Abstract
OBJECTIVES:
To examine the effects of age and race on the association of apolipoprotein E (APOE) genotypes with cognitive decline in a population sample.
DESIGN:
Longitudinal study of 18 years’ duration.
SETTING:
Biracial urban US population sample.
PARTICIPANTS:
There were a total of 5807 participants, 60% African American (AA) and 40% European American (EA).
MEASUREMENTS:
A composite cognitive function based on individual tests of episodic memory, perceptual speed, and the Mini-Mental State Examination.
RESULTS:
The frequencies of APOE ε2/ε3 (14% vs 12%), ε2/ε4 (4% vs 2%), ε3/ε4 (29% vs 22%), and ε4/ε4 (4% vs 2%) genotypes were higher among AAs than EAs. After adjusting for demographic factors, the rate of decline in global cognition was twice as high among participants with the APOE ε4/ε4 genotype compared to participants with the APOE ε3/ε3 genotype (0.097 vs 0.048 SD units [SDUs] per year; P < .0001). This doubling was not different between AAs (0.091 vs 0.045 SDUs per year) and EAs (0.118 vs 0.059 SDUs per year) (Pinteraction = .63). The APOE ε3/ε4 genotype was associated with a higher rate of decline with age (Pinteraction = .021), while the APOE ε2/ε4 genotype (Pinteraction = .016) and the APOE ε2/ε3 genotype (Pinteraction = .043) were associated with a lower rate of decline with higher age. The APOE ε2/ε2 genotype was associated with a lower rate of decline in episodic memory, while the APOE ε2/ε4 was associated with a higher rate of decline in episodic memory and perceptual speed.
CONCLUSIONS:
The association of the APOE genotypes with cognitive decline was not different between AAs and EAs. However, individuals with different APOE genotypes showed a lower or a higher rate of decline with age.
Keywords: APOE genotypes, cognitive decline, minority aging
Alzheimer disease (AD) is characterized by a severe decline in cognitive function.1–3 The apolipoprotein E (APOE) ε4 genotype is associated with an increased risk for AD4–7 and cognitive decline.8–11 The APOE ε2 genotype may reduce the risk of AD,12 slow the decline in episodic memory,13 and increase cognitive performance,14 while few studies suggest a positive association with the APOE ε2 allele increasing the risk of AD.15,16 The age-dependent effects of the APOE genotypes on cognitive decline remain unclear, although the most commonly studied genotype, the APOE ε4 genotype, is reported to have diminished association with AD after 70 years7 or peak by 75 to 78 years.17,18 However, several questions on the influence of the six APOE genotypes on cognitive decline remain unanswered.
Of particular interest are reports that the APOE ε4 genotype may not be associated with a higher risk of incident AD among African Americans (AAs),19–23 or this association might be weaker among AAs.24,25 Some studies have shown that the effect of the APOE ε4 genotype on cognitive decline may be similar among AAs and European Americans (EAs),26,27 or this association may be different during midlife,28 or for specific cognitive tests11 compared to EAs. However, the racial differences of the association of the APOE ε2 genotypes with cognitive decline have not been investigated.
The objective of this article is to examine three research questions: (1) to test the racial differences in the association of the six APOE genotypes with decline in global cognition between AAs and EAs, (2) to test if the association of the six APOE genotypes with decline in global cognition changes with age, and (3) to test the association of the APOE genotypes with individual tests of cognition.
METHODS
Study Participants
Study participants come from the Chicago Health and Aging Project (CHAP), a longitudinal population-based study of adults 65 years and older conducted between 1993 and 2012. The study procedures and sampling procedures have been described in greater detail previously.29 The CHAP study began by enrolling 78.7% of all residents older than 65 years in a door-to-door census of a geographically defined, biracial Chicago community. From 2001, community residents who attained the age of 65 years were also enrolled as successive cohorts.
Data Collection
Population interviews were performed in participants’ homes in approximately 3-year cycles. Data were collected for up to six follow-up cycles for the original cohort, and two to five follow-up cycles for successive cohorts. At the end of cycle 2 population interview, DNA samples were collected from a stratified random sample of the population of about one-sixth of all participants enrolled in the study. Toward the end of cycle 5 and all of cycle 6, DNA samples were collected from all subjects during their population interviews.
The original CHAP study consisted of 10,802 participants; we excluded participants who died without a follow-up (N = 2796), participants who declined follow-up participation (N = 171), participants with no DNA extracted (N = 1949), participants with insufficient cognitive data (N = 78), and participants without demographic data (N = 3). We compared those who died without a follow-up to those who provided two or more follow-up interviews and found that those who died had a significantly lower cognitive function at baseline (P < .0001). We also compared those who provided cognitive function tests without DNA to those with DNA extracted and found that participants with DNA had significantly better cognition than those without DNA (P < .0001). We found no differences in the years of follow-up, race/ethnicity, sex, or education between those with and without DNA.
