Abstract
Context.—
Duchenne muscular dystrophy is a rare, progressive, and fatal neuromuscular disease caused by dystrophin protein loss. Common investigational treatment approaches aim at increasing dystrophin expression in diseased muscle. Some clinical trials include assessments of novel dystrophin production as a surrogate biomarker of efficacy, which may predict a clinical benefit from treatment.
Objectives.—
To establish an immunofluorescent scanning and digital image analysis workflow that provides an objective approach for staining intensity assessment of the immunofluorescence dystrophin labeling and determination of the percentage of biomarker-positive fibers in muscle cryosections.
Design.—
Optimal and repeatable digital image capture was achieved by a rigorously qualified fluorescent scanning process. After scanning qualification, the MuscleMap (Flagship Biosciences, Westminster, Colorado) algorithm was validated by comparing high-power microscopic field total and dystrophin-positive fiber counts obtained by trained pathologists to data derived by MuscleMap. Next, the algorithm was tested on whole-slide images of immunofluorescent-labeled muscle sections from Duchenne muscular dystrophy, Becker muscular dystrophy, and control patients.
Results.—
When used under the guidance of a trained pathologist, the digital image analysis tool met predefined validation criteria and demonstrated functional and statistical equivalence with manual assessment. This work is the first, to our knowledge, to qualify and validate immunofluorescent scanning and digital tissue image-analysis workflow, respectively, with the rigor required to support the clinical trial environments.
Conclusions.—
MuscleMap enables analysis of all fibers within an entire muscle biopsy section and provides data on a fiber-by-fiber basis. This will allow future clinical trials to objectively investigate myofibers’ dystrophin expression at a greater level of consistency and detail.
Muscular dystrophies are a genetically and phenotypically heterogeneous group of muscle diseases characterized by myonecrosis, regeneration, and endomysial fibrosis leading to progressive muscle wasting.1 Duchenne muscular dystrophy (DMD) is a severe, X-linked, recessive disorder that affects approximately 1 in 3500 to 5000 live male births worldwide, making it one of the most common recessive disorders in humans.2–5 Becker muscular dystrophy (BMD) is milder and less frequent than DMD, affecting 1 in 11 500 males.4,6,7
The skeletal muscle histopathologic hallmarks of DMD are muscle fiber atrophy and hypertrophy, myonecrosis, and regeneration with progressively more severe endomysial fibrosis and fatty replacement leading to a gradual decline in muscle function.8 In the clinical trial setting, therapeutic interventions designed to restore dystrophin production and functional localization can only be directly confirmed by evaluation of appropriately sampled muscle biopsy specimens.9–11
Even though reliable dystrophin quantification is urgently needed to support efficacy findings in multiple ongoing clinical trials, assessment of the protein’s expression remains challenging.12 In recent years, the need to detect subtle changes in biomarkers of disease progression within the tissue context has spurred the development of image analysis techniques. With computational means to assess biomarkers, researchers can rapidly analyze hundreds to thousands of myofibers across entire muscle sections, aiming for an objective assessment of the tissue. Paramount for reproducible image analysis of fluorescent signals is a standardized scanning approach on properly qualified equipment by trained personnel. In addition, the importance of exposure times for optimal scanning conditions and analysis is critical. Specifically, within the context of dystrophin quantification in DMD research, many publications (when scanning conditions are disclosed) document that an exposure time was sought that put dystrophin staining intensity of control samples in the middle of the dynamic range.13 However, when the described method is used to quantify dystrophin changes in BMD and DMD samples, that approach limits the overall dynamic range for those samples, making the detection and quantification of small changes in dystrophin levels tremendously challenging.
No image analysis studies conducted before this validation study, to our knowledge, included the appropriate scanning qualification and algorithm validation that has the potential to accurately quantify small, yet meaningful, increases in dystrophin expression at the rigor required for use as outcome measures in clinical trials.13–16 Additionally, many publications do not specify whether the analysis process was overseen or guided by a pathologist experienced in evaluating immunofluorescent-labeled muscle tissues.
With this current work, we are documenting a workflow and scanning qualification process in combination with an algorithm validation that recapitulates a manual scoring paradigm. These results display functional and statistical equivalence to manual assessment of dystrophin expression at the sarcolemmal membrane. Workflow, personnel training, and documentation were performed with a rigor that mimics the intended use setting of a clinical trial. This semiautomated methodology enables quantification of muscle biomarkers for clinical trials and research studies for potential use as early surrogate endpoints in the evaluation of treatment effects and disease progression.
MATERIALS AND METHODS
Human Sample Collection
All muscle biopsies were initially obtained for routine diagnostic testing at the University of Iowa (Iowa City). Residual frozen tissue from those biopsies was subsequently accrued by the Tissue and Cell Culture Repository component of the University of Iowa’s Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (Iowa City). Biopsies in the repository are approved for research by the University of Iowa institutional review board (protocol 200510769).
Muscle Sample Sectioning
Immunofluorescent labeling was carried out on unfixed cryosections from DMD and BMD patient samples. Muscle samples that were considered to have no diagnostic abnormalities indicative of BMD, DMD, or other neuromuscular disorders were used as controls (CTRLs). The samples for this study (Table) were selected by a pathologist to represent a wide range of histopathologic disease severities across the spectrum of BMD and DMD based upon routine evaluation of hematoxylin-eosin–stained sections. Cryosections were cut at 10-lm thickness and mounted on Superfrost glass slides (Fisher Scientific, Hampton, New Hampshire). Sections were briefly air dried at room temperature, then stored 1 to 5 days at −80°C until immunostaining was performed.
| Patient and Sample Information | ||
|---|---|---|
| Sample ID | Age, y/S at Time of Biopsy | DMD Mutation Status |
| CTRL-A | 1/M | NA |
| CTRL-B | 3/M | NA |
| DMD-A | 6/M | Unknown |
| DMD-B | 6/M | Unknown |
| DMD-C | 7/M | Unknown |
| CTRL-1 | 20/M | NA |
| CTRL-2 | Young child/M | NA |
| CTRL-3 | 3/M | NA |
| CTRL-4 | 10/F | NA |
| CTRL-5 | 7/F | NA |
| BMD-1 | 53/M | Unknowna |
| BMD-2 | 14/M | Duplication of exons 21–29 |
| BMD-3 | 8/M | Duplication of exons 3–4 |
| BMD-4 | 4/M | c.2380G>A, p.E794K |
| BMD-5c | 10/F | Unknown |
| DMD-1 | 4/M | Deletion of exons 45–57 |
| DMD-2 | 7/M | Unknown |
| DMD-3 | 6/M | c.251dupT, p.L84Ffs*5 |
| DMD-4 | 6/M | Unknown |
| DMD-5 | 4/M | Stop mutation in exon 7 |
| DMD-6 | 2/M | c.1615C>T, p.R539* |
| DMD-7 | 8/M | Unknown |
| DMD-8 | 1/M | c.1615C>T, p.R539* |
| DMD-9 | 8/M | Duplication of exons 50–55 |
Abbreviations: BMD, Becker muscular dystrophy; BMD-5c, manifesting DMD mutation carrier; CTRL, control; DMD, Duchenne muscular dystrophy; NA, not applicable.
