Abstract
Hematopoietic stem cells (HSCs) reside in the bone marrow within niches that provide microenvironmental signals in the form of biophysical cues, bound and diffusible biomolecules, and heterotypic cell-cell interactions that influence HSC fate decisions. This study seeks to inform the development of a synthetic culture platforms that promote ex vivo HSC expansion without exhaustion. A library of methacrylamide-functionalized gelatin (GelMA) hydrogels is used to explore remodeling and crosstalk from murine bone marrow MSCs on the expansion and quiescence of primary murine HSCs. The use of a degradable GelMA hydrogel enables MSC-mediated remodeling, yielding dynamic shifts in the matrix environment over time. An initially low-diffusivity hydrogel combined with a 1:1 HSPC:MSC co-culture was observed to facilitate maintenance of a hematopoietic stem and progenitor cell population over 7-days. Excitingly, this platform promoted retention of a quiescent HSC population compared to HSC monocultures. Our studies reveal MSC-density dependent upregulation of MMP-9 and changes in hydrogel mechanical properties (ΔE = 2.61±0.72) suggesting MSC-mediated matrix remodeling may contribute to a dynamic culture environment. Together, we report a three-dimensional hydrogel for ex vivo HSC culture and reveal HSC expansion and quiescence is sensitive to hydrogel properties, MSC co-culture, and MSC-mediated hydrogel remodeling.
Keywords: hematopoietic stem cell, hydrogel, mesenchymal stromal cell, remodeling, differentiation
1. Introduction
Hematopoietic stem cells (HSCs) are responsible for the production of the body’s entire complement of blood and immune cells through a highly regulated differentiation hierarchy.[1, 2] HSCs lodge primarily in the bone marrow in unique tissue microenvironments termed niches, which provide a milieu of extrinsic signals in the form of biophysical cues, biomolecular gradients, and co-inhabiting niche cells that, together, promote quiescence, self-renewal, and lineage specification.[3, 4] Disruptions to the niche can lead to a loss in regulation and abnormal or unsuccessful hemostasis. The drop in HSC efficacy from niche disruption has hindered clinical use of HSC transplants (HSCTs) for the treatment of blood and immune disorders.[5] As such, there is an acute clinical need for in vitro approaches to expand or precondition donor HSCs for improved homing and engraftment to recipient bone marrow niches. A central challenge in the context of HSCT applications is a synthetic niche that provides sequences of signals to promote expansion of a hematopoietic stem and progenitor cell population without exhaustion of a subpopulation of quiescent HSCs critical for long-term hematopoiesis. Inspiration drawn from the native niche may inform development of a synthetic platform for HSC culture.[6]
In vivo, HSCs are presented with a range of microenvironment triggers that maintain hematopoietic activity,[7] including complex patterns of ligands such as laminins and fibronectin[8] and a Young’s modulus of 0.25 – 24.7 kPa.[9] In vitro, tissue engineering approaches have begun to identify critical biomaterial design parameters for HSC culture, inspired by native niche signals. Notably, fibronectin-functionalized elastic substrates that induce expansion of HSC progenitor populations[10] and maintenance of a quiescent HSC population in isolated single-cell cultures, demonstrating the potential of tissue engineering platforms.[11] However, it is becoming increasingly important to capture both direct and indirect effects of the niche in tissue engineering constructs. For example, the niche microenvironment can modulate the behavior of co-inhabiting niche cells, which then exert an influence on HSC activity via cell-cell contact (e.g., cadherin interactions) and secreted biomolecular factors.[12] Further, the extracellular matrix modulates biotransport of secreted factors through transient binding motifs (e.g. proteoglycans) or steric hindrance to diffusive and advective mass transport.[13] These complex biomolecular signaling processes offer an opportunity to develop artificial niches that regulate the production and biotransport of paracrine signals from co-encapsulated niche cells to promote culture of desired HSC subpopulations.
Expanding upon the use of conventional conditioned media experiments to interrogate the role of cell-cell signaling,[14] biomaterial designs can exploit matrix properties to create regimes of distinct cell-cell signaling mechanism (autocrine-feedback vs. paracrine signaling). In a tight matrix, with a small mesh size and low diffusivity, HSCs may experience a small radius of communication, as cell-cell communication is hindered by steric and electrostatic interactions with the matrix, enforcing a mostly autocrine feedback-rich environment. Alternatively, in a loose matrix, with a large mesh size, HSCs have a larger radius of communication, allowing for possible paracrine signaling. Culturing HSCs with hematopoietic lineage positive cells in a collagen system, we recently demonstrated improved maintenance of early progenitors in autocrine-dominated environments with low diffusivity,[15] demonstrating the potential of biomaterial designs to regulate heterotypic cell interactions.
