Abstract
The distinct clinical features of myelofibrosis (MF) have been attributed in part to dysregulated inflammatory cytokine production. Circulating cytokine levels are elevated in MF patients; a subset of which have been shown to be poor prognostic indicators. In this study, cytokine overproduction was examined in MF patient plasma and in MF blood cells ex vivo using mass cytometry. Plasma cytokines measured following treatment with ruxolitinib remained markedly abnormal, indicating that aberrant cytokine production persists despite therapeutic JAK2 inhibition. In MF patient samples, 14/15 cytokines measured by mass cytometry were found to be constitutively overproduced, with the principal cellular source for most cytokines being monocytes, implicating a non-cell-autonomous role for monocyte-derived cytokines impacting disease-propagating stem/progenitor cells in MF. The majority of cytokines elevated in MF exhibited ex vivo hypersensitivity to thrombopoietin (TPO), toll-like receptor (TLR) ligands, and/or tumor necrosis factor (TNF). A subset of this group (including TNF, IL-6, IL-8, IL-10) was minimally sensitive to ruxolitinib. All TPO/TLR/TNF-sensitive cytokines, however, were sensitive to pharmacologic inhibition of NFκB and/or MAP kinase signaling. These results indicate that NFκB and MAP kinase signaling maintain cytokine overproduction in MF, and that inhibition of these pathways may provide optimal control of inflammatory pathophysiology in MF.
Keywords: Myeloid Neoplasia, Hematopoiesis And Stem Cells
Introduction
Myeloproliferative neoplasms (MPNs) are systemic diseases of hematopoiesis. Hematopoietic clones harboring mutations in JAK2, CALR, MPL, or SH2B3 (LNK) exhibit constitutive activation of the JAK2 kinase,1–3 which leads to the phenotypically distinct MPNs: polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (MF), which can present de novo as primary myelofibrosis (PMF), or from phenotypic transformation of PV or ET (MF post-PV or post-ET). MF, in contrast to PV and ET, carries a poor prognosis, with a median survival ranging from months to years, depending on its clinical and molecular features.4,5 Its distinct clinical features include fibrosis of the bone marrow space, extramedullary hematopoiesis and consequent splenomegaly, mobilization of immature myeloid cells to the peripheral blood, and leukocytosis, frequently featuring hyperproliferation of multiple myeloid lineages, which can progress to the development of anemia and cytopenias, culminating in bone marrow failure.6 These clinical features have been hypothesized to be due in part to the overproduction of inflammatory cytokines, which is prevalent in MF. In particular, TGF-β overproduction occurs in MF and is hypothesized to be crucial for the fibrotic phenotype, as TGF-β is essential for bone marrow fibrosis induced by TPO overexpression in mice.3,7 Other cytokines, specifically circulating CXCL8/IL-8, IL-2R, IL-12, and IL-15, have been identified as negative prognostic indicators, when present at elevated levels in blood plasma.8 Furthermore, TNF (tumor necrosis factor, TNF-α) has been hypothesized as a driver of clonal dominance in MPNs, based on evidence from a JAK2 V617F mutant retroviral mouse model.9 TNF is most frequently elevated in blood plasma of MF patients versus other MPNs,9 and activates the NFκB signaling pathway, which was found to be strongly hyperactivated in hematopoietic cells from patients with JAK2 V617F mutant MF.10 Therefore, there is substantial evidence implicating overproduction of cytokines as a prominent pathophysiologic mechanism in MF. Specific roles for individual cytokines, among the multiplicity overproduced in MF patients, however, remain incompletely understood.
MF patients can only be cured by allogeneic transplantation, which is not feasible in many patients.6 Best available therapy for MF currently includes treatment with the JAK inhibitor ruxolitinib. Ruxolitinib improves constitutional symptoms and splenomegaly in MF patients. It does not, however, eradicate the malignant clone, and shows only modest (at best) benefit for survival, reduction of clonal burden, or transformation to acute myeloid leukemia (AML), which is a common outcome in MF.6,11–14 Ruxolitinib has been shown to reduce elevated levels of circulating inflammatory cytokines in MF;15 however, it has not yet been clear if circulating cytokine levels are fully rectified with long-term ruxolitinib treatment.
Prior to this study, our group investigated intracellular signaling abnormalities in MF and post-MPN secondary AML (sAML), utilizing mass cytometry (CyTOF) on samples of patient bone marrow or peripheral blood.10 Mass cytometry enables the visualization of >30 antibody-labeled cellular parameters at single cell resolution.16,17 Thereby deviations from the normal state in multiple cellular parameters can be visualized simultaneously in immunophenotypically identified cell populations.10,16–19 The study identified prevalent signaling abnormalities in MF, namely elevation of markers of activated JAK-STAT, MAP kinase/PI3 kinase, and NFκB signaling pathways. NFκB signaling hyperactivation was validated in MF patient CD34+ hematopoietic stem and progenitor cells (HSPC) from analysis of a previously published expression dataset;20 however, it was also identified in other cell populations including B and T cells.10 Due to the multi-population distribution of NFκB signaling hyperactivation in MF, we hypothesized that it is largely driven non-cell-autonomously by cytokines, including TNF, which is produced in MF patients and mouse cells transduced with JAK2 V617F mutant virus,9,10 and which is present at very low levels in healthy human blood plasma.8
In mouse model studies, Tnf was found to be essential for clonal dominance of transplanted JAK2 V617F mutant retrovirally transduced cells.9 Transplanted cells from mice lacking Tnf receptors, however, exhibited clonal advantage over normal competitors,21 suggesting Tnf is an endogenous inhibitor of normal hematopoiesis. Differential pro-malignant-clone effects of Tnf and NFκB signaling were also observed on leukemia-initiating cell (LIC) function and protection from apoptosis in AML mouse models.22,23 A mouse model with NFκB signaling hyperactivation due to elimination of the NFκB-suppressive mir-146a also exhibited fatal myeloproliferative disease, which, however, was dependent on T cell derived Il-6.24 Therefore multiple cytokines and cell populations have the potential to mediate pathophysiologic effects in MF. This makes mass cytometry a particularly advantageous tool for the study of cytokine production in MF, since both multiple cytokines and multiple cell populations can be assayed throughout patient versus healthy control hematopoiesis. The present study elucidates each of the following questions: which cytokines are recurrently overproduced in MF patients, in which cell populations are they produced, and which molecular signaling pathways are essential drivers for cytokine overproduction. Identification of the major signaling pathways driving cytokine production in MF can be a step toward improving the efficacy of best available therapy, currently with ruxolitinib.
Materials and Methods
Patient Samples
Patient and healthy donor control peripheral blood (PB) or bone marrow (BM) samples were obtained with written consent according to a protocol approved by the Washington University Human Studies Committee (WU no. 01–1014). Mononuclear cells (PBMC or BMMC) were obtained by Ficoll gradient extraction and cryopreserved according to standard procedures. Control peripheral blood samples listed with the LRS# alias (Supplementary Table S1) were extracted from leukocyte reductions system (LRS) chambers obtained from platelet donors according to protocol. Other control samples with aliases N# were extracted directly from PB or BM. Clinical and genetic information for patients studied is provided in Supplementary Table S2.
Plasma cytokine analysis
Peripheral blood plasma collected under standard protocols was stored at −80°C, from normal control subjects (N=4), MF patients Pre- and Post-ruxolitinib (N=8), other ruxolitinib-naïve MF patients (N=7), and sAML patients (N=6). Concentrations of 30 cytokines/chemokines were analyzed in duplicate using the Meso Scale Discovery platform with the V-PLEX human cytokine 30-plex kit (Meso Scale Discovery, Rockville, MD, USA). Statistical analysis was performed using GraphPad Prism.
Mass cytometry
Cell staining for mass cytometry followed either of two distinct protocols. For intracellular signaling experiments (Figure 1a, b, Figure 6d, Supplementary Figure S1; N=2 for both MF patients and healthy controls, although a more extensive set was examined in a prior study10), a protocol derived from Bendall et al., 2011,16 was used and previously described,10 with a panel of antibodies previously described.10 The intracellular cytokine protocol has been previously described, and includes live surface marker staining followed by fixation, a saponin-based cell permeabilization and intracellular cytokine staining.25,26 Antibodies used for intracellular cytokine experiments are listed in Supplementary Table S3, while the procedure is described in Supplementary Methods. Samples (N=5 for normal bone marrow, N=9 for normal peripheral blood, N=13 for MF peripheral blood) were mass-channel barcoded following antibody staining,27 and readouts were recorded on a CyTOF2 mass cytometer (Fluidigm, South San Francisco, CA, USA). Data were analyzed in Cytobank (cytobank.org). Gating for specific cell populations (as in Figure 2e–h) is shown in Supplementary Figure S2. viSNE analysis was described previously,18 and conducted using Cytobank interface. Statistical analysis was performed using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). Antibodies for supporting fluorescent flow cytometry experiments are listed in Supplementary Table S4.