Global and Individual Tests of Cognition
Cognitive function was evaluated using a battery of four tests, including two tests of episodic memory (immediate and delayed recall) derived from the East Boston Test,30,31 a test of perceptual speed (the Symbol Digits Modalities Test),32 and a test of general orientation and global cognition (the Mini-Mental State Examination [MMSE]).33 Cognitive function tests loaded on a single factor that accounted for about 75% of the variance in a factor analysis.34 Hence, a global measure of cognitive function was created based on the four tests, by averaging the four tests together after centering and scaling each to baseline mean and SD from the original cohort.
APOE Genotypes
The APOE ε4 genotypes were determined using two single-nucleotide polymorphisms (SNPs): rs7412 and rs429358.35,36 These SNPs were genotyped in each subject at the Broad Institute Center for Genotyping (Cambridge, MA, USA) using the hME Sequenom MassARRAY platform. Genotyping call rates were 100% for SNP rs7412 and 99.8% for SNP rs429358. Both SNPs were in Hardy-Weinberg equilibrium with P values of.0833 and .7925, respectively. Five indicator variables for ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε4, and ε4/ε4 genotypes, with ε3ε3 as the reference genotype, were created for analysis purposes.
Demographic Variables
Age at the time of the interview, sex (males or females), race (Hispanic ethnicity, AAs, non-Hispanic whites, or EAs), and education (measured in the number of years of schooling completed) were assessed during the baseline assessment. In our analysis, we centered baseline age at 75 years and education at 12 years and created indicator variables for males and AAs.
Statistical Analysis
Descriptive statistics were computed using means and SDs for continuous variables and percentages for categorical variables. Race-specific allelic frequencies for APOE were also estimated from the study sample. We used mixed-effects regression models to examine average levels of cognitive function and the longitudinal association of the APOE genotypes with cognitive decline stratified by race/ethnicity.37 The racial differences in average levels of cognitive function and cognitive decline for the APOE genotypes were tested using two- and three-way interactions of race with time since baseline and the indicator for the APOE genotypes. The regression models had five indicator variables for combinations of ε2, ε3, and ε4, with the ε3ε3 as the reference category, and had an interaction of the linear time since baseline, which takes the value 0 at baseline, with each of the indicator variables. This regression-based approach also included the main effect for time since baseline and interactions of this time variable with baseline age (centered at 75 years), education (centered at 12 years), sex (indicator for males), and APOE genotype indicators in our core models. The demographic variables were centered around 0, so the time since baseline variable can be interpreted as the rate of decline in cognition in ε3ε3 when all covariates are set to 0.
The analytic sample consisted of 5807 participants, of whom 2253 were selected using a stratified random sample, and the remaining 3554 participants provided DNA samples during cycles 5 and 6 of the population interviews. A subject-specific sample weight was assigned for those selected using our stratified random sample, while setting those from the population interviews to be equal to 1. Given that sample weights were design features, a model-based adjustment for sampling would provide a biased estimate of the sample variances, even though the sample means would be unbiased. To remedy this situation, we estimated bootstrap sampling weights for a mixed sampling scheme and further evaluated our bootstrap variance using a simulation study.38,39 The variances for parameter estimates in the models used 1000 bootstrap samples and used these variances to estimate the corresponding P values. All models were fitted using R version 2.15.3 using the nlm function in the lme package, and bootstrap variances were estimated with the coding program using the sampling package.40
RESULTS
The sample consisted of 5807 participants (60% AAs) with APOE genotypes. The average length of follow-up was 8.9 (SD = 4.4) years for AAs and 8.3 (SD = 4.5) years for EAs. AAs were younger and had less education compared to EAs (Table 1). AAs also scored lower on global cognition and individual tests of episodic memory, perceptual speed, and the MMSE than EAs. The overall rate of cognitive decline was 0.053 SD units (SDUs), and did not differ between AAs and EAs (Pinteraction = .18).
Table 1.
