SUMMARY
Mammals rely on a network of circadian clocks to control daily systemic metabolism and physiology. The central pacemaker in the suprachiasmatic nucleus (SCN) is considered hierarchically dominant over peripheral clocks, whose degree of independence, or tissue level autonomy, has never been ascertained in vivo. Using arrhythmic Bmal1-null mice, we generated animals with reconstituted circadian expression of BMAL1 exclusively in the liver (Liver-RE). High-throughput transcriptomics and metabolomics show that the liver has independent circadian functions, specific for metabolic processes such as the NAD+ salvage pathway and glycogen turnover. However, although BMAL1 occupies chromatin at most genomic targets in Liver-RE mice, circadian expression is restricted to ~ 10% of normally rhythmic transcripts. Finally, rhythmic clock gene expression is lost in Liver-RE mice under constant darkness. Hence, full circadian function in the liver depends on signals emanating from other clocks and light contributes to tissue-autonomous clock function.
INTRODUCTION
The mammalian circadian clock system orchestrates daily rhythms in behavior and physiology, allowing animals to anticipate environmental changes and synchronize their internal processes accordingly (Gerhart-Hines and Lazar, 2015) (Partch et al., 2014). The discovery of peripheral oscillators nearly 20 years ago changed the perspective of the field (Yamazaki et al., 2000) (Whitmore et al., 1998); (Plautz et al., 1997) (Balsalobre et al., 1998) (Carr et al., 2003). The concept of a ‘web of pacemakers’ has been largely adopted, in which suprachiasmatic nucleus (SCN) neurons control the tempo of oscillators in peripheral tissues (Schibler and Sassone-Corsi, 2002) through yet ill-defined pathways. By regulating the sleep-wake cycle, the SCN also indirectly evokes the feeding-fasting cycle, a driver of cycling transcripts in the periphery (Bechtold and Loudon, 2013).
These notions prompted the question of whether peripheral clocks may communicate with each other. Recent findings demonstrated system-wide metabolic coordination between tissues over the circadian cycle (Dyar et al., 2018). Also, genetic ablation of the clock in pancreatic beta cells disrupts clock-dependent insulin signaling in the liver (Lee et al., 2013) (Sadacca et al., 2011) (Yamajuku et al., 2012), while lung adenocarcinoma distally rewires circadian homeostasis in the liver (Masri et al., 2016). Thus, identifying the interplay between peripheral clocks will provide critical information on physiology and disease.
The liver clock controls cyclic metabolism and adapts to changes in nutritional regimes (Stokkan et al., 2001) (Eckel-Mahan et al., 2013). Liver-specific clock ablation causes loss of circadian transcription and ultimately loss of oscillation in glucose, lipid and oxidative pathways (Lamia et al., 2008) (Zhang et al., 2010). Nevertheless, some cyclic transcripts persist in the absence of a functioning clock, suggesting alternative mechanisms contributing to fluctuations in the liver (Kornmann et al., 2007). The presence of a circadian clock in virtually all cells begs the question of its dependence on external cyclic signals (Panda et al., 2002).
Tissue-specific clock ablation has been instrumental in identifying the functions of peripheral clocks, however they have not allowed assessment of their degree of autonomy. We have developed a mouse model in which the liver clock is reconstituted in an otherwise BMAL1-deficient animal (Liver-RE). This model is a tool to study whether and to what extent a peripheral clock operates independently from all other clocks. We demonstrate that the liver is intrinsically capable of clock function even in absence of functioning clocks in all other tissues. The liver clock retains a degree of autonomy at the tissue level, restricted to specific genes and metabolic pathways. Remarkably, the genome-wide capacity and specificity of reconstituted BMAL1 to bind chromatin is similar to that of wild type mice, demonstrating that external clock-dependent inputs are required to elicit a full circadian program. Lastly, lack of circadian rhythms in liver-RE mice maintained under constant darkness reveals a critical role of the light-dark cycle on tissue-autonomous function.
RESULTS
Reconstitution of liver clock function
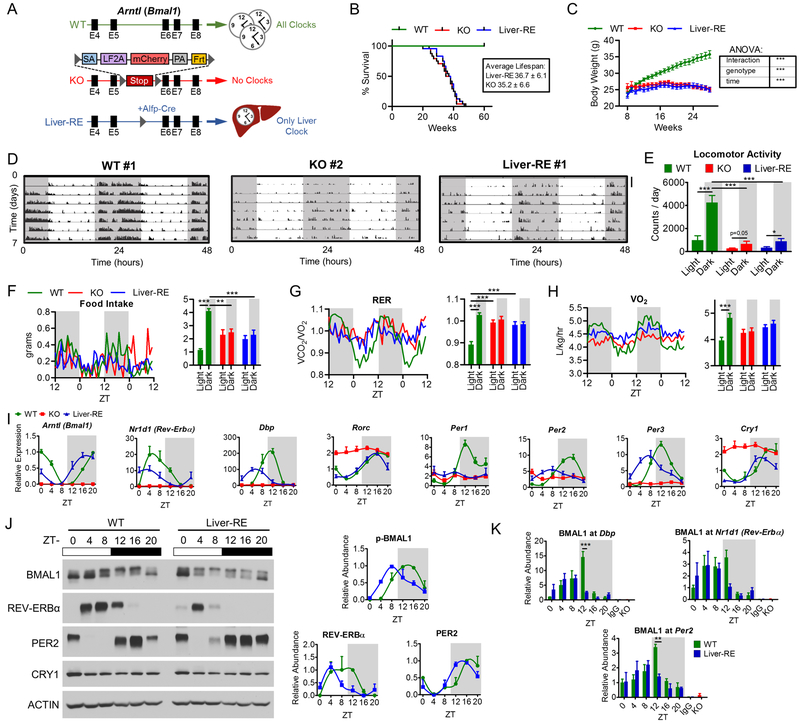
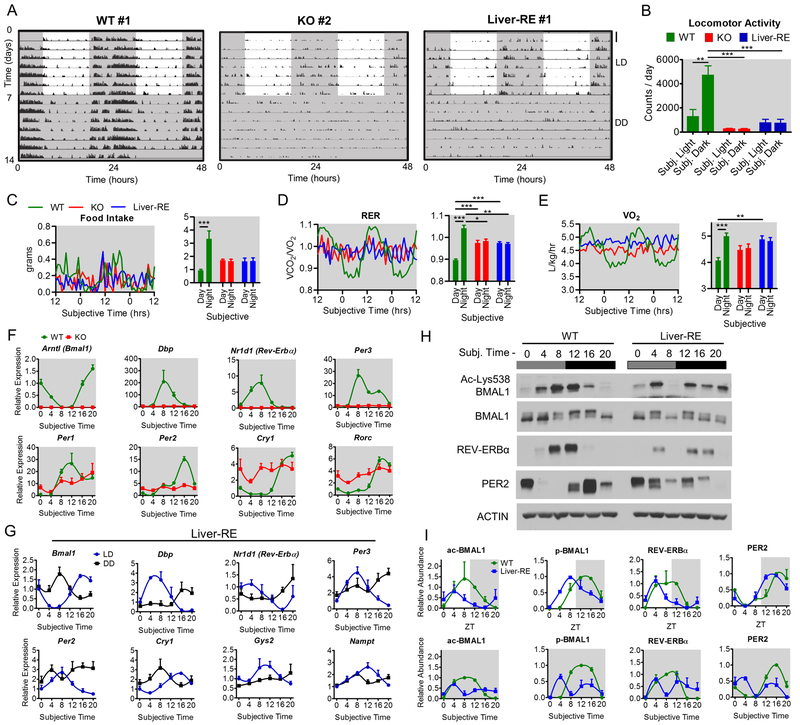
Using a conditional gene trap approach, we generated Bmal1-stop-FL mice (KO) that are fully devoid of BMAL1 and display analogous phenotypes as Bmal1−/− mice, such as reduced lifespan and attenuated weight gain (Fig. 1A–C) (Kondratov et al., 2006). Subsequent crossings with mice expressing cre recombinase under the Alfp promoter generated mice with hepatocyte-specific expression of Bmal1 (Fig. S1A). The unique feature of this model is that the reintroduced Bmal1 gene is under control of the endogenous molecular clockwork (Fig. 1). The number of BMAL1-positive nuclei is comparable in WT and Liver-RE (Fig. S1B). Thus, system-wide functional clocks are not required to direct liver BMAL1 expression. Yet, both KO and Liver-RE mice have reduced lifespan and bodyweight, demonstrating the importance of an intact circadian network (Fig. 1B–C).
Fig. 1. Reconstitution of the liver clock using Bmal1-Stop-FL mice.
A) Scheme of genetic reconstitution of the liver clock. See also Fig. S1A–B. B) Survival curves, WT n=25; KO n=24; Liver-RE n=24. C) Progression of body weight, WT n=27; KO n=39; Liver-RE n=27. Two-way ANOVA, ***=p<0.001. D-E) Locomotor activity in LD cycle. D) Representative trace, duplicated over 2 days for visualization; scale bar = 30 counts. E) Group quantification, n=5. Two-way ANOVA, *=p<0.05; ***=p<0.001. See also Fig. S1C. F-H) Metabolic cage assessment of mice in LD. Traces of group averages (left) and light phase (ZT0–12)/dark phase (ZT12–24) averages (right). RER – respiratory exchange ratio. WT n=8; KO n=5; Liver-RE n=7. Two-way ANOVA, **=p<0.01; ***=p<0.001. See also Fig. S1D–E. I) Gene expression in liver; n=4. See also Fig. S1F. J) Protein levels in liver whole-cell extracts. Right, quantification of n=3. See also Fig. S1G. J) ChIP-qPCR for BMAL1 recruitment to promoters, n=4. IgG and KO negative controls are from ZT20. Two-way ANOVA, **=p<0.01; ***=p<0.001.
Next, we examined the contribution of behavioral rhythmicity under ad libitum feeding and 12:12 hr LD cycle. Whereas WT mice exhibit robust rhythmic locomotor activity, KO and Liver-RE are heterogeneous for a light masking effect, in which light inhibits locomotion (Fig. 1D, S1C). This is in line with previous studies on Bmal1−/− and brain region-specific Bmal1 null mouse models (Bunger et al., 2000) (Izumo et al., 2014). Of 5 Liver-RE mice tested, 3 displayed robust masking (i.e. ~80% of activity during dark phase, similar to WT), 1 did not (no difference between light and dark) and 1 had intermediate phenotype (Fig. 1D, S1C). Overall, dark phase activity was markedly reduced in KO and Liver-RE compared to WT (Fig. 1E). Food is a prominent zeitgeber for the liver (Asher and Sassone-Corsi, 2015). In WT mice, we confirmed a clear pattern of heightened feeding during the dark phase (ZT12–24), diurnal cycles of oxygen consumption, respiratory exchange ratio and energy expenditure (Fig. 1F–H, S1D). In contrast, KO and Liver-RE feed equally between light and dark phases and do not display the accompanying metabolic cycles (Fig. 1F–H, S1D). Notably, body weight (8–12 weeks of age) and total caloric intake did not differ between genotypes (Fig. 1C and S1E). Thus, Liver-RE mice behave comparably to KO mice.