As a source of niche-biomolecules, mesenchymal stromal cells (MSCs) show significant promise for the development of HSC culture platforms. MSCs co-localize with HSCs in the perivascular regions and central marrow,[16, 17] and mediate HSC activation and quiescence via cell-cell contact and diffusive signaling[4, 11, 17, 18] through production of factors such as CXCL12,[19] IL-6,[2, 20] and TPO.[21] Biomaterial systems offer the opportunity to identify heterotypic HSC-MSC cultures that support HSCs without exogenic factors. However, while the matrix can directly influence MSC-HSC interactions, MSCs are also equipped to remodel the matrix via protein deposition and enzymatic degradation,[22, 23] leading to non-static material properties, and a feedback loop termed dynamic reciprocity.[23, 24]As a result, it is important to consider initial properties of a biomaterial system, the dynamic shifts in these properties and their effect on soluble cell signaling. As a result, HSC-MSC biomaterial culture platforms offer an essential opportunity to define a role for dynamic reciprocity as a design paradigm in artificial niches.
The objective of this study was to employ a library of three-dimensional methacrylamide-functionalized gelatin (GelMA) hydrogels to describe the influence of hydrogel poroelastic (stiffness, diffusion) properties, in conjunction with MSC-secreted biomolecular (paracrine) signals and matrix remodeling, on the expansion, differentiation, and quiescence of primary murine HSCs (Figure 1). We report myeloid lineage and HSC quiescence events in response to increasing concentrations of MSCs and matrix remodeling. Together this work represents an integrated HSC culture platform that leverages concepts of dynamic biotransport of paracrine signals as an essential biomaterial design paradigm.
Figure 1. Hematopoietic cell-cell interactions in a dynamic biomaterial landscape.
A) Hematopoietic lineage hierarchy. Long-term repopulating HSCs and Short-term repopulating HSCs are grouped as a singular HSC population. HSCs and Multipotent Progenitors comprise the HSC progenitor, HSPCs, population. Common Myeloid Progenitors and Lin+ cells make up the Terminal cell population. B) Autocrine and paracrine signaling can be tuned by adjusting seeding density and altering biotransport properties. In an HSC-only culture, an increase in mesh size leads to a diffusive regime that increases paracrine signaling. In an HSC-MSC co-culture, small mesh constrains secreted biomolecules and leads to a mostly autocrine signaling. However, as mesh size is increased, autocrine feedback is reduced and paracrine signaling begins to dominate cell-cell interactions.
2. Experimental Section
2.1. Material characterization
2.1.1. GelMA and hydrogel fabrication
Gelatin (#G2500, Sigma, St. Louis, MO) was functionalized with methacrylamide groups following a previously described protocol.[25, 26] In brief, methacrylate anhydride (MA) was added dropwise to a gelatin and PBS solution before quenching, dialysis, and lyophilization. The gelatin:MA ratio was tuned to control the degree of functionalization (DOF), quantified by H1 NMR.[26] Methacrylamide-functionalized gelatin (GelMA) hydrogels were formed from 20μL GelMA precursor suspension with lithium acylphosphinate (LAP) photoinitiator (PI) in circular molds (5mm dia.), and exposed to 7.14 mW/cm2 UV light for 30 seconds.[27, 28]
2.1.2. Determination of poroelastic properties
A library of 18 acellular hydrogels were fabricated from 4, 5, and 7.5% GelMA at 35, 50, and 85% DOF, with 0.05 or 0.1% (w/v) PI. The Young’s modulus was determined using an Instron 5943 in unconfined compression (Instron, Norwood, MA). Briefly, a 0.005N preload was applied followed by 0.1mm/min compression until 20% compression was reached. The Young’s modulus was taken as the slope of a linear fit applied to 10% of the stress vs. strain data, with an offset of 0 or 5%, using Origin Statistical Software (Northampton, MA).[29] The moduli of hydrogels containing 1×105 or 1×106 MSCs/mL were traced over seven days, using the same compression protocol.