Diagnostic immunostaining was interpreted by the reviewing pathologist as strongly suggestive of the presence of an in-frame deletion.
Immunofluorescent Labeling
Slides were brought to room temperature and dual-labeled with anti-dystrophin at 1:25 (MANDYS106, clone 2C6, obtained directly from Glenn Morris, Wolfson Centre for Inherited Neuromuscular Disease, Oswestry, United Kingdom) and anti-merosin at 1:400 (ab11576, clone 4H8, lot GR95776–14; Abcam, Cambridge, United Kingdom) antibodies. Briefly, sections were incubated for 60 minutes at ambient temperature with 100 μL of a primary antibody cocktail diluted in phosphate-buffered saline. After incubation, the slides were washed twice for 5 minutes with phosphate-buffered saline then incubated with 100 μL of a detection cocktail that consisted of Alexa Fluor 594 goat anti-mouse immunoglobulin (Ig)G2a (A21135, lot 1366503; Molecular Probes, Eugene, Oregon) and Alexa Fluor 488 rabbit anti-rat IgG (H + L) (A21210, lot 53122A; Molecular Probes), each at 1:400 in phosphate-buffered saline for 30 min. After incubation, the slides were washed twice for 5 minutes with phosphate-buffered saline. Coverslips were mounted with ProLong Gold (ThermoFisher Scientific, Waltham, Massachusetts).
Slide Scanning and Manual Image Annotation
Slides were scanned at ×20 in the fluorescein isothiocyanate (FITC) and tetramethylrhodamine (TRITC) detection channels on a 3DHISTECH Pannoramic MIDI fluorescent scanner (PANMIDI; PerkinElmer, Waltham, Massachusetts) at optimal exposure times and following established protocols (see supplemental digital content containing Figures 1 and 2 at www.archivesofpathology.org in the February 2019 table of contents). Scanned images were manually annotated within ImageScope (Leica Biosystems, Buffalo Grove, Illinois) before analysis. One large inclusion annotation was drawn around the muscle tissue intended for analysis. Smaller exclusion annotations were placed around areas omitted from analysis, eg, tissue folds, staining artifacts, regions of fibrofatty tissue, areas of necrosis, among others. All annotations were performed by a trained technician and reviewed and approved by a pathologist.
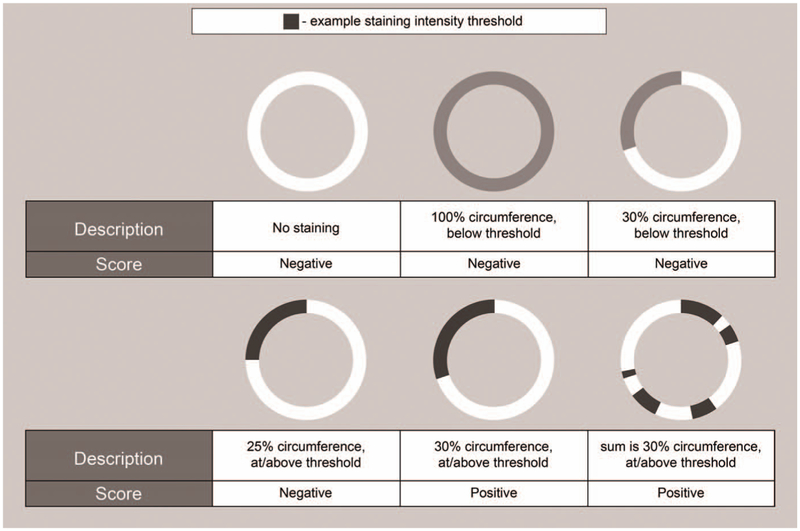
Figure 1.
Schematic examples of the manual-scoring paradigm. Schematic ring structures each represent a muscle fiber with its membrane. Different shades of gray represent different staining intensities (white, no staining). The examples cover a range of fiber staining with their corresponding scores. All scores are independent of fiber size or shape.
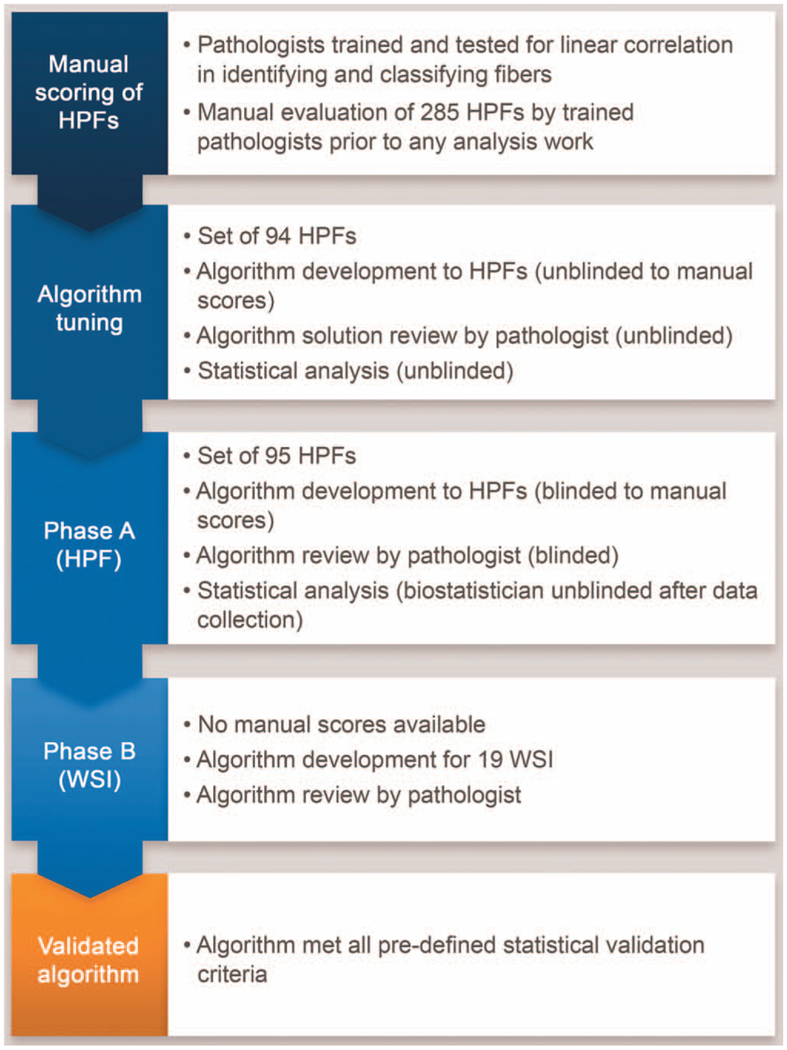
Figure 2.
