Abstract
Upon RANKL binding, RANK promotes osteoclast formation through the recruitment of TNF receptor-associated factors (TRAFs). In vitro assays identified two RANK intracellular motifs that bind TRAFs: PVQEET560–565 (Motif 2) and PVQEQG604–609 (Motif 3), which potently mediate osteoclast formation in vitro. To validate the in vitro findings, we have generated knockin (KI) mice harboring inactivating mutations in RANK Motifs 2 and 3. Homozygous KI (RANKKI/KI) mice are born at the predicted Mendelian frequency and normal in tooth eruption. However, RANKKI/KI mice exhibit significant more trabecular bone mass than age- and sex-matched heterozygous KI (RANK+/KI) and wild-type (RANK+/+) counterparts. Bone marrow macrophages (BMMs) from RANKKI/KI mice do not form osteoclasts when they are stimulated with M-CSF and RANKL in vitro. RANKL is able to activate the NF-ĸB, ERK, p38 and JNK pathways in RANKKI/KI BMMs, but it cannot stimulate c-Fos or NFATc1 in the RANKKI/KI cells. Previously, we showed that RANK signaling plays an important role in Porphyromonas gingivalis (Pg)-mediated osteoclast formation by committing BMMs into the osteoclast lineage. Here, we show that RANKL-primed RANKKI/KI BMMs are unable to differentiate into osteoclasts in response to Pg stimulation, indicating that the two RANK motifs are required for Pg-induced osteoclastogenesis. Mechanistically, RANK Motifs 2 and 3 facilitate Pg-induced osteoclastogenesis by stimulating c-Fos and NFATc1 expression during the RANKL pretreatment phase as well as rendering c-Fos and NFATc1 genes responsive to subsequent Pg stimulation. Cell penetrating peptides (CPPs) conjugated with RANK segments containing Motif 2 or 3 block RANKL- and Pg-mediated osteoclastogenesis. The CPP conjugates abrogate RANKL-stimulated c-Fos and NFATc1 expression, but do not affect RANKL-induced activation of NF-ĸB, ERK, p38, JNK or Akt signaling pathway. Taken together, our current findings demonstrate that RANK Motifs 2 and 3 play pivotal roles in osteoclast formation in vivo and mediate Pg-induced osteoclastogenesis in vitro.
Keywords: Osteoclastogenesis, RANKL, RANK Signaling, Porphyromonas gingivalis
Introduction
Osteoclasts, the body’s bone-resorbing cells, are multinucleated cells differentiating from mononuclear precursors of the monocyte/macrophage lineage. The bone-resorbing cell plays critical roles in skeletal development and continues to participate in adult bone remodeling upon skeletal maturation (1,2). Consequently, abnormal osteoclast formation and/or activity lead to the development of bone diseases including osteoporosis and periodontal bone loss (3,4). Osteoclast differentiation requires two key factors: the macrophage colony-stimulating factor (M-CSF) and RANKL(1). RANKL drives osteoclast differentiation through activating its receptor RANK (1), which was identified as a member of the TNF receptor family (5,6).
TNF receptor family members lack intrinsic enzymatic activity and, hence, initiate intracellular signaling typically through the recruitment of TRAFs (7–9). We previously identified three mouse RANK intracellular motifs: PFQEP369–373 (Motif 1), PVQEET560–565 (Motif 2) and PVQEQG604–609 (Motif 3), which were capable of promoting osteoclastogenesis in vitro (10) (Supplemental Fig. 1). These RANK motifs interact with distinct TRAFs, which then facilitate the formation of signaling complexes leading to the activation of NF-ĸB, JNK, ERK, p38, Akt and NFATc1 (11–13). Notably, RANK Motifs 2 and 3 exhibit higher capacity to promote osteoclastogenesis than Motif 1; mutational inactivation of both motifs dramatically impairs the ability of RANK to mediate osteoclastogenesis in vitro (10,14). Additionally, RANK cytoplasmic domain possesses another functional motif (IVVY535−538), which does not bind TRAFs. This TRAF-independent motif stimulates the osteoclast lineage commitment via up-regulating the expression of NFATc1 (15–17), a master transcription factor of osteoclastogenesis (18). Importantly, our subsequent work has shown that RANK Motifs 2 and 3 collaborate with the IVVY motif to promote osteoclast formation through mediating the lineage commitment (14).
Alveolar bone loss is a major complication associated with periodontitis (19). Porphyromonas gingivalis (Pg) is one of major microbes implicated in the development of both periodontitis and periodontal bone loss (20). We previously showed that Pg plays a dual role in osteoclast differentiation (21). Specifically, Pg enhances osteoclastogenesis from RANKL-primed BMMs, but it inhibits osteoclast formation from non-primed cells, revealing that RANKL-induced lineage commitment is critical for Pg-induced osteoclast formation. Furthermore, we found that Pg is unable to induce the expression of NFATc1 and c-Fos (21), two key transcriptional factors for osteoclast differentiation (18,22). Intriguingly, Pg suppresses RANKL-induced expression of c-Fos and NFATc1 in BMMs. Nevertheless, Pg instead enhances c-Fos and NFATc1 expression in RANKL-pretreated BMMs (21). Thus, Pg modulates osteoclastogenesis by modulating RANKL-mediated expression of c-Fos and NFATc1.
The functional role of RANK Motif 2 and Motif 3 in osteoclastogenesis was identified in vitro using a chimeric receptor approach. Moreover, the precise mechanism by which RANKL primes BMMs to assist Pg to induce osteoclastogenesis remains unclear. In this work, we further investigate the in vivo role of RANK Motifs 2 and 3 in osteoclastogenesis by generating knockin (KI) mice bearing inactivating mutations in RANK Motifs 2 and 3. In addition, using BMMs from the KI mice we examine whether RANK Motifs 2 and 3 play roles in RANKL-mediated priming of BMMs that is important for Pg-induced osteoclastogenesis.
Materials and Methods
Chemicals, reagents and bacteria
Chemicals were bought from Sigma (St. Louis, MO, US) unless indicated otherwise. Cell permeable peptide (CPP)-conjugated peptides were synthesized by ChinaPeptides Company (Shanghai, China). Recombinant GST-RANKL was purified as previously described (23). Mouse M-CSF was prepared from CMG14–12 cells, an M-CSF-producing cell line (24). Antibodies against IĸBα (4812S), phospho-IĸBα (2859S), p44/42ERK (4695S), phospho-p44/42ERK (#9101S), JNK (9252S), phospho-JNK (9251S), p38 (#9212), phospho-p38 (9215S), Akt (9272S), phospho-Akt (#9271S), and c-Fos (2250S) were obtained from Cell Signaling Technology (Beverly, MA, US). Antibodies against NFATc1 (sc-7294) and RANK (sc-34249) were from Santa Cruz Biotechnology (Santa Cruz, CA, US). Alexa Fluor-488 phalloidin (A12379) was bought from Life Technologies (Carlsbad, CA, US), and Hoechst-33258 (sc-391053) was from Santa Cruz Biotechnology. Pg strain ATCC33277 was grown in enriched trypticase soy agar plates at 37 °C under the conditions of 10% H2, 5% CO2, and 85% N2 as previously described (21). Bacteria were collected and washed with phosphate-buffered saline (PBS). The bacterial colony forming units were measured as the optical density at 580 nm.
Generation of KI mice harboring inactivating mutations in RANK Motifs 2 and 3
The targeting vector for generating knockin mice was constructed on a pBluescript-SK plasmid. The targeting vector was electroporated into C57BL/6N-tac embryonic stem cells (PRX-B6N Primogenix, Laurie, MO, US). Transfected embryonic stem cells were positively selected with G418 followed by negative selection with ganciclovir. Surviving clones were examined by Southern blot analysis. Positive clones were then injected into blastocysts from albino C57BL/6J to generate RANK+/KI/RANK+/+ chimeras. Embryonic stem electroporation and blastocyst injection were performed at the UAB Transgenic and Genetically Engineered Models Facility. Chimeric male founder animals were bred to albino C57BL/6J females, and black-coated offspring were assessed by PCR to determine transmission of the targeted alleles. The LoxP-flanked NEO cassette was then excised by breeding to a Cre transgene mouse line (B6.FVB-Tg(EIIa-cre)C5379Lmgd/J, JAX Mice #003427). RANK+/KI siblings were mated to produce RANK+/+, RANK+/KI, and RANKKI/KI mice. Mice were generated following the regulations of the Institutional Animal Care and Use Committee (IACUC) at the University of Alabama at Birmingham (UAB) (animal protocol approval number: IACUC-20300). The mice were maintained and used for experiments according to the regulations of UAB IACUC (animal protocol approval number: IACUC-07854).