Stress-relaxation indentation tests were performed on acellular hydrogels using MFP-3D AFM (Asylum Research, Goleta, CA), with a 4.5μm spherical tip (Novascan, Ames, IA). The generated force-curve was used to determine the shear modulus and diffusion coefficient of water. Specimens were indented to 0.5, 1, or 2μm, and held for 5 seconds. This process has been described elsewhere; in brief, the instantaneous force was normalized to the far-field force and fit to a finite element analysis model by modulating the characteristic time and material properties (shear modulus, diffusion).[30, 31]
2.1.3. Mesh size estimate
The mesh size was determined using swelling ratios and is a function (1) of the polymer volume fraction and the mean-squared end-to-end distance of the gelatin chain.[32–34]
| (1) |
It was assumed that the gelatin chains were long enough to neglect chain-end effects, and that the molecular weight of the repeat unit, Mr, was the averaged molecular weight of the amino acid composition. The mass swelling ratio was determined by the ratio of dry to wet hydrogel weight following crosslinking (relaxed state) or after 24hrs in PBS (equilibrium state). Hydrogels were weighed before and after lyophilization to obtain the mass swelling ratio, and then transformed to the volume fraction of gelatin (2), using the solvent density (ρ(s) = 1.01g/cm3)[35] and gelatin density (ρ(p) = 1.345g/cm3).[36, 37]
| (2) |
The molecular weight between crosslinks, Mc, was estimated using the Flory-Rehner equation adapted by Bray and Merrill for use in polymers crosslinked in solvent (3).[33, 38] The applicability of the modified equation for specific concentration regimes was confirmed by Peppas and Merrill [32]. The Mc is a function of material properties such as the molar weight of the solvent, V1, the specific volume of the polymer, , and the solvent-polymer interaction, X1, also known as Flory’s Chi parameter. It is also a function of the system parameters such as the volume fraction of the polymer in its relaxed state and in the equilibrium state, v2r and v2s, and the number average molecular weight before crosslinking, Mn (see Table S1 for an explanation of variables).
| (3) |
Flory’s characteristic ratio,Cn, can be approximated by a constant as the number of monomers goes to infinity, limn→∞ Cn = C∞. For a stiff chain, this can be approximated using the worm-like or Kratky-Porod persistence model using the persistence length, lp, the monomer unit length, l, and modified for proteins.[39]
| (4) |
2.2. Hematopoietic stem and progenitor cell isolation
All work involving primary cell extraction was conducted under approved animal welfare protocols (Institutional Animal Care and Use Committee, University of Illinois at Urbana-Champaign). Primary murine hematopoietic stem and progenitor cells (HSPC) were extracted from the femur and tibia bone marrow of C57BL/6 female mice, age 4 – 8 weeks (The Jackson Laboratory). The bones were gently washed with PBS + 5%FBS and filtered with a 40μm sterile filter.[15, 27] The suspension was lysed with ACK lysis buffer (Invitrogen, Carlsbad, CA), and then rinsed with PBS/FBS. Initial HSPC isolation was performed with EasySep™ Mouse Hematopoietic Progenitor Cell Enrichment Kit and Magnet (#19756, #18000, Stemcell Technologies, CA). A secondary isolation further enriched the HSPC population by collecting the Lin− Sca-1+ c-kit+ (LSK) fraction (SMethods 1) using a BD FACS Aria II flow cytometer.[40, 41] The LSK faction was collect in PBS + 25% FBS on ice and immediately used.
2.3. Mesenchymal stem cell isolation
Following gentle crushing with a pestle and mortar, MSCs were isolated from murine femur and tibia bones following commercially available defined protocols of MesenCult™ Proliferation Kit with MesenPure™ (#05512 Stemcell Technologies, CA). The MSCs were allowed to expand for two weeks prior to collection.