Sample Characteristics of 5807 Participants in a Biracial Population Sample
| AAs Only |
EAs Only |
All Participants |
||
|---|---|---|---|---|
| Characteristics | Measures | (N = 3495) | (N = 2312) | (N = 5807) |
| Demographic | Age, mean (SD), ya | 71.1 (5.8) | 74.5 (7.4) | 72.5 (6.7) |
| Education, mean (SD), ya | 11.8 (3.3) | 14.1 (3.2) | 12.8 (3.5) | |
| Female sex, No. (%) | 2235 (64) | 1423 (62) | 3658 (63) | |
| Cognitive test results, mean (SD)b | Global cognition | 0.217 (0.716) | 0.561 (0.667) | 0.354 (0.717) |
| Memory score | 0.253 (0.854) | 0.532 (0.794) | 0.352 (0.839) | |
| Perceptual speed | 0.192 (0.874) | 0.846 (0.831) | 0.452 (0.915) | |
| MMSE | 0.205 (0.704) | 0.445 (0.582) | 0.301 (0.668) | |
| APOE genotypes, No. (%) | ε2/ε2 | 34 (<1) | 14 (<1) | 48 (<1) |
| ε2/ε3 | 500 (14) | 282 (12) | 782 (14) | |
| ε2/ε4 | 154 (4) | 49 (2) | 203 (4) | |
| ε3/ε3 | 1642 (47) | 1412 (61) | 3054 (53) | |
| ε3/ε4 | 1017 (29) | 517 (22) | 1534 (26) | |
| ε4/ε4 | 148 (4) | 38 (2) | 186 (3) | |
Abbreviations: AA, African American; APOE, apolipoprotein E; EA, European American; MMSE, Mini-Mental State Examination.
Age and education were significantly different between AAs and EAs (P < .001).
The global cognitive function, memory score, perceptual speed, and MMSE were also significantly different between AAs and EAs (P < .0001).
The Frequency of APOE Genotypes
From Table 1, less than 1% of AAs and EAs had the APOE ε2/ε2 genotype. The frequencies of APOE ε2/ε3, ε2/ε4, ε3/ε4, and ε4/ε4 genotypes were higher among AAs compared to EAs, while the frequency of the APOE ε3/ε3 genotype was lower among AAs compared to EAs, by almost 14%.
Association of APOE Genotypes With Global Cognitive Function
The average baseline level and the rate of decline in global cognition are shown in Table 2. After adjusting for demographic characteristics, the APOE ε3/ε4 and ε4/ε4 genotypes were associated with a lower level of global cognition compared to those with the ε3/ε3 genotype. In particular, participants with the ε4/ε4 genotype had average baseline cognition 0.221 SDUs lower than those with the ε3/ε3 genotype.
Table 2.
Demographic Adjusted Average Level and Rate of Change in Global Cognition Among AAs and EAsa
| AAs Only |
EAs Only |
|
|---|---|---|
| APOE Genotype | (N = 3495) | (N = 2312) |
| Average Level of Baseline Cognition | ||
| ε3/ε3 | 0.118 (0.017)b | 0.565 (0.018)b |
| ε2/ε2 vs ε3/ε3 | −0.101 (0.098) | 0.093 (0.152) |
| ε2/ε3 vs ε3/ε3 | 0.045 (0.029) | 0.025 (0.036) |
| ε2/ε4 vs ε3/ε3 | −0.007 (0.048) | −0.008 (0.081) |
| ε3/ε4 vs ε3/ε3 | −0.018 (0.023) | −0.098 (0.029) |
| ε4/ε4 vs ε3/ε3 | −0.185 (0.049)b | −0.315 (0.091)b |
| Rate of Decline in Cognition | ||
| ε3/ε3 | 0.046 (0.003)b | 0.059 (0.003)b |
| ε2/ε2 vs ε3/ε3 | −0.032 (0.013)c | −0.010 (0.027) |
| ε2/ε3 vs ε3/ε3 | −0.004 (0.004) | −0.010 (0.006) |
| ε2/ε4 vs ε3/ε3 | 0.009 (0.006) | 0.026 (0.013)d |
| ε3/ε4 vs ε3/ε3 | 0.018 (0.003)b | 0.031 (0.005)b |
| ε4/ε4 vs ε3/ε3 | 0.045 (0.009)b | 0.059 (0.013)b |
Abbreviations: AA, African American; APOE, apolipoprotein E; EA, European American.
Data are given coefficient (SE). Race-specific models adjusted for main effects of age, sex, and education, and interaction of linear time since baseline with each of the demographic variable. Each of the race-specific models also consist of five indicator variables for APOE genotypes, with APOE ε3ε3 as the reference category.
P < .0001.
P < .05.
P < .001.