Genotyping the KI mice by PCR
Genomic DNA was isolated from the tail tip of each mouse, and processed using 100 μl lysis buffer (50 mM KCl, 10 mM Tris, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20) freshly supplemented with 0.1 mg/ml Proteinase K (LS004222; Worthington Biochemical Corporation, Lakewood, JN, US). DNA samples were then subjected to PCR amplification in a 20 μl reaction mixture consisting of 10 μl DreamTaq Green PCR Master Mix (K1082; Thermo Scientific, Waltham, MA, US), 0.5 μl forward primer, 0.5 μl reverse primer, 2 μl sample DNA, and 7 μl DNAase-free water. The reaction was initiated at 95 ℃ for 4 minutes, followed by 35 cycles of 95 ℃ for 30 seconds, 59 ℃ for 30 seconds and 72 ℃ for 35 seconds, completed at 72 ℃ for 5 minutes. The PCR products were examined in 2.5% agarose gel stained with ethidium bromide (80 V, 30 minutes), and visualized by UV light. PCR primers are 5’-ATGCACGGCCACTACCACTAAGGCTT-3’ (forward) and 5’-AGATTTAGTGGCCAAGTACCTGCCAC-3’ (reverse).
X-ray and micro-CT (μCT) analyses of bone
Excised mouse femurs were positioned on the middle shelf of the X-ray cabinet and scanned at 18 kV. A Faxitron Specimen Radiography system (MX-20, DC4; Faxitron X-ray Corporation, Tucson, AZ, US) was used to obtain digital X-ray images. Femur samples were also analyzed with the Scanco μCT40 desktop cone-beam micro-CT scanner (Scanco Medical AG, Brüttisellen, Switzerland). To obtain data of the whole bone, femurs were placed horizontally in 20-mm-diameter scanning holders, while inverted in 12-mm-diameter scanning holders in order to perform the analyses of cortical and trabecular parameters. The following settings were set up: 70 kVp, 114 μA, 200 ms (integration time), 10 μm voxel size for whole bone, and 6 μm voxel size for cortical or trabecular bone. Scans were then performed and specifically, for cortical bone, femurs were scanned at the midshaft for a scan of 25 slices, whereas for trabecular bone, the scans began 2.5 mm from the growth plate and had 200 slices (6 μm). The region of interest (ROI) was drawn on each of the slices accordingly, and bones were thresholded at 548.8 mgHA/ccm (whole bone), 1033 mgHA/ccm (cortical bone), or 548.8 mgHA/ccm (trabecular bone). The 3-D reconstruction was performed using the outlined slices (μCT Ray v3.8). Data were generated for bone volume (BV), total volume (TV), BV/TV, bone density, trabecular number (Tb.N), trabecular separation (Tb.Sp), trabecular thickness (Tb.Th) and cortical thickness (Ct.Th).
Static and dynamic histomorphometric analyses
Lumbar vertebrae and femurs of mice were dissected and fixed in 70% ethanol for at least 24 hours. Then all samples were processed for infiltration and plastic embedding. The fully plasticized sample sections were cut in the coronal planes at 5 μm thickness, and stained with Goldner’s Trichrome. The osteoclast surface (OcS/BS), osteoblast surface (ObS/BS), osteoclast number (N.Oc/BS), and osteoblast number (N.Ob/BS) were quantified (Bioquant Osteo 2018 software, Nashville, TN, US). For dynamic histomorphometric analysis, intraperitoneal injections of calcein (30 mg/kg; C0875; Sigma) were performed 6 and 2 days before euthanasia. Thick sections (10 μm) with calcein labeling were analyzed under the fluorescent microscope (Olympus BX51) with the U-MF2 filter set. The mineralizing surface (MS/BS), mineral apposition rate (MAR), and bone formation rate (BFR/BS, BFR/BV) were calculated (Bioquant Osteo 2018 software, Nashville, TN, US). All measurements were based on the recommendation of the ASBMR Histomophometry Nomenclature Committee (25).
In vitro osteoclastogenesis and bone resorption assays
Murine bone marrow cells were collected from femurs and tibiae of 6- to 8-week-old WT or RANKKI/KI mice. Then cells were cultured overnight in tissue culture dishes with α-MEM consisting of 10% heat-inactivated fetal bovine serum (α−10 medium) to remove stromal cells. The next day, non-adherent cells were obtained and subject to Ficoll-Hypaque density gradient centrifugation purifications. Purified cells were counted and plated in tissue culture plates at density indicated in individual assays. BMMs were enriched by one- or two-day M-CSF treatment (40 ng/ml) prior to the addition of RANKL (100 ng/ml). Incubation of cell cultures was performed in a humidified 37 °C, 7% CO2 incubator. Culture plates were stained for tartrate-resistant acid phosphatase (TRAP) activity using a Leukocyte Acid Phosphatase kit (387A-1KT; Sigma). The method of quantifying osteoclasts was to count TRAP-positive multinucleated cells (MNCs). Bovine cortical bone slices (thickness: 0.25–0.5 mm) were autoclaved and plated in 24-well plates. BMMs (1 × 105 cells/well) were then seeded and cultured under the conditions of individual assays. Next, bone slices were sonicated to remove cells, fixed with 4% paraformaldehyde (PFA), treated with 0.3% H2O2, and washed with PBS. The resorption of bone slices was examined using scanning electronic microscopy.
Immunofluorescence analysis of in vitro osteoclastogenesis
The procedures of actin ring and nucleic staining were performed as described previously (26). First, BMMs (1 × 105 cells/well) were seeded and cultured under the conditions of individual assays. Cells were then fixed for 20 minutes at room temperature with 4% PFA and washed 2 times with PBS. After fixation, cells were penetrated with 0.1%−0.2% Triton X-100 for 10 minutes. DNA-specific fluorescent Hoechst 33258 was used for nucleic staining (5–10 minutes), and Alexa Fluor 488-labeled phalloidin for actin ring staining (15 minutes). Cells were viewed by an inverted fluorescence microscopy (ECLIPSE TE2000-U; Nikon, Tokyo, Japan). An EXFO X-Cite 120 lamp was the light source. Images were obtained and analyzed by the software QCapture Pro (Version 5.1.1.14; Q Imaging, Media Cybernetics, Rockville, MD, US).
Western blot
BMMs were washed and lysed at indicated time points as described in individual experiments. For c-Fos or NFATc1 analysis, cells were washed with ice-cold PBS 2 times and lysed in lysis buffer containing 10× Cell Lysis Buffer (#9803S; Cell Signaling Technology), 100× Protease Inhibitor Cocktail (P8340; Sigma). For the analysis of Akt, IĸBα, p44/42ERK, JNK, p38, and the phosphorylated form of these antibodies, lysis buffer consisted of 10× Cell Lysis Buffer, 100× Protease Inhibitor Cocktail, 100× Phosphatase Inhibitor Cocktail 1 (P2850; Sigma), and 100× PHOSPHATASE INHIBITOR COCKTAIL II (P5726; Sigma). The protein extracts were boiled in the presence of 4× Laemmli Sample Buffer (161–0747; Bio-Rad, Hercules, CA, US) for 5–10 minutes, and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were then transferred to nitrocellulose membranes (1620112; Bio-Rad), blocked in Odyssey Blocking Buffer (927–40000; LI-COR Biosciences, Lincoln, NE, US) for 1 hour, and incubated with primary antibodies at 4 °C overnight. Membranes were then incubated with dye-labeled second antibodies (LI-COR Biosciences) and were visualized by the Image Studio software (LI-COR Biosciences). Alternatively, membranes were blocked in 5% nonfat dry milk in TBST for 1 hour and incubated with primary antibodies overnight at 4 °C. Subsequently, membranes were washed 3 times with TBST. After 1 hour of incubation with secondary antibodies, membranes were washed with TBST and blots were detected by the HRP Substrate kit from Millipore (WBKLS0100; Billerica, MA, US).
Statistical analysis
All results are representative of mean ± standard deviation (S.D.) of three independent replicates. Student’s t test or one-way ANOVA was used to determine differences among different groups. Statistical significance was defined as p value < 0.05.