2.4. HSC and MSC culture in GelMA hydrogels
HSPCs or MSCs in a suspension were added to a GelMA solution at seeding densities of 1×105 HSPCs/mL and 1×105 or 1×106 MSCs/mL to create ratios of 1:0 (monoculture), 1:1,1:10 (HSPC:MSC). Following the same crosslinking conditions as outlined in Methods 2.1.1, hydrogel-cell constructs were cultured in 48-well plates at 37°C (5% CO2) for 7 days in 300μL StemSpan™ SFEM (#09650 Stemcell Technologies) supplemented with 100 ng/mL SCF (Peprotech) and 0.1% Pen Strep (Gibco), with media changes every 2 days.[10]
2.5. Analysis of differentiation patterns
Differentiation patterns were analyzed via Fluorescence-Assisted Cytometry (FACs), using a BD LSR Fortessa (BD Biosciences, San Jose, CA). Cell-seeded hydrogels were degraded with 100 Units of Collagenase Type IV (Worthington Biochemical, Lakewood, NJ) for 30 minutes at 37°C, and gently mixed via pipetting. The cells were re-suspended in PBS + 5% FBS and stained with a cocktail of antibodies (SMethods 1.) Cells were classified as Long-Term repopulating HSCs (LT-HSCs: CD34lo CD135lo Lin− Sca1+ c-kit+)[42, 43]; Short-Term repopulating HSCs (ST-HSCs: CD34hi CD135lo LSK)[42, 43]; Multipotent Progenitors (MPPs: CD34+ CD135+ LSK)[43, 44]; lineage-specified Common Myeloid Progenitors (CMPs: Lin− c-kit+ Sca1− CD34+ CD16/32−).[41, 44, 45] Individual cell populations are reported as percentages of the total hematopoietic cell population in each culture, with terminal cells classified as non-LSK cells. For some analyses, LT-HSC and ST-HSC were grouped together as a common HSC population.
2.6. Cell cycle analysis
Conditions identified as optimal in maintaining early progenitors were chosen for subsequent cell cycle analysis. Cells in 1:0 Low and 1:1 High (HSPC:MSC) conditions were cultured and isolated at day 7. Experimental replicates (n=10/condition) were performed; each replicate contained three separately cultured hydrogels (ngels=30/condition). Isolated cells were live/dead stained with LIVE/DEAD™ Fixable Red Dead Cell Stain Kit (#L34971, ThermoFisher) following the standard protocol, and LT-HSC, ST-HSC, and MPP populations identified via antibody cocktail (Methods 2.5). Stained cells were fixed and permeabilized with Foxp3 / Transcription Factor Staining Buffer Set (#00-5523-00, eBioscience), and stained with BV605-conjugated Ki-67 (1:100 dilution) and DAPI (#D21490, ThermoFisher). Quiescent cells (DAPI2n Ki-67−) from each hematopoietic population were subsequently analyzed via Flow Cytometry.
2.7. MSC-mediated remodeling profiles
Matrix remodeling associated gene expression profiles were examined for MSC-seeded hydrogel specimens via QuantiStudio 7 Flex real-time PCR (Applied Biosystems). Frozen hydrogels isolated at days 0, 1, 4, and 7 during culture were crushed using a pestle, with mRNA extracted using RNeasy Plant Mini Kit (Qiagen, Germany). mRNA to cDNA was accomplished using the QuantiTect Rev. Transcription kit (Qiagen) and prepped for RT-PCR using PIPETMAX (Gilson, Middleton, WI). The CT of each gene/sample was performed in duplicate. Results were analyzed using the comparative CT method (2−ΔΔCT). Primer specificity and efficiency were validated prior to comparative CT analysis.[46] Matrix deposition genes examined were collagen, type I, alpha 1 (Col1a1), and tissue inhibitors of metalloprotease TIMP-1, TIMP-2, and TIMP-3. Matrix degradation genes examined were matrix metalloproteases MMP-2 and MMP-9, also known as Gelatinase-A and Gelatinase-B.[47] Gene expression was normalized to the housekeeper glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and then compared against the gene expression at day 0 of each condition (Figure S1,2). Primer information can be found in Table S2.
2.8. Statistical Analysis
Power analysis was used to confirm all sample sizes were large enough to maintain a maximum Type I and II error of α=0.05 and β=0.20.[48] Significance was tested with Origin Statistical Software using 2-way ANOVA at each time point, with stiffness and seeding density as independent variables; normality of the data was determined using the Shapiro-Wilkes test,[49] and homoscedasticity determined using Levene’s Test.[50] Quiescence significance was determined with the nonparametric Mann-Whitney test (2-sample) at the 0.05 significance level. Young’s modulus outliers were removed using Dixon’s Q-test at significance level 0.05.[51] Means are reported with their associated standard deviations.
2.9. Data Sharing Statement
For original data, please contact the corresponding author (BACH). All flow cytometry files have been uploaded to flowrepository.org (FR-FCM-Z2ZU; FR-FCM-Z2ZV).