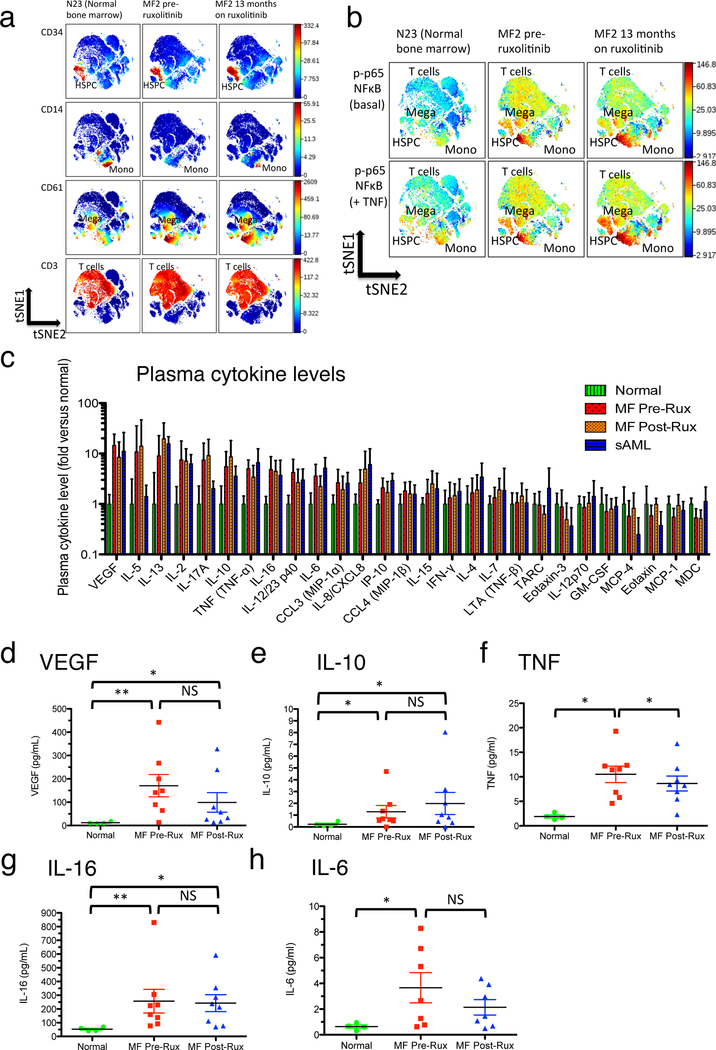
Figure 1. Persistence of NFκB activation and plasma cytokine elevations in myelofibrosis despite ruxolitinib.
a. Mass cytometry surface marker labeling to identify cell populations in viSNE.18 viSNE plots show (top to bottom) labeling for CD34+ (HSPC), CD14+ (monocytes), CD61+ (megakaryoblasts), and CD3+ (T cells) populations. Samples are (left to right) normal control bone marrow N23, MF2 (JAK2 V617F mutant MF post ET) blood immediately prior to commencing ruxolitinib therapy, and MF2 blood after 13 months of ruxolitinib therapy. b. Mass cytometry labeling of p-p65/RELA for samples illustrated in a. Upper panels are basal unstimulated p-p65/RELA. Lower panels are p-p65/RELA following 15 minute ex vivo incubation with 20ng/mL TNF. c. Plasma cytokine levels observed in patient samples, normalized to levels observed in healthy controls. Error bars indicate mean +/− 95% CI. Cytokines are positioned left to right by rank of highest to lowest fold elevation in pre-ruxolitinib MF plasma versus healthy control (measured values shown in Supplementary Table S5). Bars are normalized to mean of levels in healthy controls (N=4; Supplementary Table S1), including zero values, but cytokines undetected in normal plasmas are excluded (Supplementary Table S5). Patient sample sizes are MF Pre- and Post-ruxolitinib, N=8; sAML, N=6. d-h: Individual plasma cytokine measurements (mean of two replicates per individual from MSD array), comparing normal control (Normal), MF patients prior to commencing ruxolitinib therapy (MF Pre-Rux), and the same patients on ruxolitinib (MF Post, Rux; see Supplementary Table S2B for durations of ruxolitinib treatment). Error bars = mean +/− SEM. Significance was determined by Mann-Whitney U-test for disease versus normal comparisons, and by paired T-test for pre- versus post-ruxolitinib comparisons: *, P<0.05; **, P<0.01. d. VEGF. e. IL-10. f. TNF. g. IL-16. h. IL-6.
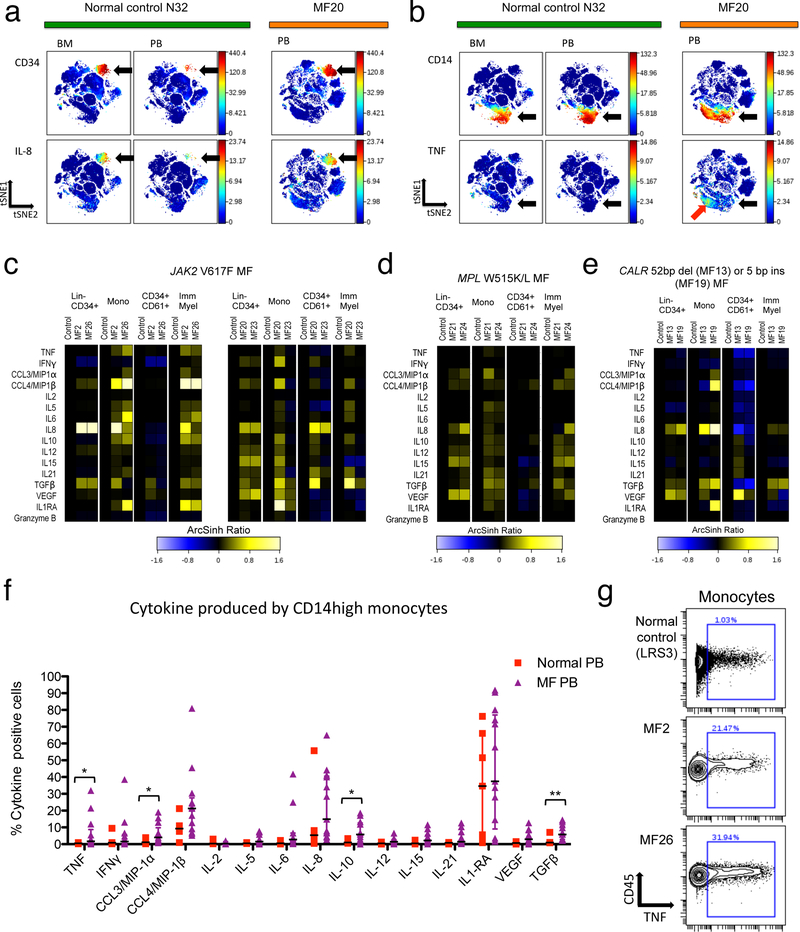
Figure 2. Cytokine overproduction in MF myeloid cell populations.
a-d: viSNE analysis comparing normal control N32 bone marrow, N32 blood, and blood from MF20 (JAK2 V617F mutant PMF). a. CD34 (upper panels) and CXCL8/IL-8 (lower panels) are shown, with arrows indicating HSPC. b. CD14 (upper panels) and TNF (lower panels) are shown, with black arrows indicating monocytes and red arrow indicating a subset of monocytes expressing TNF in MF20 blood. c-e. Heat maps showing median staining for 15 cytokines plus granzyme B, normalized to normal control blood levels (left column of each panel) on ArcSinh ratio scale.16 Heat map panels show (left to right) blood from two MF patients versus one healthy control (from an individual experiment) for the cell populations: Lin-CD34+ HSPC, CD14+ monocytes (Mono), CD34+CD61+ megakaryoblasts, and CD11b+CD11c+CD34- immature myeloid cells (Imm Myel). c. JAK2 V617F mutant MF patients MF2, MF26, MF20, and MF23 versus controls. d. MPL mutant patients MF21 and MF24 versus controls. e. CALR mutant patients MF13 and MF19 versus controls. f. Percent of monocytes manually gated as clearly positive for expression of each of the 15 cytokines. Plots show samples for normal peripheral blood (N=7) and MF blood (N=13: 7 JAK2 V617F mutant, 4 CALR mutant, 2 MPL W515L/K mutant). Error bars indicate median +/− interquartile range. Significance was determined by Mann-Whitney U-test: *, P<0.05; **, P<0.01. g. Biaxial plots illustrating positive versus negative gating (as used in f and Supplementary Figure S7) for TNF from a normal blood control sample and two JAK2 V617F mutant MF patients.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed on a published dataset of gene expression in sorted PMF (N=42) and control CD34+ (N=15) cells.20 Published expression data was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/ series GSE53482). GSEA was performed using software from Broad Institute (http://software.broadinstitute.org/gsea/index.jsp), as described previously.10 Overexpression, in MF versus healthy control CD34+ cells, was ranked by GSEA score: Score = (μPMF μcontrol)/(SDPMF + SDcontrol), where μ=mean expression value among probes in microarray and SD=standard deviation of expression values for each gene.
Data Sharing Statement
For original data, contact Stephen Oh, stoh@wustl.edu. Mass cytometry data will be made public on Cytobank.org upon publication.
Results
NFκB hyperactivation and multi-cytokine overproduction in MF patients persist despite therapeutic ruxolitinib.