Next, we asked whether the hepatic clock oscillates independently of other clocks and in the absence of a feeding-fasting cycle. The molecular clock components of Liver-RE oscillated similarly to WT, albeit with slightly dampened amplitude and advanced timing (Fig. 1I–K, S1F–G). Exemplified by Arntl (Bmal1), Nr1d1 (Rev-Erbα), Per3, Cry1 and Dbp, the expression peaks and troughs were restored for core clock genes, with the exception of Per1 (Fig. 1I). Importantly, we observed a phase advancement of clock gene expression in Liver-RE mice that was also present at the protein level (see REV-ERBα and PER2; Fig. 1J). BMAL1 phosphorylation, which is indicative of its transcriptional activity (Tamaru et al., 2009), was also present at ZT8 in Liver-RE as compared to ZT12 in WT mice (Fig. 1J). Correspondingly, the peak of BMAL1 recruitment to Dbp, Rev-Erbα, and Per2 promoters was also phase advanced at ZT8 compared to ZT12 (Fig. 1K). Thus, under physiological conditions, oscillations within the liver are independent from other body clocks, revealing tissue-level autonomy.
Metabolomics reveals autonomous functional output of liver clock
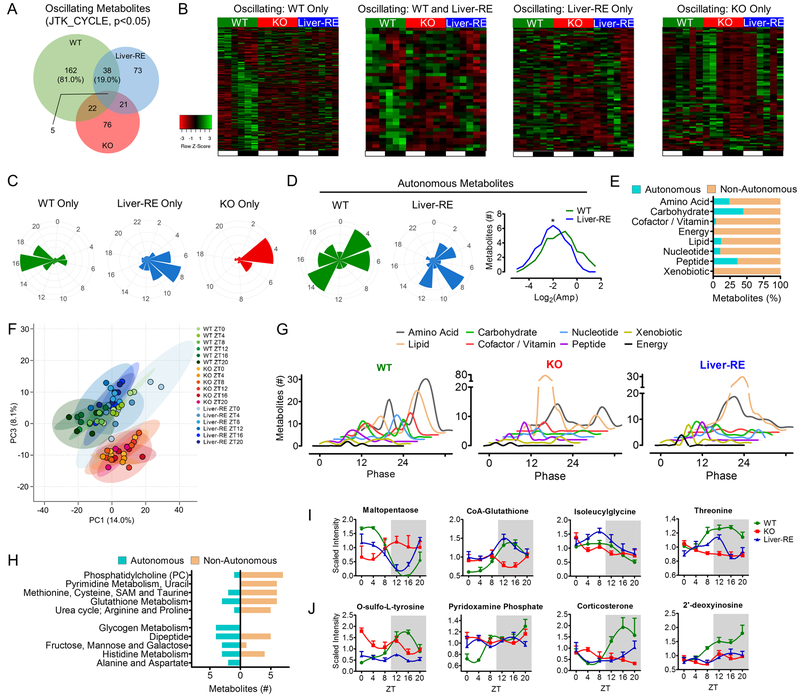
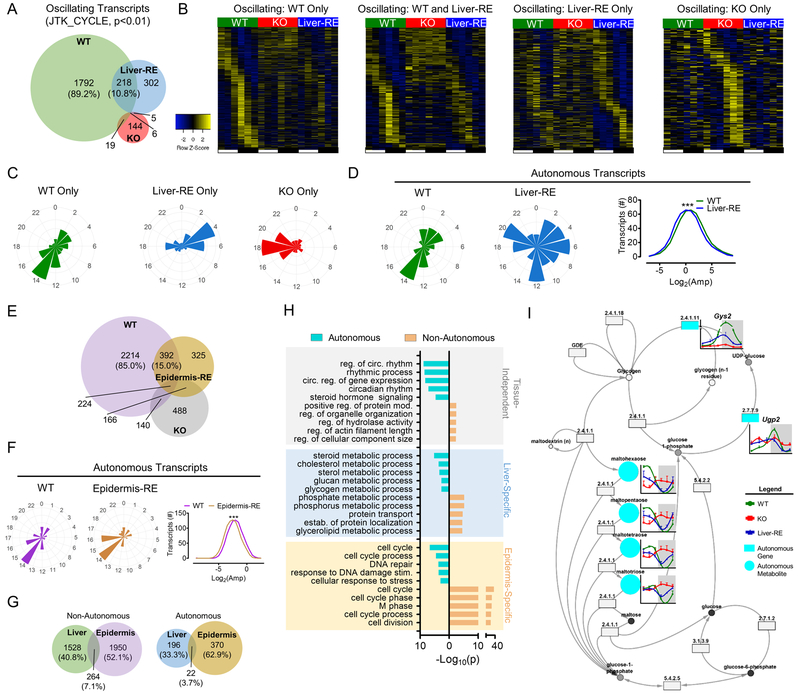
We generated liver metabolite profiles of WT, KO and Liver-RE mice over the diurnal cycle by ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS). In total, 757 annotated metabolites were identified, quantified and analyzed for Bmal1-dependent features (Table S1). In WT liver, 200 Bmal1-dependent metabolites (26.4%) of diverse chemical classes oscillated, in line with previous reports (Fig. 2A–C, S2A) (Hughes et al., 2010) (Krishnaiah et al., 2017) (Eckel-Mahan et al., 2013) (Agostinelli et al., 2016). Strikingly, 38 (19%) of WT oscillatory metabolites retained oscillation in Liver-RE mice (Fig. 2A–D). We defined this group of metabolites as autonomous, without influence of a feeding-fasting cycle or other clocks. Most of these metabolites are peptides, amino acids and carbohydrates and are phase advanced (60.5%) and reduced in amplitude (79.0%) as compared to WT (Fig. 2A–E, S2B). This parallels the advanced timing and dampened amplitude of core clock transcription (Fig. 1). Another 73 metabolites oscillated exclusively in Liver-RE, suggesting that under normal conditions external inputs might dampen oscillation of a subset of liver metabolites (Fig. 2A–C, S2C). Notably, only 5 metabolites oscillated independently of BMAL1 across all genotypes (Fig. 2A). Thus, reconstitution of the hepatic clock drives a specific portion of metabolism independently.
Fig. 2. Diurnal metabolome reveals autonomous metabolic output.
A) Overlap of oscillating metabolites from liver obtained at 6 time-points over 12 hr LD cycle. Percentages are of the total number of Bmal1-dependent metabolites oscillating in WT, n=4. See also Fig. S2A. B) Phase sorted heatmaps of distinct groups of oscillating metabolites. C) Polar histogram plots of peak phase for oscillating metabolites. D) Polar histogram plots of peak phase for autonomously oscillating metabolites (left) and amplitude histogram (right). See also S2B. E) Chemical classification of oscillating metabolites, presented as % of total oscillating for that class. See also S2C–D. F) Principal component analysis of metabolites at each time-point. G) Histograms showing the peak phase of oscillating metabolites in each class. H) Metabolic process or pathway for oscillating metabolites (top 5), see also Table S1. I-J) Examples of autonomously (I) and non-autonomously (J) oscillating metabolites. See also S2E.
Principal component analysis (PCA) across all ZTs clustered metabolite profiles of Liver-RE and WT away from KO mice (Fig. 2F), confirming partial restoration of diurnal hepatic metabolism in Liver-RE. WT livers display distinct light and dark phase peaks for oscillating metabolites (ZT4–8 and ZT16, respectively), especially for amino acids, lipids, nucleotides and carbohydrates (Fig. 2G) (Eckel-Mahan et al., 2012). BMAL1 ablation led to loss of the majority of WT peaks and introduction of newly oscillating species of lipids and amino acids with a prominent peak at ZT4 (Fig. 2G) (Adamovich et al., 2014). Liver-RE partially reintroduced phase peaks during the day, particularly in carbohydrates and xenobiotics, but not the nighttime peaks of nucleotides, cofactors and vitamins (Fig. 2G). In addition, Liver-RE display only one peak for lipids and amino acids which spans ZT4–8 to ZT20, as well as 73 uniquely oscillating metabolites clustering into one peak between ZT4 and ZT12 (Fig. 2C, 2G, S2C).
Next, we sought to define the metabolic processes driven by the autonomous hepatic clock. Carbohydrate and amino acid metabolic processes are prominently represented in the 38 autonomous metabolites, including those involved in glycogen, fructose, mannose and galactose metabolism (Fig. 2H, 2I and Fig. 4). Several metabolic sub-pathways contained metabolites that oscillated autonomously and non-autonomously, such as dipeptides, histidine, methionine, cysteine, SAM, taurine and other amino acids (Fig. 2H–J). Although the total amino acid pool oscillated in both WT and Liver-RE, Liver-RE mice had significantly lower total levels of amino acids as observed in KO (Fig. S2D), likely reflecting defective BMAL1-regulated protein metabolism in tissues such as skeletal muscle or intestine (Jeyaraj et al., 2012) (Mukherji et al., 2013). Redox-related metabolites were restored in Liver-RE; specifically, 3’-dephospho-CoA-glutathione, CoA-glutathione and S-methylglutathione regained oscillation concomitant with partially restored balance of oxidized and reduced glutathione (Fig. 2I, S2E). NAD+ was restored at 3 of 6 ZTs, as FAD at ZT12 and ZT16 (Fig. S2E; Fig. 5). The other 81% of WT metabolites not oscillating in Liver-RE were central to liver function, including xenobiotic detoxification, a vast array of lipids for synthesis, oxidation and membranous function, purine and pyrimidine nucleotides, cofactors and vitamin metabolites (Fig. 2E, H and J).
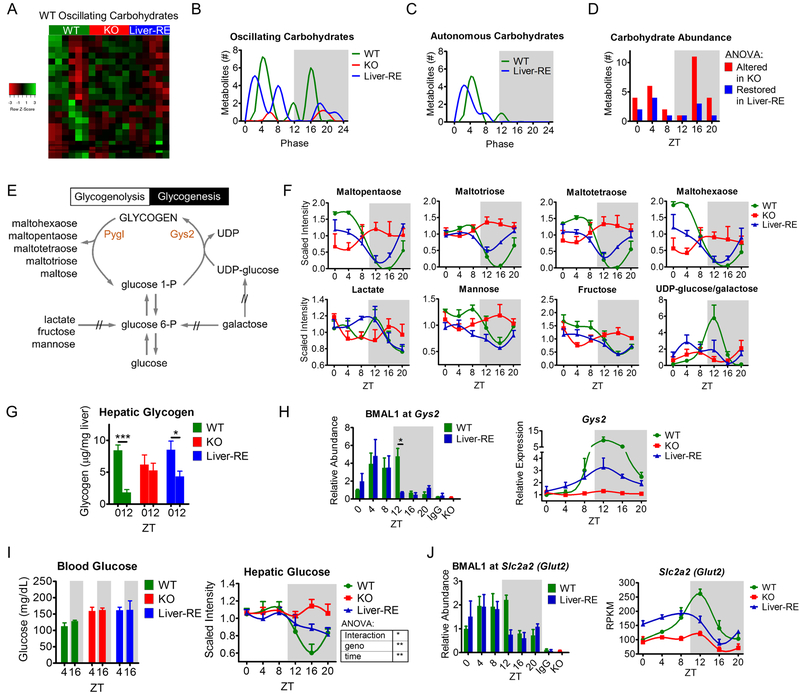
Fig. 4. Liver clock drives diurnal glycogen metabolism.