3. Results
3.1. Characterization of GelMA hydrogels
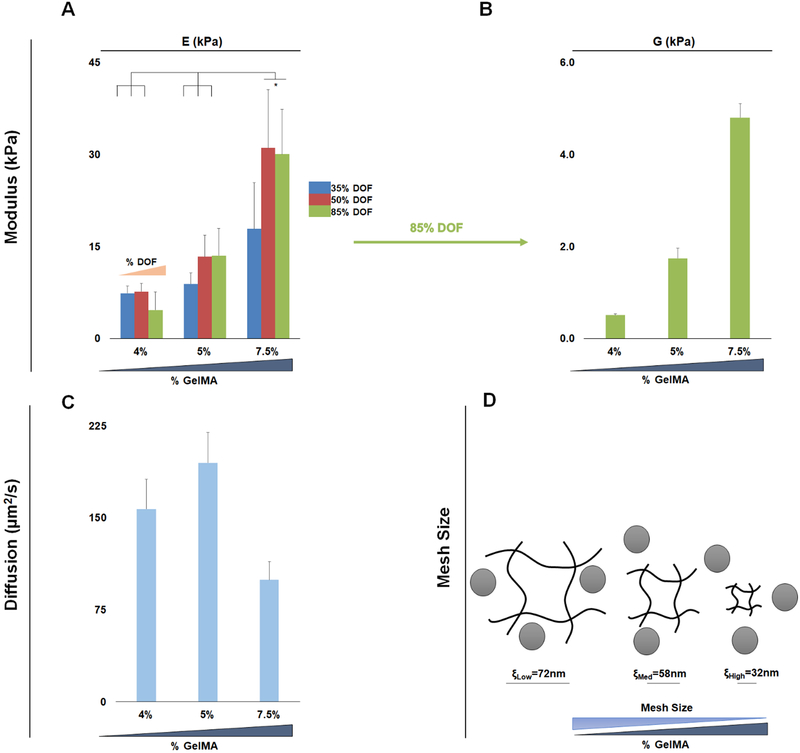
The poroelastic parameters of a library of hydrogels were examined as a function of GelMA wt%, degree of methacrylamide functionalization (DOF), and photoinitiator (PI) concentration. The Young’s modulus (E) of the library spanned a range of marrow-mimetic values (2.67±1.39 to 32.23±10.7kPa), and showed strong dependence on wt% GelMA and DOF, with minimal influence of PI (0.05, 0.1%) (Figure S3).Concentrating on a subset of hydrogels fabricated with 0.1% PI, these hydrogels displayed compressive moduli ranging from 4.67±3.20kPa (35% DOF, 4wt% GelMA) to 30.06±12.35kPa (85% DOF, 7.5wt% GelMA) (Figure 2A). We subsequently examined (via indentation) poroelastic properties of this library of hydrogels, observing ranges of shear moduli (G; 0.36±0.03 to 6.05±0.50kPa) and diffusion coefficient of water (Dw; 75.86±11.84 to 211.13±51.84 μm2/s) (Figure S4 and S5).
Figure 2. Mechanical characterization of the library of methacrylamide-functionalized hydrogels. (n=6).
A) Young’s modulus of 9 hydrogel conditions, keeping the PI constant at 0.1% B) Shear modulus data from the Low, Med, and High hydrogel variants used for cell culturing, keeping DOF and PI constant at 85% DOF and 0.1%PI. C) Diffusion of water in the Low, Med, and High variants exhibits two regimes, Low and Med (4, 5% GelMA) have a higher diffusion, whereas High (7.5% GelMA) has ~50% reduction in diffusion D) Mesh size estimates for the Low, Med, and High variants decreases with increasing GelMA content. There is a trend of Low and Med exhibiting similar mesh and diffusion, while High exhibits a significantly (p<0.05) lower biotransport regime.
From this library of hydrogels, we identified a subset of three hydrogels (85% DOF, 0.1%PI, and 4, 5, 7.5%GelMA) that spanned a range of mechanical properties associated with the native bone marrow niche: Low, ELow = 4.67±3.20; Med, EMed = 13.54±4.81; High, EHigh = 30.06±12.35kPa (Figure 2A). This subset also displayed an increasing shear modulus (GLow = 0.51±0.03; GMed = 1.74±0.22; GHigh = 4.80±0.3kPa respectively) (Figure 2B). Low and Med hydrogels displayed similar water diffusion characteristics (157.19±24.45, 195.04±24.66 μm2/s) whereas the High hydrogel variant had a significantly reduced Dw (99.48±15.11 μm2/s) (Figure 2C). Employing mesh size (ξ) calculations, we see a decreasing mesh size associated with increasing GelMA content: ξLow=72, ξMed=58, ξHigh=32nm (Figure 2D).