NFκB pathway hyperactivation was previously identified as a prevalent signaling abnormality in MF and post-MPN sAML.10 NFκB activity in MF frequently exhibited a pan-hematopoietic pattern, in which nearly all hematopoietic cell populations observed ex vivo by mass cytometry exhibited levels of the activated NFκB subunit p65/RELA (phospho-S529) above those found in their healthy counterparts.10 In MF patients from the prior study,10 elevated levels of phospho-p65/RELA were visualized using the cell clustering algorithm viSNE (visualization of Stochastic Nearest-neighbor Embedding; Figure 1a, b, Supplementary Figure S1).18 A pan-hematopoietic distribution of activated NFκB subunit p65/RELA was observed, suggesting a non-cell-autonomous etiology, potentially mediated by TNF and other cytokines. In patients treated with ruxolitinib, pan-hematopoietic phospho-p65/RELA was observed to persist (Figure 1a, b, Supplementary Figure S1). Hypersensitivity of p65/RELA phosphorylation in response to TNF also persisted in HSPC, monocytes, and megakaryoblasts (Figure 1b).
Plasma cytokine levels were assayed in MF patients prior to and on therapy with ruxolitinib, in parallel with normal controls and post-MPN sAML patients (Figure 1c). Among assayed cytokines, five were significantly elevated in pre-ruxolitinib MF patient versus normal plasma, namely VEGF, IL-10, TNF, IL-16, and IL-6 (Figure 1d–h; Supplementary Table S5 for P values). Of these five cytokines, three (VEGF, IL-10, and IL-16) remained significantly elevated in post-ruxolitinib samples, while the other two (TNF and IL-6) still showed a trend toward elevation rather than approximating reversion to normal (Figure 1d–h). A prior study showed reduced levels of multiple cytokines in plasma of MF patients after 28 days of ruxolitinib in vivo.15 In the present study, patients on prolonged treatment with ruxolitinib (Supplementary Table S2b, S5) exhibited only modest overall reductions in levels of any cytokine assayed from pre-ruxolitinib values. Levels of cytokines elevated in MF plasma, whether before or after in vivo ruxolitinib, were notably similar to those in sAML plasma, despite the high leukemic cell burden in the latter disease (Figure 1c).
MF patient elevated cytokines derive from distinct hematopoietic cell populations
An intracellular cytokine labeling protocol was employed to identify cytokine-producing cellular populations in MF patient blood samples, as quantified by mass cytometry. A panel of antibodies including surface markers for immunophenotyping, antibodies to 15 cytokines, and the activated cytotoxic T and NK cell marker Granzyme B, was utilized to identify cellular loci of cytokine expression (Supplementary Table S3). MF patient samples consisting of peripheral blood mononuclear cells (PBMC) were compared with control samples from healthy bone marrow and peripheral blood (BMMC and PBMC, Figure 2). Both PBMC and BMMC were deemed necessary as controls due to the presence of mobilized CD34+ HSPC and other immature cells in MF PBMC, which are very rare in normal PBMC, while normal BMMC may nonetheless underrepresent mature cell populations present in normal and MF PBMC. Cell populations were identified using surface marker labeling with viSNE18 or by gating schemata for Lin-CD34+ HSPC, monocytes, megakaryoblasts, and immature myeloid cells (Supplementary Figure S2–S4). Megakaryoblasts analyzed were specifically an early CD34+CD61+ population (Supplementary Figure S3), similar to those described as megakaryocyte progenitors in published studies.28,29
In comparison with normal BMMC and PBMC samples, individual cytokines were overproduced in MF PBMC in multiple cell populations. In MF, HSPC are mobilized to the peripheral blood, and these were found to express high levels of IL-8/CXCL8 (Figure 2a; Supplementary Figure S5–S7). TNF, in contrast, was mainly expressed in MF monocytes, and minimally expressed in unstimulated normal control BM or PB samples (Figure 2b). HSPC and myeloid cells from MF patients reliably overproduced several to multiple cytokines compared to in-experiment controls, as was observed for MF patients with JAK2, MPL, or CALR mutations (Figure 2c–e; Supplementary Figure S7).
For the majority of the cytokines studied, the largest portion of signal in unstimulated samples derived from monocytes, in both MF patients and controls (Figure 2f). Across the MF patients studied versus controls, significant elevations in peripheral blood monocytes were observed for the cytokines TNF, CCL3/MIP-1α, IL-10, and intracellular latent TGFβ (Figure 2f). Additional cytokines, notably IL-6, IL-8/CXCL8, and CCL4/MIP1β, were significantly overproduced in immature myeloid cells, B cells, and/or CD8+ effector T cells from MF patients (Supplementary Figure S6). In monocytes and immature myeloid cells, the cytokines IL-6, IL-8/CXCL8, and CCL4/MIP1β, were frequently coexpressed with TNF, but not with TGFβ (Supplementary Figure S8). Contrarily, IL-12, IL-15, and VEGF, were more frequently coexpressed with TGFβ, while HSPC-expressed IL-8/CXCL8 was not preferentially coexpressed with either TNF or TGFβ (Supplementary Figure S8). Among monocyte subclasses, the strongest cytokine producers were the “classical” CD14+CD16- monocytes, although the less abundant CD14+CD16+ “inflammatory” monocytes, and CD16+CD14- “non-classical” monocytes and myeloid cells, all exhibited similar profiles of cytokine expression (Supplementary Figure S9).30
Myeloid cytokines are inducible by TPO, TNF, and TLR ligands
To identify signaling pathways driving cytokine production in MF, samples of MF or control PBMC were stimulated with ligands activating signaling pathways previously shown to be hyperactivated in MF HSPC and myeloid cells.10 The JAK-STAT and NFκB signaling pathways were chosen for study due to their previously identified hyperactivation in MF, and their sensitivity to stimulation by identified, distinct ligands. The cytokine TPO signals through the JAK-STAT signaling pathway, which is hyperactivated in human MF, and indeed roughly 5–10% of human MF patients harbor activating mutations in the TPO receptor gene MPL, while the MPL receptor is itself constitutively activated by binding the neomorphic C-terminus of mutant CALR protein, which is present in the ~30% of human MF patients harboring CALR mutations.1,31 TNF activates the NFκB pathway (but not the JAK-STAT pathway),16 and this activation is hypersensitive in MF HSPC.10
Incubation with TNF induced a limited set of cytokines: IL-8/CXCL8 in Lin-CD34+ cells and monocytes, plus IL-6, CCL4/MIP1β, and IL-1RA in monocytes (Figure 3a, b). The NFκB pathway can also be activated by Toll-like receptors (TLRs), whose expression has been shown to be upregulated in myelodysplastic syndromes.32,33 Gene-set enrichment analysis (GSEA) of a published dataset of gene expression in MF versus normal CD34+ HSPC20 revealed that members of the S100 family of endogenous TLR ligands are among the most overexpressed genes in MF HSPC (Figure 3c, d). The TLR agonists PAM3CSK4 and R848 (resiquimod) activate TLR1/2 and TLR7/8 respectively. Therefore R848, and PAM3CSK4 were chosen, along with TPO, to test inducibility of cytokines in a set of MF PBMC samples including patients harboring JAK2, MPL, and CALR mutations. The JAK inhibitor ruxolitinib was included, along with stimulations, as a potential suppressor of cytokine induction.
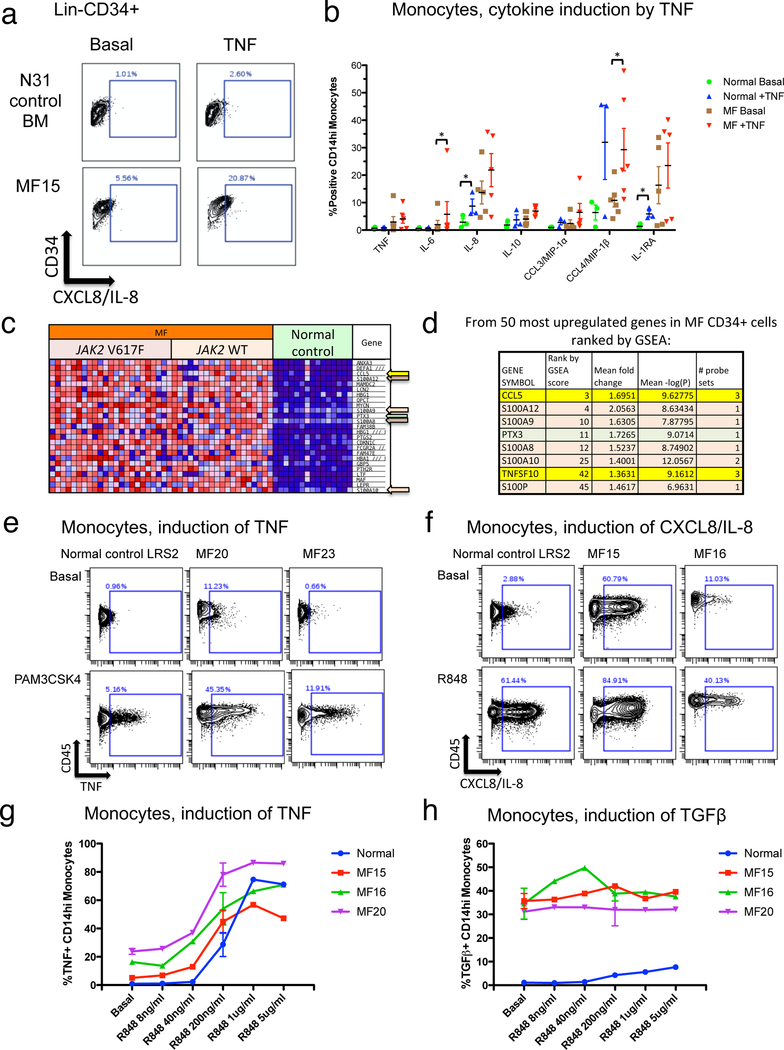
Figure 3. NFκB activating ligands induce a set of MF overproduced cytokines.