A) Phase-sorted heatmap showing all oscillating carbohydrates in WT livers. B-C) Peak phases of all oscillating and autonomously oscillating carbohydrates. D) Differential abundance at each ZT (as percent of total metabolites altered in KO). ANOVA with Fisher’s LSD, altered in KO = WT vs KO – FDR<0.05; restored in Liver-RE = WT vs KO – FDR<0.05, WT vs Liver-RE – FDR>0.05, Liver-RE vs KO – FDR<0.05. E) Simplified schematic of hepatic glycogen metabolism. Rate-limiting enzymes are orange, Pygl – glycogen phosphorylase, liver form; Gys2 – glycogen synthase 2 (liver). // - indicates multiple steps. F) Examples of glycogen-related metabolites. G) Hepatic glycogen content at indicated ZTs, as μg/mg tissue, n=4. Student’s t-test was performed to determine a diurnal difference within each genotype, *=p<0.05; ***=p<0.001. H) Left - BMAL1 recruitment to Gys2 promoter region; two-way ANOVA, *=p<0.05. Right – Gys2 expression validated by qPCR. I) Left – blood glucose measurements at indicated ZTs, n=3–6. Right – hepatic glucose levels, Two-Way ANOVA – *=p<0.05; **=p<0.01. J) Left - BMAL1 recruitment to Slc2a2 (Glut2) promoter. Two-way ANOVA, p>0.05. Right – Slc2a2 (Glut2) expression (RNA-sequencing).
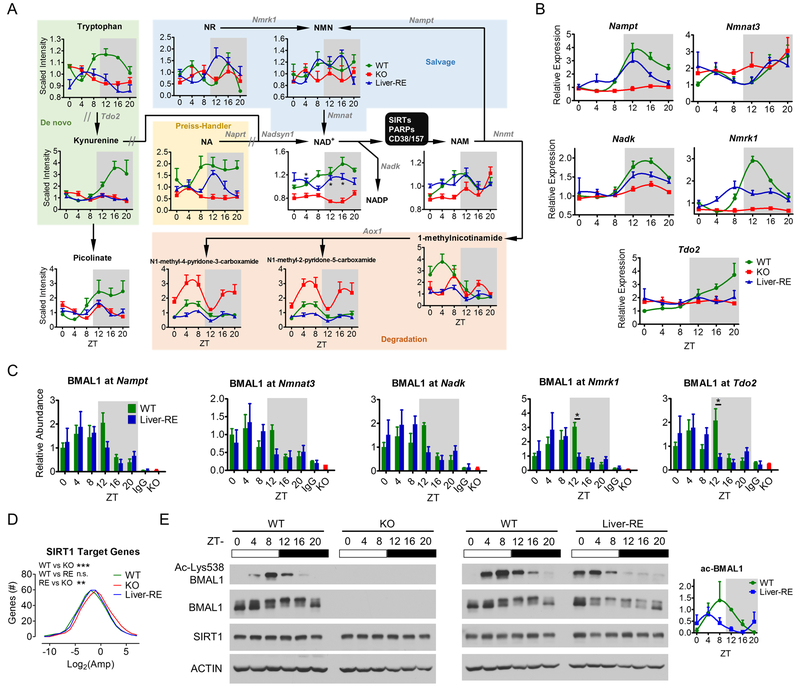
Fig. 5. Clock regulation of hepatic NAD+ metabolism.
A) Schematic of NAD+ metabolism showing the effect of organism-wide clock deficiency and reconstitution of liver clock. One-way ANOVA, *=p<0.01. Metabolite names are black: NR, nicotinamide riboside; NMN, nicotinamide mononucleotide; NA, nicotinate; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide. Enzymes are gray: Nmrk1 – nicotinamide riboside kinase 1; Nampt – nicotinamide phosphoribosyltransferase; Nmnat – nicotinamide mononucleotide adenylyltransferase; Tdo2 – tryptophan 2,3-dioxygenase; Naprt – nicotinate phosphoribosyltransferase; Nadsyn1 – glutamine-dependent NAD+ synthetase 1; Nnmt – NAM N-methyltransferase; Nadk - NAD+ kinase; Aox1 – aldehyde oxidase 1. // - multiple enzymatic steps. B) Key enzymes of NAD+ metabolism validated by qPCR. See also Fig. S4A. C) BMAL1 recruitment to NAD+-related genes, IgG and KO negative controls at ZT20. Two-way ANOVA, *=p<0.05. D) Amplitude of circadian SIRT1 target genes. One-way ANOVA, n.s.=not significant; **=p<0.01; ***=p<0.001. E) Representative western blot from liver whole-cell extracts, n=3. BMAL1 and ACTIN blots are the same as Fig. 1 and S1.
Peripheral clocks drive a subset of tissue-specific transcriptional output
Transcriptomics by RNA sequencing revealed 18,079 coding transcripts, of which 2,010 are Bmal1-dependent transcripts oscillating in WT (Fig. 3A–D, S3A, Table S2 and S3). Of these, 218 (10.8%) also oscillated in Liver-RE mice (Fig. 3A–D). A small group of genes (302) oscillated exclusively in Liver-RE (Fig. 3A–D) and 144 genes oscillated exclusively in KO mice. Only 6 genes oscillated independently of BMAL1 in WT, KO and Liver-RE mice (Fig. 3A–D). Principal component analysis (PCA) of all ZTs revealed Liver-RE profiles cluster more closely with WT than KO, suggesting restoration of gene expression (Fig. S3B). Differential analysis at each ZT found that 29.5–54.3% of gene expression was restored (Fig. S3C), suggesting BMAL1 may also act to maintain overall levels of hepatic gene expression.
Fig. 3. Autonomous transcriptional output of peripheral clocks is tissue-specific.
(A) Overlap of oscillating liver transcripts in a 12 hr LD cycle. Percentages are of the total number of Bmal1-dependent transcripts oscillating in WT; n=3. See also Fig. S3A. B) Phase sorted heatmaps of distinct groups of transcripts. C) Polar histogram plots of peak phase for oscillating transcripts. D) Polar histogram plots of peak phase for autonomously oscillating transcripts (left) and amplitude histogram (right). See also S3D. E) Overlap of oscillating transcripts in epidermis of WT, KO and epidermis reconstituted (Epidermis-RE) mice (Welz et al.). Percentages are of the total number of oscillating transcripts in WT, JTK – p<0.01; n=3–4. F) Polar histogram plots of peak phase and amplitude histogram for autonomously oscillating transcripts in epidermis. See also Fig. S3F. G) Overlap of oscillating transcripts between Liver-RE and Epidermis-RE. Percentages are of the total number of oscillating transcripts. H) GO Biological Process enrichment analysis of oscillating transcripts, grouped by tissue-specificity and autonomy (top 5), see also Table S4. I) Example of cohesively autonomous pathway – glycogen metabolism (KEGG). Metabolites = circles; genes = rectangles, EC number is given for specific enzymatic activity. See also Fig. S3G.
Consistent with previous reports (Koike et al., 2012) (Eckel-Mahan et al., 2013), two peak phases exist for oscillating genes, one during the early daytime between ZT0 and ZT4 and one containing the majority of transcripts during the early nighttime around ZT12 to ZT16 (Fig. 3B–D). In Liver-RE, 92.2% of transcripts were phase advanced, resulting in a dampening of the early nighttime peak at ZT12 to ZT16 and a broadened phase distribution overall (Fig. 3D, S3D). Additionally, 78.4% of the autonomous genes were reduced in amplitude (opposed to 21.6% increased) compared to WT (Fig. S3D). Based on phase advanced core clock profiles, these genes are likely directly regulated by the autonomous branch of the clock. Lastly, transcription factor binding site motif analysis of genes oscillating in WT only, WT and Liver-RE, or Liver-RE mice only, revealed that all groups were enriched for canonical liver TFs such as LXR, RXR and PPARα (Fig. S3E).
Importantly, autonomous oscillations are also present in epidermis-specific Bmal1 reconstituted mice (Epidermis-RE) (Welz et al. 2019; Fig. 1 and 2). 2606 genes oscillate in epidermis in a Bmal1-dependent manner, of which 392 (15.0%) also oscillate in Epidermis-RE (p<0.01) (Fig. 3E). Similar to liver, 73.2% of autonomous genes displayed reduced amplitude (Fig. 3F, S3F). However, in contrast to liver, half (50.0%) were in phase with WT including core clock components (Fig. 3F, S3F). Comparing the overlap of oscillating transcripts in liver and epidermis, only 22 (3.7%) autonomous genes are common (Fig. 3G), 9 (40.9%) being core clock genes. Non-autonomous (oscillating in WT only) genes also showed a small degree of overlap, with 264 (7.1%) genes in both tissues (Fig. 3G). Aside from the circadian rhythm class, GO enrichment analysis revealed steroid hormone signaling as the only autonomous pathway enriched in both liver and epidermis (Fig. 3H). Common, non-autonomous pathways, which include protein modification process, hydrolase activity and actin filament length, may depend on clocks in other tissues. Liver-specific autonomous pathways were those central to metabolism such as cholesterol, triglyceride, glycogen metabolic process and oxidation-reduction (Fig. 3H, Table S4), consistent with oscillating metabolites (Fig. 2H–J). Top non-autonomous pathways included phosphate and phosphorus metabolic process, protein transport and localization (Fig. 3H, Table S4). In contrast, autonomous and non-autonomous pathways overlapped in the epidermis, being enriched for cell cycle and DNA damage/repair (Fig. 3H).
To determine functional coherence of metabolome and transcriptome, we performed integrated pathway enrichment analysis using IMPaLA (Kamburov et al., 2011). In WT, gene expression and metabolites were highly coherent in more than 14 pathways (Fig. S3G). Livers of KO mice exhibit no coherence, whereas Liver-RE mice show coherence for 3 pathways including metabolism, metabolism of carbohydrates and transport of small molecules (Fig. S3G). As an example, see restoration of the glycogen pathway (Fig. 3I). Thus, the hepatic clock restores coherence for select pathways while a full clocks network is needed for complete rhythmic output.
The hepatic clock drives diurnal glycogen metabolism
Glucose homeostasis is clock-controlled in several tissues (Kalsbeek et al., 2014). Still, the extent to which the liver relies on other clocks for proper glucose handling remains unclear. The feeding-fasting cycle generates a diurnal rhythm in dietary glucose influx such that circulating carbohydrates are plentiful during the dark/active phase and lacking during the light/resting phase. Consequently, the liver maintains blood glucose levels within a narrow, constant range. Of 44 measured carbohydrates, 21 (47.7%) oscillated in WT and exhibited two peak phases, one during the light phase at ZT4–8 and another during the dark at ZT16–20 (Fig. 4A–C). BMAL1 ablation abolished rhythmicity in all but 1 of these metabolites and altered the total abundance of 28 carbohydrates across all ZTs (Fig. 4A–C). Remarkably, Liver-RE retained rhythmicity of 9 (42.9%) of the metabolites and restored the total abundance of 12 (57.1%) carbohydrates across all ZTs (Fig. 4C and D). As a result, Liver-RE mice regained the light-phase carbohydrate peak, with advanced timing (Fig. 4C).