3.2. MSC-mediated remodeling of GelMA hydrogels
As MSCs can interact and remodel gelatin-based hydrogels, we quantified changes in the Young’s modulus of the Low, Med, and High hydrogels as a function of culture time (0 – 7 days) and MSC seeding density (0, 1×105, 1×106 MSCs/mL), with results reported as absolute values (Figure 3A) and normalized against the initial properties (day 0) of each variant (Figure 3B). Notably, no observable trends in modulus change were observed in either Low and Med hydrogel variants. However, the High hydrogel (7.5%GelMA; 85% DOF; 0.1%PI) displayed significant remodeling with the highest seeding density (1×106 MSCs/mL). Here, biosynthetic remodeling was observed, characterized by significant increases in Young’s modulus (ΔE = 2.61±0.72), twice that of other conditions (ΔE = 1.13±0.45, acellular; ΔE = 1.25±0.13, 1×105 MSCs/mL) (Figure 3B).
Figure 3. Bulk Young’s modulus of Low, Med, and High hydrogels cultured over 7 days with seeding densities of 0, 1×105, and 1×106 MSCs/mL (1:0, 1:1, 1:10 HSPC:MSC). (n=3–6).
A) There is minimal change in Young’s modulus over time in any hydrogel and seeding conditions. The high hydrogel shows a significant increase (p<0.05) from initial seeding to day 7 in the presence of a high concentration of the highest density of MSCs, 1×106 MSCs/mL. B) The Young’s modulus was normalized to the initial value, day 0, of the specific seeding and hydrogel variant to show fold change. This highlights that only the High with the highest concentration of MSCs (1×106 MSCs/mL) experiences significant increase (p<0.05) in bulk Young’s modulus over time.
While shifts in bulk hydrogel properties were not always significant, analysis of gene expression profiles overtime identifies significant changes in remodeling-associated genes as early as day 1. MMP9 expression increased significantly (6-fold) across all conditions on day 1, with the largest fold-change occurring in Low hydrogels at the highest seeding density (10.8±1.93 fold change; Figure 4A,B). The MMP-activity repressor, TIMP-1, showed modest upregulation at day 1 (1.3-fold, 1×105 MSCs/mL; 1.5-fold, 1×106 MSCs/mL) for Low, Med, and High hydrogels; however, by day 4, all matrix-additive genes (TIMP-1 TIMP-2, TIMP-3, and Col1(a1)) were downregulated (Figure 4A,B)
Figure 4. MSC-mediated remodeling signatures.
Relative gene expression of matrix-associated genes normalized to day 0 expression, shown on a log2 scale, with downregulation shown in red and upregulation shown in blue. Gray values for TIMP-2 show non-determinant values, indicative of low yields of expression. Matrix degradation genes analyzed were matrix metalloproteases MMP-2 and MMP-9. Matrix additive genes were the tissue inhibitors of metalloproteases TIMP-1, TIMP-2, and TIMP-3, along with the matrix protein collagen, type 1, alpha 1. (n=3–6). A) Gene expression of hydrogels seeded with low density of MSCs 1×105 MSCs/mL (1:1 HSPC:MSC) (Low, Med, and High). B) Gene expression of hydrogels seeded with high density of MSCs 1×106 MSCs/mL (1:10 HSPC:MSC) (Low, Med, and High).
3.3. Culture system dependent hematopoietic differentiation patterns
The differentiation and expansion of primary murine HSPCs were examine in GelMA hydrogels, seeded at a dilute concentration (2000 HSPCs/20μL solution) to avoid direct cell-cell contact. Lineage analysis at day 7 of the HSPC monocultures reveals a significant dependence on hydrogel biophysical properties, with decreasing percentages of HSCs within increasing GelMA wt% (Figure 5). Not unexpectedly, decreases in early progenitor population maintenance led to an increase in later progenitors’ patterns (CMPs and terminal). The CMP population made up 4.95±1.02% of the hematopoietic cell population in the Low hydrogel, but was significantly larger in the High hydrogel, 13.72±2.93% (Figure 5B), with similar trends observed in terminal myeloid population (Figure S6).