Schematic overview of the algorithm validation process. Abbreviations: HPF, high-power field; WSI, whole-slide image.
Selection of High-Power Fields for Analysis
To enable manual pathology review, high-power field (HPF) images were generated from the whole slide scans of samples CTRL-1 through 5, BMD-1 through 5, and DMD-1 through 9. The GridMap (Flagship Biosciences, Westminster, Colorado) algorithm plugin for ImageScope was used to generate 15 HPF annotations (size, 750 × 750 μm) selected at random locations throughout the annotated tissue of each slide resulting in a total of 285 HPF images. These grid annotations were transferred to the single-channel TRITC or FITC images, and images were exported that captured the content of each HPF.
Manual Scoring
Four board-certified pathologists performed the manual scoring of samples. All pathologists participated in a training session to gain proficiency in (1) applying the scoring paradigm, and (2) the use of ImageJ software (National Institutes of Health, Bethesda, Maryland) for manual scoring.
After training and before scoring samples, all pathologists were required to demonstrate linear correlation for both total fiber count and positive fiber count (minimum requirement, Pearson r ≥ 0.75) to a predefined gold standard per a fifth experienced pathologist by scoring a training set of images under standardized conditions described below. All pathologists obtained a linear correlation greater than 0.98 for the total number of fibers and greater than 0.94 for the dystrophin-positive fibers.
Manual Scoring Paradigm
For merosin labeling, any labeled structure that was identified by the pathologist as a muscle fiber was counted in ImageJ as a muscle fiber. Fibers cut off by the edge of the HPF were counted as muscle fibers if both a piece of the membrane and a region of the sarcoplasm that the membrane encompasses were visible and unambiguously identifiable as muscle fibers. This procedure yielded total fiber counts for each HPF evaluated.
For dystrophin labeling, myofibers at or above a predefined labeling intensity threshold were counted as positive if that labeling was 30% or more of the individual fiber’s total circumference (Figure 1). Visually identifying membrane staining at or above the intensity threshold was included in pathologist training and testing.
Manual Scoring Process
The 285 HPFs generated (described above) were separated into their individual FITC and TRITC channels, resulting in 570 HPFs total. Those HPFs were split evenly among the 4 pathologists, with matching FITC and TRITC images assigned to the same pathologist for manual evaluation of total fiber numbers (by FITC) or dystrophin-positive fibers (by TRITC). To ensure the greatest amount of comparability between the individual pathologist’s assessments, all pathologists evaluated HPFs individually in an enclosed and darkened room under standardized conditions. All pathologists used the same computer, software, and monitors to evaluate HPFs, within the same room, and with the same controlled lighting conditions. Room access and usage was restricted and documented. Pathologists only had access to the HPFs they were assigned. They evaluated those images per the scoring paradigm described above and saved the counts generated in ImageJ to the computer. Those data were considered the raw data of the pathologist’s scores, including both a total fiber count and a positive fiber count.
Tissue Image Analysis: MuscleMap
The MuscleMap (MM) algorithm was specifically developed for the analysis of muscle fiber biomarkers and used for dystrophin quantification in the current application. For this study, the algorithm defined fibers based on both morphometrics and the FITC labeling for merosin. Based on the FITC signal, a membrane mask was generated allowing quantification of dystrophin per TRITC labeling in membrane areas only. Primary data outputs included, but are not limited to, mean membrane intensity for each fiber in a biopsy, cumulative percentage membrane-intensity curves for all fibers in the biopsy, and classification of individual fibers as either positive or negative based on pathologist-chosen dystrophin intensity and membrane completeness thresholds (ie, the percentage of the membrane circumference that displayed dystrophin labeling above the intensity threshold). Those pathologist-chosen settings were set at the beginning of the study and maintained on all samples.
MM Validation Study Design
The MM algorithm was validated in installation qualification, operational qualification, and performance qualification testing processes. Briefly, the algorithm was (1) tuned with unblinded HPFs, (2) tested for concordance with the pathologists on blinded HPFs, and (3) run on whole slide scans and reviewed by a pathologist in a fashion that mimicked the use of the validated process in future clinical trials and research studies (Figure 2).
Algorithm Development and Tuning Phase
Of the dual-labeled HPFs manually scored (n = 285), 94 of 285 (33%) were selected via stratified, random sampling from the study samples for this phase (CTRLs [24 of 94; 26%], BMDs [26 of 94; 28%], and DMDs [44 of 94; 47%]). Sampling was stratified to ensure equitable representation of CTRL, BMD, and DMD samples. Algorithms were developed to correctly identify fibers based on merosin-labeling to generate a total fiber count. In addition, algorithms were tuned to adequately classify fibers as “positive” or “negative” for dystrophin labeling, in a fashion that recapitulated the manual scoring paradigm. That entailed uniform settings for all samples for fiber classification based on dystrophin staining. To ensure accurate myofiber identification across samples, myofiber-detection parameters were adjusted on a per-HPF basis. The manual scoring results were available for final algorithm tuning to ensure that detection of positive fibers closely mirrored manual evaluation results. Algorithms were reviewed, and algorithm results were collected for statistical analysis to ensure that the developed algorithm solutions would meet basic validation criteria. Finalized parameters of positive-fiber classification (including staining-intensity thresholds) were uniform across samples and carried forward without further changes.