Results
Generation and characterization of KI mice bearing inactivating mutations in RANK Motifs 2 and 3
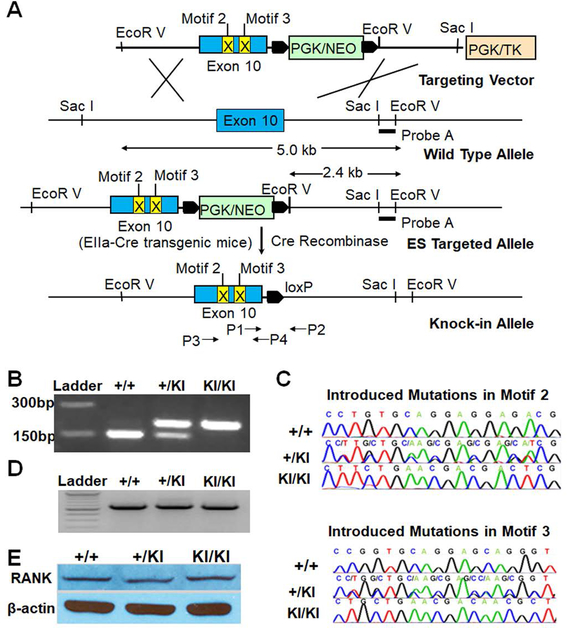
Using standard molecular techniques, we developed a targeting vector to convert WT RANK allele to mutated allele bearing nucleotide substitutions in Exon 10 which blocked the function of Motifs 2 and 3 in our previous study (10) (Fig. 1A). The targeting vector was electroporated into C57BL/6N embryonic stem cells and selected with G418 and gancyclovir. Surviving embryonic stem cell colonies were screened by Southern blots with Probe A to identify correct clones (Fig. 1A). The introduction of an EcoR V site enabled the discrimination of the targeted allele (2.4 kb) from WT allele (5.0 kb) (Fig. 1A). Embryonic stem cell clones were microinjected into albino blastocysts to produce chimeric founder mice that were subsequently bred to albino C57BL/6J females. Black-coated offspring were analyzed by PCR to establish germline transmission. The PGK/neo cassette was then excised by breeding to Cre transgenic mice (Fig. 1A). The remaining LoxP site provided a unique insert for identifying the KI allele by PCR (Fig. 1B) using primers P1 and P2. Introduced mutations were confirmed by sequencing PCR products (Fig. 1C) amplified by primers P3 and P4. The mutations did not alter mutant RANK expression (Fig. 1D–E). Also, we sequenced the transcript to confirm that the LoxP site did not affect splicing and absence of other unwanted mutations in the transcript in addition to the introduced mutations.
Fig. 1.
Generation, genotyping and validation of the KI mice. (A) The targeting strategy. (B) Genotyping by PCR using primers P1 and P2 shown in Fig. 1A. This pair of primers yields 160 bp PCR products from WT allele. The presence of a LoxP site in the KI allele results in 200 bp products. (C) Sequencing results of PCR products amplified using P3 and P4 shown in Fig. 1A and genomic DNA isolated from RANK+/+, RANK+/KI and RANKKI/KI mice. (D) Semi-quantitative RT-PCR analysis of RANK mRNA levels in BMMs from RANK+/+, RANK+/KI and RANKKI/KI mice. The assay was independently repeated twice. (E) Western blot analysis of RANK protein levels in BMMs from RANK+/+, RANK+/KI and RANKKI/KI mice. The assay was independently repeated twice.
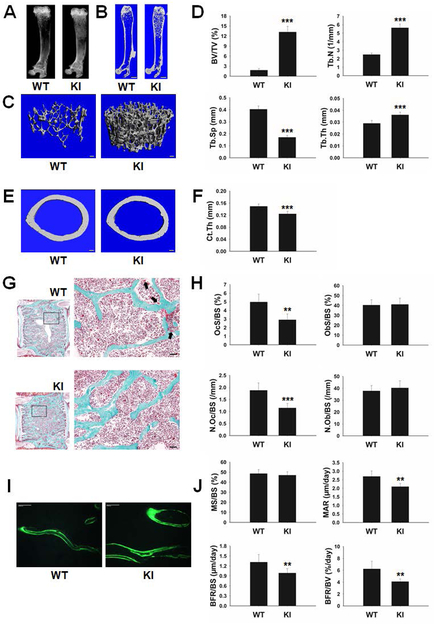
Homozygous KI (RANKKI/KI) mice were born at the predicted Mendelian frequency, normal in tooth eruption, and similar in size to WT mice. X-ray and μCT analysis of whole femurs from 8-week-old female WT and RANKKI/KI mice showed more trabecular bone in RANKKI/KI mice than WT controls (Fig. 2A–B). The 3-D μCT reconstruction of trabecular bone further confirmed the dramatic difference in trabecular bone among these genotypes (Fig. 2C). Statistical analysis of the 3-D μCT data indicates that RANKKI/KI mice exhibited significant differences in BV/TV, Tb.N, Tb.Sp and Tb.Th in comparison to WT mice (Fig. 2D). Moreover, RANKKI/KI mice showed a significant reduction in cortical bone thickness versus WT mice (Fig. 2E–F).
Fig. 2.
Characterization of bone phenotypes of 8-week-old female WT and KI mice. X-ray (A) and μCT (B) analysis of femurs from 8-week-old WT and RANKKI/KI (KI) mice. Scale bar = 1 mm. (C) Representative images of μCT 3-D reconstruction of the trabecular region. Scale bar = 100 μm. (D) Statistical analysis of μCT 3-D data (WT, n=6; KI, n=6). ***, p < 0.001. (E) Transverse μCT images of cortical bone at the midshaft of femurs. Scale bar = 100 μm. (F) Statistical analysis of the cortical bone thickness (WT, n=6; KI, n=5). ***, p < 0.001. (G) Representative images of static histomorphometric analyses of lumbar vertebrae. Osteoclasts are indicated by black arrows. Scale bar = 50 μm. (H) The quantification of static histomorphometric parameters OcS/BS, ObS/BS, N.Oc/BS, and N.Ob/BS (WT, n=5; KI, n=6). **, p < 0.01; ***, p < 0.001. (I) Representative images of dynamic histomorphometric analyses of femurs. Scale bar = 50 μm. (J) The quantification of dynamic histomorphometric parameters MS/BS, MAR, BFR/BS, and BFR/BV (WT, n=6; KI, n=7). **, p < 0.01.
Static histomorphometric analysis revealed significant lower percentage of bone surface occupied by osteoclasts (OcS/BS) and significant reduced osteoclast number per bone surface (N.Oc/BS) in RANKKI/KI mice compared to those in WT mice (Fig. 2G–H). These data indicate that the two RANK motifs play important roles in osteoclast differentiation in vivo.
In contrast, no significant difference in osteoblast parameters (ObS/BS and N.Ob/BS) was observed between RANKKI/KI and WT mice (Fig. 2G–H). Interestingly, dynamic histomorphometric analysis demonstrates that MAR, BFR/BS and BFR/BV, but not MS/BS, in RANKKI/KI mice were lower than those in WT mice (Fig. 2I–J). These findings demonstrate that while the mutation of RANK Motifs 2 and 3 does not affect osteoblast differentiation or survival, it leads to impaired bone formation and mineralization.
RANKKI/KI BMMs fail to form osteoclasts due to impaired c-Fos and NFATc1 expression
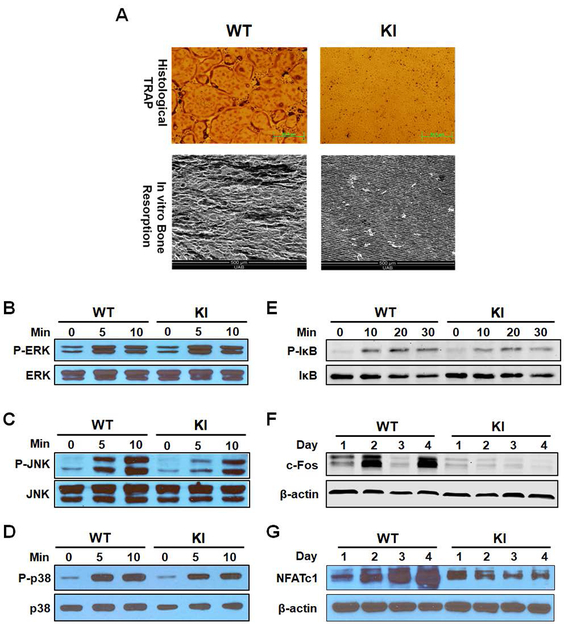
We then performed in vitro assays using BMMs from WT and RANKKI/KI mice. When RANKKI/KI BMMs were stimulated with M-CSF and RANKL, they did not differentiate into osteoclasts (top panel, Fig. 3A). Consistent with osteoclast formation data, no resorption pits were observed in bone resorption assays with BMMs from RANKKI/KI mice (bottom panel, Fig. 3A). In addition, we found that RANKL was still able to activate the ERK, JNK, p38 and NF-κB pathways in RANKKI/KI BMMs (Fig. 3B–E). The data indicate that the dual mutation of RANK Motifs 2 and 3 does not considerably influence the activation of NF-κB and MAPK pathways since Motif 1 is still intact in the mutant RANK and is able to activate these pathways. More importantly, the data have also ruled out the concern that the bone phenotypes in RANKKI/KI mice are due to adverse effects of the inactivating mutations on the 3-D structure of RANK. However, the dual mutation of Motifs 2 and 3 dramatically reduced the capacity of RANK to up-regulate c-Fos and NFATc1 expression (Fig. 3F–G). Consistent with the previous finding that c-Fos transcriptionally activates NFATc1 (27), we found that the impaired c-Fos expression is associated with decreased NFATc1 mRNA levels in RANKKI/KI (Supplemental Fig. 2). These findings indicate that Motifs 2 and 3 promote osteoclast formation by inducing c-Fos and NFATc1 expression.
Fig. 3.
