., Pelletier E., Mangenot S., Kuypers M. M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464 543–550. 10.1038/nature08883 [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Shima S., van de Pas-Schoonen K. T., Kahnt J., Medema M. H., Op den Camp H. J., et al. (2008). Denitrifying bacteria anaerobically oxidize methane in the absence of archaea. Environ. Microbiol. 10 3164–3173. 10.1111/j.1462-2920.2008.01724.x [DOI] [PubMed] [Google Scholar]
- Fritsch J., Scheerer P., Frielingsdorf S., Kroschinsky S., Friedrich B., Lenz O., et al. (2011). The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479 249–253. 10.1038/nature10505 [DOI] [PubMed] [Google Scholar]
- Greening C., Biswas A., Carere C. R., Jackson C. J., Taylor M. C., Stott M. B., et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10 761–777. 10.1038/ismej.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C., Constant P., Hards K., Morales S. E., Oakeshott J. G., Russell R. J., et al. (2015). Atmospheric hydrogen scavenging: from enzymes to ecosystems. Appl. Environ. Microbiol. 81 1190–1199. 10.1128/AEM.03364-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczár T., Csáki R., Bodrossy L., Murrell J. C., Kovács K. L. (2002). Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch. Microbiol. 177 167–172. 10.1007/s00203-001-0372-4 [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Hanson T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon M. F., Hu S., Shi Y., Imelfort M., Keller J., Hugenholtz P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500 567–570. 10.1038/nature12375 [DOI] [PubMed] [Google Scholar]
- Hou S., Makarova K. S., Saw J. H. W., Senin P., Ly B. V., Zhou Z., et al. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum verrucomicrobia. Biol. Direct 3:26. 10.1186/1745-6150-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T., Jensen S., Reigstad L. J., Larsen O., Birkeland N.-K. (2008). Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105 300–304. 10.1073/pnas.0704162105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalms J., Schmidt A., Frielingsdorf S., Utesch T., Gotthard G., von Stetten D., et al. (2018). Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase. Proc. Natl. Acad. Sci. U.S.A. 115 E2229–E2237. 10.1073/pnas.1712267115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. P., Anthony C., Murrell J. C. (2005). Insights into the obligate methanotroph Methylococcus capsulatus. Trends Microbiol. 13 195–198. 10.1016/j.tim.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Wieczorek A., Mohammadi S. S., Francoijs K. J., Stunnenberg H. G., et al. (2011). Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193 4438–4446. 10.1128/JB.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem A. F., Pol A., Wieczorek A. S., Jetten M. S. M., Op den Camp H. J. M. (2012). Metabolic regulation of “Ca. Methylacidiphilum fumariolicum” SolV cells grown under different nitrogen and oxygen limitations. Front. Microbiol. 3:266 10.3389/fmicb.2012.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke S., Bousquet P., Ciais P., Saunois M., Canadell J. G., Dlugokencky E. J.,, et al. (2013). Three decades of global methane sources and sinks. Nat. Geosci. 6 813–823. 10.1038/ngeo1955 [DOI] [Google Scholar]
- Knittel K., Boetius A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63 311–334. 10.1146/annurev.micro.61.080706.093130 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N., Allen J. F., Martin W. (2010). How did LUCA make a living? Chemiosmosis in the origin of life. Bioessays 32 271–280. 10.1002/bies.200900131 [DOI] [PubMed] [Google Scholar]
- Lubitz W., Ogata H., Rüdiger O., Reijerse E. (2014). Hydrogenases. Chem. Rev. 114 4081–4148. 10.1021/cr4005814 [DOI] [PubMed] [Google Scholar]
- Mohammadi S., Pol A., van Alen T. A., Jetten M. S. M., Op den Camp H. J. M. (2017). Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11 945–958. 10.1038/ismej.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga Y., Yamanaka S., Ishizaki A., Hirose Y. (1978). Growth characteristics and cell composition of Alcaligenes eutrophus in chemostat culture. Agric. Biol. Chem. 42 439–444. 10.1271/bbb1961.42.439 [DOI] [Google Scholar]