Hepatic glycogen is synthesized during feeding and degraded during fasting to maintain blood glucose (Fig. 4E) (Doi et al., 2010). In WT, oligosaccharides of glycogen breakdown accumulate during the light phase, when glycogenolysis is maximal (Fig. 4F). Substrates for glycogen synthesis such as glucose, fructose, mannose and lactate also display this diurnal pattern, utilized for glycogenesis during the dark phase (Fig. 4F). This is reflected in hepatic glycogen content, with peak and trough at ZT0 and ZT12, respectively (Fig. 4G). In contrast to KO, Liver-RE recapitulated the WT pattern of glycogen metabolites and displayed diurnal difference in hepatic glycogen (Fig. 4G). BMAL1 is rhythmically recruited to the Glycogen synthase 2 (Gys2) promoter in both WT and Liver-RE, eliciting cyclic expression of this rate-limiting enzyme (Doi et al., 2010) (Hatanaka et al., 2010)(Fig. 4H). Thus, diurnal glycogen regulation is directly under hepatic clock control.
Despite loss of rhythmic metabolism, Bmal1−/− mice have only slightly elevated blood glucose, presumably because the lack the feeding-fasting cycle, receive a steady influx of dietary glucose (Rudic et al., 2004). Yet, with an intact feeding-fasting cycle, liver-specific Bmal1−/− mice show fasting hypoglycemia, presumably due to inadequate hepatic glucose export through the BMAL1 target Slc2a2 (Glut2) (Lamia et al., 2008). As expected, there is no overt difference in resting blood glucose between WT, KO and Liver-RE (Fig. 4I). However, hepatic glucose is cyclic in WT, abolished in KO, and partially restored in Liver-RE (Fig. 4I), paralleling rhythmic BMAL1 recruitment to Glut2 promoter and cyclic expression (Fig. 4J). As BMAL1 ablation leads to a loss of SCN and pancreatic clock-driven hormonal signals (Lee et al., 2013) (Bechtold and Loudon, 2013), we deduce that the hepatic clock autonomously governs glucose concentration and export. Yet, as amplitudes are dampened in Liver-RE mice, inputs external to the liver likely provide robustness to this process.
Clock regulation of hepatic NAD+ metabolism
Based on the restoration of redox pathways in Liver-RE, we postulated that redox feedback is an autonomous feature of the hepatic clock. The clock and NAD+ metabolism are interlocked (Bellet et al., 2013) (Ramsey et al., 2009) (Nakahata et al., 2009). Our investigation of the metabolome revealed restoration of NAD+ levels in Liver-RE. Several key NAD+ metabolites also displayed telling patterns of abundance across WT, KO and Liver-RE livers (Fig. 5A). Levels of de novo biosynthesis metabolites such as tryptophan, kynurenine, and nicotinate are reduced in KO, and remained so in Liver-RE compared to WT. In contrast, NAD+ levels are significantly reduced in KO yet markedly restored in Liver-RE at ZT4, ZT12 and ZT16 (Fig. 5A). The NAM catabolism products N1-Methyl-2-pyridone-5-carboxamide (2py) and N1-Methyl-4-pyridone-3-carboxamide (4py) are elevated in KO compared to WT, indicative of an abnormal increase in the NAM clearance pathway. Strikingly, 2py and 4py are normalized to WT levels in Liver-RE, signifying proper salvage pathway function and recycling of NAM to NAD+ (Fig. 5A). Thus, the autonomous hepatic clock drives the NAD+ salvage pathway and partially restores NAD+ oscillation. On the other hand, de novo NAD+ biosynthesis requires a full circadian system.
Expression of genes in the NAD+ salvage such as Nampt, Nmrk1 and Nadk is rhythmic in WT and Liver-RE, whereas expression of Tdo2 (tryptophan-kynurenine pathway) and Aox1 (NAM clearance pathway) is constant (Fig. 5B, S4A). These rhythmically expressed genes also remain direct targets of BMAL1 in Liver-RE. Indeed, BMAL1 is still rhythmically recruited to the promoters of these genes, with advanced phase at ZT4-ZT8 compared to ZT8-ZT12 as in WT (Fig. 5C). This suggests that the NAD+ salvage pathway is an autonomous transcriptional target of the hepatic clock.
Restoration of Nampt expression and NAD+ levels indicates a functioning clock-NAD+-SIRT1 feedback loop in Liver-RE. The NAD+-dependent deacetylase SIRT1 is known to regulate a distinct set of rhythmic genes through interplay with BMAL1 at genomic targets. Liver-specific Sirt1 ablation enhances amplitude or promotes de novo oscillation of thousands of genes (Masri et al., 2014). To assess Bmal1-dependence of SIRT1 function we compared SIRT1 target genes in WT, KO and Liver-RE. Amplitude of SIRT1 circadian target genes is significantly increased in KO, suggesting disruption of SIRT1 function (Fig. 5D). The amplitude of these genes are reduced back to WT levels in Liver-RE, indicative of restored SIRT1 function (Fig. 5D). A circadian function of SIRT1 is the rhythmic deacetylation of BMAL1(Nakahata et al., 2009) (Hirayama et al., 2007). WT livers display rhythmic Lys538 BMAL1 acetylation that peaks at ZT8-ZT12 (Fig. 5E) (Nakahata et al., 2008). Remarkably, BMAL1 rhythmic acetylation is restored in Liver-RE. As for BMAL1 phosphorylation and clock protein accumulation (Fig. 1), acetylation is phase advanced by approximately 4 hours compared to WT (Fig. 5E). Thus, the autonomous branch of the hepatic clock is sufficient to support SIRT1 circadian function.
Autonomous recruitment of BMAL1 to chromatin
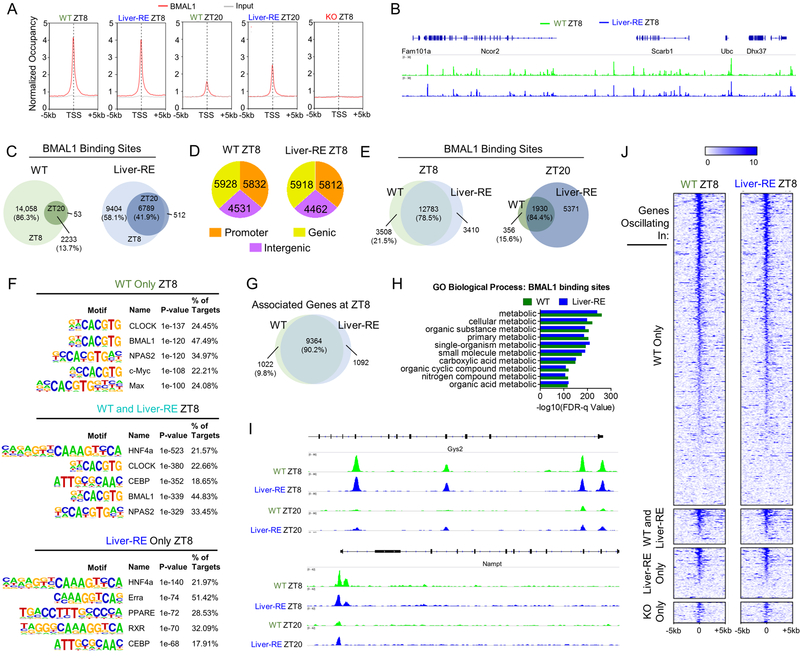
We investigated whether BMAL1 recruitment to chromatin (Trott and Menet, 2018) in WT and Liver-RE would parallel the differences in circadian transcriptional profiles. Using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq), we determined BMAL1 genome-wide occupancy at ZT8 and ZT20 (Fig. 6; see also Fig. S5A–B and Table S5). In contrast to transcriptomics results, BMAL1 binding throughout the genome exhibited highly similar patterns in both genotypes (Fig. 6A–B). We identified 16,291 and 16,193 bona fide BMAL1 binding sites in WT and Liver-RE, respectively, at ZT8 (Fig. 6C). These sites were similarly distributed at promoter, genic and intergenic regions (Fig. 6D, S5C). Indicative of rhythmic binding for both genotypes, significantly fewer sites were identified at ZT20 (WT – 2,286; Liver-RE – 7,301) (Fig. 6C).
Fig. 6. BMAL1 recruitment to chromatin in the liver.
A) Normalized occupancy of BMAL1 genome-wide. TSS – transcription start site. See also Fig. S5A–B. B) Example section of genome. C) Overlap of binding sites between ZT8 and ZT20. Percentages are of the total number of sites at ZT8. D) Genomic distribution of binding sites. See also Fig. S5C. E) Overlap of binding sites in WT and Liver-RE. Percentages of total number of sites in WT at that ZT. F) Motif enrichment analysis at ZT8. See also Fig. S5G and Table S6. G) Overlap of genes targeted in WT and Liver-RE at ZT8. H) GO enrichment analysis of all binding sites in WT or in Liver-RE. “Process” was removed from terms for clarity. See also Table S6. I) Example of binding peaks for WT and Liver-RE at autonomously oscillating genes. See also Fig. S5D–F, J) Heatmap of BMAL1 occupancy at the distinct groups of oscillating genes. 0 = transcription start site. See also Fig. S5H.
Of WT sites, 78.5% (ZT8) and 84.4% (ZT20) were restored in Liver-RE (Fig. 6E, S5D–E). These included 99 (86.8%) of 114 peaks corresponding to core clock genes, (Fig. S5F). Notably, an additional 3,508 and 3,410 sites were exclusive to WT and Liver-RE, respectively (Fig. S5D and E). However, only 28.8% (WT) and 29.5% (Liver-RE) of genes exclusively rhythmic in that particular genotype were associated with these unique sites. Motif analysis confirmed E-box sequences as highly enriched for both genotypes (Fig. 6F, Table S6). The REV-ERB motif was also enriched in shared WT and Liver-RE peaks at both ZT8 and ZT20 (Table S6). To further probe potential cooperation with REV-ERBα, as its binding displays significant overlap with BMAL1, we compared our results with previously published REV-ERBα cistromes (Bugge et al., 2012) (Cho et al., 2012) (Zhang et al. 2015). The proportion of REV-ERBα binding sites associated with autonomously or non-autonomously oscillating genes was similar, suggesting that genomic occupancy of REV-ERBα alone may not explain transcript oscillation in Liver-RE (Fig. S5G).
Upon assigning genes to BMAL1 binding loci, we found that 90.2% of WT BMAL1-target genes were targeted in Liver-RE (Fig. 6G, Table S5). GO analysis revealed nearly identical enrichment for WT and Liver-RE of metabolic pathways (Fig. 6H, Table S6). Of the autonomously regulated genes (Fig. 3), 93.8% of WT binding sites were restored in Liver-RE. This is exemplified by BMAL1 binding at Gys2 and Nampt in WT and Liver-RE (Fig. 6I). A Heat-map of BMAL1 binding for rhythmic genes illustrates disconnect with gene rhythmicity in Liver-RE. Of the 2,010 Bmal1-dependent genes in WT, 1,377 (68.5%) have at least one corresponding BMAL1 peak in WT (Fig. 6J, S5H). However, though BMAL1 still binds 1276 (92.7%) of those 1,377 genes in Liver-RE, their rhythmicity was not restored. Thus, despite substantial BMAL1 chromatin recruitment, the clock in Liver-RE mice fails to drive the bulk of rhythmic expression.
Analysis of Liver-RE mice under constant conditions
In constant darkness (DD), WT mice continue to display rhythmic activity, feeding behavior and cycles of VO2, RER and energy expenditure (Fig. 7A–E, S6A–C, S1). In KO and Liver-RE mice, locomotor activity remained markedly reduced, yet the heterogeneous light masking effect was lost in DD, resulting in equal activity between subjective day and night (Fig. 7A–B). As in LD, rhythmic feeding behavior and accompanying metabolic cycles were absent (Fig. 7C–E, S6A–C).