The rate of decline in global cognition was about twofold higher in participants with the APOE ε4/ε4 genotype compared to participants with the ε3/ε3 genotype (0.097 vs 0.048 SDUs per year). This higher rate of decline in cognition was similar among AAs and EAs (Pinteraction = .63). The rate of decline in global cognition was greater by about 50% in participants with the APOE ε3/ε4 genotype compared to those with the ε3/ε3 genotype (0.070 vs 0.048 SDUs per year). Although this association appeared to be weaker among AAs, no statistical difference was found between AAs and EAs (Pinteraction = .09). The rate of decline in global cognition was greater by about 25% in participants with the APOE ε2/ε4 genotype compared to participants with the ε3/ε3 genotype (0.060 vs 0.048 SDUs per year). This difference appeared to be larger among EAs than AAs (0.026 vs 0.009 SDUs per year), but not statistically significant (Pinteraction = .27).
The presence of the APOE ε2/ε2 genotype was associated with a lower rate of decline in global cognition by about 60% compared to participants with the APOE ε3/ε3 genotype (0.020 vs 0.048 SDUs per year; Pinteraction = .032). Although this association appeared to be weaker among EAs than AAs, no racial differences were observed (P = .47). The APOE ε2/ε3 genotype was not associated with the rate of decline in global cognition (Pinteraction = .08).
Age-Dependent Association of APOE Genotypes With Global Cognitive Function
In general, we found no statistically significant racial differences in the association of the APOE genotypes with rate of decline in global cognition, although some of this association appeared to be weaker among AAs. The average age was higher among EAs compared to AAs for the APOE genotypes (Supplementary Table 1).
The APOE ε2/ε3 genotype was associated with a lower rate of decline in global cognition among those older than 75 years than among those younger than 75 years compared to the ε3/ε3 genotype (Pinteraction = .045) (Table 3). In contrast, the APOE ε3/ε4 genotype was associated with a higher rate of decline in global cognition among those older than 75 years than among those younger than 75 years compared to the ε3/ε3 genotype (Pinteraction = .021). The APOE ε2/ε4 genotype was associated with a higher rate of decline among those younger than 75 years and a lower rate of decline among those older than 75 years (Pinteraction = .016). The association of the APOE ε2/ε2 genotype did not appear to be modified by age (Pinteraction = .89).
Table 3.
Average Level and Rate of Decline in Global Cognition, by Age Groupsa
| Aged <75 y |
Aged >75 y |
||
|---|---|---|---|
| APOE Genotype | (N = 4158) | (N = 1651) | PInteraction |
| Average Level of Baseline Cognition | |||
| ε3/ε3 | 0.509 (0.023)b | 0.713 (0.041)b | <.0001 |
| ε2/ε2 vs ε3/ε3 | −0.036 (0.088) | −0.098 (0.184) | .47 |
| ε2/ε3 vs ε3/ε3 | 0.027 (0.024) | 0.042 (0.051) | .92 |
| ε2/ε4 vs ε3/ε3 | 0.016 (0.042) | −0.127 (0.103) | .11 |
| ε3/ε4 vs ε3/ε3 | −0.014 (0.019)c | −0.141 (0.043)c | .011 |
| ε4/ε4 vs ε3/ε3 | −0.164 (0.043)b | −0.469 (0.054)b | .002 |
| Rate of Decline in Cognition | |||
| ε3/ε3 | 0.040 (0.003)b | 0.053 (0.007)b | <.0001 |
| ε2/ε2 vs ε3/ε3 | −0.028 (0.013)d | −0.030 (0.030) | .89 |
| ε2/ε3 vs ε3/ε3 | −0.004 (0.003) | −0.014 (0.009) | .043 |
| ε2/ε4 vs ε3/ε3 | 0.019 (0.006)d | −0.023 (0.019) | .016 |
| ε3/ε4 vs ε3/ε3 | 0.019 (0.003)b | 0.038 (0.008)b | .021 |
| ε4/ε4 vs ε3/ε3 | 0.046 (0.006)b | 0.059 (0.023)d | .87 |
Abbreviation: APOE, apolipoprotein E.
Data are given as coefficient (SE). The model for interaction test included main effects of age, sex, race, and education, and interaction of linear time since baseline with each of the demographic variables. Age-specific models adjusted for main effects of age, sex, and education, and interaction of linear time since baseline with each of the demographic variables.
P < .0001.
P < .001.
P < .05.