In vitro characterization of osteoclast formation/function and RANK signaling in BMMs from WT and homozygous KI (RANKKI/KI) mice. (A) BMMs were plated in 24-well plates (1 × 105 cells/well) and treated with 40 ng/ml M-CSF and 100 ng/ml RANKL for 4 days. Then cultures were stained for tartrate-resistant acid phosphatase (TRAP) activity (top panel). Scale bar = 50 μm. BMMs were plated on bovine cortical bone slices placed in 24-well plates (1 × 105 cells/well) and treated with 40 ng/ml M-CSF and 100 ng/ml RANKL for 7 days. Bone slices were harvested for scanning electron microscopy analysis (bottom panel). Scale bar = 500 μm. Assays were performed in triplicates and repeated twice. (B-E) Activation of ERK, JNK, p38 and NF-κB/IκB in WT and RANKKI/KI BMMs by RANKL was assessed as phosphorylation of ERK, JNK, and p38 and IκB using Western blot analysis with antibodies against phospho-JNK, phospho-ERK, phospho-p38 or phospho-IκB. Min: minute. The assays were independently repeated twice. (F-G) BMMs were treated with 40 ng/ml M-CSF and 100 ng/ml RANKL for 1, 2, 3, or 4 days, and harvested for Western blot analysis of c-Fos and NFATc1 expression.
We next determined whether the ectopic expression of the constitutively active form of NFATc1 (CA-NFATc1) in RANKKI/KI BMMs restores the impaired osteoclastogenesis. To this end, WT and RANKKI/KI BMMs were infected with virus expressing GFP or CA-NFATc1, and the expression of CA-NFATc1 was then assessed by Western blot analysis (Supplemental Fig. 3A). The infected cells were also used for osteoclastogenesis assays in the absence or presence of RANKL. While the ectopic expression of CA-NFATc1 in WT BMMs was unable to promote osteoclastogenesis in the absence of RANKL (top row in the left panel, Supplemental Figure 3B and 3C), it enhanced osteoclastogenesis in the presence of 50 ng/ml or 100 ng/ml RANKL (two bottom rows in the left panel, Supplemental Figure 3B and 3C). This finding suggests that, in addition to NFATc1, RANK also initiates an unidentified signaling pathway(s) which is critical for osteoclastogenesis in response to RANKL stimulation. The ectopic expression of CA-NFATc1 in RANKKI/KI BMMs also failed to promote osteoclastogenesis without RANKL (top row in the right panel, Supplemental Figure 3B and 3C). This further suggests that RANK initiates an unidentified signaling pathway(s) required for osteoclastogenesis. Notably, the ectopic expression of CA-NFATc1 in RANKKI/KI BMMs did not enhance osteoclastogenesis in the presence of 50 ng/ml or 100 ng/ml RANKL (two bottom rows in the right panel, Supplemental Figure 3B and 3C). This finding suggests that the two RANK motifs are responsible for initiating the unidentified signaling pathway(s).
RANKL-primed RANKKI/KI BMMs do not form osteoclasts in response to Pg stimulation
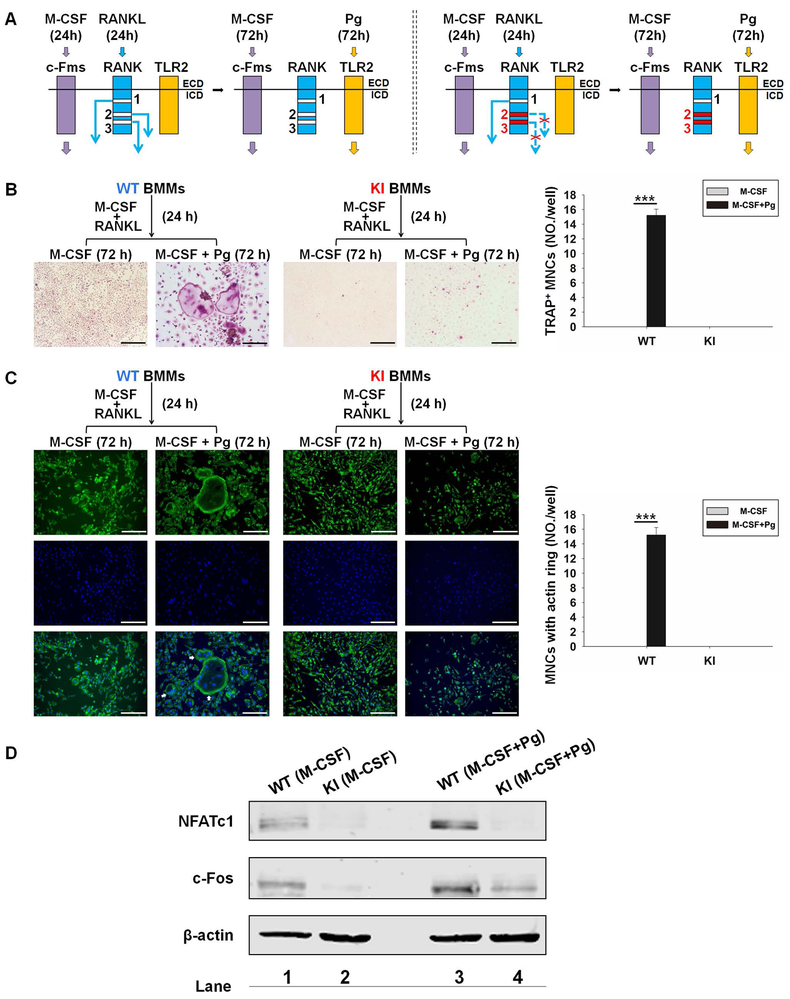
We previously showed that RANK signaling is essential for Pg-mediated osteoclast differentiation by committing BMMs into the osteoclast lineage (21). Using BMMs from WT and RANKKI/KI mice, we investigated the potential involvement of RANK Motifs 2 and 3 in Pg-induced osteoclast formation. To this end, we followed the experimental procedures shown in Fig. 4A. WT and KI BMMs were treated with M-CSF and RANKL for 1 day to prime BMMs to the osteoclast lineage. Then, the RANKL-primed cells were treated for 3 days with M-CSF and Pg to determine their capacity to differentiate osteoclasts with Pg stimulation. It is noted that while all the three RANK motifs were functional and activated their downstream signaling pathways in WT BMMs during the 24-hour pretreatment, in KI BMMs only Motif 1 was functional and initiated its signaling pathways (Fig. 4A). Our data demonstrated that while RANKL-primed WT BMMs formed osteoclasts when stimulated with Pg in the presence of M-CSF, KI BMMs formed no osteoclast (Fig. 4B). Given that multinucleation and actin ring formation are two key characteristics of osteoclastogenesis (28,29), we further examined these two phenotypical features in Pg-induced osteoclastogenesis. As shown in Fig. 4C, osteoclasts differentiated from RANKL-primed WT BMMs followed by Pg stimulation contained multiple nuclei and formed large actin ring. RANKL-primed KI BMMs remained mononuclear and lacked typical actin ring. The findings show that RANKL-primed RANKKI/KI BMMs are unable to differentiate into osteoclasts when treated with Pg stimulation, indicating that RANK Motifs 2 and 3 are required for Pg-induced osteoclastogenesis.
Fig. 4.
RANK Motifs 2 and 3 play a role in Pg-induced osteoclastogenesis by modulating NFATc1 and c-Fos expression. (A) The experimental procedures. Left panel: when WT BMMs were pretreated with M-CSF (40 ng/ml) plus RANKL (100 ng/ml) for 1 day, c-Fms signaling and all RANK signaling pathways (Motifs 1, 2 and 3) were activated. During subsequent 3-day Pg treatment (m.o.i = 50), c-Fms signaling and TLR2 signaling were activated. Right panel: in contrast, in KI BMMs only RANK Motif 1 signaling was functional during the RANKL pretreatment. KI, RANKKI/KI; ECD, extracellular domain; ICD, intracellular domain. (B) Representative images (left panel) of osteoclastogenesis assays as described in Fig. 4A. Quantification of osteoclastogenesis (right panel). (C) Actin ring and nuclear staining. The culture of BMMs was performed as in Fig. 4A. Cells were then stained for actin ring formation and nucleation. Osteoclasts are indicated by white arrows (left panel). Quantification of osteoclastogenesis (right panel). For Fig. 4B and 4C, scale bar = 200 μm. Data are mean ± S.D. of three independent experiments. ***, p < 0.001. (D) Western blot analysis. WT or KI BMMs were pretreated with M-CSF (40 ng/ml) plus RANKL (100 ng/ml) for 1 day and then with M-CSF, or M-CSF plus Pg (m.o.i.= 50) for 1 day. The expression of NFATc1 and c-Fos was assessed by Western blot analysis using β-actin as a loading control. The results are representative of three independent experiments.