Algorithm Testing Phase A
For this phase of the algorithm validation, 95 dual-labeled HPFs (CTRLs [30 of 95; 32%], BMDs [23 of 95; 24%], and DMDs [42 of 95; 44%]) were selected via stratified random sampling to be analyzed via MM without prior knowledge of the manual scoring results. Those HPFs were different from those used during algorithm development and tuning, although HPFs from individual patients contributed to both tuning and testing sets. Algorithms were developed and adjusted on a per-HPF basis to correctly identify individual myofibers based on merosin labeling. For classification of dystrophin-positive fibers, parameters and thresh-olds were carried forward from the previous tuning phase without any modifications. Algorithms were reviewed by a pathologist, and algorithm results were collected for statistical analysis to test whether algorithm solutions would meet the basic validation criteria.
Algorithm Testing Phase B
After MM passed phase A, the algorithms were tested on whole slide images (WSIs) of the same sample set. In this scenario, no manual scoring data were available for direct data comparison. Instead, pathology evaluation was performed in a fashion that mimicked the use of the validated process in future clinical trials and research studies. The WSIs were manually annotated (described in Materials and Methods) to define the region of analysis. Algorithm solutions were developed and adjusted as described in testing phase A. Both the annotations and the algorithm performance were visually reviewed and approved by a pathologist. Minimum passing criteria for algorithm performance during pathologist’s review were (1) correct identification of approximately 75% of myofibers, and (2) correct classification of fibers as dystrophin-positive or dystrophin-negative for approximately 75% of detected fibers. Percentages were visually estimated. If an algorithm solution did not meet those criteria, no data were generated for that WSI. To meet MM validation criteria, pathologist-approved algorithm solutions were required for at least 75% of all WSI samples (ie, 15 out of 19 samples [79%]).
Interrun Comparison
To demonstrate interrun reproducibility by MM, 10 dual-labeled HPFs (CTRLs [2 of 10; 20%], BMDs [4 of 10; 40%], and DMDs [4 of 10; 40%]) were analyzed with the same algorithm parameters on 5 separate days. Algorithm results were extracted for statistical analysis.
Statistical Analysis
All statistical analyses were performed with R (version 3.3.2, R Development Core Team, Vienna, Austria).17 Statistical significance was set at the usual α = .05 level.
Comparison of Manual Scoring to MM Scoring on HPFs
Equivalence between MM and manual scoring was evaluated by comparing the rate ratio between the 2 counting modalities to a null hypothesis using a 2 one-sided test procedure. If MM and manual counting modalities were equivalent, the rate ratio between the 2 would be 1.0. Therefore, the null hypothesis for the 2 one-sided tests was that the rate ratio (the modality-fixed effect) was both (1) less than three-quarters or 0.75, and (2) greater than four-thirds or 1.33. The alternative hypothesis—that both MM and manual counting had equivalent rates—was that the rate ratio was both (1) greater than or equal to three-quarters or 0.75, and (2) less than or equal to four-thirds or 1.33. That hypothesis was set to establish the equivalence margin; that range represents the clinically meaningful deviation allowable for the MM algorithm relative to the manual counting. The confidence level was set at 95%, which corresponds to calculating a 90% CI for the 2 one-sided test framework, and limits were calculated by profiling the likelihood function. A rate ratio in which the confidence limits fell within the bounds of the equivalence margin would indicate that the MM algorithm and manual counting were statistically equivalent.
Interrun Comparison of the Algorithm on HPFs
An analysis of variance was conducted on total and positive fibers to estimate the amount of variance attributable to different runs of MM. The residual variance was the variance that was not accounted for by the subject factor. That estimates how variable the MM would be when run on separate days with the exact same parameter settings. Acceptable residual variance was equal to 2% or less of total variance.
RESULTS
MM Validation on HPFs
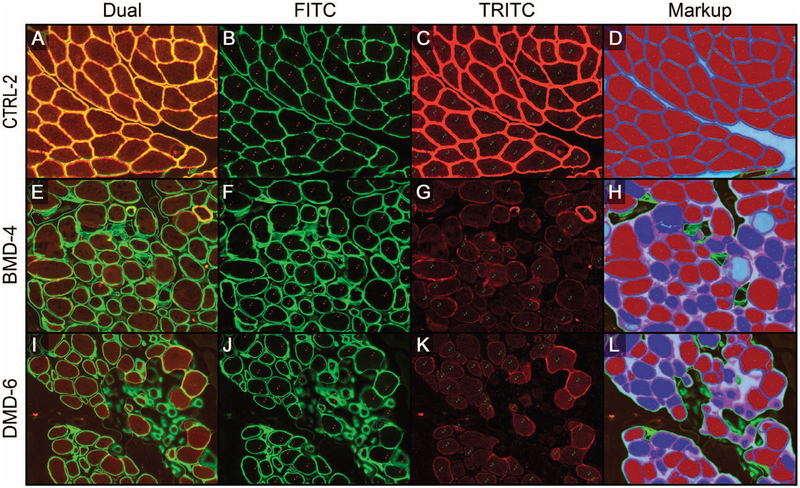
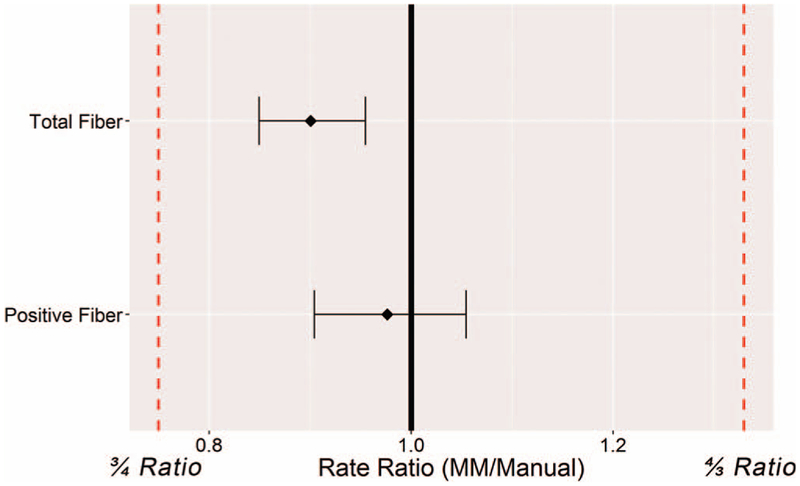