- Murrell J. C., Jetten M. S. M. (2009). The microbial methane cycle. Environ. Microbiol. Rep. 1 279–284. 10.1111/j.1758-2229.2009.00089.x [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Herringa J. (2000). T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Olson J. W., Maier R. J. (2002). Molecular hydrogen as an energy source for Helicobacter pylori. Science 298 1788–1790. 10.1126/science.1077123 [DOI] [PubMed] [Google Scholar]
- Op den Camp H. J. M., Islam T., Stott M. B., Harhangi H. R., Hynes A., Schouten S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic verrucomicrobia. Environ. Microbiol. Rep. 1 293–306. 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- Pol A., Heijmans K., Harhangi H. R., Tedesco D., Jetten M. S., Op den Camp H. J. M. (2007). Methanotrophy below pH 1 by a new verrucomicrobia species. Nature 450 874–878. 10.1038/nature06222 [DOI] [PubMed] [Google Scholar]
- Power J. F., Carere C. R., Lee C. K., Wakerley G. L. J., Evans D. W., Button M., et al. (2018). Microbial biogeography of 925 geothermal springs in New Zealand. Nat. Comm. 9:2876. 10.1038/s41467-018-05020-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer C., Bommer M., Hennig S. E., Jeoung J. H., Dobbek H., Lenz O. (2016). Structure of an Actinobacterial-Type [NiFe]-Hydrogenase Reveals Insight into O2-Tolerant H2 Oxidation. Structure 24 285–292. 10.1016/j.str.2015.11.010 [DOI] [PubMed] [Google Scholar]
- Semrau J. D., DiSpirito A. A., Vuilleumier S. (2011). Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol. Lett. 323 1–12. 10.1111/j.1574-6968.2011.02315.x [DOI] [PubMed] [Google Scholar]
- Semrau J. D., DiSpirito A. A., Yoon S. (2010). Methanotrophs and copper. FEMS Microbiol. Rev. 34 496–531. 10.1111/j.1574-6976.2010.00212.x [DOI] [PubMed] [Google Scholar]
- Sharp C. E., Op den Camp H. J. M., Tamas I., Dunfield P. F. (2013). “Unusual members of the PVC superphylum: the methanotrophic verrucomicrobia genus “Methylacidiphilum”,” in Planctomycetes: Cell Structure, Origins and Biology, ed. Fuerst J. A. (New York, NY: Humana Press; ), 211–227. 10.1007/978-1-62703-502-6_9 [DOI] [Google Scholar]
- Sharp C. E., Smirnova A. V., Graham J. M., Stott M. B., Khadka R., Moore T. R., et al. (2014). Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 16 1867–1878. 10.1111/1462-2920.12454 [DOI] [PubMed] [Google Scholar]
- Sharp C. E., Stott M. B., Dunfield P. F. (2012). Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303. 10.3389/fmicb.2012.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura Y., Yoon K. S., Nishihara H., Higuchi Y. (2011). Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479 253–256. 10.1038/nature10504 [DOI] [PubMed] [Google Scholar]
- Søndergaard D., Pedersen C. N. S., Greening C. (2016). HydDB: a web tool for hydrogenase classification and analysis. Sci. Rep. 6:34212. 10.1038/srep34212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. H., Dalton H. (1982). Role of ribulose-1,5-bisphosphate carboxylase/oxygenase in Methylococcus capsulatus (Bath). J. Gen. Microbiol. 128 2927–2935. 10.1099/00221287-128-12-2927 [DOI] [Google Scholar]
- Taylor S. C., Dalton H., Dow C. S. (1981). Ribulose-1,5-bisphosphate carboxylase/oxygenase and carbon assimilation in Methylococcus capsulatus (Bath). J. Gen. Microbiol. 122 89–94. 10.1099/00221287-122-1-89 [DOI] [Google Scholar]
- van Teeseling M. C., Pol A., Harhangi H. R., van der Zwart S., Jetten M. S., Op den Camp H. J. M., et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 80 6782–6791. 10.1128/AEM.01838-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais P. M., Billoud B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107 4206–4272. 10.1021/cr050196r [DOI] [PubMed] [Google Scholar]
- Voulgarakis A., Naik V., Lamarque J.-F., Shindell D. T., Young P. J., Prather M. J., et al. (2013). Analysis of present day and future OH and methane lifetime in the ACCMIP simulations. Atmos. Chem. Phys. 13 2563–2587. 10.5194/acp-13-2563-2013 [DOI] [Google Scholar]
- Ward L., Taylor M. W., Power J. F., Scott B. J., McDonald I. R., Stott M. B. (2017). Microbial community dynamics in inferno crater lake, a thermally fluctuating geothermal spring. ISME J. 11 1158–1167. 10.1038/ismej.2016.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.