a. Biaxial graphs showing production of CXCL8/IL-8 in the Lin-CD34+ population basally (left) or following 4-hour incubation with 20ng/mL TNF (right). Bone marrow from healthy control N31 (above) is compared to blood from MF patient MF15 (JAK2 V617F mutant MF post ET, below). The positive gate shows cells with CXCL8/IL-8 levels above ~99% of the basal control population. b. Cytokine induction by TNF in monocytes. Column graph shows the percent of cells identified as expressing a given cytokine (from biaxial flow plots; Supplementary Figure S7), both basally and following 4-hour incubation with 20ng/mL TNF. Data are shown from both normal blood control samples (N=3) and MF patient samples (N=6; 4 JAK2 V617F mutant, 2 CALR mutant, Supplementary Table S2). Error bars = mean +/− SEM. Statistical significance is shown between basal and TNF-treated samples where identified (*, P<0.05) by Wilcoxon sign-rank test. c. Heat map of gene expression profiling (GEP) analysis, showing the top 25 most upregulated genes in MF versus normal bone marrow CD34+ cells, ranked by gene set enrichment analysis (GSEA) score (“Materials and Methods”). d. TLR and NFκB signaling related genes from among the 50 most upregulated genes in MF CD34+ cells ranked by GSEA.10,20 Genes denoted by arrows in c are listed (those in the ranking range of 1–25, plus two additional genes in the ranking range of 26–50), with corresponding GSEA rank, mean fold change (MF/control), and mean –log(P). These include S100 family endogenous TLR ligands (peach highlight), inflammatory cytokines that are targets of NFκB signaling10 (yellow highlight), and the inflammatory TLR/NFκB target PTX3 (green highlight). e. Biaxial plots showing production of TNF in monocytes basally (top) and after 4-hour incubation with 50ng/mL PAM3CSK4 (bottom). Monocytes from normal control blood are shown along with those from JAK2 V617F mutant MF patients MF20 and MF23. f. Biaxial plots showing production of CXCL8/IL-8 basally (top) and after 4-hour incubation with 5μg/mL R848 (bottom). Monocytes from normal control blood are shown along with those from JAK2 V617F mutant MF patients MF15 and MF16. g. Dose-response showing induction of TNF in response to R848. h. Dose-response showing induction of TGFβ in response to R848. Error bars in g-h = mean +/− SD.
Ex vivo production of several myeloid-produced cytokines, including TNF (Figure 3e, f), exhibited hypersensitivity to stimulation by TLR ligands in MF patients versus normal controls. Dose-response experiments demonstrated that monocytes from JAK2 mutant MF patients were more sensitive to low doses of R848 than normal control monocytes (Figure 3g, Supplementary Figure S10). Cytokines inducible by R848 and PAM3CSK4 included those inducible by TNF (Supplementary Figure S11). Another set of cytokines basally overproduced in MF, notably including TGFβ, VEGF, and IFNγ, were not induced by TNF or TLR ligands (Figure 3h, Supplementary Figure S10).
Incubation with TPO induced cytokines including IL-8, TNF, and CCL4/MIP-1β, in Lin-CD34+ cells (Figure 4a, Supplementary Figure S12). An overview of cytokine production by viSNE, however, revealed that TPO induced cytokine production outside of CD34+ cells as well, with the induced cells being predominantly identifiable as CD14+ monocytes (Figure 4b). Cytokine induction by TPO in both MF and control monocytes could be entirely suppressed by co-incubation with the JAK inhibitor ruxolitinib (Figure 4c).
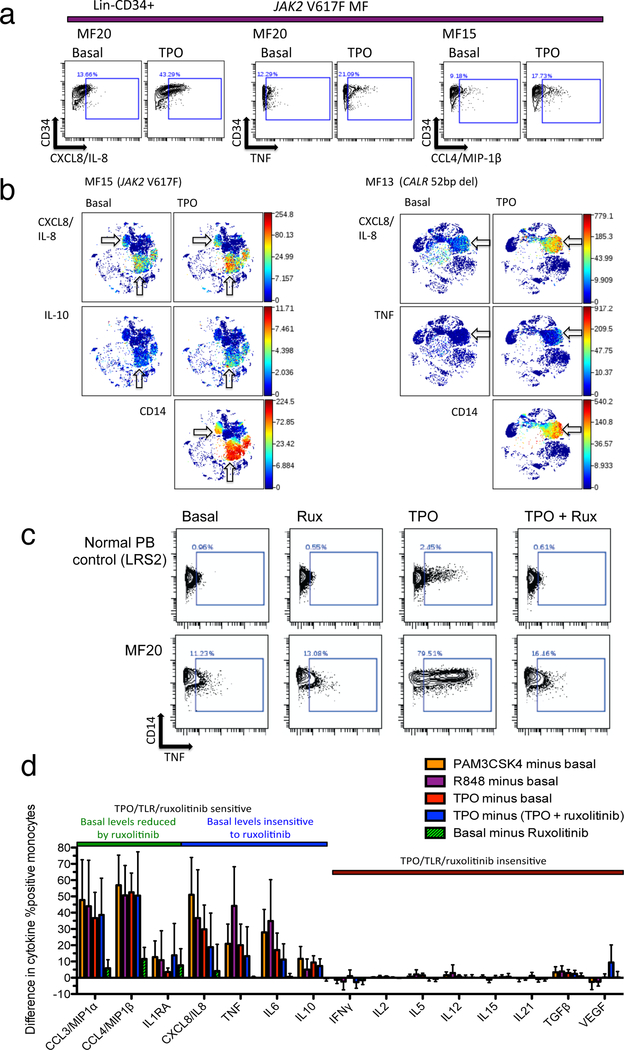
Figure 4. Differential myeloid cytokine induction by TPO and inhibition by ruxolitinib.
a. Biaxial graphs show cytokine production in Lin-CD34+ cells from JAK2 V617F mutant MF patients basally and after 4-hour incubation with 10ng/mL TPO. The positive gate corresponds to cytokine levels above 99% of the healthy control basal population from the same experiment (Supplementary Figure S12). b. viSNE plots show cytokine levels basally and induced by TPO in JAK2 V617F mutant MF patient MF15 (left) and CALR mutant MF patient MF13 (right). Arrows indicate cell populations producing cytokines CXCL8/IL-8, IL-10, and TNF. Bottom panels show CD14 labeling of monocytes. c. Biaxial graphs show TNF in monocytes from healthy control LRS2 (above) and JAK2 V617F mutant MF patient MF20 (below) following 4-hour incubation conditions (left to right): basal, 5μM ruxolitinib, 10ng/mL TPO, TPO plus ruxolitinib. The positive gate corresponds to cytokine levels above 99% of the healthy control basal population. d. Sensitivity of MF overproduced cytokines to TPO, TLR ligands and ruxolitinib, shown as bar graph with differences in MF patient values (% positive, as in c) according to the following comparisons: PAM3CSK4, R848, or TPO minus basal, TPO minus (TPO + ruxolitinib), and basal minus ruxolitinib. The latter two comparisons illustrate inhibition of cytokine either TPO-induced or basal cytokine production by ruxolitinib. Error bars show mean +/− 95% CI. Colored bar above graph denotes separation of cytokines into groups based on reduction of basal cytokine levels by ruxolitinib (green versus blue) and sensitivity to TPO and TLR ligand stimulation and ruxolitinib inhibition (green/blue versus brown).
MF overproduced cytokines were separable into three groups based on their responses to TPO and ruxolitinib in monocytes. The first group, composed of CCL3/MIP-1α, CCL4/MIP-1β, and IL1RA (Figure 4d, Supplementary Figure S13), showed TPO induction and reduction of both basal and TPO-induced levels by ruxolitinib. The second group, including TNF, IL-6, IL-8, and IL-10, showed induction by TPO but no reduction of basal levels by ruxolitinib. The third group, including TGFβ, VEGF, and IFNγ, did not respond to either TPO or ruxolitinib. Cytokines that were induced by TLR ligands were invariably also induced by TPO, while those that were insensitive to TLR ligands were also insensitive to TPO and ruxolitinib.