Before the capturing of whole slide scans, the fluorescent scanner was qualified for reproducibility through a detailed performance qualification of the fluorescent scanning procedure (see supplemental material). As outlined above, randomly selected HPFs (Figures 3, A through L) were generated from all dual-labeled slides (Figure 3, A, E, and I). The FITC (Figure 3, B, F, and J) and TRITC (Figure 3, C, G, and K) color channels were separated and manually scored. Evaluation of FITC images resulted in a total fiber count per image and the TRITC image evaluation yielded the number of dystrophin-positive fibers present in each HPF. These data were compared with total fiber and positive fiber counts generated by the MM algorithms (markup examples are shown in Figure 3, D, H, and L). After that comparison yielded satisfactory statistical results (the algorithm development and tuning phase; statistical analysis data not shown), the process was repeated on HPFs for which the team was blinded to the manual evaluation data (algorithm testing phase A). Statistical analysis demonstrated equivalence between the manual evaluation and the digital image analysis of HPFs by meeting the predefined validation criteria in which ratios and confidence limits were between 0.75 and 1.33 (Figure 4). In general, MM algorithms slightly under-counted the total number of fibers (rate ratio, 0.901; 95% CI, 0.850–0.955), whereas the percentage of positive fibers was close to the data derived from the manual evaluation (rate ratio, 0.976; 95% CI, 0.904–1.055). The undercounting of total fibers was likely due to pathologists being instructed to count all fibers at the edge of the HPF during the manual evaluation, even if partially cut off by the field of view. During digital-image analysis, however, the MM algorithm did not include all such edge fibers in its analysis, but only those fibers in which most of the fiber was present in the HPF. That situation would not be present in the intended use setting in which WSIs were being evaluated.
Figure 3.
Representative images of algorithm-testing phase A. The high-power field (HPF) examples for each category were controls (CTRL [A–D]), Becker muscular dystrophy (BMD [E–H]), and Duchenne muscular dystrophy (DMD [I–L]). Each row of images shows the dual-stained, merosin/dystrophin-labeled HPF (A, E, and I), the isolated FITC channel (green) displaying merosin labeling of all fibers, including the ImageJ markup of manual counts (red) of all fibers [B, F, and J], the isolated tetramethylrhodamine (TRITC) channel (red) displaying dystrophin labeling of fibers, including the ImageJ markup of manual counts (green) of dystrophin-positive fibers [C, G, and K], and MuscleMap (MM) markups [D, H, and L], which show fiber classification based on dystrophin quantification (blue outline with red center, positive fiber; blue outline with blue center, negative fiber; and teal line, manual annotation) (original magnification ×10 [A through L]).
Figure 4.
Statistical analysis of algorithm testing phase A (on high-power fields [HPFs]). Rate ratios of the total and positive fiber counts were within the required 3/4 and 4/3 ratios. The means of MuscleMap (MM) counts are less than the manual-count standard, resulting in a rate ratio <1.0, indicating that MM tends to undercount fibers compared with the manual assessment results.
MM Validation on WSIs (Algorithm Testing Phase B)
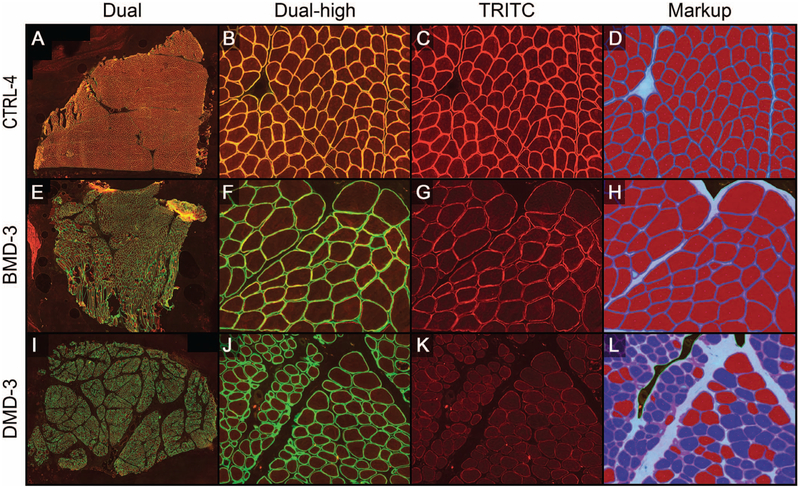
After MM met the validation criteria for the HPF analysis stage (phase A), scans of the entire muscle section (Figure 5, A through L) were then analyzed. Dual-stained, dystrophin-labeled slide images were captured (Figure 5, A, B, E, F, I, and J). Algorithm solutions previously used on HPFs were tuned for optimal fiber identification on WSIs, whereas dystrophin quantification and fiber classification parameters were not changed from the previous phase. To test MM performance in a workflow that closely resembled the intended future use of the tool, no manual scores were generated on WSIs before image analysis. Algorithm markups were reviewed by a pathologist and required to meet predetermined validation criteria for accurate total fiber classification and adequate classification and enumeration of positive fibers recapitulating the manual scoring paradigm. To that end, algorithm markups were reviewed (Figure 5, D, H, and L) in comparison to isolated TRITC channel images (Figure 5, C, G, and K). The reviewing pathologist classified an algorithm as passed or failed based on the aforementioned criteria. When feasible, failed algorithms were further tuned to improve total fiber detection and reevaluated by the reviewing pathologist. At least 75% of the WSIs analyzed were required to pass pathologist’s review to meet the predetermined validation criteria. All samples (19 of 19; 100%) passed the pathologist’s review. Depending on the size of the native biopsy, 1375 to 17 618 fibers were analyzed per sample (CTRLs, 1375–6332 fibers; BMDs, 1816–5921 fibers; and DMDs, 2149–17 618 fibers).
Figure 5.