Figure 5. Analysis of cell populations at day 7 (n=6).
A) The lineage hierarchy of hematopoietic stem cells, with long-term and short-term hematopoietic stem cells grouped as HSCs (blue), Multipotent Progenitors (orange), and Common Myeloid Progenitors (lavender). B) The relative population of HSC (blue), MPP (orange), CMP (lavender) are shown for single-culture and co-culture at 1:1 and 1:10 (HSPC:MSC), in the Low, Med, and High variants. The height of each subcolumn reflects how much of the total cell population is made up of the specified cell type. The blue columns (HSC) show that the highest percentage of HSCs are found in the single-culture Low variant (1:0), and in the 1:1 co-culture High variant. C) The percentage of HSCs that make up the total hematopoietic cell population, arranged in order of increasing MSC density (1:0, 1:1, 1:10 HSPC:MSC), and in order of increasing hydrogel stiffness (Low, Med, and High). The largest percent of HSC is found in the High variant at a 1:1 seeding ratio and is significantly higher (p<0.05) than all other conditions.
The behavior of heterotypic cultures of HSPCs and MSCs were examined at dilute seeding densities to reduce direct cell-cell contact to study the role of soluble biochemical interactions between cells. Co-cultures led to important shifts in HSC differentiation patterns (Figure 5). Unlike HSPC monoculture where lower wt% GelMA hydrogels promoted the highest percentage of early progenitors, the addition of MSCs in a 1:1 (HSPC:MSC) ratio reversed that trend. Rather, increased HSC maintenance in heterotypic cultures was observed in the High hydrogel variant with HSCs comprising 11.11±3.18% of the hematopoietic population after 7 days, significantly higher than HSPC:MSC cultures in Med and Low hydrogel variants (Figure 5B,C). Notably, we observe retention of 50.2%, 22.08%, and 18.50% (High, Med, and Low; Figure 6) of the day 1 HSC populations, with higher HSC maintenance associated with a reduced expansion of a differentiated myeloid population (77.54±5.83% of overall hematopoietic population at day 7 for the High hydrogel variant; 82.70±6.09%, Med; 91.50±4.67%, Low; Figure S6). However, when HSPCs were co-cultured with a ten-fold higher density of MSCs (1:10 HSPC:MSC, 1×106 MSCs/mL), hematopoietic differentiation towards the myeloid lineage was enhanced (Figure 5), with significantly reduced HSC retention and differentiated myeloid cell populations representing more than 99% of the hematopoietic cells regardless of hydrogel biophysical properties (Figure S6).
Figure 6. Relative fold change of cell population from day 1 to day 7.
A) Color-coded hematopoietic lineage hierarchy B) Fold change of HSC population. Darker blue indicates higher maintenance of HSC (highest maintenance in High variant at 1:1 seeding). C) Fold change of MPP population. Darker orange indicates higher maintenance of MPP (highest in High variant at 1:1 seeding). D) Fold change of CMP population. Darker purple indicates higher maintenance of CMP (highest in Med variant at 1:1 seeding). E) Fold change of Terminal population. Darker gray indicates higher maintenance of Terminal population (highest in Med variant at 1:1 seeding).
3.4. Culture dependent shifts in HSC quiescence
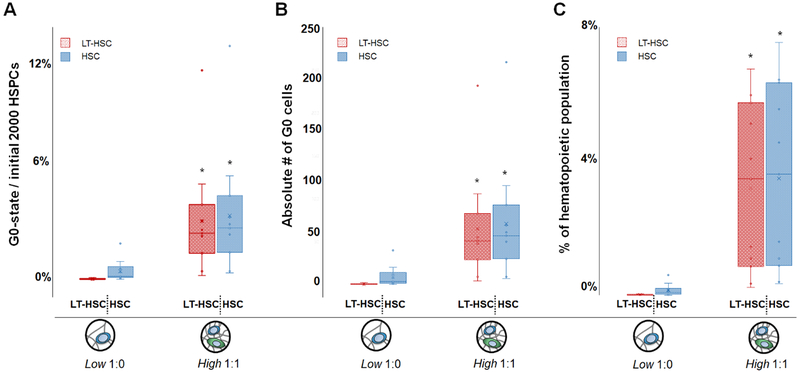
Given the observed differences in HSC retention, we subsequently examined cell cycle status for a subset of culture conditions that maintained the largest HSC population in the absence (Low 1:0) or presence (High 1:1) of MSCs. Here we compare the effect of MSC inclusion within the hydrogel environment on HSC quiescence, a key parameter for long-term culture success. Notably, inclusion of MSCs in a low diffusive environment (High 1:1) led to a significant increase in the number of quiescent (G0-state) hematopoietic stem cells. Inclusion of MSCs led to a 129-fold larger quiescent LT-HSC subpopulation and a 7.7-fold for the overall HSC population versus monocultures of HSCs in a highly diffusive environment (Low 1:0) (Figure 7). Excitingly, inclusion of a hydrogel matrix for HSC culture promoted enhanced quiescence compared to conventional liquid cultures of HSCs or HSC-MSC mixtures (Figure S7).