Fig. 7. Characterization of Liver-RE mice under constant conditions.
(A-B) Locomotor activity in 12:12 hr light-dark (LD) and dark-dark (DD) cycles. A) Representative actogram (double plotted for visualization, LD same as in Fig. 1 for comparison), scale bar = 30 counts. B) Group quantification, n=5. Two-way ANOVA, **=p<0.01; ***=p<0.001. See also Fig. S1B; C-E) Metabolic cage assessment of mice at 3–4 days in DD. Left – traces of group averages; Right – subjective light phase (CT0–12)/subjective dark phase (CT12–24) values. RER = respiratory exchange ratio. For food intake, n=4–6. Otherwise, WT n=8; KO n=5; Liver-RE n=7. Two-way ANOVA, *=p<0.05; **=p<0.01; ***=p<0.001. See Fig. S6A–B. F) Clock genes expression after 7 days of DD. See also Fig. S6D, G) Direct comparison of LD and DD conditions in Liver-RE; n=3–4. See Fig. S6E–F. H) Liver whole-cell extracts from DD. I) Quantification of core clock proteins in LD (top, as in Fig. 1) and DD (bottom) for n=3.
After 7 days in DD, the WT molecular clock oscillated robustly, in stark contrast to Liver-RE mice (Fig. 7F–G, S6D–E). Cosinor analysis did not identify significant oscillations (20–26 hrs) for clock gene transcripts, including Bmal1, Dbp, Rev-erbα, Cry1, Per2 and Per3, as well as autonomous output genes Nampt and Gys2 (Fig. 7G, S6E–F). Analysis of protein accumulation yielded similar results; 24 hr oscillations of acetyl-BMAL1, phospho-BMAL1, PER2 and REV-ERBα were lost (Fig. 7H–I, S6F).
SCN lesion or absence of the SCN clock in DD leads to labile oscillations in livers of individual mice, postulated to have widely dispersed phases (Tahara et al., 2012) (Husse et al., 2014) (Izumo et al., 2014) (Yoo et al., 2004). Thus, synchrony is lost between animals and variability within groups increases. Clock gene expression patterns of Liver-RE mice in DD appear consistent between animals, albeit with disrupted rhythmicity. In fact, of all clock genes, only Per2 was statistically more variable in DD compared to LD (p<0.01, Fig. S6F). Thus, reconstituted livers are still coherent between animals in DD, but do not exhibit circadian oscillations. Similar results were obtained with the reconstituted epidermis clock in DD (Welz et al. 2019; Fig. 3).
DISCUSSION
The cell-autonomous nature of individual cells has been established (Pando et al., 2002) (Nagoshi et al., 2004) (Maywood et al., 2006) (Yoo et al., 2004). In vivo, the discovery that metabolic pathways exhibit cyclic coordination and communication (Dyar et al. 2018) suggests that clocks function as an organized network. Yet, whether peripheral clocks operate as tissue-autonomous oscillators remains unclear. We used a mouse model that reconstitutes only the hepatic clock in an otherwise clock deficient animal (Liver-RE) to demonstrate autonomy of the liver clock and identify possible mechanisms of integration into the local clockwork. Based on the accompanying study (Welz et al. 2018) these findings likely apply to other peripheral clocks.
While the degree of reconstitution of the core clock components in the Liver-RE model is remarkable, only a fraction of circadian function is achieved. At the molecular level, BMAL1 displays cyclic post-translational modifications characteristic of transcriptional potential, is properly recruited to chromatin at target genes and drives rhythmic gene expression. Yet, reconstituted livers were only capable of recapitulating ~10% of transcript and ~20% of metabolite oscillations. Thus, other clocks appear to distally influence BMAL1 liver function through signals that might operate via specialized signaling pathways (Dang et al., 2016) (Koike et al., 2012).
Loss of detectable circadian oscillations under DD in Liver-RE suggests that the light-dark cycle synchronizes the hepatic clock when all other clocks are disrupted. Lesioning of the SCN results in arrhythmicity of various hepatic clock genes in both LD and DD (Reddy et al., 2007) (Akhtar et al., 2002). Yet, SCN-specific ablation of Bmal1, which leaves intact anatomical structure and neuronal connections, shows that liver clock genes still oscillate in LD but after 2–3 days in DD oscillation dampens (Husse et al., 2014). Hence, light can entrain peripheral clocks independently of the SCN clock but still through SCN-dependent mechanism(s) (Husse et al., 2014) (Izumo et al., 2014). Light may be acting through several pathways to affect liver function. Light activates the adrenal gland, inducing corticosterone and liver Per1 expression (Ishida et al., 2005) (Son et al., 2008). This pathway, however, is unlikely to impact Liver-RE as it relies on a functioning SCN clock and/or adrenal clock (Oster et al., 2006) and the WT oscillation of hepatic corticosterone is not restored in Liver-RE mice. The masking effect of light on locomotor activity under LD could indirectly affect clock gene drive and/or synchrony in the liver (Redlin and Mrosovsky, 1999). Yet, Liver-RE mice are heterogeneous for this behavior and thus masking is unlikely to fully explain their diurnal profile. Intriguingly, sympathetic innervation via the PVN appears to support hepatic oscillation of Per1, Per2 and Bmal1 (Terazono et al., 2003), thus neuronal connections in light-transducing pathways from brain to liver may be involved.
Studies using bioluminescent clock gene reporters demonstrate that tissues retain the capacity to oscillate ex vivo (Izumo et al., 2014) and indicate in vivo that peripheral clocks of individual mice still oscillate in the absence of a functional SCN, yet their phases become rapidly dispersed (Tahara et al., 2012) (Yoo et al., 2004). Per2:LUC oscillation following SCN lesion is present in 50% of livers with dampened amplitude (Tahara et al., 2012). Similarly, Bmal1:LUC peripheral expression wanes a week after SCN lesion in mice fed ad libitum (Saini et al., 2013). One explanation for the discrepancy between group data versus individual animals could be that monitoring a single clock gene does not fully inform on clock function. Many hepatic clock genes are differentially regulated by systemic signals. Indeed, Per2 is regulated by feeding-fasting signals, temperature, cyclic AMP and serum-borne signals, in addition to the clock (Travnickova-Bendova et al., 2002) (Kornmann et al., 2007). Given that nearly all clock genes in Liver-RE displayed similar variability in LD and DD (Fig. 7; S7), light-dark cycle appears critical for establishing liver circadian oscillations. The core clock and its tissue-autonomous output appear to be supported by SCN clock-independent effects of light.
Our metabolome profiling revealed semi-autonomous regulation of NAD+ metabolism. Clock regulation of the NAD+ salvage pathway is well characterized (Ramsey et al., 2009) (Nakahata et al., 2008) (Asher et al., 2008) (Nakahata et al., 2009). In the absence of inputs from other clocks and feeding-fasting cycle, NAD+ levels are restored sufficiently to drive SIRT1 circadian function. Thus, the NAD+ salvage pathway is dependent on the autonomous liver clock, whereas NAD+ de novo biosynthesis relies on extra-hepatic clocks. One explanation is the misregulation of BMAL1-dependent amino acids metabolism in the gut or skeletal muscle results in lowered biosynthetic substrates (Jeyaraj et al., 2012) (Mukherji et al., 2013), such as was observed for tryptophan in Liver-RE mice. Interestingly, hepatic de novo biosynthesis of NAM from tryptophan and subsequent shuttling via the blood stream is the major route through which extra-hepatic tissues generate NAD+ (Liu et al., 2018). Therefore, NAD+ biosynthetic pathways and their metabolites might link to the clock system systemically. Indeed, hepatic clock reconstitution does not reverse the reduced lifespan of BMAL1−/− mice, despite restoration of hepatic NAD+ salvage pathway, a key feature of caloric restriction-induced clock reprogramming in aged mice (Sato et al., 2017).
Diurnal liver glucose metabolism depends on many clock-mediated signals including SCN-driven feeding-fasting cycle, autonomic innervation, pancreatic insulin and glucagon secretion, release of leptin and other adipokines from WAT, post-prandial glucose uptake by skeletal muscle and rhythmic expression of rate-limiting liver enzymes (Akhtar et al., 2002) (Doi et al., 2010) (Kalsbeek et al., 2014). However, despite an environment where presumably most of these signals are disrupted, the clock in Liver-RE is sufficient to restore significant carbohydrate oscillations, specifically glycogen metabolism. This parallels a partial rescue of diurnal hepatic glucose content, which reflects export into the circulation. In contrast to carbohydrates, the vast majority of oscillating lipid and xenobiotic metabolites were not restored in Liver-RE. Interestingly, these metabolic pathways are targets of auxiliary clock loops and might indicate disrupted coupling to these output arms. For example, REV-ERBα is a major circadian lipid regulator in the liver, directly targeting lipid homeostasis in cooperation with HNF6 (Bugge et al. 2012) (Zhang et al., 2016). D-box binding PAR bZIP transcription factors, such as DBP, TEF and HLF, regulate the temporal detoxification of xenobiotics (Gachon et al., 2006). In Liver-RE, however, expression of REV-ERBα targets is restored, along with a comparable proportion of REV-ERBα sites between autonomous and non-autonomous genes. Likewise, Dbp, Tef and Hlf expression in Liver-RE was comparable to WT. Collectively, autonomous clock output alone does not appear to drive rhythmic abundance of hepatic lipid and xenobiotic species, again indicating that systemic signals from other clocks contribute to circadian liver homeostasis.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paolo Sassone-Corsi (psc@uci.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice were bred and housed at the Barcelona Science Park, Spain, animal facilities in accordance with European Union and Spanish regulations. Animal care and experimental use was approved by the government of Catalonia, Spain, in line with national and local legislation. An additional cohort of mice was housed and utilized for the collection of experimental data at the University of California – Irvine vivarium in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at UC-Irvine. Animal experiments were designed and conducted with consideration of the ARRIVE guidelines, the details of which were as follows. Bmal1-stop-FL mice and subsequent crossings were generated as described in the “Generation of BMAL1-stop-FL Mice” section below. Male and female littermates were used for lifespan and body weight experiments. Unless otherwise noted, female mice were used for all other experiments, group housed on a 12 hr light-dark cycle. Experiments were conducted with 8–12 week-old mice prior to the phenotypic onset of abnormalities.
METHOD DETAILS
Generation of Bmal1-stop-FL Mice
A stop cassette, containing a splice acceptor (SA), a LF2A sequence, mCherry, poly-A tail (PA) and an Frt sequence, is flanked by loxP sites and was inserted between exon 5 and 6 of the Arntl (Bmal1) allele (as seen in Figure 1A). These Bmal1-stop-FL mice were generated on the C57BL/6 background and crossed with Alfp-cre mice, which were backcrossed to the C57BL/6 background for at least ten generations. Cre-mediated recombination specifically in Alfp-cre positive hepatocytes results in excision of the stop sequence, loss of mCherry expression and expression of Bmal1, leaving behind one intronic loxP. Mice heterozygous for Bmal1-stopFL and Alfp-cre were crossed to generate experimental mice: WT – Bmal1wt/wt, Alfp-cre−/tg; KO – Bmal1stopFL/stopFL, Alfp-cre−/−; Liver-RE – Bmal1stopFL/stopFL, Alfp-cre−/tg.