Representative images of algorithm testing phase B (whole-slide images). Example images for control (CTRL [A–D]), Becker muscular dystrophy (BMD [E–H]), and Duchenne muscular dystrophy (DMD [I–L]) samples. Each row of images shows the dual-stained merosin/dystrophin-labeled high-power fields (HPF) at low magnification [A, E, and I], at higher magnification [dual-high magnification B, F, and J], the isolated tetramethylrhodamine (TRITC) channel displaying dystrophin labeling of fibers [C, G, and K], as well as MuscleMap (MM) markups [D, H, and L], which show fiber classification based on dystrophin quantification (blue outline with red center, positive fiber; blue outline with blue center, negative fiber) (original magnifications ×1 [A, E, and I] and ×10 [B through D, F through H, and J through L).
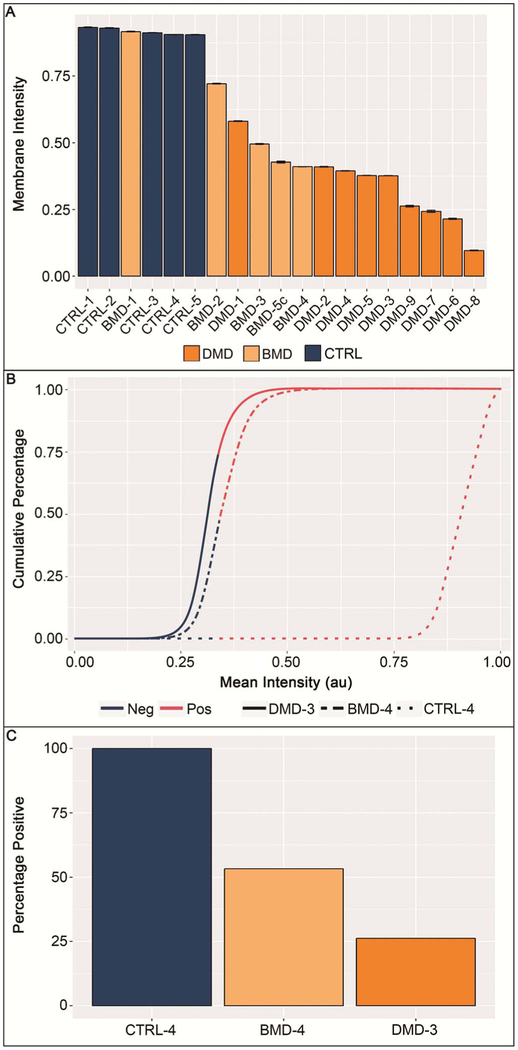
Although not part of the validation procedure, the average membrane staining intensity for all samples was analyzed (Figure 6, A through C). As expected, staining intensity (Figure 6, A) was similar between the CTRL and BMD sample sets, as well as between the BMD and DMD sample sets, which is representative of the variability of dystrophin expression among patients with BMD. However, the general order of samples visually separated DMD and CTRL samples into their respective diagnostic categories. Additionally, CTRL and DMD samples showed notable differences in mean staining intensity, indicating not only that sample labeling and analysis performance were in concordance with existing general disease knowledge but also that the optimized exposure times yielded desired results on the validation cohort with differentiation of individual DMD samples by staining intensity, whereas imaging CTRL samples was at near-saturation levels. Similar characteristics were revealed when cumulative histogram data were evaluated (Figure 6, B), displaying data from the same samples as shown in Figure 5. Most fibers stained in the CTRL samples displayed saturated signals near 1.0 arbitrary units (au), and 6332 of 6332 (100%) of fibers detected in that sample (CTRL-4) had a mean fiber intensity greater than 0.34 au, the threshold for positivity. DMD-3 and BMD-4 had 1284 of 4899 (26.2%) and 3155 of 5921 (53.3%) of fibers, with mean fiber intensities above that threshold, respectively (Figure 6, C).
Figure 6.
Analysis of staining intensity. Average membrane staining intensity per slide/sample displayed for all samples. A, Duchenne muscular dystrophy (DMD) and control (CTRL) samples displayed intensities that formed 2 distinct groups, with a slight overlap of both groups with Becker muscular dystrophy (BMD) samples. All CTRL samples show staining intensities at the upper limit of the dynamic range. Error bars are standard errors of the mean. B, Cumulative distribution histogram data of 3 representative samples (as displayed in Figure 5). Curves show that, although 100% of fibers stain at a high intensity for CTRL samples, the curve profiles of DMD and BMD samples are substantially different, with more fibers being negative or at lower staining intensities compared with CTRLs. The red portion of the lines indicate staining intensity above predefined staining-intensity threshold (0.34 arbitrary units [au]); dark blue portions indicate staining intensity below the predefined staining-intensity threshold. C, The percentage of positive fibers, as determined by MuscleMap, are shown for the same 3 samples displayed in Figure 5. Abbreviations: Neg, negative; Pos, positive.
Evaluation of interrun performance of MM was part of the predefined validation plan. To that end, 10 HPFs were analyzed on 5 consecutive days. Resulting data met the acceptable residual variance of less than 2%. In fact, only one sample showed a minimal variation (<0.1% residual variance across all samples) of total fiber counts, whereas no variations were observed in the number of positive fibers (data not shown).
In summary, all predetermined performance criteria were met, resulting in the qualification and validation of a scanning and digital image analysis process, respectively, which allows for semiautomated analysis of myofibers and their dystrophin content under the direct supervision of a trained pathologist.
DISCUSSION
In this article, we describe a strategy of equipment and procedure qualification, as well as algorithm validation, that aimed to produce standardized, quantitative, and reproducible results in a fashion that met the rigor required for measurements of dystrophin treatment effects in a clinical trial setting.
Reproducible fluorescent imaging is challenging, and the lack of standards or reference material makes it difficult to compare data generated from different microscopes or to monitor equipment performance over time.18,19 Therefore, we used an automated fluorescent scanning approach and fluorescent test slides that were not subject to photo-bleaching to ensure that slide scans were consistent over time and image capture was performed in a standardized fashion. As shown by our data, exposure time can have a profound effect on measured parameters such as staining intensity (see Supplemental Materials).
As a first step, we recommend that the performance of the fluorescent microscope/scanning system be monitored before the capture of any study samples to ensure reproducible conditions at the optimal exposure. The exposure settings used here were chosen to bring DMD samples into an appropriate dynamic range for dystrophin quantification. Those procedures, when performed at the start of the scanning of a batch of slides, ensure that the light intensity, optical path, and image capture are functioning correctly before study specimens are evaluated. The process and equipment tested in this study passed all qualification parameters.