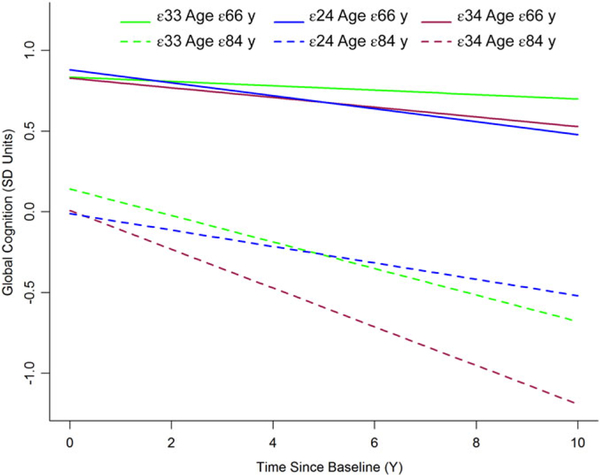
The rate of cognitive decline for APOE genotypes for the 10th and 90th percentiles of the age distribution is shown in Figure 1. The solid lines show the cognitive decline at 66 years of age, while the dashed line shows the cognitive decline at 84 years of age. The APOE ε2/ε4 genotype declines faster in earlier ages (solid blue line) and slower in later ages (dashed blue line), while the APOE ε3/ε4 genotype declines faster in later years (dashed red line) and slower in earlier ages (solid red line).
Figure 1.
Age-dependent rate of cognitive decline in specific apolipoprotein E (APOE) genotypes over the duration of the study. The age distributions are based on the lower (10th) and upper (90th) percentiles of the age distributions. Only APOE genotypes with significant differences are shown here.
Association of APOE Genotypes With Individual Tests of Cognition
The APOE ε4/ε4 genotype was associated with significantly lower episodic memory, perceptual speed, and MMSE scores than participants with the ε3/ε3 genotype (Table 4).
Table 4.
Average Level and Rate of Decline in Individual Cognitive Tests of Episodic Memory, Executive Function, and MMSE in the CHAP Biracial Samplea
| APOE Genotype | Episodic Memory | Executive Function | MMSE |
|---|---|---|---|
| Average Level of Baseline Cognition | |||
| ε3/ε3 | 0.435 (0.019)b | 0.706 (0.021)b | 0.379 (0.016)b |
| ε2/ε2 vs ε3/ε3 | −0.073 (0.104) | −0.084 (0.100) | −0.019 (0.085) |
| ε2/ε3 vs ε3/ε3 | 0.021 (0.028) | 0.059 (0.027)c | 0.043 (0.023) |
| ε2/ε4 vs ε3/ε3 | −0.009 (0.052) | −0.012 (0.049) | −0.018 (0.042) |
| ε3/ε4 vs ε3/ε3 | −0.078 (0.022)d | −0.016 (0.021) | −0.022 (0.018) |
| ε4/ε4 vs ε3/ε3 | −0.275 (0.054)b | −0.148 (0.054)c | −0.176 (0.044)b |
| Rate of Decline in Cognition | |||
| ε3/ε3 | 0.033 (0.003)b | 0.056 (0.002)b | 0.054 (0.003)b |
| ε2/ε2 vs ε3/ε3 | −0.050 (0.015)d | −0.001 (0.012) | −0.003 (0.014) |
| ε2/ε3 vs ε3/ε3 | −0.008 (0.004)c | 0.002 (0.003) | −0.008 (0.004)c |
| ε2/ε4 vs ε3/ε3 | 0.016 (0.007)c | 0.016 (0.006)d | 0.003 (0.007) |
| ε3/ε4 vs ε3/ε3 | 0.019 (0.003)b | 0.016 (0.003)b | 0.024 (0.003)b |
| ε4/ε4 vs ε3/ε3 | 0.046 (0.008)b | 0.034 (0.006)b | 0.058 (0.007)b |
Abbreviations: APOE, apolipoprotein E; CHAP, Chicago Health and Aging Project; MMSE, Mini-Mental State Examination.
Data are given as coefficient (SE). Models adjusting for main effects of age, sex, race, and education, and interaction of linear time since baseline with each of the demographic variables.
P < .0001.
P < .05.
P < .001.
The APOE ε3/ε4 and ε4/ε4 genotypes were associated with a higher rate of decline in all three cognitive tests, episodic memory, perceptual speed, and the MMSE, compared to participants with the APOE ε3/ε3 genotype. The APOE ε2/ε2 genotype was associated with a lower rate of decline in episodic memory compared to participants with the APOE ε3/ε3 genotype (0.017 vs 0.033 SDUs per year, a difference of 0.050 SDUs per year). The APOE ε2/ε3 genotype showed a lower rate of decline in episodic memory and MMSE scores compared to participants with the APOE ε3/ε3 genotype. The APOE ε2/ε4 genotype was associated with a higher rate of decline in episodic memory and perceptual speed scores compared to participants with the APOE ε3/ε3 genotype.