Metabolic Cage Data Acquisition
Indirect calorimetry was performed with negative flow Oxymax-CLAMS (Columbus Instruments, Columbus, OH) hardware system cages. Mice were singly housed and given a 24-hour acclimation period to the metabolic cage. Throughout the experiment, standard chow and water were available ad libitum. Measurements of VO2, VCO2, food intake, water intake and feeding activity were taken every 10 minutes for up to 8 consecutive days at a room temperature. Mice were put under a 12 hr light-dark cycle for 3–4 days and then released into constant darkness for another 3–4 days. Respiratory exchange ratio (RER = VCO2/VO2) was calculated by the accompanying Oxymax software.
Locomotor Activity
Locomotor activity of individually housed mice was measured using optical beam motion detection (Starr Life Sciences). Data were collected using Minimitter VitalView 5.0 data acquisition software and analysed using Clocklab (Actimetrics).
Western Analyses
Whole cell tissue lysates were obtained for western blot analysis. Frozen tissue was homogenized in RIPA lysis buffer (50 mM tris-HCl [pH 8], 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2 and 1% NP-40) supplemented with 500 μM PMSF (serine protease inhibitor), 20 mM NaF (phosphatase inhibitor), Protease Inhibitor Cocktail (Roche, Basel, Switzerland), 10 mM nicotinamide (Sirtuin inhibitor) and 330 nM TSA (Class I and II HDAC inhibitor). Samples were lysed for 30 min on ice with periodic mixing, sonicated (5 sec on, 5 sec off, 4 cycles) and centrifuged at 13,200 rpm for 15 min at 4°C and the supernatant was collected. Protein concentration was determined by the Bradford method using Protein Assay Dye (BioRad) 20–60 μg protein from each sample was separated on 6% or 8% gels by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and blocked with 5% instant non-fat dry milk in TBS-T (0.1% Tween-20, TBS) for 2 hours at room temperature. Primary antibodies were diluted in 5% milk or 5% BSA TBS-T and incubated with membranes overnight at 4°C (BMAL1, Abcam – ab93806; acetyl-BMAL, EMD Millipore – AB15396; Rev-Erbα, Cell Signaling – 13418; Cry1, Bethyl Labs – A302–614; Per2, Alpha Diagnostic – PER21-A; SIRT1, EMD Millipore – 07–131; ACTIN, Abcam – ab3280). Following HRP-conjugated secondary antibody incubation (Mouse IgG-HRP conjugate, EMD Millipore – AP160P; Rabbit IgG-HRP linked, EMD Millipore – 12–348) for 1 hour at room temperature, blots were visualized with Immobilon Western chemiluminescent HRP substrate (Millipore, Burlington, MA) and developed on HyBlot CL autoradiography film (Denville Scientific, Holliston, MA).
RNA Extraction
Total RNA was extracted from liver tissue by homogenization in TRIzol (Invitrogen, Carlsbad, CA). Following RNA, DNA and protein layer separation with chloroform, RNA was precipitated with a standard isopropanol and ethanol procedure. Final pelleted and washed RNA was resuspended in RNase-free water and quantified using the NanoDrop (Thermo Fisher, Waltham, MA).
Real-Time Quantitative PCR
Real-Time Quantitative PCR was carried out with SsoAdvanced Universal SYBR Green Supermix (BioRad) on an Applied Biosystems Quant Studio 3 (Thermo Fisher). Delta CT’s were calculated and further normalized to 18S rRNA values for each sample. Primer sequences are listed in Table S7. For gene expression, complimentary DNA (cDNA) was synthesized from 1 μg RNA by reverse transcription with iScript cDNA Synthesis Kit (BioRad, Hercules, CA). For ChIP experiments, purified DNA was added directly to the PCR mix.
RNA-Sequencing
Total RNA was extracted (see RNA Extraction) and monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios for 260/280nm and 260/230nm. Library construction was performed according to the Illumina TruSeq Total RNA stranded protocol. The input quantity for total RNA was 1ug and rRNA was depleted using ribo-zero rRNA gold removal kit (human/mouse/rat). The rRNA depleted RNA was chemically fragmented for three minutes. First strand synthesis used random primers and reverse transcriptase to make cDNA. After second strand synthesis the ds cDNA was cleaned using AMPure XP beads and the cDNA was end repaired and then the 3’ ends were adenylated. Illumina barcoded adapters were ligated on the ends and the adapter ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The concentration for denaturation was 2nM and the final concentration for clustering was 200pM. The multiplexed libraries were sequenced by the Genomics High-Throughput Facility at the University of California – Irvine on six lanes using single end 100 cycles chemistry for the HiSeq 4000. The version of HiSeq control software was HCS 3.4.0.38 with real time analysis software, RTA 2.7.7.
Metabolomics
Metabolomics data was generated by Metabolon, Inc. (Durham, NC) as previously described previously (Evans et al., 2009). Sample preparation was carried out with the automated MicroLab STAR system from Hamilton Company (Reno, NV) in the presence of recovery standards for QC. Biochemicals were extracted and analyzed by ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) in four ways: 1) acidic positive ion conditions (water and methanol), optimized for hydrophilic compounds; 2) acidic positive ion conditions (water, methanol and acetonitrile), optimized for hydrophobic compounds; 3) basic negative ion conditions; 4) negative ionization. Raw data was processed with software developed at Metabolon, Inc. (Dehaven et al., 2010). Metabolites were identified by comparison to a library of purified standards with retention time/index (RI), mass to charge ratio (m/z) and chromatographic data (MS/MS spectral data). Corresponding peaks were quantified using area-under-the-curve and normalized by amount of starting material (tissue).
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as follows. All buffers prior to sonication were supplemented with 500 μM PMSF (serine protease inhibitor), 20 mM NaF (phosphatase inhibitor), Protease Inhibitor Cocktail (Roche), 10 mM nicotinamide (Sirtuin inhibitor) and 330 nM TSA (Class I and II HDAC inhibitor). 150 ± 10 mg of liver tissue was homogenized in 4 ml PBS and cross-linked with 2 mM disuccinimidyl glutarate (DSG, and 1 mM MgCl2) for 40 min at room temperature (RT). Samples were further cross-linked with 1% formaldehyde for 6 min at RT then cross-linking was stopped by 125 mM glycine incubation for 10 min at RT. Samples were centrifuged at 3000 rpm for 5 min at 4°C. Pellets were washed with PBS, resuspended in 1 ml cell lysis buffer (5mM HEPES [pH 8.0], 85 mM KCl, 0.5% NP-40) and incubated for 10 min at 4°C. Samples were then centrifuged at 3000 rpm for 5 min at 4°C. Pellets were resuspended in 600 μl SDS lysis buffer (50 mM tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS) and incubated on ice for 20 min. Sonication was achieved with the Bioruptor (Diagenode, Denville, NJ), 8 cycles of 30 sec on / 30 sec off. Samples were then centrifuged at 13,200 rpm for 10 min at 4°C and supernatants were diluted 1:5 with dilution buffer (50 mM tris-HCl [pH 8.0], 167 mM NaCl, 1.1% Triton X-100, 0.11% Na deoxycholate). Next, 100 μl of this chromatin was saved for input DNA quantification and an additional 1 ml was pre-cleared with 50 μl of BSA-blocked Protein G-PLUS Agarose, 50% slurry (Santa Cruz, Santa Cruz, CA) for 2 hours at 4°C. Samples were then incubated with antibodies (BMAL1, Abcam – ab93806; Rabbit IgG, Santa Cruz Biotech. – sc-2027) overnight at 4°C. The next day, 50 μl of BSA-blocked Protein G Sepharose, 50% slurry (Sigma, St. Louis, MO), was added to the samples for 3 hours at 4°C with rocking. Beads were then washed in the following manner: 1X in low-salt RIPA (150 mM NaCl, RIPA: 50 mM tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% SDS, 0.1% Na deoxycholate); 1X in high-salt RIPA (500 mM NaCl, RIPA); 1X in LiCl wash (0.25 M LiCl, 10 mM tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 0.5% NP-40, 0.5% Na deoxycholate); and 2X with TE wash (10 mM tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). Samples were reverse cross-linked by overnight incubation at 65°C in 200 μl direct elution buffer (10 mM tris-HCl [pH 8.0], 300 mM NaCl, 5 mM EDTA [pH 8.0], 0.5% SDS). The next series of sample treatments was as follows: 1) 2 μl of 2 mg/ml RNase A for 30 min at 37°C; 2) 1 μl of 10 mg/ml Proteinase K for 1 hour at 55°C; 3) addition of 2 μl of 20 mg/ml glycogen. DNA was then extracted twice with a standard phenol/chloroform/isoamyl alcohol procedure. Lastly, DNA was precipitated with ethanol and resuspended in TE buffer (pH 8.0).
ChIP-Sequencing
Library construction was performed using the PrepX Illumina ChipSeq kit and the PrepX Complete ILMN DNA barcodes on the Apollo324 System. The DNA quantity was measured by Qubit DNA High Sensitivity and the fragment size was measured by Agilent Bioanalyzer 2100 High Sensitivity DNA chip assay. The input amount of DNA was 1.5ng. The ends of the DNA were repaired and adenylated. The reaction mixture was then cleaned up using AMPure XP magnetic beads and Illumina barcoded adapters were ligated onto the blunt-end/adenylated product. The adapter ligated product was cleaned using AMPure XP beads and then amplified for adapter ligated products using 12 cycles of PCR. The resulting library was cleaned with AMPure XP beads and quantified by qPCR with Kapa Sybr Fast universal for Illumina Genome Analyzer kit. The library size was determined by analysis using the Bioanalyzer 2100 DNA High Sensitivity Chip. The library sequenced on the HiSeq 4000 Sequencer. Sequencing was performed using single read flowcell chemistry and 100 cycles with additional cycles for the index read. The HiSeq real time analysis software (RTA 2.7.7) converted the images into intensities and base calls. Post-processing of the run to generate the FASTQ files was performed at the Institute for Genomics and Bioinformatics (UCI IGB).
Hepatic Glycogen Content
Hepatic glycogen was quantified with the Glycogen Assay Kit II (colorimetric) (Abcam, ab169558) according to the manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Circadian Rhythmicity Determination
For time series datasets, rhythmic transcripts or metabolites were detected with the non-parametric JTK_CYCLE algorithm incorporating a period of 24 hours. Output measures including amplitude and phase of rhythmic features were also determined. Transcripts/metabolites which displayed no detectable expression/abundance in more than six biological replicates were filtered out before rhythmicity was tested. Genes were considered rhythmic over the circadian cycle if their permutation-based, adjusted p-value was <0.01. Metabolites were evaluated with a p-value of <0.05 given the inherent variability of metabolite quantification across mice and the n-value of this experiment. Additional detection of rhythmicity for transcripts and metabolites was conducted using a different, independently generated algorithm – BIO_CYCLE – to support JTK_CYCLE results.
Bioinformatics
RNA-seq
The reads from each replicate experiment were separately aligned to the reference genome assembly mm10 and corresponding transcriptome using the short-read aligner ELAND v2e (Illumina). Reads uniquely aligned to known exons or splice junctions extracted from the RefSeq (O’Leary et al., 2016) database with no more than two mismatches were included in the transcriptome. Reads uniquely aligned, but with more than two mismatches were removed from analyses. Similarly, reads matching several locations in the reference genome were removed from analysis. Gene expression levels were directly computed from the read alignment results for each replicate. Standard RPKM values (Mortazavi et al., 2008) (reads per kilobase of exon model per million mapped reads) were extracted for each gene covered by the sequencing data and each replicate used in this study. 18,079 annotated genes in RefSeq were found expressed in at least one of the replicate experiments and were considered during the next steps of the analysis. Genes systematically observed with no expression in all replicates were no longer considered in this study.