Semiquantitative assessment of fluorescent staining, including that of dystrophin staining, has been performed for years. Although the manual assessment by a pathologist is generally considered the gold standard for tissue analysis, it can be flawed.12,20,21
Especially when viewing fluorescent staining, phenomena that influence perception of contrast and staining intensity can have a substantial effect on the pathologist’s ability to reliably and reproducibly assess samples (reviewed by Aeffner et al22). Digital image analysis can aid in avoiding effects on data that are created by limitations of human vision.22
Instead of relying on manual assessment by a pathologist alone, the semiautomated digital image analysis approach validated in this work aimed at providing a tool for dystrophin quantification at the myofiber membrane that can generate more-accurate, reproducible, and fully quantitative data.
Two key aspects are required for successful validation of an image analysis tool and the process described herein: (1) successful identification of muscle fibers, and (2) successful classification of dystrophin-positive muscle fibers in a fashion that recapitulates the manual scoring paradigm. Therefore, both of those aspects of MM were evaluated independently.
Total fiber counts were based on the outlining of all viable myofibers (diseased or not) via merosin immunostaining and dystrophin-positive classification was established by quantifying the dystrophin fluorescence in regions identified by merosin. Fiber counting “by hand” is not only tedious and error prone, it is also cost and time prohibitive, and it is unrealistic to expect it to be performed on entire muscle biopsy sections that can contain as many as 10 000 fibers. Therefore, most techniques previously published on this topic have relied on the analysis of a few, selected HPFs, which may not be representative of the entire tissue section. Even when digital solutions are employed to mitigate those shortcomings, only selected HPFs are assessed. The HPF selection process itself is also prone to bias and risks not capturing biologic heterogeneity.
The HPF evaluation was followed by the evaluation of the algorithm on WSIs, with review by a trained pathologist. Through scanning the entire biopsy section, MM captured up to 17 000 fibers in the sample set presented here. Imaging a tissue section in its entirety eliminates the bias that manual selection of HPFs would introduce into any analysis. In addition, capturing a dual-labeled tissue section eliminates variability that could be introduced by single-labeled slides in consecutive sections. Here, the detection of merosin staining (by FITC) was used to create a virtual mask to quantify dystrophin (by TRITC) in membrane-associated areas only. Definition of membrane areas via merosin allowed not only for collection of fiber-by-fiber data on dystrophin expression and other fiber parameters but also for inclusion of membrane staining completeness into the digital scoring paradigm. The data presented herein show that MM passed validation criteria on WSIs and pathologist quality control assessment of algorithm performance. This formally demonstrated functional and statistical equivalence to manual assessments and counts when MM was used as described above.
Although other published methods aim to exclude revertant fibers from the overall analysis,12 the currently described method generates data for every fiber identified via merosin staining. With assessment on a per-fiber basis, revertant fibers can be included in the overall data set. Postprocessing of data can have various aims, for example (1) analyzing revertant fibers separately, (2) analyzing revertant fibers together with all other fibers, or (3) excluding revertant fibers from further analysis. In addition, because this method uses merosin labeling to outline every individual myofiber, morphological data on a fiber-by-fiber basis can be generated (such as fiber area, fiber diameter, fiber circumference, among others) and analyzed individually, in aggregates, or in subgroups (eg, dystrophin-positive versus dystrophin-negative fibers).
Despite the benefits of semiautomated image analysis methods, including MM, there are several scenarios that can challenge their application in the intended-use setting. For example, we found that tissue mounting in an orientation that allowed for cross-sections of myofibers at their shortest diameter was paramount to accurate fiber identification and measurement of fiber morphometric parameters. In addition, to demonstrate the methods described herein, the MANDYS106 antibody was used to stain for dystrophin. However, MM can be used to analyze a variety of different antibodies selected for staining. Antibodies binding specific epitopes should be chosen in the context of the patients’ specific DMD mutation to ensure binding sites are appropriately expressed.
Differences in dystrophin expression between BMD and DMD, between individual patients within each disease category, and between individual fibers within a single section are consistent with earlier reports in the literature.11 That includes the observation of trace amounts of dystrophin expression in myofibers of patients with DMD,23 as well as the prevalence of revertant fibers.15 The number of revertant fibers as well as trace dystrophin expression within a patient has been shown not to change over time.15 Together with the practice of comparing pretreatment and on-treatment samples from the same patient for treatment effects, stable expression of revertant fibers eliminates further potential variables from clinical trial data evaluation.
The results generated by MM, when considered in the context of other methodologies (eg, Western blot) performed on the same biopsy sample for assessing dystrophin protein content, provides complementary confidence in the amount of dystrophin actually present. This is especially true when considering requirements to measure low levels of dystrophin in untreated patient samples in the clinical setting. The goal is to accurately determine changes in the dystrophin protein content of disease muscle after therapeutic interventions designed to restore dystrophin protein production.
In summary, we describe the qualification and validation of a scanning and digital image analysis process that allows for semiautomated identification of muscle fibers and semiautomated classification of dystrophin-labeled myofibers for samples from patients with DMD and BMD using the MM image analysis tool. This approach has the potential of greatly improving the clinical trial process for DMD drug candidates that aim at increasing dystrophin expression. This approach provides a validated mechanism for assessing treatment effects on a molecular and cellular level, using the entirety of each muscle biopsy. Moving forward, use of MM in clinical trials will provide a rigorous test of this digital image analysis approach within the context of quantification of biomarker changes from drug treatment.
Supplementary Material
Acknowledgments
We acknowledge the University of Iowa’s Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for supplying all biopsy samples used in this study (U54-NS053672). We also thank the following people for their contributions to this work: Terese Nelson, BS (University of Iowa) and Elie Schuchardt, BS, Sara K. Whitney, BA, Stefan Pieterse, BA, Nicholas Landis, BS, and Karen Ryall, PhD (all at Flagship Biosciences).