DISCUSSION
Our findings suggest that AAs and EAs with the APOE ε3/ε4 and APOE ε4/ε4 genotypes had similar rates of decline in global cognition. About 33% of AAs had the APOE ε3/ε4 and ε4/ε4 genotypes, while only 24% of EAs had these combinations. This difference in allelic frequencies might translate to a larger burden of decline in global cognition in AAs due to higher APOE ε4 frequency. The APOE ε4 allele findings largely support previous studies that the APOE ε3/ε4 and APOE ε4/ε4 genotypes were associated with cognitive decline,8–11 but no significant racial differences with cognitive decline were observed.26,27
The APOE ε2/ε4 genotype association with cognitive decline decreased with age, with faster decline in those younger than 75 years and slower decline in those older than 75 years. The APOE ε2/ε3 genotype was associated with cognitive decline that increased with age. These associations have not been reported in previous literature. The presence of the APOE ε2 genotype has been reported to be associated with reduced risk of AD12 and slower decline in episodic memory.13 However, our findings suggest that the protective associations are stronger among those with two copies of the APOE ε2 genotype, mostly for slower decline in episodic memory, which did not change with age. The association of the APOE ε3/ε4 allele with cognitive decline increased with age. This finding is contrary to findings that report this association with AD to decrease with age.17,18
The APOE ε2/ε2 and APOE ε2/ε3 genotypes showed slower decline in episodic memory, whereas the APOE ε2/ε4, APOE ε3/ε4, and APOE ε4/ε4 genotypes showed increased decline in episodic memory compared to those with the APOE ε3/ε3 genotype. Several studies have shown the association of the APOE ε4 genotype with domains of cognitive decline.10,11,13 The APOE ε2/ε4 genotype was associated with increased decline in perceptual speed, while the APOE ε2/ε3 was associated with slower decline in MMSE.
This study is based on a large population-based sample of AAs and EAs from Chicago. AAs were younger, had fewer years of education, and had lower baseline scores on global and individual tests of cognition. To adjust for these differences, we used a random-effects model with subject-specific intercept and slopes, and age and education centered to a common reference, 75 years for age and 12 years for education, to examine the association of APOE genotypes with average baseline cognition and the rate of cognitive decline separately in AAs and EAs. Our age-dependent analysis showed that the effect of APOE genotypes, especially the APOE ε4 genotypes, was more pronounced in those older than 75 years, hence making our overall estimates somewhat conservative. In a sensitivity analysis, we also controlled for several adverse health measures, using a summary measure for stroke, hypertension, myocardial infarction, cancer, diabetes, and hip fracture, health measures such as depressive symptoms, and body mass index. The results of this sensitivity analysis did not change the main findings.
Several studies classified individuals with one or more APOE ε4 alleles into a single group. However, the dose-effect response to the presence of one or more copies of APOE ε4 genotypes is evident for cognitive decline. The progression of AD entails a higher degree of decline in cognitive function over time for those with two copies of the APOE ε4 genotype. The allelic frequency of the APOE ε4/ε4 genotype was twice as large among AAs as EAs; interestingly, several studies have reported a lack of association of the APOE ε4 genotype with clinically diagnosed AD.19–22 The APOE ε2/ε2 genotype carriers had slower cognitive decline, but comprised less than 1% of the population, and the slower decline for those with the APOE ε2/ε3 is only observed in those older than 75 years.
Several strengths and limitations of our study need to be discussed. Some of the strengths include a longitudinal study performed over 18 years with multiple cohorts, a large sample of the participants genotyped, nearly 60% AAs, and cognitive test battery collected uniformly over the duration of the study. Several limitations also need to be noted: a partially random sample of individuals that might lead to a selection bias, geographical sample restricted to urban areas in Chicago makes generalizability a concern, and small sample size for the rare APOE genotype (ε2/ε2) makes reproducibility in future studies of high significance. A high mortality rate might bias our racial comparisons. However, average cognition of participants who died before follow-up did not differ between AAs and EAs. Also, the association of the APOE genotypes with mortality does not differ by race.41 A joint model for cognitive decline and time to mortality did not find any noticeable differences in the association of the APOE genotypes with cognitive decline. However, average cognitive level was lower among those who died without a follow-up, especially among those with the APOE ε4 allele.
Our findings suggest that the APOE ε3/ε4 and the ε4/ε4 genotypes are associated with a faster cognitive decline in both AAs and EAs equally. The APOE ε2/ε2 allele is associated with slower decline in the episodic memory. The association of the APOE ε2/ε4 with faster cognitive decline decreases with age, while the association of the APOE ε2/ε3 with slower cognitive decline increases with age. The association of the APOE ε4 genotype with faster cognitive decline increases with age. The APOE genotypes were associated with cognitive decline, but these associations were not different between AAs and EAs. Future studies focusing on identifying potential health and social factors that might impact the association of APOE genotypes with cognitive decline could be of potential interest for understanding health disparities and designing better preventative strategies.