Comparison of liver and epidermis was carried out using the transcriptome dataset from Welz et al. 2019 (Fig. 2). GO Biological Process (FAT) enrichment analysis was conducted using The Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 (Huang da et al., 2009). Differential analysis of gene expression at individual ZTs was carried out using Cyber-T (Kayala and Baldi, 2012); ANOVA with Tukey-HSD post-hoc test was used to determine significance – altered in KO (WT vs KO = p<0.01) and restored in RE (KO vs RE = p<0.01, WT vs RE = n.s.).
ChIP-seq
Raw reads were mapped to mouse reference genome NCBI build 37 (mm9) by bowtie v1.1.0, allowing up to 1 mismatch. To adjust the difference of library size, uniquely mapped reads of each sample were down-sampled to the lowest reads count among all samples including input (Figure S6). Then the ChIP-seq peaks were called by MACS v1.4.2 with a cutoff of p ≤1e−8, and clonal reads were automatically removed by MACS. The peaks overlapped with any “negative peaks” (peaks are called by swapping the ChIP and input) were discarded. As the ChIP-seq profile of two replicates are highly correlated (Pearson’s R≥0.935, Figure S6), data from replicate 1 were applied in downstream analysis. Called peaks from KO negative control liver (59) were removed from WT and Liver-RE analysis since there are non-specific peaks. For peak overlap analysis, peaks from WT and Liver-RE of same time point were merged together as consensus regions. Then peaks of each sample which have at least 50% overlap with the consensus regions were defined as overlapped peaks. The ChIP-seq reads density was determined by deepTools v2.3.4, and then the average binding profile was visualized using R v3.2.3. The “Find Motifs Genome” function of HOMER (v4.8) (Heinz et al., 2010) was applied for motif searching at BMAL1 binding peaks with the option of “-size 400”, which allows identification of motifs at the 400bp region centered by peak midpoint. Binding peaks were assigned associated genes using the GREAT tool (McLean et al., 2010), which also performed GO Biological Process enrichment analysis from WT and Liver-RE peaks at ZT8. Annotation of genes nearest to each peak in REV-ERBα ChIP-seq datasets was carried out by Peak Assignment and Profile Search Tool (PAPST) software.
Metabolomics
General metabolite chemical classification (lipid, carbohydrate, etc.) and subpathway (fatty acid synthesis, polyunsaturated fatty acid, etc.) was determined by a combination of literature evidence and cross-reference to Metabolon’s internal database. Statistical analysis, including principal component analysis (PCA) and ANOVA, was carried out with MetaboAnalyst 3.0 (Xia and Wishart, 2016) using pre-normalized peak area-under-the-curve values for each metabolite, which were then log transformed and auto-scaled. At each ZT, ANOVA with Fisher’s LSD (FDR<0.1) determined metabolites altered in KO (WT vs KO = p<0.05) and restored in Liver-RE (KO vs Liver-RE = p<0.05, WT vs Liver-RE = n.s.). Full lists of metabolites and metrics can be found in Table S1. Outliers were removed using the Grubbs’ test (ESD method).
Integrated Pathway and Enzyme-Metabolite Pairs analysis
Pathway analysis integrating gene and metabolite enrichment was performed using the Integrated Molecular Pathway Level Analysis (IMPaLA) tool (Kamburov et al., 2011). This analysis returned separate enrichment p-values for genes and metabolites. Pathways were considered significantly coherent if enriched with ≥3 genes (p<0.01) and ≥2 metabolites (p<0.01).
Statistics
For each experiment, the number of biological replicates, statistical test, significance threshold and visual representation information (i.e. mean, SEM, etc. of graphs) can be found in the figure legends or main text. Inclusion/exclusion criteria, when appropriate, is reported in the figure legends. Complex statistical analyses are described within their corresponding methods section. Unless otherwise stated, data were analyzed in Prism 5.0 (GraphPad). Sample size was determined by referencing literature standards recently reported for circadian studies (Hughes et al. 2017).
DATA AND SOFTWARE AVAILABILITY
Raw RNA-sequencing data was deposited in Gene Expression Omnibus (GEO): GSE117134. ChIP-sequencing and metabolomics data are provided as supplemental tables: Table S5; Table S1.
Supplementary Material
Table S1. Metabolomics Dataset and Analysis - Related to Figure 2
Table S2. Transcriptome Dataset - Related to Figure 3
Table S3. Transcriptome JTK Analysis - Related to Figure 3
Table S4. GO Analysis of Liver-RE and Epidermis-RE - Related to Figure 3
Table S5. ChIP-seq Binding Peak Lists - Related to Figure 6
Table S6. ChIP-seq Motif Enrichment and GO Analysis - Related to Figure 6
Table S7. Primer Sequences - Related to STAR Methods
Figure S1. Related to: Figure 1. Reconstitution of the liver clock using BMAL1-Stop-FL mice.
(A) Western blot for BMAL1 in various tissues from WT, KO and Liver-RE mice demonstrating the tissue-specificity of Alfp-cre in the BMAL1-Stop-FL model. (B) Left – BMAL1 immunostaining of liver sections co-stained with nuclear marker DRAQ7, representative maximum intensity Z projections of 15μm tissue sections are displayed. Right – quantification of 6 random fields of view analyzed per animal, between 1574 and 1825 nuclei per genotype; n=3. (C) Locomotor activity traces for individual mice (double plotted for visualization), 7 days in LD followed by 7 days in DD. (D) Food intake for individual mice over a 2 day period in LD. Energy expenditure group averages over 2 days (left) and quantification of light and dark periods (right); Two-way ANOVA, **=p<0.01; ***=p<0.001. (E) Daily caloric intake of mice subjected to metabolic cages. (F) qPCR-determined expression of clock genes in the liver; n=4. (G) Representative western blot of core clock proteins from liver whole-cell extracts for WT and KO mice; n=3.
Figure S2. Related to: Figure 2. The diurnal metabolome reveals the autonomous metabolic output of the liver clock
(A) Venn diagram showing the overlap of oscillating metabolites using the BIO_CYCLE algorithm, p<0.05. (B) Pie charts of the phase and amplitude of autonomously oscillating metabolites in Liver-RE compared to WT. (C) Percentage of oscillating metabolites of each super-classification in each distinct group. (D) Sum of the raw intensity values for the 20 common amino acids over the diurnal cycle. Two-way ANOVA, statistics are presented on the graph. (E) Examples of redox metabolites over the diurnal cycle in each genotype. FAD – Flavin adenine dinucleotide. One-way ANOVA, *=p<0.01.
Figure S3. Related to: Figure 3. The autonomous transcriptional output of peripheral clocks is tissue-specific
(A) Venn diagram showing the overlap of oscillating transcripts using the BIO_CYCLE algorithm, p<0.01. (B) Principal component analysis of transcriptome profiles at each time-point for WT, KO and Liver-RE. (C) Differential expression at each ZT was determined using Cyber-T. Data presented as percent of total number of transcripts altered in KO. ANOVA with Tukey-HSD, altered in KO = WT vs KO – p<0.01; restored in Liver-RE = WT vs KO – p<0.01, WT vs Liver-RE – p>0.01, Liver-RE vs KO – p<0.01. (D) Pie chart showing the phase and amplitude of autonomously oscillating transcripts in Liver-RE compared to WT. (E) Enrichr was used to probe the enrichment of oscillating transcripts in each distinct group for enrichment of transcription factor target genes from published mouse liver ChIP-seq datasets (chEA). (F) Pie chart showing the phase and amplitude of autonomously oscillating transcripts in Epidermis-RE compared to WT. See also Welz et al. 2019 – Figure 1 and 2. (G) Integrated pathway enrichment analysis for oscillating metabolite and oscillating transcripts in each genotype. Dashed lines are significance cutoffs. Pathway names for WT are shown in gray and black for Liver-RE to highlight coherent, autonomously oscillating pathways.
Figure S4. Related to: Figure 5. Clock regulation of hepatic NAD+ metabolism is autonomous and non-autonomous.
(A) Expression of other NAD+ metabolism-related enzymes and Sirt1 from RNA-sequencing.
Figure S5. Related to: Figure 6. Inherent BMAL1 recruitment to chromatin in the liver.
(A) ChIP-sequencing sample reads and mapping metrics. (B) Correlation of the two biological replicates for each genotype and time point. (C) Pie chart of the genomic distribution of binding sites within promoter, genic or intergenic regions in WT or Liver-RE at ZT20. (D-F) Examples of peaks found only in WT, only in Liver-RE, or near core clock genes. (G) Proportion of published REV-ERBα binding sites (GSE number listed) associated with autonomously or non-autonomously oscillating genes. (H) BMAL1 occupancy at distinct groups of oscillating genes, for comparison of all groups and time points. RE = Liver-RE. TSS = transcription start site.
Figure S6. Related to: Figure 7. Characterization of Liver-RE mice under constant conditions.
(A) Food intake for individual mice over a 2-day period in constant darkness. (B) Energy expenditure group averages over 2 days (top) and quantification of subjective light and dark periods (bottom). (C) Daily average caloric intake of mice subjected to metabolic cages under constant darkness. (D) qPCR-determined expression of clock genes in the liver for WT and KO mice, n=3–4. (E) Direct comparison of LD and DD conditions (7 days) of qPCR-determined expression of clock genes in the liver for Liver-RE mice. (F) Cosinor analysis of clock gene mRNA and protein in LD and DD. Single factor ANOVA determined the variance of each gene and Levene’s test was used to compare variances between LD and DD.
ACKNOWLEDGEMENTS
P.S.W generated Liver-RE mice. We thank all members of the Sassone-Corsi lab for discussions and support. We thank Melanie Oakes, Seung-Ah Chung and Valentina Ciobanu of Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (CA-62203) at UC-Irvine (NIH – 1S10RR025496–01; 1S10OD010794–01; 1S10OD021718–01). The European Research Council (ERC), Spanish Ministry of Economy and Development and IRB-Barcelona (recipient of Severo Ochoa Award of Excellence from MINECO [Government of Spain]) supported the lab of S.A.B. P.S.W. was supported by EMBO long-term fellowship and Ayudas Juan de la Cierva fellowship (MINECO). V.M.Z. received financial support by “la Caixa” INPhINIT Fellowship Grant for Doctoral studies at Spanish Research Centers of Excellence, “la Caixa” Foundation, Barcelona (ID 100010434, code LCF/BQ/IN17/11620018). P.S.-C. lab is supported by NIH grants R21DK114652, R21AG053592 and a Challenge Grant of the Novo Nordisk Foundation (NNF-202585). W.L. lab is supported by R01 CA193466 and R01 HG007538. K.B.K is supported by NIH-NINDS, T32 5T32NS045540. K.K. was supported by a fellowship from Japan Society for the Promotion of Science (JSPS). P.B. lab was supported by the grants NIH GM123558 and DARPA D17AP00002 with computing support by Yuzo Kanomata.