RANK Motifs 2 and 3 play a role in Pg-induced osteoclastogenesis by mediating NFATc1 and c-Fos expression
Two transcription factors (c-Fos and NFATc1) are critically involved in osteoclastogenesis (18,22), and RANK promotes osteoclast formation in part by stimulating NFATc1 expression via c-Fos (30). Previously, we showed that Pg is not able to induce the expression of NFATc1 or c-Fos in normal BMMs (21). Nonetheless, 24-hour RANKL pretreatment increases c-Fos and NFATc1 expression and subsequent Pg stimulation further enhances RANKL-induced expression of the two transcriptional factors, indicating that RANK signaling assists Pg-induced osteoclastogenesis by modulating the expression of c-Fos and NFATc1. To address whether RANK Motifs 2 and 3 specifically play a role in Pg-induced osteoclastogenesis by regulating the expression of c-Fos and NFATc1, WT and KI cells were stimulated for 24 hours with M-CSF and RANKL and then treated for 1 day with M-CSF alone or M-CSF plus Pg before the assessment of c-Fos and NFATc1 expression. 24-hour RANKL pretreatment induced the expression of c-Fos and NFATc1 in WT BMMs (lane 1, Fig. 4D), but not in KI BMMs (lane 2, Fig. 4D). Moreover, subsequent Pg stimulation of RANKL-pretreated WT BMMs enhanced the expression of c-Fos and NFATc1 (lane 1 vs. 3, Fig. 4D). In contrast, succeeding stimulation of RANKL-pretreated KI BMMs by Pg increased c-Fos expression but not NFATc1 expression (lane 2 vs. 4, Fig. 4D). However, Pg-induced c-Fos expression in KI BMMs was lower than that in WT BMMs. These data demonstrate that RANK Motifs 2 and 3 facilitate Pg-induced osteoclast differentiation by up-regulating c-Fos and NFATc1 expression during the RANKL pretreatment phase as well as rendering c-Fos and NFATc1 genes responsive to subsequent Pg stimulation.
CPP conjugated with RANK fragments containing Motif 2 or 3 inhibit osteoclastogenesis and the expression of c-Fos and NFATc1
To further study the function of RANK Motifs 2 and 3 in osteoclastogenesis, we developed CPP conjugated with RANK fragments containing Motif 2 or 3 and examined their effect on osteoclastogenesis. We reasoned that these CPP conjugates could competitively bind to TRAFs and thus prevent these RANK motifs from initiating downstream signaling (Supplemental Fig. 4A–B). Specifically, we synthesized CPP (Hph-1) conjugated with 10-a.a RANK fragment containing WT Motif 2 (M2–10), inactivated Motif 2 (iM2–10), WT Motif 3 (M3–10), inactivated Motif 3 (iM3–10) (Supplemental Fig. 4C). These CPP conjugates were tagged with FITC at their N-termini. Hph-1 is a protein-transduction domain from a human transcriptional factor (31). The mutations introduced to inactivate these motifs were identified in our previous study (10). To examine the capability of the CPP conjugates to transduce BMMs and transduction kinetics, BMMs were incubated with iM2–10 (Supplemental Fig. 4D), M2–10 (Supplemental Fig. 4E), iM3–10 (Supplemental Fig. 4F), or M3–10 (Supplemental Fig. 4G) for different lengths of time. The uptake of these CPP conjugates by BMMs was detected (distributed mainly in the cytoplasm) in most of cells at 2 hours and reached the plateau at 6 hours (Supplemental Fig. 4D–G).
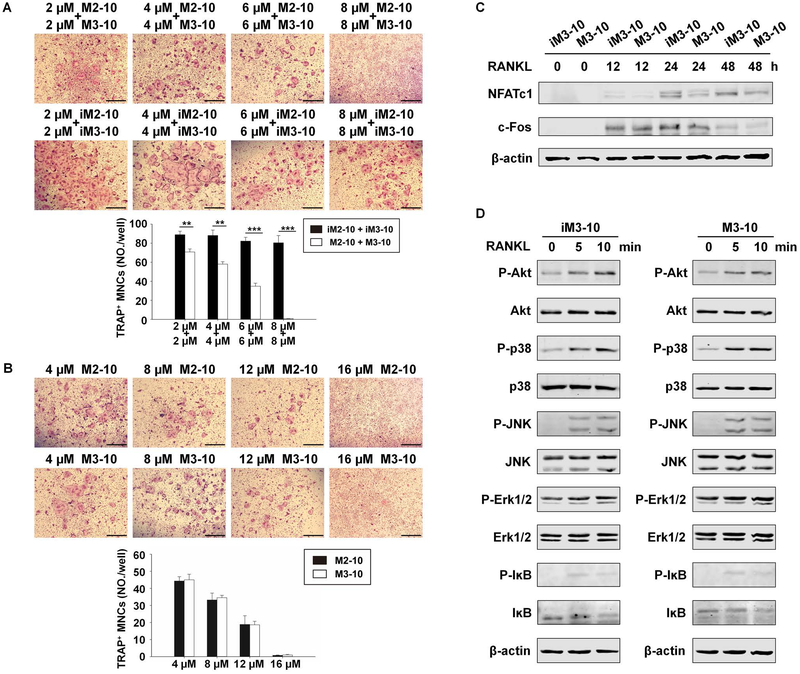
Having demonstrated that the CPP conjugates can efficiently transduce BMMs, we then investigated their effect on osteoclastogenesis. BMMs were treated with M-CSF and RANKL in the presence of 2 μM M2–10 plus 2 μM M3–10, 4 μM M2–10 plus 4 μM M3–10, 6 μM M2–10 plus 6 μM M3–10, or 8 μM M2–10 plus 8 μM M3–10 for 4 days (top panel, Fig. 5A). As control, the same set of assays were performed with iM2–10 and iM3–10 (middle panel, Fig. 5A). The data showed that a combination of M2–10 and M3–10 dose-dependently inhibited osteoclastogenesis (bottom panel, Fig. 5A). Notably, 8μM M2–10 plus 8μM M3–10 almost completely blocked osteoclastogenesis (Fig. 5A).
Fig. 5.
M2–10 and M3–10 inhibit osteoclastogenesis and the expression of c-Fos and NFATc1. (A) Osteoclastogenesis assay with a combination of M2–10 and M3–10. After two-day incubation with M-CSF (40 ng/ml) in 24-well plates, WT BMMs were treated with M-CSF (40 ng/ml) plus different doses of CPP conjugates for 7 hours. RANKL (100 ng/ml) was then added and the cultures were continued for 4 days prior to TRAP staining. Representative areas of different culture conditions are shown (top and middle panel). TRAP-positive MNCs per well were counted (bottom panel). (B) Osteoclastogenesis assay with M2–10 or M3–10. The assays were performed as in Fig. 5A. Representative areas of TRAP staining (top and middle panel) and quantification of osteoclastogenesis (bottom panel) were shown. For A and B, Scale bar = 500 μm. Each condition had three replicates. Data are means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (C) Western blot analysis. WT BMMs were cultured with M-CSF (40 ng/ml) for 2 days, continued with M-CSF (40 ng/ml) plus iM3–10 (12 μM) or M-CSF plus M3–10 (12 μM) for 7 hours before the treatment with RANKL (100 ng/ml) for 0, 12, 24 or 48 hours. The expression of NFATc1 and c-Fos was determined by Western blot. β-actin was used as a loading control. (D) WT BMMs were treated with 12 μM iM3–10 or M3–10 for 24 hours in the presence of M-CSF (40 ng/ml), followed by the stimulation of RANKL (100 ng/ml) for 0, 5, or 10 minutes. Activation of the NF-ĸB, Akt, and MAPK (ERK, JNK, and p38) pathways was assessed as phosphorylation of IĸBα, ERK, JNK, and p38 using Western blot with specific antibodies. The immunoblots were stripped and reprobed with antibodies against IĸBα, Akt, JNK, ERK, and p38. Data are representative of three independent experiments.
As shown in Supplemental Fig. 1, RANK Motif 2 shares high similarity in amino acid sequence with Motif 3. This raises a possibility that M2–10 is not only able to block Motif 2 function but can also inhibit Motif 3 function, and M3–10 can do the same. To investigate this possibility, BMMs were treated with M-CSF and RANKL in the presence of 4 μM M2–10, 8 μM M2–10, 12 μM M2–10, or 16 μM M2–10 for 4 days (top panel, Fig. 5B). In addition, the same set of assays were performed with M3–10 (middle panel, Fig. 5B). The capacity of 16 μM M2–10 or M3–10 to inhibit osteoclastogenesis (Fig. 5B) was similar to that of the combination of 8 μM M2–10 plus 8 μM M3–10 (Fig. 5A). Moreover, the inhibitory capacity of M2–10 was similar to that of M3–10 (Fig. 5B). These findings demonstrate that M2–10 not only can block Motif 2 function but is also able to inhibit Motif 3 function, and M3–10 can do the same.