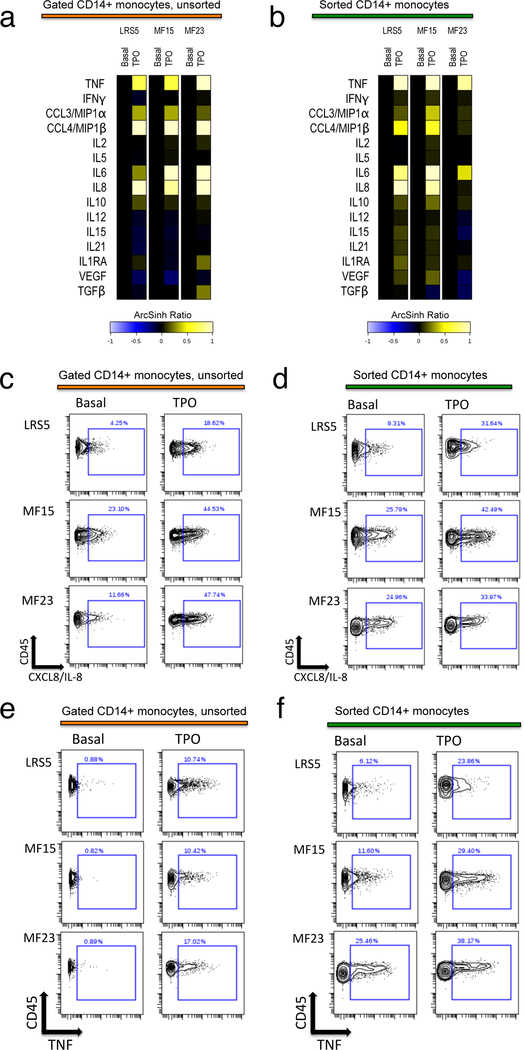
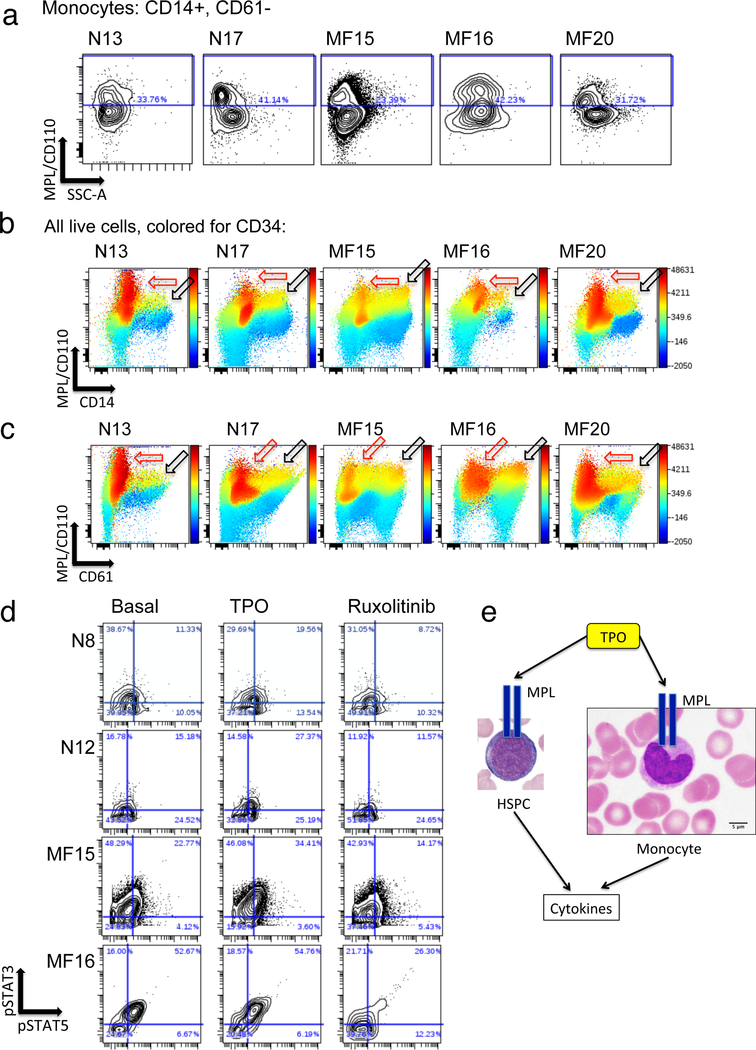
TPO stimulation of FACS-sorted CD14+ monocytes resulted in nearly identical cytokine responses to those observed in monocytes in ex vivo PBMC samples (Figure 5). These findings indicate that the effect of TPO on monocytes was likely direct, mediated via monocyte expressed MPL, which was identified on a subset of monocytes from both MF and normal control samples (Figure 6a–c). MPL expressing monocytes and megakaryoblasts also expressed low to intermediate levels of CD34, suggesting they were a relatively immature subset within the total CD14+ and CD61+ populations (Figure 6b, c). These CD34 low to intermediate expressing cells were 22–34% of monocytic lineage cells in both normal bone marrow and MF blood samples. In the megakaryocytic lineage, they represented 45–50% of normal bone marrow CD61+CD14- cells, and 54–62% of the equivalent cells in MF blood samples. The MPL/CD110 antibody utilized for this experiment, clone 1.6.1 (originally developed by Immunex Corporation), was identified independently as being strongly specific for MPL among multiple human MPL/CD110 antibodies tested, and able to sort xenotransplantable human hematopoietic stem cells (HSC) from within the labeled CD34+CD38- cell population.34 Monocytes from MF patients were observed to harbor high basal levels of phosphorylated STAT3 (pSTAT3) and STAT5 (pSTAT5), consistent with JAK2 hyperactivation, which could be further elevated by TPO, consistent with MPL expression, and suppressed by ruxolitinib (Figure 6d, e).
Figure 5. Direct induction of monocyte expressed cytokines by TPO.
A, B: Heat maps comparing induction of CD14+ monocyte expressed cytokines by TPO, in monocytes within cryopreserved and stimulated PBMC cultures (a) versus isolated flow-sorted CD14+ monocytes from the identical individuals (b). Original cryopreserved samples used for a, b were identical, and treatment with TPO was simultaneous (see Supplementary Methods). Heat maps illustrate the 90th percentile level of TPO-induced versus basal unstimulated cytokine expression, in ArcSinh ratio scale. PBMC from control LRS5 and JAK2 V617F mutant MF patients MF15 and MF23 were utilized. c-f: Biaxial contour plots show cytokine-positive gating in monocytes (Y axis = CD45, X axis = cytokine) for IL-8/CXCL8 (c, d) and TNF (e, f); comparing unsorted monocytes from PBMC cultures (c, e) versus isolated flow-sorted monocytes (d, f), from control LRS5 and JAK2 V617F mutant MF patients MF15 and MF23. Percent of monocytes gated as cytokine positive is denoted by rectangle gate.
Figure 6. Evidence for MPL expression on monocytic and megakaryocytic lineage cells.
a. Expression of CD110/MPL (Y axis) visualized by fluorescent flow cytometry versus side scatter (X axis) in CD14+ monocytes from two normal control bone marrow samples (N13 and N17) and blood from three JAK2 V617F mutant MF patients (MF15, MF16, MF20). CD14+ monocytes were also gated as negative for 7AAD (viability marker), CD3, CD19, CD16, and CD61 (see Supplementary Methods). b, c: Visualization of all live cells from the same same experiment as a, potted by CD110/MPL (Y axis) versus CD14 (b) or CD61 (c), and colored by intensity of labeling for CD34. Note highest levels of CD110/MPL expression in CD34+ high expressing cells (red outlined arrows) and high levels in CD34+/intermediate cells also expressing CD14 (black outlined arrows in b) or CD61 (black outlined arrows in c). d. Biaxial contour plots illustrate phosphorylation of STAT3 (Y axis) and STAT5 (X axis) in gated monocytes as assayed by mass cytometry. Plots are shown for normal bone marrow controls N8 and N12 and JAK2 V617F mutant MF patients MF15 and MF16. Conditions shown (left to right) are basal unstimulated, 15 min with 10ng/mL TPO, and 1h with 5μM ruxolitinib. The quadrant gate shows the percent of cells identified as positive for pSTAT3 and/or pSTAT5. Basal STAT3,5 phosphorylation (compare MF15 and MF16 versus N8 and N12) may be a consequence of constitutive JAK2 kinase activity.10 e. Schematic illustrating the hypothesis that cytokine expression is induced by TPO in monocytes as in HSPC, directly via the TPO receptor MPL.
In addition to ruxolitinib, other JAK inhibitors were used to verify whether resistance to suppression by JAK2 inhibition was a consistent feature of MF overexpressed cytokines. Basal cytokine production was compared to cytokine levels after incubation with 5μM ruxolitinib, momelotinib (JAK1/JAK2/JNK1/TBK1/IKBKE inhibitor),35–38 itacitinib (JAK1 inhibitor),39 pacritinib (JAK2/FLT3 inhibitor), or fedratinib (JAK2/FLT3 inhibitor).40 Itacitinib showed little if any reduction of cytokine production, while momelotinib, pacritinib, and fedratinib showed profiles similar to ruxolitinib, but with possibly greater potency, in reducing basal levels of CCL4/MIP-1β, IL-8/CXCL8 and IL1RA (Supplementary Figure S14).