Footnotes
DECLARATION OF INTERESTS
J.M.K is an employee of Metabolon, Inc. (no role in the study design)
REFERENCES
- Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, and Asher G (2014). Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 19, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinelli F, Ceglia N, Shahbaba B, Sassone-Corsi P, and Baldi P (2016). What time is it? Deep learning approaches for circadian rhythms. Bioinformatics 32, i8–i17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, and Kyriacou CP (2002). Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12, 540–550. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, and Schibler U (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328. [DOI] [PubMed] [Google Scholar]
- Asher G, and Sassone-Corsi P (2015). Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, and Schibler U (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Bechtold DA, and Loudon AS (2013). Hypothalamic clocks and rhythms in feeding behaviour. Trends Neurosci 36, 74–82. [DOI] [PubMed] [Google Scholar]
- Bellet MM, Nakahata Y, Boudjelal M, Watts E, Mossakowska DE, Edwards KA, Cervantes M, Astarita G, Loh C, Ellis JL, et al. (2013). Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl Acad Sci U S A 110, 3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, and Lazar MA (2012). Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev 26, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, and Bradfield CA (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AJ, Johnston JD, Semikhodskii AG, Nolan T, Cagampang FR, Stirland JA, and Loudon AS (2003). Photoperiod differentially regulates circadian oscillators in central and peripheral tissues of the Syrian hamster. Curr Biol 13, 1543–1548. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. (2012). Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F, Sun X, Ma X, Wu R, Zhang D, Chen Y, Xu Q, Wu Y, and Liu Y (2016). Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat Commun 7, 12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R, Oishi K, and Ishida N (2010). CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J Biol Chem 285, 22114–22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, and Sassone-Corsi P (2013). Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, and Sassone-Corsi P (2012). Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A 109, 5541–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, and Schibler U (2006). The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4, 25–36. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, and Lazar MA (2015). Circadian metabolism in the light of evolution. Endocr Rev 36, 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. (2010). Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol 30, 5636–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, and Sassone-Corsi P (2007). CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, and Kornacker K (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J, Leliavski A, Tsang AH, Oster H, and Eichele G (2014). The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J 28, 4950–4960. [DOI] [PubMed] [Google Scholar]
- Ishida A, Mutoh T, Ueyama T, Bando H, Masubuchi S, Nakahara D, Tsujimoto G, and Okamura H (2005). Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab 2, 297–307. [DOI] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, and Takahashi JS (2014). Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj D, Scheer FA, Ripperger JA, Haldar SM, Lu Y, Prosdocimo DA, Eapen SJ, Eapen BL, Cui Y, Mahabeleshwar GH, et al. (2012). Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab 15, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A, la Fleur S, and Fliers E (2014). Circadian control of glucose metabolism. Mol Metab 3, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Cavill R, Ebbels TM, Herwig R, and Keun HC (2011). Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27, 2917–2918. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, and Antoch MP (2006). Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev 20, 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, and Schibler U (2007). System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, et al. (2017). Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab 25, 1206. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, and Weitz CJ (2008). Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105, 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moulik M, Fang Z, Saha P, Zou F, Xu Y, Nelson DL, Ma K, Moore DD, and Yechoor VK (2013). Bmal1 and beta-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced beta-cell failure in mice. Mol Cell Biol 33, 2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Su X, Quinn WJ 3rd, Hui S, Krukenberg K, Frederick DW, Redpath P, Zhan L, Chellappa K, White E, et al. (2018). Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell Metab 27, 1067–1080 e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, Jacks T, and Sassone-Corsi P (2016). Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell 165, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, and Hastings MH (2006). Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol 16, 599–605. [DOI] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, and Chambon P (2013). Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, and Schibler U (2004). Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, and Sassone-Corsi P (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, and Sassone-Corsi P (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, and Eichele G (2006). The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab 4, 163–173. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, and Hogenesch JB (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. [DOI] [PubMed] [Google Scholar]
- Pando MP, Morse D, Cermakian N, and Sassone-Corsi P (2002). Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110, 107–117. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, and Takahashi JS (2014). Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, and Kay SA (1997). Independent photoreceptive circadian clocks throughout Drosophila. Science 278, 1632–1635. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, and Hastings MH (2007). Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45, 1478–1488. [DOI] [PubMed] [Google Scholar]
- Redlin U, and Mrosovsky N (1999). Masking of locomotor activity in hamsters. J Comp Physiol A 184, 429–437. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, and Fitzgerald GA (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadacca LA, Lamia KA, deLemos AS, Blum B, and Weitz CJ (2011). An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Liani A, Curie T, Gos P, Kreppel F, Emmenegger Y, Bonacina L, Wolf JP, Poget YA, Franken P, et al. (2013). Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev 27, 1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, and Sassone-Corsi P (2017). Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 170, 664–677 e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, and Sassone-Corsi P (2002). A web of circadian pacemakers. Cell 111, 919–922. [DOI] [PubMed] [Google Scholar]
- Son GH, Chung S, Choe HK, Kim HD, Baik SM, Lee H, Lee HW, Choi S, Sun W, Kim H, et al. (2008). Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci U S A 105, 20970–20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, and Menaker M (2001). Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Tahara Y, Kuroda H, Saito K, Nakajima Y, Kubo Y, Ohnishi N, Seo Y, Otsuka M, Fuse Y, Ohura Y, et al. (2012). In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, and Sassone-Corsi P (2009). CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat Struct Mol Biol 16, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terazono H, Mutoh T, Yamaguchi S, Kobayashi M, Akiyama M, Udo R, Ohdo S, Okamura H, and Shibata S (2003). Adrenergic regulation of clock gene expression in mouse liver. Proc Natl Acad Sci U S A 100, 6795–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, and Sassone-Corsi P (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A 99, 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott AJ, and Menet JS (2018). Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet 14, e1007156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, and Sassone-Corsi P (1998). Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci 1, 701–707. [DOI] [PubMed] [Google Scholar]
- Yamajuku D, Inagaki T, Haruma T, Okubo S, Kataoka Y, Kobayashi S, Ikegami K, Laurent T, Kojima T, Noutomi K, et al. (2012). Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep 2, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, and Tei H (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. (2010). Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16, 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Damle M, Guan D, Li Z, Kim YH, Gannon M, and Lazar MA (2016). HNF6 and Rev-erbalpha integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev 30, 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Metabolomics Dataset and Analysis - Related to Figure 2
Table S2. Transcriptome Dataset - Related to Figure 3
Table S3. Transcriptome JTK Analysis - Related to Figure 3
Table S4. GO Analysis of Liver-RE and Epidermis-RE - Related to Figure 3
Table S5. ChIP-seq Binding Peak Lists - Related to Figure 6
Table S6. ChIP-seq Motif Enrichment and GO Analysis - Related to Figure 6
Table S7. Primer Sequences - Related to STAR Methods
Figure S1. Related to: Figure 1. Reconstitution of the liver clock using BMAL1-Stop-FL mice.
(A) Western blot for BMAL1 in various tissues from WT, KO and Liver-RE mice demonstrating the tissue-specificity of Alfp-cre in the BMAL1-Stop-FL model. (B) Left – BMAL1 immunostaining of liver sections co-stained with nuclear marker DRAQ7, representative maximum intensity Z projections of 15μm tissue sections are displayed. Right – quantification of 6 random fields of view analyzed per animal, between 1574 and 1825 nuclei per genotype; n=3. (C) Locomotor activity traces for individual mice (double plotted for visualization), 7 days in LD followed by 7 days in DD. (D) Food intake for individual mice over a 2 day period in LD. Energy expenditure group averages over 2 days (left) and quantification of light and dark periods (right); Two-way ANOVA, **=p<0.01; ***=p<0.001. (E) Daily caloric intake of mice subjected to metabolic cages. (F) qPCR-determined expression of clock genes in the liver; n=4. (G) Representative western blot of core clock proteins from liver whole-cell extracts for WT and KO mice; n=3.
Figure S2. Related to: Figure 2. The diurnal metabolome reveals the autonomous metabolic output of the liver clock
(A) Venn diagram showing the overlap of oscillating metabolites using the BIO_CYCLE algorithm, p<0.05. (B) Pie charts of the phase and amplitude of autonomously oscillating metabolites in Liver-RE compared to WT. (C) Percentage of oscillating metabolites of each super-classification in each distinct group. (D) Sum of the raw intensity values for the 20 common amino acids over the diurnal cycle. Two-way ANOVA, statistics are presented on the graph. (E) Examples of redox metabolites over the diurnal cycle in each genotype. FAD – Flavin adenine dinucleotide. One-way ANOVA, *=p<0.01.
Figure S3. Related to: Figure 3. The autonomous transcriptional output of peripheral clocks is tissue-specific
(A) Venn diagram showing the overlap of oscillating transcripts using the BIO_CYCLE algorithm, p<0.01. (B) Principal component analysis of transcriptome profiles at each time-point for WT, KO and Liver-RE. (C) Differential expression at each ZT was determined using Cyber-T. Data presented as percent of total number of transcripts altered in KO. ANOVA with Tukey-HSD, altered in KO = WT vs KO – p<0.01; restored in Liver-RE = WT vs KO – p<0.01, WT vs Liver-RE – p>0.01, Liver-RE vs KO – p<0.01. (D) Pie chart showing the phase and amplitude of autonomously oscillating transcripts in Liver-RE compared to WT. (E) Enrichr was used to probe the enrichment of oscillating transcripts in each distinct group for enrichment of transcription factor target genes from published mouse liver ChIP-seq datasets (chEA). (F) Pie chart showing the phase and amplitude of autonomously oscillating transcripts in Epidermis-RE compared to WT. See also Welz et al. 2019 – Figure 1 and 2. (G) Integrated pathway enrichment analysis for oscillating metabolite and oscillating transcripts in each genotype. Dashed lines are significance cutoffs. Pathway names for WT are shown in gray and black for Liver-RE to highlight coherent, autonomously oscillating pathways.
Figure S4. Related to: Figure 5. Clock regulation of hepatic NAD+ metabolism is autonomous and non-autonomous.
(A) Expression of other NAD+ metabolism-related enzymes and Sirt1 from RNA-sequencing.
Figure S5. Related to: Figure 6. Inherent BMAL1 recruitment to chromatin in the liver.
(A) ChIP-sequencing sample reads and mapping metrics. (B) Correlation of the two biological replicates for each genotype and time point. (C) Pie chart of the genomic distribution of binding sites within promoter, genic or intergenic regions in WT or Liver-RE at ZT20. (D-F) Examples of peaks found only in WT, only in Liver-RE, or near core clock genes. (G) Proportion of published REV-ERBα binding sites (GSE number listed) associated with autonomously or non-autonomously oscillating genes. (H) BMAL1 occupancy at distinct groups of oscillating genes, for comparison of all groups and time points. RE = Liver-RE. TSS = transcription start site.
Figure S6. Related to: Figure 7. Characterization of Liver-RE mice under constant conditions.
(A) Food intake for individual mice over a 2-day period in constant darkness. (B) Energy expenditure group averages over 2 days (top) and quantification of subjective light and dark periods (bottom). (C) Daily average caloric intake of mice subjected to metabolic cages under constant darkness. (D) qPCR-determined expression of clock genes in the liver for WT and KO mice, n=3–4. (E) Direct comparison of LD and DD conditions (7 days) of qPCR-determined expression of clock genes in the liver for Liver-RE mice. (F) Cosinor analysis of clock gene mRNA and protein in LD and DD. Single factor ANOVA determined the variance of each gene and Levene’s test was used to compare variances between LD and DD.
Data Availability Statement
Raw RNA-sequencing data was deposited in Gene Expression Omnibus (GEO): GSE117134. ChIP-sequencing and metabolomics data are provided as supplemental tables: Table S5; Table S1.