This study was supported by Sarepta Therapeutics. Dr Moore was supported in part by grant U54-NS053672 from the National Institutes of Health (Bethesda, Maryland), the Iowa Wellstone Muscular Dystrophy Cooperative Research Center (Iowa City). Dr Moore had a fee-for-services contract with Sarepta Therapeutics and Flagship Biosciences; Drs Aeffner, Faelan, Black, Wilson, Kanaly, Rudmann, Lange, Young, and Milici and Mr Moody were full-time or part-time employees at Flagship Biosciences; Drs Charleston and Frank and Messrs Dworzak, Piper, and Ranjitkar were full-time employees and shareholders of Sarepta Therapeutics; and Drs Faelan, Lange, Young, and Milici had stock options with Flagship Biosciences during production of this work. Dr Milici is the author of the patent that pertains to this work: US 9,784,665—Methods for Quantitative Assessment of Muscle Fibers in Muscular Dystrophy (MuscleMap). Drs Aeffner, Black, and Rudmann and Mr Moody were employees at the time of the study but are no longer with Flagship Biosciences; Dr Aeffner maintains stock in Flagship Biosciences. Drs Charleston and Frank and Messrs Piper and Dworzak are full-time employees of Sarepta Therapeutics and have stock options in the company. Mr Ranjitkar was with Sarepta Therapeutics at the time of the study; he is no longer, but maintains stock in Sarepta Therapeutics.
Footnotes
Supplemental digital content is available for this article at archivesofpathology.org in the February 2019 table of contents.
References
- 1.Mah JK, Korngut L, Fiest KM, et al. A systematic review and meta-analysis on the epidemiology of the muscular dystrophies. Can J Neurol Sci. 2016;43(1): 163–177. [DOI] [PubMed] [Google Scholar]
- 2.Emery AE. Population frequencies of neuromuscular diseases, II—amyotrophic lateral sclerosis (motor neurone disease). Neuromuscul Disord. 1991;1(5):323–325. [DOI] [PubMed] [Google Scholar]
- 3.Mendell JR, Shilling C, Leslie ND, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71(3):304–313. [DOI] [PubMed] [Google Scholar]
- 4.Nigro V, Piluso G. Spectrum of muscular dystrophies associated with sarcolemmal-protein genetic defects. Biochim Biophys Acta. 2015;1852(4):585–593. [DOI] [PubMed] [Google Scholar]
- 5.Romitti PA, Zhu Y, Puzhankara S, et al. Prevalence of Duchenne and Becker muscular dystrophies in the United States [published correction appears in Pediatrics. 2015;135(5):945]. Pediatrics. 2015;135(3):513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushby KM, Gardner-Medwin D, Nicholson LV, et al. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy, II: correlation of phenotype with genetic and protein abnormalities. J Neurol. 1993;240(2):105–112. [DOI] [PubMed] [Google Scholar]
- 7.Flanigan KM, Dunn DM, von Niederhausern A, et al. ; United Dystrophinopathy Project Consortium. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30(12):1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, Kunkel LM. The pathogenesis and therapy of muscular dystrophies. Annu Rev Genomics Hum Genet. 2015;16:281–308. [DOI] [PubMed] [Google Scholar]
- 9.Flanigan KM. Duchenne and Becker muscular dystrophies. Neurol Clin. 2014;32(3):671–688, viii. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson LV, Johnson MA, Bushby KM, Gardner-Medwin D. Functional significance of dystrophin positive fibres in Duchenne muscular dystrophy. Arch Dis Child. 1993;68(5):632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholson LV, Johnson MA, Gardner-Medwin D, Bhattacharya S, Harris JB. Heterogeneity of dystrophin expression in patients with Duchenne and Becker muscular dystrophy. Acta Neuropathol. 1990;80(3):239–250. [DOI] [PubMed] [Google Scholar]
- 12.Taylor LE, Kaminoh YJ, Rodesch CK, Flanigan KM. Quantification of dystrophin immunofluorescence in dystrophinopathy muscle specimens. Neuropathol Appl Neurobiol. 2012;38(6):591–601. [DOI] [PubMed] [Google Scholar]
- 13.Beekman C, Sipkens JA, Testerink J, et al. A sensitive, reproducible and objective immunofluorescence analysis method of dystrophin in individual fibers in samples from patients with duchenne muscular dystrophy. PLoS One. 2014; 9(9):e107494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arechavala-Gomeza V, Kinali M, Feng L, et al. Immunohistological intensity measurements as a tool to assess sarcolemma-associated protein expression [published correction appears in Neuropathol App Neurobiol. 2014; 40(4):515]. Neuropathol Appl Neurobiol. 2010;36(4):265–274. [DOI] [PubMed] [Google Scholar]
- 15.Arechavala-Gomeza V, Kinali M, Feng L, et al. Revertant fibres and dystrophin traces in Duchenne muscular dystrophy: implication for clinical trials. Neuromuscul Disord. 2010;20(5):295–301. [DOI] [PubMed] [Google Scholar]
- 16.Kinali M, Arechavala-Gomeza V, Feng L, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8(10):918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 18.Dixon A, Heinlein T, Wolleschenky R. Need for standardization of fluorescence measurements from the instrument manufacturer’s view, In: Resch-Genger U, (ed). Standardization and Quality Assurance in Fluorescence Measurements, II—Bioanalytical and Biomedical Applications. Vol 6 Berlin, Germany: Springer-Verlag; 2008. pp 3–24. [Google Scholar]
- 19.Resch-Genger U, Hoffmann K, Nietfeld W, et al. How to improve quality assurance in fluorometry: fluorescence-inherent sources of error and suited fluorescence standards. J Fluoresc. 2005;15(3):337–362. [DOI] [PubMed] [Google Scholar]
- 20.Conway C, Dobson L, O’Grady A, Kay E, Costello S, O’Shea D. Virtual microscopy as an enabler of automated/quantitative assessment of protein expression in TMAs. Histochem Cell Biol. 2008;130(3):447–463. [DOI] [PubMed] [Google Scholar]
- 21.Sirota RL. Defining error in anatomic pathology. Arch Pathol Lab Med. 2006;130(5):604–606. [DOI] [PubMed] [Google Scholar]
- 22.Aeffner F, Wilson K, Martin NT, et al. The gold standard paradox in digital image analysis: manual versus automated scoring as ground truth. Arch Pathol Lab Med. 2017;141(9):1267–1275. [DOI] [PubMed] [Google Scholar]
- 23.Voit T, Stuettgen P, Cremer M, Goebel HH. Dystrophin as a diagnostic marker in Duchenne and Becker muscular dystrophy: correlation of immunofluorescence and western blot. Neuropediatrics. 1991;22(3):152–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.