Inhibition of NFκB and MAP kinase signaling reduces MF myeloid cytokine production.
The observation that only a subset of MF elevated myeloid cytokines exhibited reductions in basal levels by ruxolitinib suggested that the residual basal cytokine overproduction was not directly dependent on ongoing JAK2 kinase activity. Signaling pathways potentially responsible include NFκB, which is activated by TLR ligands and hyperactive in MF,10,32 and MAP kinases, which include a subset activated by TNF and thereby potentially hyperactive in MF, as well as in MDS and AML.9,22,23,41–43 A set of inhibitors was therefore chosen to assess sensitivity of basally elevated cytokine production in MF to blockade of these signaling pathways. These included pevonedistat (MLN4924), a NEDD8-activating enzyme (NAE) inhibitor, which blocks NFκB activation by preventing the NEDDylation and consequent degradation of IκBα.44,45 Also tested were the MEK inhibitor trametinib,46 the p38 MAPK inhibitor VX-745,47 and JNK inhibitor 8 (JNKi8).48,49
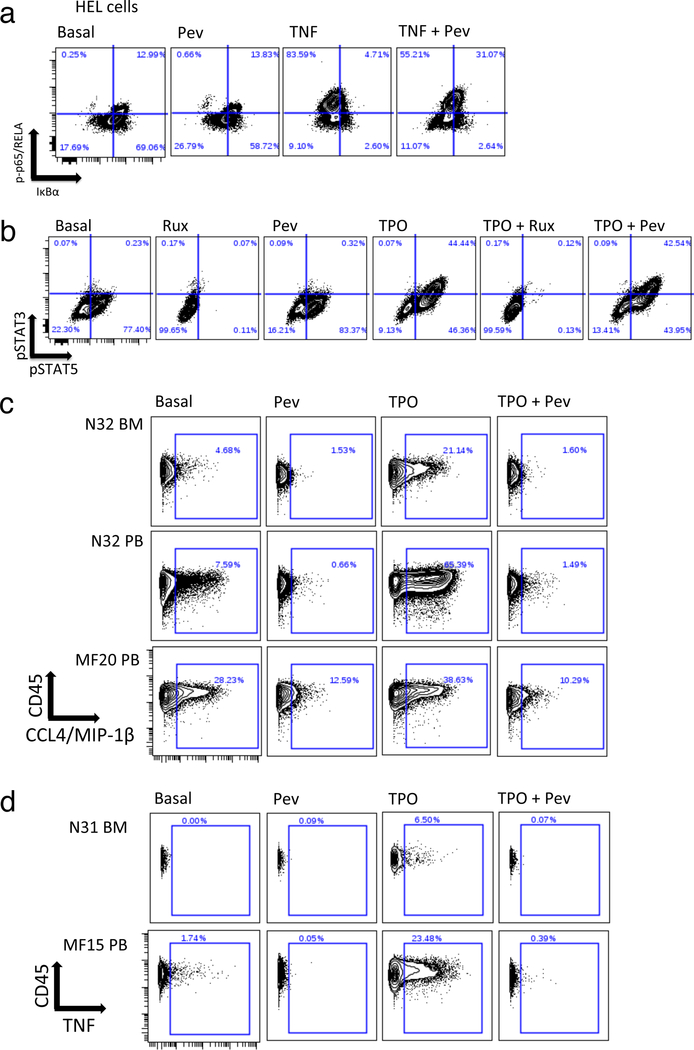
In JAK2 mutant HEL cells, pevonedistat was shown to inhibit degradation of IκBα (Figure 7a), without affecting levels of p-p65/RELA or pSTAT3/5 (Figure 7a, b). In MF monocytes, TPO/TNF/TLR ligand inducible cytokines exhibited inhibition by pevonedistat of in vitro cytokine induction (Figure 7c, d; Supplementary Figure S15). Basally elevated MF cytokines not inducible by TPO, TNF, or TLRs, such as IFNγ and TGFβ, were also unaffected by pevonedistat (Supplementary Figure S15).
Figure 7. Inhibition of TPO induced cytokine production by pevonedistat.
a, b: Responses to pevonedistat versus other stimuli in HEL cells. a. Biaxial plots showing p-p65/RELA (Y axis) versus total IκBα (X axis) in the following conditions (left to right): basal, 1h incubation with 1μM pevonedistat, 15 min incubation with 20ng/mL TNF, 1h incubation with 1μM pevonedistat followed by 15 min incubation with 20ng/mL TNF. Quadrant gate illustrates degradation of IκBα and phosphorylation of p65/RELA on S529. b. Biaxial plots showing phosphorylation of STAT3 (Y axis) and STAT5 (X axis) in the following conditions (left to right): basal, 1h incubation with 5 μM ruxolitinib, 1h incubation with 1μM pevonedistat, 15 min incubation with 10ng/mL TPO, 1h incubation with 5 μM ruxolitinib followed by 15 min incubation with 10ng/mL TPO, 1h incubation with 1μM pevonedistat followed by 15 min incubation with 10ng/mL TPO. c. Biaxial plots showing CCL4/MIP-1β (X axis) versus CD45 (Y axis) in control (N32 BM and PB) versus MF20 (JAK2 V617F mutant PMF) CD14+ blood monocytes. Conditions shown (left to right) correspond to the following 4-hour incubations: basal, 1μM pevonedistat, 10ng/mL TPO, 1μM pevonedistat plus 10ng/mL TPO. d. Biaxial plots showing TNF (X axis) versus CD45 (Y axis) in control (N31 BM) versus MF15 (JAK2 V617F mutant post ET, blood) CD14+ monocytes. The following 4-hour incubations are shown (left to right): basal, 10ng/mL TPO, 1μM pevonedistat, 1μM pevonedistat plus 10ng/mL TPO.
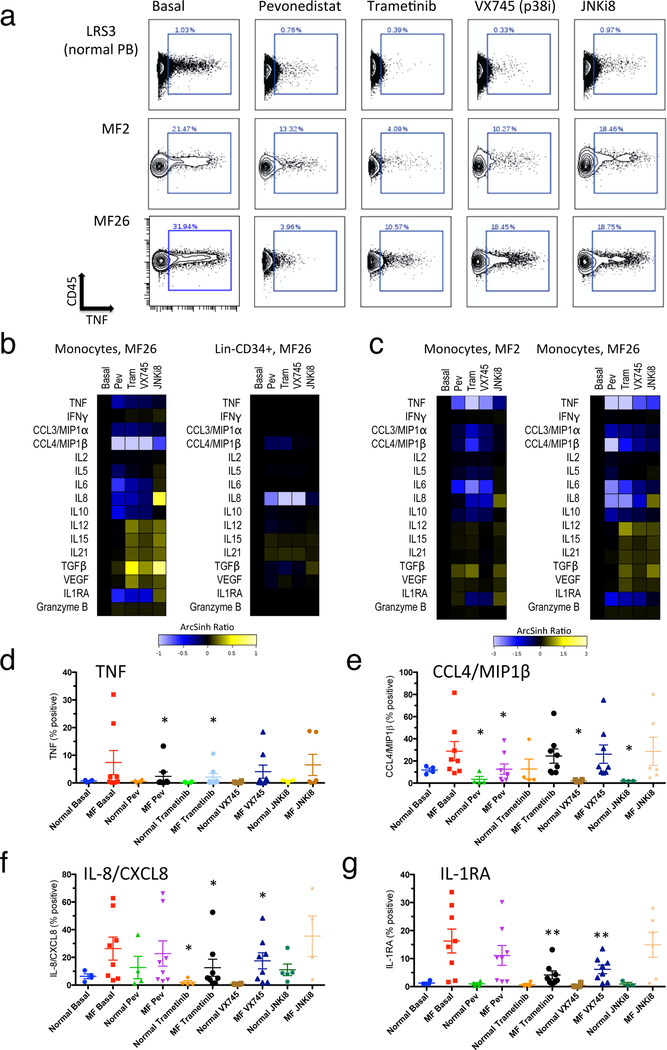
Individual patients, with the most pronounced basal elevations of TPO/TNF/TLR ligand inducible cytokines, exhibited partial reductions of median basal cytokine elevations with all of the inhibitors pevonedistat, trametinib, VX-745, and JNKi8 (Figure 8a; Supplementary Figure S16, S17). This included strong inhibition of CXCL8/IL-8 in Lin-CD34+ cells and of CCL4/MIP1β in monocytes, with pevonedistat, trametinib, and VX-745 (Figure 8b). Particular cytokines were, contrarily, induced by these inhibitors: including CXCL8/IL-8 induced by JNKi8, and TGFβ by trametinib and JNKi8, in monocytes (Figure 8b). The highest cytokine expressing cells, represented by the 90th percentile, showed a broader range of cytokines inhibited by pevonedistat, trametinib, and VX-745, including inhibition of TNF, IL-6, IL-10, CXCL8/IL-8, CCL3/MIP-1α, CCL4/MIP-1β, and IL1RA, in monocytes (Figure 8c–g; Supplementary Figure S17).