Supplementary Material
Table S1. Mean Age (SD) of Participants, by APOE Genotypes and Race
ACKNOWLEDGMENTS
The authors thank the Chicago Health and Aging Project research team and study participants for their time, effort, and commitment.
Financial Disclosure: Funding was received from the National Institutes on Aging Grants R01AG051635 and RF1AG057532.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Conflicts of Interest: The authors have no conflicts of interest to report.
REFERENCES
- 1.Dixon RA, de Frias CM. The Victoria Longitudinal Study: from characterizing cognitive aging to illustrating changes in memory compensation. Aging Neuropsych Cog. 2004;11:346–376. [Google Scholar]
- 2.Berg S. Aging, behavior, and terminal decline. In: Birren JE, Schaie KW, eds. Handbook of the Psychology of Aging. 4th ed New York, NY: Academic Press; 1996. [Google Scholar]
- 3.Daviglus ML, Bell CC, Berrettini W, et al. NIH state-of-the-science conference statement: preventing Alzheimer’s disease and cognitive decline. NIH Consens State Sci Statements. 2010;27:1–30. [PubMed] [Google Scholar]
- 4.Greyshock N, Guyton JR, Sebastian S, et al. In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews® [Internet]APOE p.Leu167del-Related Lipid Disorders. Seattle, WA: University of Washington, Seattle; 2014:1993–2017. Available at: https://www.ncbi.nlm.nih.gov/books/NBK208534/. Accessed September 15, 2017. [Google Scholar]
- 5.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci. 1993;90: 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saunders AM, Strittmatter WJ, Schemchel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. [DOI] [PubMed] [Google Scholar]
- 7.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer’s disease. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 8.Knopman DS, Mosley TH, Catellier DJ, Coker LH. Atherosclerosis risk incommunities study brain MRI study. Fourteen-year longitudinal study of vascular risk factors, APOE genotype, and cognition: the ARIC MRI study. Alzheimers Dement. 2009;5:207–214. [DOI] [PubMed] [Google Scholar]
- 9.Bretsky P, Guralnik JM, Launer L, Albert M, Seeman TE. MacArthur studies of successful aging. The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur studies of successful aging. Neurology. 2003;60:1077–1081. [DOI] [PubMed] [Google Scholar]
- 10.Packard C, Westendorp R, Stott D, et al. Association between apolipoprotein E4 and cognitive decline in elderly adults. J Am Geriatr Soc. 2007;55: 1777–1785. [DOI] [PubMed] [Google Scholar]
- 11.Barnes LL, Arvanitakis Z, Yu L, Kelly J, De Jager PL, Bennett DA. APOE and change in episodic memory in blacks and whites. Neuroepidemiology. 2013;40:211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late-onset Alzheimer’s disease. Nat Genet. 1994;7: 180–184. [DOI] [PubMed] [Google Scholar]
- 13.Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon 2 genotype and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002;73:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staehelin HB, Perrig-Chiello P, Mitrache C, et al. Apolipoprotein E genotypes and cognitive functions in healthy elderly persons. Acta Neurol Scand. 1999;100:53–60. [DOI] [PubMed] [Google Scholar]
- 15.Sorbi S, Nacmias B, Forleo P, et al. APOE allele frequencies in Italian sporadic and familial Alzheimer’s disease. Neurosci Lett. 1994;177:100–102. [DOI] [PubMed] [Google Scholar]
- 16.Van Duijn CM, de Knijff P, Wehnert A, et al. The apolipoprotein E ε2 allele is associated with an increased risk of early-onset Alzheimer’s disease and a reduced survival. Ann Neurol. 1995;37:605–610. [DOI] [PubMed] [Google Scholar]
- 17.Sando SB, Melquist S, Cannon A, et al. APOE epsilon 4 lowers age at onset and is a high-risk factor for Alzheimer’s disease: a case-control study from Central Norway. BMC Neurol. 2008;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frisoni GB, Manfredi M, Geroldi C, et al. The prevalence of apoE-epsilon4 in Alzheimer’s disease is age dependent. J Neurol Neurosurg Psychiatry. 1998;65:103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang M-X, Stern Y, Marder K, et al. The APOE-ε4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–755. [DOI] [PubMed] [Google Scholar]