To determine the minimal length of sequences capable of blocking RANK motif function, we further developed CPP conjugates containing shorter RANK fragments (8 a.a or 6 a.a): M2–8 vs. iM2–8, M3–8 vs. iM3–8 (Supplemental Fig. 5A), and M2–6 vs. iM2–6, M3–6 vs. iM3–6 (Supplemental Fig. 5B). To examine their blocking capacity, we repeated the assays in Fig. 5 with M2–8, iM2–8, M3–8 and iM3–8 (Supplemental Fig. 5C), or with M2–6, iM2–6, M3–6 and iM3–6 (Supplemental Fig. 5D). The data showed that M2–8/M3–8 and M2–6/M3–6 had lower blocking capacity than M2–10/M3–10 (Supplemental Fig. 5C–D). Hence, we used M2–10 and/or M3–10 for the remaining studies.
Next, we determined the effect of M3–10 on RANKL-mediated c-Fos and NFATc1 expression. To this end, WT BMMs were pretreated with M3–10 or iM3–10 for 7 hours and followed by RANKL stimulation for 0, 12, 24, or 48 hours (Fig. 5C). c-Fos and NFATc1 expression was then analyzed by Western blot. Compared to iM3–10, M3–10 reduced the level of NFATc1 and c-Fos by 74.41% and 53.75% at day 1. Moreover, the expression of NFATc1 and c-Fos in the M3–10 group decreased by 46.27% and 44.71% at day 2 than the iM3–10 group (Fig. 5C). The results demonstrate that M3–10 exhibits higher capacity to suppress the expression of c-Fos and NFATc1 than iM3–10, indicating that the CPP conjugate inhibits osteoclastogenesis by suppressing RANKL-induced expression of c-Fos and NFATc1. Moreover, we determined the effect of M3–10 on the activation of the NF-ĸB, Akt, and MAPKs (JNK, ERK, and p38) pathways by RANKL. WT BMMs were pretreated with M3–10 or iM3–10 overnight and were then treated with RANKL for 0, 5, or 10 minutes prior to cell lysis for assessing the activation of the NF-ĸB, Akt, and MAPK pathways. Our data show that M3–10 does not affect the activation of the NF-ĸB, Akt, or MAPK pathway by RANKL (Fig. 5D).
Addition of M2–10 and M3–10 during RANKL pretreatment abrogates the capacity of BMMs to differentiation into osteoclasts
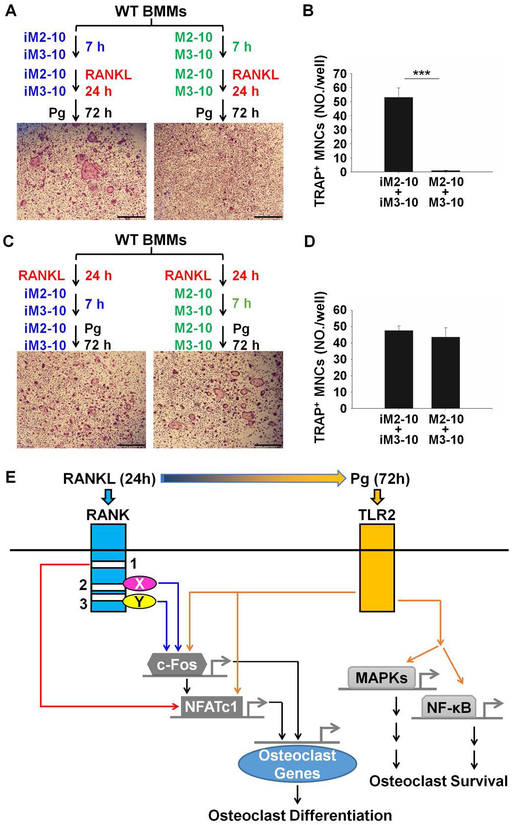
Finally, we addressed whether the CPP conjugates can block Pg-induced osteoclastogenesis. More specifically, we determined if the addition of M2–10 and M3–10 during RANKL pretreatment affects the capacity of BMMs to differentiate into osteoclasts with Pg stimulation. As shown in Fig. 6A, WT BMMs were pretreated with 8 μM iM2–10 plus 8 μM iM3–10 or 8 μM M2–10 plus 8 μM M3–10 for 7 hours and RANKL was then added to the cultures. 24 hours after the addition of RANKL, RANKL was washed out and the cultures were further stimulated with Pg for 3 days. While BMMs pretreated with RANKL in the presence of iM2–10 plus iM3–10 formed osteoclasts in response to subsequent Pg stimulation, the cells pretreated with RANKL in the presence of M2–10 plus M3–10 failed to do so (Fig. 6A–B). Potentially, the failure of BMMs pretreated with RANKL in the presence of M2–10 plus M3–10 to differentiate into osteoclasts in response to Pg treatment partially results from the effect of M2–10 and M3–10 that were present inside BMMs and could not be removed during the washing process. To address this possibility, we performed an additional experiment shown in Fig. 6C; WT BMMs were pretreated with RANKL for 24 hours. After removal of RANKL, 8 μM iM2–10 plus 8 μM iM3–10 or 8 μM M2–10 plus 8 μM M3–10 were added to the cultures. 7 hours later, Pg was added and the cultures were maintained for 3 days. The results showed that the cultures with M2–10 and M3–10 formed osteoclasts compared to those with iM2–10 and iM3–10, indicating that the addition of M2–10 and M3–10 after RANKL pretreatment had no effect on osteoclastogenesis (Fig. 6C–D). Taken together, these findings indicate that the addition of M2–10 and M3–10 during RANKL pretreatment abrogates the capacity of BMMs to differentiate into osteoclasts in response to Pg treatment.
Fig. 6.
M2–10 and M3–10 block Pg-induced osteoclastogenesis. (A) M2–10 and M3–10 block RANKL-mediated priming during RANKL pretreatment that is required for Pg-induced osteoclastogenesis. WT BMMs were pretreated 8 μM M2–10 plus 8 μM M3–10, or 8 μM iM2–10 plus 8 μM iM3–10, in the presence of M-CSF (40 ng/ml) for 7 hours and RANKL (100 ng/ml) was then added. 1 day later, RANKL was removed and the cultures were continued with M-CSF (40 ng/ml) plus Pg (m.o.i. = 50) for 3 days. The cultures were then stained for TRAP activity. A representative area of the cultures from each condition is shown. Scale bar = 500 μM. (B) Quantification of assays in Fig. 6A. Data are expressed as mean ± S.D. of three independent experiments. ***, p < 0.001. (C) The addition of M2–10 and M3–10 after RANKL pretreatment had no effect on Pg-induced osteoclastogenesis. The culture of BMMs was similar to that described for A except that M2–10 and M3–10 were added after RANKL pretreatment. Scale bar = 500 μM. (D) Quantification of assays in Fig. 6C. Data are expressed as mean ± S.D. of three independent experiments. ***, p < 0.001. (E) The proposed model for the molecular mechanism by which RANKL pretreatment facilitates Pg-induced osteoclastogenesis.
Discussion
RANK is a member of the TNF receptor family (32). Given that TNF receptor family members normally initiate intracellular signaling via the recruitment of various TRAFs through their specific cytoplasmic motifs (33), early studies on RANK signaling were primarily focused on characterizing RANK cytoplasmic regions that interact with TRAF proteins using various in vitro biochemical assays (6,34–37). However, these biochemical studies generated conflicting results, leading to a concern regarding the physiological significance of these in vitro interaction findings to osteoclast formation and activity. To generate more physiologically relevant findings, we developed a chimeric receptor to define the RANK intracellular motif(s) that mediate osteoclast differentiation and function, which are important functional readouts (11). This receptor comprises mouse TNFR1 external domain conjugated with the mouse RANK transmembrane and intracellular domains. Given that TNFR1 and RANK belong to the TNF receptor family and both are activated by ligand-triggered trimerization, we reasoned that the chimera can be activated by TNF to activate RANK signaling involved in the osteoclast differentiation and activity. This chimeric receptor approach enabled the identification of three motifs that mediate osteoclast differentiation and function in vitro (10) (Supplemental Fig. 1). Nonetheless, these functional data are in vitro findings obtained from an artificial study system (i.e., the chimeric receptor), therefore it is critical to validate the in vitro findings using an in vivo mouse model. Since Motif 2 and Motif 3 are more active than Motif 1 in regulating osteoclastogenesis in vitro (10), we investigated in vivo function of RANK Motifs 2 and 3 by generating KI mice bearing inactivating mutations in these two motifs. We found that, similar to RANK KO mice (38,39), our KI mice have an increase in trabecular bone mass and a decline in osteoclast numbers. In addition, it is noted that the KI mice exhibit impaired bone formation and mineralization (Fig. 2I–J), a phenotype also seen in RANK KO mice (39). The impaired bone formation and mineralization in the KI mice likely result from the coupling of bone formation to bone resorption during bone remodeling (3). However, our KI mice do not have abnormalities such as retarded growth, shortened long bones and defective tooth eruption seen in RANK KO mice. This difference is likely due to the fact that RANK Motif 1 in the KI mice is still functional and able to mediate a certain degree of bone resorption. In summary, the KI mice have provided firm evidence that RANK Motifs 2 and 3 play a crucial role in osteoclastogenesis in vivo.