Figure 8. Reduction of basally elevated MF cytokine levels by signaling inhibitors.
a. Biaxial plots showing TNF (X axis) versus CD45 (Y axis) in CD14+ monocytes from healthy control (LRS3 PB, top row) and JAK2 V617F mutant MF patients MF2 and MF26 (lower rows). The following 4-hour incubations are shown (left to right): basal, 1μM pevonedistat, 10μM trametinib, 1μM VX-745, 1μM JNKi8. b-c: Heat maps showing staining for 15 cytokines plus granzyme B, in ArcSihn ratio scale with values from inhibitor-treated cells normalized to basal (left column of each heat map). Columns left to right in each heat map represent 4-hour incubations conditions as in a. Abbreviations Pev = pevonedistat, Tram = trametinib. b. Median cytokine levels in CD14+ monocytes (left) and Lin-CD34+ cells (right) from MF26 (JAK2 V617F mutant MF post PV). c. 90th percentile cytokine levels in CD14+ monocytes from JAK2 V617F mutant MF patients MF2 and MF26. d-g: Statistical representation of suppression of basal cytokine production by signaling inhibitors in monocytes. Percent of monocytes identified as expressing each cytokine shown (from biaxial plots, as in a) from healthy control (N=4) or MF patient (N=8; 6 JAK2 V617F mutant and 2 CALR mutant) blood samples. Results are shown for the cytokines TNF (d), CCL4/MIP1β (e), IL-8/CXCL8 (f) and IL-1RA (g). Error bars = mean +/− SEM. Statistical significance is shown between basal and inhibitor treated samples where identified by Wilcoxon sign-rank test (*, P<0.05; **, P<0.01).
Discussion
Previous studies have established that (1) multiple cytokines are abnormally produced in MF patients,15 and can be associated with poor survival;8 that in mouse models of MPNs (either JAK2 or MPL mutant), multiple cytokines are also overproduced, including non-cell-autonomously by nonmalignant cells;50 and that TNF specifically is required for clonal dominance in a JAK2 mutant mouse model.9 The present study establishes that (1) ruxolitinib therapy broadly reduces but does not rectify cytokine overproduction in human MF; (2) ruxolitinib does not reliably eliminate widespread NFκB hyperactivation in MF blood cells; (3) monocytes are the cell population most responsible for cytokine overproduction in MF blood; (4) MF overproduced cytokines divide into subsets sensitive to induction by TPO and TLR ligands and either sensitive or insensitive to ruxolitinib, or, alternatively, insensitive to all three conditions; and (5) basally elevated levels of the TPO/TLR/ruxolitinib sensitive cytokines can be partially reduced by inhibitors of NFκB, MEK, p38-MAPK, and JNK. Collectively, these findings indicate that cytokine overproduction in MF myeloid cells is driven by multiple signaling pathways beyond JAK-STAT. Notably, these implicated signaling pathways can all be activated by TNF, and have been implicated in both malignant cell survival in AML22,23 and myeloproliferation in stress hematopoiesis.51
Our prior studies of intracellular signaling in MF revealed prevalent hyperactivation of NFκB and MAP kinase, as well as JAK-STAT, signaling.10 NFκB hyperactivation has since been identified in the MPL mutant retroviral mouse model;52 therefore this aberrant signaling is a consequence of the primary molecular defect. Cytokine elevations in this mouse model have been attributed to STAT3 and epigenetic reprogramming.50,52 In this study, a set of MF elevated cytokines, including TNF, was observed to be driven partly by each of the NFκB, MAP kinase, and JAK-STAT pathways. The observation that TPO induced production of these cytokines was blocked by pevonedistat as effectively as by ruxolitinib implies that an intact NFκB pathway is necessary for maximal production of these cytokines in MF, which therefore suggests a potential benefit from NFκB and/or MAP kinase inhibition in MF therapy.
Contrarily, a set of MF overproduced cytokines, including IFNγ and TGF-β, was refractory to all stimuli or inhibitors tested. Therefore, ongoing expression of these cytokines does not appear to be reliant on JAK-STAT, MAP kinase, or NFκB signaling. Since TGF-β has been implicated in bone marrow fibrosis, and IFNγ responsive genes have been associated with stress hematopoiesis53 and thrombocytosis,54 it is plausible that signaling other than JAK-STAT, MAP kinase, or NFκB, will need to be targeted for optimal therapeutic benefit in MF. It is possible, however, that inhibitors of JAKs and MAP kinases could also successfully inhibit signaling downstream of IFNγ and TGF-β.
While the JAK inhibitors momelotinib, pacritinib, and fedratinib showed some ability to reduce basal MF overproduced cytokines beyond that observed with ruxolitinib, this may be attributed to their inhibitory actions on other signaling molecules besides JAK2: JNK1, TBK1, and IKBKE (the latter two upstream of NFκB), in the case of momelotinib,35,38 and a combination of IRAK1 for pacritinib, and MAP kinases downstream of active FLT3, for both pacritinib and fedratinib.40,55 Itacitinib, in contrast, showed less reduction of MF cytokine production than ruxolitinib. In clinical studies, momelotinib therapy led to reductions in plasma cytokine levels similar to those observed for ruxolitinib,56,57 while itacitinib and fedratinib did not.39,58 Itacitinib, however, was shown to reduce a few specific molecules, including RANTES and CD40 ligand,39 suggesting there may be an important role for JAK1 in production of particular cytokines, as well as the essential role of JAK1 in sensitivity to type I interferons and IL-3.53
Most importantly, the consequences of cytokine overproduction in MF disease pathogenesis remain relatively unexplored. As observed here, MF cytokine overproduction is reduced but not abrogated by ruxolitinib therapy. This is likely relevant for disease features that remain refractory to ruxolitinib, including malignant clonal burden and marrow fibrosis. Cytokines are expected to have fibrogenic, proliferative, and pro-malignant-survival effects in MF. TNF, however, is known to promote pro-apoptotic as well as anti-apoptotic signaling:22,23,59 this pro-apoptotic signaling is in fact upregulated in MF,10 and harnessing it while inhibiting anti-apoptotic signaling is a plausible therapeutic strategy.59 Among the most upregulated cytokines observed in MF, IL-8/CXCL8 was previously shown to be a pro-survival and proliferative factor for MDS and AML blasts, in which it is also frequently upregulated,60 as well as being a poor prognostic indicator in MF.8 Whether individual cytokines, or signaling pathways activated by multiple cytokines, are more relevant targets for MF therapy, remains to be clarified by future studies.
Supplementary Material
Key Points.
Ruxolitinib therapy does not fully rectify in vivo cytokine overproduction in myelofibrosis.
Cytokine overproduction in myelofibrosis is driven by NFκB and MAP kinase signaling pathways, as well as JAK-STAT.
Acknowledgements
This work was supported by NIH grants K08HL106576 (Oh), R01HL134952 (Oh), and T32HL007088 (Engle, Fisher). This research was also supported by an American Cancer Society Postdoctoral Fellowship (Fisher). This work was also supported by a Doris Duke-Damon Runyon Clinical Investigator Award (Oh) and a Challenge Grant from the MPN Research Foundation (Oh). Technical support was provided by the Alvin J Siteman Cancer Center Tissue Procurement Core Facility, Flow Cytometry Core, and Immunomonitoring Laboratory, which are supported by NCI Cancer Center Support Grant P30CA91842. The Immunomonitoring Laboratory is also supported by the Andrew M and Jane M Bursky Center for Human Immunology and Immunotherapy Programs. We thank K. Luber for assistance with patient samples, C. Holley and the staff of the Alvin J Siteman Cancer Center Flow Cytometry Core for assistance with cell sorting, D. Bender and C. Wilson for assistance with plasma cytokine assays, R. Lin and R. Betz for assistance with mass cytometry experiments, and M. Fulbright and E. De Togni for providing results of pharmacologic assays not presented here, relevant for determining doses of inhibitors for cell treatments used. We also thank M. Fulbright for laboratory management.
Footnotes
Disclosure of Conflicts of Interest
The authors report no conflicts of interest.