- 20.Hendrie HC, Hall KS, Hui S, et al. Apolipoprotein E genotypes and Alzheimer’s disease in a community study of elderly African Americans. Ann Neurol. 1995;37:118–120. [DOI] [PubMed] [Google Scholar]
- 21.Evans DA, Bennett DA, Wilson RS, et al. Incidence of Alzheimer disease in abiracial urban community. Arch Neurol. 2003;60:185–189. [DOI] [PubMed] [Google Scholar]
- 22.Osuntokun BO, Sahota A, Ogunniyi AO, et al. Lack of an association between apolipoprotein E ε4 and Alzheimer’s disease in elderly Nigerian. Ann Neurology. 1995;38:463–465. [DOI] [PubMed] [Google Scholar]
- 23.Weuve J, Barnes LL, de Leon CFM, et al. Cognitive aging in black and white Americans: cognition, cognitive decline, and incidence of Alzheimer disease dementia [published online August 31, 2017]. Epidemiology. 2018;29: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maestery G, Ottman R, Stern Y, et al. Apolipoprotein E and Alzheimer’s disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37:254–259. [DOI] [PubMed] [Google Scholar]
- 25.Mayeux R, Stern K, Ottman R, et al. The apolipoprotein ε4 allele in patients with Alzheimer’s disease. Ann Neurol. 1993;34:752–754. [DOI] [PubMed] [Google Scholar]
- 26.Sawyer K, Sachs-Ericsson N, Preacher KJ, Blazer DG. Racial differences in the influence of the APOE epsilon 4 allele on cognitive decline in a sample of community-dwelling older adults. Gerontology. 2009;55:32–40. [DOI] [PubMed] [Google Scholar]
- 27.Fillenbaum GG, Landerman LR, Blazer DG, Saunders AM, Harris TB, Launer LJ. The relationship of APOE genotype to cognitive functioning in older African-American and Caucasian community residents. J Am Geriatr Soc. 2001;49:1148–1155. [DOI] [PubMed] [Google Scholar]
- 28.Blair CK, Folsom AR, Knopman DS, Bray MS, Mosley TH, Boerwinkle E. Atherosclerosis Risk in Communities (ARIC) Study Investigators. APOE genotype and cognitive decline in a middle-aged cohort. Neurology. 2005; 64:268–276. [DOI] [PubMed] [Google Scholar]
- 29.Rajan KB, Wilson RS, Weuve J, Barnes LL, Evans DA. Cognitive impairment18 years prior to clinical diagnosis of Alzheimer’s disease dementia. Neurology. 2015;85:898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert M, Smith LA, Scherr PA, et al. Use of brief cognitive tests to identify individuals in the community with clinically diagnosed Alzheimer’s disease. Int J Neurosci. 1991;57:167–178. [DOI] [PubMed] [Google Scholar]
- 31.Wilson RS, Bennette DA, Bienias JL, et al. Cognitive activity and incident AD in a population-based sample of older adults. Neurology. 2002;59:1910–1914. [DOI] [PubMed] [Google Scholar]
- 32.Smith A Symbol Digit Modalities Test Manual-Revised. Los Angeles, CA: Western Psychological; 1982. [Google Scholar]
- 33.Folstein MF, Folstein SE, McHugh TR. “Mini-Mental State”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 34.Wilson RS, Bennett DA, Beckett LA, et al. Cognitive activity in older persons from a geographically defined population. J Gerontol B Psychol Sci Soc Sci. 1999;54:155–160. [DOI] [PubMed] [Google Scholar]
- 35.Hixson J, Vernier DO. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with Hha l. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 36.Wenham PR, Price WH, Blundell G. Apolipoprotein E genotyping by one-stage PCR. Lancet. 1991;337:1158–1159. [DOI] [PubMed] [Google Scholar]
- 37.Diggle PJ, Heagerty P, Liang KY, Zeger SL. Analysis of Longitudinal Data.2nd ed New York, NY: Oxford University Press; 2004. [Google Scholar]
- 38.Preston J Rescaled bootstrap for stratified multistage sampling. Survey Methodology. 2009;35:227–234. [Google Scholar]
- 39.Canty AJ, Davison AC. Resampling-based variance estimation for laborforce surveys. Statistician. 1999;48:379–391. [Google Scholar]
- 40.R: A Language and Environment for Statistical Computing: Version 2.15.2. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 41.Rajan KB, Barnes LL, Wilson RS, et al. Racial differences in the association between apolipoprotein E risk alleles and overall and total CVD cardiovascular mortality over 18 years. J Am Geriatr Soc. 2017;65:2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mean Age (SD) of Participants, by APOE Genotypes and Race