Earlier studies showed that the ectopic expression of CA-NFATc1 promoted osteoclastogenesis in the absence of RANKL (40,41). Thus, we determined whether the ectopic expression of CA-NFATc1 in RANKKI/KI BMMs could restore the impaired osteoclastogenesis. Our data indicate that the ectopic expression of CA-NFATc1 in both WT and RANKKI/KI BMMs was unable to promote osteoclastogenesis without RANKL. This discrepancy is likely due to different types of cells used; we used primary BMMs while RAW264.7 cells were used in the previous investigations (40,41). Importantly, our findings suggest that NFATc1 is not the only important downstream effector. This notion is consistent with a recent study which has revealed that, in addition to NFATc1, other pathways such as the MYC/estrogen receptor-related receptor α (ERRα) axis, also participate in mediating osteoclastogenesis (42). This observation has opened a new avenue for future studies to further investigate the molecular mechanism by which RANK Motifs 2 and 3 regulate osteoclastogenesis.
Pg is an important factor in the development of periodontal bone loss (20). Our previous study has demonstrated that RANK signaling is required for Pg-induced osteoclastogenesis (21). Lipopolysaccharide (LPS) has been shown to be a key factor causing periodontal bone loss (4). Our initial study showed that LPS blocked osteoclast differentiation from normal osteoclast precursors but enhanced osteoclastogenesis when osteoclast precursors were pretreated with RANKL in vitro (43). We further demonstrated that Pg exerts similar effects on osteoclastogenesis; Pg blocks osteoclast differentiation from normal osteoclast precursors but it enhances osteoclast formation from RANKL-primed precursors (21). A subsequent and independent study confirmed that Pg-induced periodontal bone loss is RANKL dependent (44). These findings indicate that Pg-induced osteoclastogenesis requires prior priming of osteoclast precursors by RANKL. This raises a question of whether RANK Motifs 2 and 3 are implicated in the priming of osteoclast precursors that is required for Pg-induced osteoclastogenesis. Using BMMs from the KI mice, we show that these two RANK motifs play a crucial role in Pg-induced osteoclastogenesis.
c-Fos and NFATc1 are two key transcription factors for osteoclastogenesis (18,22), and RANKL promotes osteoclastogenesis primarily by inducing the expression of NFATc1 via c-Fos (30). Whereas Pg is unable to induce the expression of NFATc1 and c-Fos in normal BMMs, 24-hour RANKL pretreatment increases c-Fos and NFATc1 expression and subsequent Pg stimulation further enhances RANKL-induced expression of the two factors (21), indicating that RANK signaling supports Pg-induced osteoclastogenesis by modulating c-Fos and NFATc1 expression. In this study, we have further demonstrated that RANK Motifs 2 and 3 are involved in regulating the expression of c-Fos and NFATc1 that is required for Pg-mediated osteoclastogenesis. Specifically, RANK Motifs 2 and 3 play a role in Pg-induced osteoclastogenesis by up-regulating c-Fos and NFATc1 expression during the RANKL priming phase as well as rendering c-Fos and NFATc1 genes responsive to subsequent Pg stimulation.
We further investigated the role of RANK Motifs 2 and 3 in osteoclastogenesis using CPP conjugated with RANK fragments containing Motif 2 or 3. Our initial assay revealed that the combination of CPP conjugated with RANK fragments containing Motif 2 (M2–10) and CPP conjugated with RANK fragments containing Motif 3 (M3–10) at 8 μM each effectively inhibited osteoclastogenesis (Fig. 5A). Motif 2 and Motif 3 are very similar in amino acid sequence, raising a possibility that M2–10 is able to block the function of both Motif 2 and Motif 3, and the same is probably true for M3–10. To address this possibility, we further investigated and compared the capacity of 16 μM M2–10 or M3–10 to inhibit osteoclastogenesis with that of the combination of 8 μM M2–10 plus 8 μM M3–10. We found that the inhibitory effect of 16 μM M2–10 or M3–10 on osteoclastogenesis (Fig. 5B) was quite similar to that of the combination of 8 μM M2–10 plus 8 μM M3–10 (Fig. 5A). It is noted that we previously demonstrated that inactivation of a single motif (Motif 2 or 3) had no considerable inhibitory effect on osteoclastogenesis (10). Taken together, these findings support the notion that M2–10 can block the function of both Motif 2 and Motif 3, and M3–10 can do the same. The CPP conjugates inhibit osteoclastogenesis by suppressing RANKL-induced expression of c-Fos and NFATc1. In contrast, the CPP conjugates do not affect the activation of the NF-ĸB, Akt, or MAPK signaling pathways (Fig. 5D). Additionally, the addition of the CPP conjugates during RANKL pretreatment is sufficient to suppress the differentiation of RANKL-primed BMMs into osteoclasts in response to subsequent Pg stimulation. These studies provide additional evidence supporting the role of RANK Motifs 2 and 3 in osteoclastogenesis.
Based on our published and current work, we propose the following model to explain how RANKL priming supports Pg-mediated osteoclastogenesis (Fig. 6E). RANKL promotes osteoclastogenesis by stimulating the expression of NFATc1 and c-Fos as well as inducing the activation of NF-ĸB and MAPK pathways (30). Whereas NFATc1 and c-Fos cooperatively regulate gene expression during osteoclast differentiation (22,45), NF-ĸB and MAPK pathways primarily mediate the function and survival of osteoclasts (11). We previously showed that Pg is unable to induce the expression of NFATc1 and c-Fos, but it can activate NF-ĸB and MAPK pathways in BMMs (21). During the RANKL pretreatment, c-Fos expression is up-regulated by the signaling pathways activated by RANK Motifs 2 and 3 (Fig. 6E). In collaboration of Motif 1-mediated signaling pathways, c-Fos in turn stimulates the expression of NFATc1. Thus, upon the completion of the RANKL pretreatment, the level of c-Fos and NFATc1 is sufficient to promote osteoclastogenesis. Moreover, the RANKL priming also renders c-Fos and NFATc1 genes responsive to Pg and this enables Pg to further enhance c-Fos and NFATc1 expression through TLR2 during the Pg treatment phage (Fig. 6E). Upon Pg binding, TLR2 can also activate NF-ĸB and MAPK pathways (21), which collectively mediate the function and survival of osteoclasts (11), in the absence of RANKL. Hence, RANKL pretreatment and subsequent Pg stimulation together are able to induce both the expression of NFATc1 and c-Fos and the activation of NF-ĸB and MAPK pathways to promote osteoclast differentiation (Fig. 6E).
The validation of the in vivo role of RANK Motifs 2 and 3 in osteoclastogenesis in combination of in vitro blocking experiments with the CPP conjugates support the promising potential of RANK Motifs 2 and 3 to serve as new therapeutic targets for bone disorders such as osteoporosis and periodontal bone loss. The RANKL/RANK system was recognized as an attractive new antiresorptive drug target for osteoporosis shortly after its discovery and Amgen subsequently developed a humanized anti-RANKL antibody (denosumab) (46), which was approved in 2010 for treating postmenopausal osteoporosis (47). However, denosumab causes several side effects: increased risk of serious infections (48,49), osteonecrosis of the jaw (50), and atypical femur fracture (51). In particular, the increased risk of serious infections and osteonecrosis of the jaw are likely due to the suppressive effect of denosumab on the immune system function since RANKL regulates dendritic cell biology (32), T cell development (32), and B cell differentiation (52). Thus, a better therapeutic targeting strategy would be to target RANK signaling pathways that are involved in osteoclast formation, but not in the immune system function. The current study has revealed that RANK containing inactivating mutations in Motifs 2 and 3 is still able to activate the NF-κB and MAPK signaling pathways, which regulate immune cell development and function (53–55). Thus, specifically targeting these two RANK motifs is less likely to affect the immune system function. This is particularly important for treating periodontal bone loss because the normal immune system function needs to be preserved for combating bacterial infection.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (No. AR47830 to X.F.), a grant from the National Institute of Dental and Craniofacial Research (No. DE026465 to P.Z.), and the National Scholarship for Building High Level University from Chinese Ministry of Education (CSC No. 201706240241 to Y.L.). The University of Alabama at Birmingham Transgenic & Genetically Engineered Models facility (RAK) is supported by the National Institutes of Health (grant numbers P30 CA13148, P30 AR048311, P30 DK074038, P30 DK05336 and P60 DK079626).