References
- 1.Elf S, Abdelfattah NS, Chen E, Perales-Paton J, Rosen EA, Ko A, et al. Mutant Calreticulin Requires Both Its Mutant C-terminus and the Thrombopoietin Receptor for Oncogenic Transformation. Cancer Discov 2016; 6(4): 368–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD Jr., , et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood 2010; 116(6): 988–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 2017; 129(6): 667–79. [DOI] [PubMed] [Google Scholar]
- 4.Zhou A, Oh ST. Prognostication in MF: from CBC to cytogenetics to molecular markers. Best Pract Res Clin Haematol 2014; 27(2): 155–64. [DOI] [PubMed] [Google Scholar]
- 5.Tefferi A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, et al. CALR vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia 2014; 28(7): 1472–7. [DOI] [PubMed] [Google Scholar]
- 6.Iurlo A, Cattaneo D. Treatment of Myelofibrosis: Old and New Strategies. Clin Med Insights Blood Disord 2017; 10: 1179545X17695233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chagraoui H, Komura E, Tulliez M, Giraudier S, Vainchenker W, Wendling F. Prominent role of TGF-beta 1 in thrombopoietin-induced myelofibrosis in mice. Blood 2002; 100(10): 3495–503. [DOI] [PubMed] [Google Scholar]
- 8.Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2011; 29(10): 1356–63. [DOI] [PubMed] [Google Scholar]
- 9.Fleischman AG, Aichberger KJ, Luty SB, Bumm TG, Petersen CL, Doratotaj S, et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood 2011; 118(24): 6392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher DAC, Malkova O, Engle EK, Miner CA, Fulbright MC, Behbehani GK, et al. Mass cytometry analysis reveals hyperactive NF Kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia 2017; 31(9): 1962–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervantes F, Pereira A. Does ruxolitinib prolong the survival of patients with myelofibrosis? Blood 2017; 129(7): 832–7. [DOI] [PubMed] [Google Scholar]
- 12.Verstovsek S, Gotlib J, Mesa RA, Vannucchi AM, Kiladjian JJ, Cervantes F, et al. Long-term survival in patients treated with ruxolitinib for myelofibrosis: COMFORT-I and -II pooled analyses. J Hematol Oncol 2017; 10(1): 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verstovsek S, Mesa RA, Gotlib J, Gupta V, DiPersio JF, Catalano JV, et al. Long-term treatment with ruxolitinib for patients with myelofibrosis: 5-year update from the randomized, double-blind, placebo-controlled, phase 3 COMFORT-I trial. J Hematol Oncol 2017; 10(1): 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deininger M, Radich J, Burn TC, Huber R, Paranagama D, Verstovsek S. The effect of long-term ruxolitinib treatment on JAK2p.V617F allele burden in patients with myelofibrosis. Blood 2015; 126(13): 1551–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. The New England journal of medicine 2010; 363(12): 1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332(6030): 687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. Journal of immunological methods 2010; 361(1–2): 1–20. [DOI] [PubMed] [Google Scholar]
- 18.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology 2013; 31(6): 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV Jr., Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nature biotechnology 2011; 29(10): 886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood 2014; 124(13): e21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pronk CJ, Veiby OP, Bryder D, Jacobsen SE. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. The Journal of experimental medicine 2011; 208(8): 1563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S, et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. The Journal of clinical investigation 2014; 124(2): 528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volk A, Li J, Xin J, You D, Zhang J, Liu X, et al. Co-inhibition of NF-kappaB and JNK is synergistic in TNF-expressing human AML. The Journal of experimental medicine 2014; 211(6): 1093–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao JL, Rao DS, O’Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife 2013; 2: e00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin D, Gupta S, Maecker HT. Intracellular Cytokine Staining on PBMCs Using CyTOF Mass Cytometry. Bio Protoc 2015; 5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Gorman WE, Kong DS, Balboni IM, Rudra P, Bolen CR, Ghosh D, et al. Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J Autoimmun 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zunder ER, Finck R, Behbehani GK, Amir el AD, Krishnaswamy S, Gonzalez VD, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 2015; 10(2): 316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyawaki K, Iwasaki H, Jiromaru T, Kusumoto H, Yurino A, Sugio T, et al. Identification of unipotent megakaryocyte progenitors in human hematopoiesis. Blood 2017; 129(25): 3332–43. [DOI] [PubMed] [Google Scholar]
- 29.Psaila B, Barkas N, Iskander D, Roy A, Anderson S, Ashley N, et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol 2016; 17: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011; 118(5): e16–31. [DOI] [PubMed] [Google Scholar]
- 31.Hobbs GS, Rampal RK. Clinical and molecular genetic characterization of myelofibrosis. Current opinion in hematology 2015; 22(2): 177–83. [DOI] [PubMed] [Google Scholar]
- 32.Monlish DA, Bhatt ST, Schuettpelz LG. The Role of Toll-Like Receptors in Hematopoietic Malignancies. Front Immunol 2016; 7: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei Y, Dimicoli S, Bueso-Ramos C, Chen R, Yang H, Neuberg D, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia 2013; 27(9): 1832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocault Petit L, Fleury M, Clay D, Larghero J, Vanneaux V, Souyri M. Monoclonal antibody 1.6.1 against human MPL receptor allows HSC enrichment of CB and BM CD34(+)CD38(−) populations. Experimental hematology 2016; 44(4): 297–302 e1. [DOI] [PubMed] [Google Scholar]
- 35.Barbie TU, Alexe G, Aref AR, Li S, Zhu Z, Zhang X, et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. The Journal of clinical investigation 2014; 124(12): 5411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S, Imamura Y, Jenkins RW, Canadas I, Kitajima S, Aref A, et al. Autophagy Inhibition Dysregulates TBK1 Signaling and Promotes Pancreatic Inflammation. Cancer Immunol Res 2016; 4(6): 520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov 2014; 4(4): 452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardanani A, Lasho T, Smith G, Burns CJ, Fantino E, Tefferi A. CYT387, a selective JAK1/JAK2 inhibitor: in vitro assessment of kinase selectivity and preclinical studies using cell lines and primary cells from polycythemia vera patients. Leukemia 2009; 23(8): 1441–5. [DOI] [PubMed] [Google Scholar]
- 39.Mascarenhas JO, Talpaz M, Gupta V, Foltz LM, Savona MR, Paquette R, et al. Primary analysis of a phase II open-label trial of INCB039110, a selective JAK1 inhibitor, in patients with myelofibrosis. Haematologica 2017; 102(2): 327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griesshammer M, Sadjadian P. The BCR-ABL1-negative myeloproliferative neoplasms: a review of JAK inhibitors in the therapeutic armamentarium. Expert Opin Pharmacother 2017; 18(18): 1929–38. [DOI] [PubMed] [Google Scholar]
- 41.Grosjean-Raillard J, Ades L, Boehrer S, Tailler M, Fabre C, Braun T, et al. Flt3 receptor inhibition reduces constitutive NFkappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Apoptosis : an international journal on programmed cell death 2008; 13(9): 1148–61. [DOI] [PubMed] [Google Scholar]
- 42.Grosjean-Raillard J, Tailler M, Ades L, Perfettini JL, Fabre C, Braun T, et al. ATM mediates constitutive NF-kappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene 2009; 28(8): 1099–109. [DOI] [PubMed] [Google Scholar]
- 43.Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001; 98(8): 2301–7. [DOI] [PubMed] [Google Scholar]
- 44.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 2010; 115(18): 3796–800. [DOI] [PubMed] [Google Scholar]
- 45.Khalife J, Radomska HS, Santhanam R, Huang X, Neviani P, Saultz J, et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia 2015; 29(10): 1981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess MR, Hwang E, Firestone AJ, Huang T, Xu J, Zuber J, et al. Preclinical efficacy of MEK inhibition in Nras-mutant AML. Blood 2014; 124(26): 3947–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duffy JP, Harrington EM, Salituro FG, Cochran JE, Green J, Gao H, et al. The Discovery of VX-745: A Novel and Selective p38alpha Kinase Inhibitor. ACS Med Chem Lett 2011; 2(10): 758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szczepankiewicz BG, Kosogof C, Nelson LT, Liu G, Liu B, Zhao H, et al. Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J Med Chem 2006; 49(12): 3563–80. [DOI] [PubMed] [Google Scholar]
- 49.Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, et al. Discovery of potent and selective covalent inhibitors of JNK. Chem Biol 2012; 19(1): 140–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov 2015; 5(3): 316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizrahi K, Askenasy N. Physiological functions of TNF family receptor/ligand interactions in hematopoiesis and transplantation. Blood 2014; 124(2): 176–83. [DOI] [PubMed] [Google Scholar]
- 52.Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, et al. Dual Targeting of Oncogenic Activation and Inflammatory Signaling Increases Therapeutic Efficacy in Myeloproliferative Neoplasms. Cancer cell 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleppe M, Spitzer MH, Li S, Hill CE, Dong L, Papalexi E, et al. Jak1 Integrates Cytokine Sensing to Regulate Hematopoietic Stem Cell Function and Stress Hematopoiesis. Cell stem cell 2017; 21(4): 489–501 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, et al. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer cell 2010; 18(5): 524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staudt D, Murray HC, McLachlan T, Alvaro F, Enjeti AK, Verrills NM, et al. Targeting Oncogenic Signaling in Mutant FLT3 Acute Myeloid Leukemia: The Path to Least Resistance. Int J Mol Sci 2018; 19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta V, Mesa RA, Deininger MW, Rivera CE, Sirhan S, Brachmann CB, et al. A phase 1/2, open-label study evaluating twice-daily administration of momelotinib in myelofibrosis. Haematologica 2017; 102(1): 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pardanani A, Laborde RR, Lasho TL, Finke C, Begna K, Al-Kali A, et al. Safety and efficacy of CYT387, a JAK1 and JAK2 inhibitor, in myelofibrosis. Leukemia 2013; 27(6): 1322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pardanani A, Gotlib JR, Jamieson C, Cortes JE, Talpaz M, Stone RM, et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2011; 29(7): 789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heaton WL, Senina AV, Pomicter AD, Salama ME, Clair PM, Yan D, et al. Autocrine Tnf signaling favors malignant cells in myelofibrosis in a Tnfr2-dependent fashion. Leukemia 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schinke C, Giricz O, Li W, Shastri A, Gordon S, Barreyro L, et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood 2015; 125(20): 3144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.