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
References
- 1.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337. [DOI] [PubMed] [Google Scholar]
- 2.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–84. [DOI] [PubMed] [Google Scholar]
- 3.Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves DT, Li J, Cochran DL. Inflammation and uncoupling as mechanisms of periodontal bone loss. J Dent Res. 2011;90(2):143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa N, Kinosaki M, Yamaguchi K, et al. RANK Is the Essential Signaling Receptor for Osteoclast Differentiation Factor in Osteoclastogenesis. Biochemical and Biophysical Research Communications. 1998;253(2):395–400. [DOI] [PubMed] [Google Scholar]
- 6.Hsu H, Lacey DL, Dunstan CR, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96(7):3540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hehlgans T, Pfeffer K. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology. 2005;115(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nature reviews immunology. 2003;3(9):745. [DOI] [PubMed] [Google Scholar]
- 9.Jules J, Ashley JW, Feng X. Selective targeting of RANK signaling pathways as new therapeutic strategies for osteoporosis. Expert Opinion on Therapeutic Targets. 2010;14(9):923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Xu D, Yang H, et al. Functional identification of three receptor activator of NF-κB cytoplasmic motifs mediating osteoclast differentiation and function. Journal of Biological Chemistry. 2004;279(52):54759–69. [DOI] [PubMed] [Google Scholar]
- 11.Feng X Regulatory roles and molecular signaling of TNF family members in osteoclasts. Gene. 2005;350(1):1–13. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Wang S, Wei S, Sun L, Feng X. Receptor activator of NF-kappaB (RANK) cytoplasmic motif, 369PFQEP373, plays a predominant role in osteoclast survival in part by activating Akt/PKB and its downstream effector AFX/FOXO4. J Biol Chem. 2005;280(52):43064–72. [DOI] [PubMed] [Google Scholar]
- 13.Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunological Reviews. 2009;231(1):241–56. [DOI] [PubMed] [Google Scholar]
- 14.Jules J, Wang S, Shi Z, Liu J, Wei S, Feng X. The IVVY Motif and Tumor Necrosis Factor Receptor Associated Factor (TRAF) Sites in the Cytoplasmic Domain of the Receptor Activator of Nuclear Factor kappa B (RANK) Cooperate to Induce Osteoclastogenesis. Journal of Biological Chemistry. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Wang S, Liu W, Liu J, Feng X. A novel receptor activator of NF-kappaB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J Biol Chem. 2006;281(8):4678–90. [DOI] [PubMed] [Google Scholar]
- 16.Jules J, Zhang P, Ashley JW, et al. Molecular basis of requirement of receptor activator of nuclear factor kappaB signaling for interleukin 1-mediated osteoclastogenesis. J Biol Chem. 2012;287(19):15728–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HK, Kang HR, Jung E, Kim TE, Lin JJ, Lee SY. Early estrogen-induced gene 1, a novel RANK signaling component, is essential for osteoclastogenesis. Cell Res. 2013;23(4):524–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayanagi H, Kim S, Koga T, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. [DOI] [PubMed] [Google Scholar]
- 19.Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. [DOI] [PubMed] [Google Scholar]
- 20.Mysak J, Podzimek S, Sommerova P, et al. Porphyromonas gingivalis: Major Periodontopathic Pathogen Overview. Journal of Immunology Research. 2014;2014:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Liu J, Xu Q, et al. TLR2-dependent modulation of osteoclastogenesis by Porphyromonas gingivalis through differential induction of NFATc1 and NF-κB. Journal of Biological Chemistry. 2011:jbc. M110. 198085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–40. [DOI] [PubMed] [Google Scholar]
- 23.McHugh KP, Hodivala-Dilke K, Zheng MH, et al. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105(4):433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15(8):1477–88. [DOI] [PubMed] [Google Scholar]
- 25.Dempster DW, Compston JE, Drezner MK, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jules J, Shi Z, Liu J, Xu D, Wang S, Feng X. The receptor activator of NF-kappa B (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-alpha (TNF)-mediated osteoclastogenesis. Journal of Biological Chemistry. 2010:jbc. M110. 149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takayanagi H The role of NFAT in osteoclast formation. Ann N Y Acad Sci. 2007;1116:227–37. [DOI] [PubMed] [Google Scholar]
- 28.Jimi E, Nakamura I, Duong LT, et al. Interleukin 1 Induces Multinucleation and Bone-Resorbing Activity of Osteoclasts in the Absence of Osteoblasts/Stromal Cells. Experimental Cell Research. 1999;247(1):84–93. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Xie Y, Du Q-S, et al. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J Cell Biol. 2003;160(4):565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo K, Galson DL, Zhao C, et al. NFAT rescues osteoclastogenesis in precursors lacking c-Fos. Journal of Biological Chemistry. 2004. [DOI] [PubMed] [Google Scholar]
- 31.Choi J-M, Ahn M-H, Chae W-J, et al. Intranasal delivery of the cytoplasmic domain of CTLA-4 using a novel protein transduction domain prevents allergic inflammation. Nature Medicine. 2006;12:574. [DOI] [PubMed] [Google Scholar]
- 32.Anderson DM, Maraskovsky E, Billingsley WL, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390(6656):175–9. [DOI] [PubMed] [Google Scholar]
- 33.Arch RH, Thompson CB. Lymphocyte survival--the struggle against death. Annu Rev Cell Dev Biol. 1999;15:113–40. [DOI] [PubMed] [Google Scholar]
- 34.Darnay BG, Haridas V, Ni J, Moore PA, Aggarwal BB. Characterization of the intracellular domain of receptor activator of NF-kappaB (RANK). Interaction with tumor necrosis factor receptor-associated factors and activation of NF-kappab and c-Jun N-terminal kinase. J Biol Chem. 1998;273(32):20551–5. [DOI] [PubMed] [Google Scholar]
- 35.Wong BR, Josien R, Lee SY, Vologodskaia M, Steinman RM, Choi Y. The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor. J Biol Chem. 1998;273(43):28355–9. [DOI] [PubMed] [Google Scholar]
- 36.Darnay BG, Ni J, Moore PA, Aggarwal BB. Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif. J Biol Chem. 1999;274(12):7724–31. [DOI] [PubMed] [Google Scholar]
- 37.Galibert L, Tometsko ME, Anderson DM, Cosman D, Dougall WC. The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-kappaB, a member of the TNFR superfamily. J Biol Chem. 1998;273(51):34120–7. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Sarosi I, Yan XQ, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci U S A. 2000;97(4):1566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dougall WC, Glaccum M, Charrier K, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13(18):2412–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirotani H, Tuohy NA, Woo JT, Stern PH, Clipstone NA. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem. 2004;279(14):13984–92. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda F, Nishimura R, Matsubara T, et al. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J Clin Invest. 2004;114(4):475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae S, Lee MJ, Mun SH, et al. MYC-dependent oxidative metabolism regulates osteoclastogenesis via nuclear receptor ERRalpha. J Clin Invest. 2017;127(7):2555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Wang S, Zhang P, Said-Al-Naief N, Michalek SM, Feng X. Molecular mechanism of the bifunctional role of lipopolysaccharide in osteoclastogenesis. Journal of Biological Chemistry. 2009;284(18):12512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X, Lin X, Yu X, et al. Infection-Associated Periodontal Bone Resorption Is Dependent on Receptor Activator of NF-κB Ligand. Infection and Immunity. 2013;81(5):1502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40(2):251–64. [DOI] [PubMed] [Google Scholar]
- 46.Lacey DL, Boyle WJ, Simonet WS, et al. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nat Rev Drug Discov. 2012;11(5):401–19. [DOI] [PubMed] [Google Scholar]
- 47.Rizzoli R, Yasothan U, Kirkpatrick P. Denosumab. Nat Rev Drug Discov. 2010;9(8):591–2. [DOI] [PubMed] [Google Scholar]
- 48.Anastasilakis AD, Toulis KA, Goulis DG, et al. Efficacy and safety of denosumab in postmenopausal women with osteopenia or osteoporosis: a systematic review and a meta-analysis. Horm Metab Res. 2009;41(10):721–9. [DOI] [PubMed] [Google Scholar]
- 49.Toulis KA, Anastasilakis AD. Increased risk of serious infections in women with osteopenia or osteoporosis treated with denosumab. Osteoporos Int. 2010;21(11):1963–4. [DOI] [PubMed] [Google Scholar]
- 50.Boquete-Castro A, Gomez-Moreno G, Calvo-Guirado JL, Aguilar-Salvatierra A, Delgado-Ruiz RA. Denosumab and osteonecrosis of the jaw. A systematic analysis of events reported in clinical trials. Clin Oral Implants Res. 2016;27(3):367–75. [DOI] [PubMed] [Google Scholar]
- 51.Shane E, Burr D, Abrahamsen B, et al. Atypical subtrochanteric and diaphyseal femoral fractures: second report of a task force of the American Society for Bone and Mineral Research. J Bone Miner Res. 2014;29(1):1–23. [DOI] [PubMed] [Google Scholar]
- 52.Kong YY, Yoshida H, Sarosi I, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397(6717):315–23. [DOI] [PubMed] [Google Scholar]
- 53.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–79. [DOI] [PubMed] [Google Scholar]
- 54.Huang G, Shi LZ, Chi H. Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine. 2009;48(3):161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6(7):532–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.