Figure 7. Comparison of LT-HSC and HSC quiescence after 7 days in conditions that led to the greatest maintenance of an HSC population when cultured in the absence (Low 1:0 HSPC:MSC) vs. presence (High 1:1 HSPC:MSC) of MSCs (gels=30, n=10).
A) Fraction of quiescent LT-HSC or HSC, shown as a percentage of the initial 2000 HSPCs seeded per hydrogel condition. B) Absolute number of quiescent (G0) LT-HSC and HSCs within each sample. C) Percentage of quiescent cells calculated vs. the total number of hematopoietic lineage positive cells after 7 days in culture as a percentage of the overall hematopoietic lineage positive cell population. *: significantly different (p < 0.05) versus the quiescence in high diffusivity hydrogel containing HSCs only (Low 1:0).
4. Discussion
Synthetic niche analogs for ex vivo culture of HSCs have begun to uncouple the complex signals that are inherent to the native HSC niche. Traditional efforts have demonstrated that changes in matrix composition, mechanics, and selective inclusion of growth factors can substantially alter HSC maintenance and differentiation patterns.[27, 52] More recently, the concept of soluble cell-cell signaling and its role on HSC lineage specification have been examined, primarily in liquid (unhindered-diffusion) experimental systems. Csaszar et al. demonstrated that HSC-secreted cytokines can alter ex vivo expansion of HSCs within a bioreactor,[53] while Müller et al. demonstrated that individual HSCs cultured in microcavities (autocrine feedback) led to increased HSC quiescence.[54] Efforts described here that create heterotypic cell cultures within a biomaterial matrix expand upon this work by providing an avenue to modulate kinetics of signaling modes, i.e. paracrine vs autocrine, via the poroelastic properties of the hydrogel matrix itself. We previously reported the potential of this concept in a collagen hydrogel, showing paracrine signals from Lin+ hematopoietic cells enhancing myeloid specification, while autocrine feedback in diffusion-limited collagen environments promoted HSC maintenance.[15]
Importantly, while hydrogels facilitate studies of mechanical and heterotypic cell signaling on HSC activity, it may also be sensitive to cell-mediated remodeling. Concepts of dynamic reciprocity, often employed in the context of wound remodeling or the tumor microenvironment,[55] offer a novel avenue to design HSC culture platforms. Therefore, here we developed a culture system that takes advantage of paracrine signaling and matrix remodeling from niche-associated MSCs. Our use of GelMA enables robust control over mechanical cues, transport of soluble factors, and possesses cleavage sites for matrix metalloproteinases produced by MSCs.[26] The library of hydrogels spans a range of mechanical properties inspired by the native HSC niche,[9] and advanced poroelastic characterization methods[30] allow us to examine dynamic relationships between MSCs, the local matrix architecture, and soluble cell signaling. Co-cultures of HSPCs with MSCs at varying ratios (1:0, 1:1, 1:10 HSPC:MSC) demonstrate that HSC activity is modulated by combinations of initial hydrogel properties, MSC-mediated remodeling, and the balance of autocrine feedback versus MSC-generated paracrine signals. The novelty of this work is the systematic approach to demonstrate the coordinated effects of matrix poroelastic properties, cell mediated remodeling, and heterotypic cell-cell interactions on HSC expansion vs. quiescence.
Drawing insight from the marrow niche, the GelMA hydrogels were tuned to match the range of moduli observed in vivo (Figure 2). Of the three hydrogels chosen for cellular studies (Low, Med, High), the mesh size and diffusion suggests that the High hydrogel supports a small radius of cell-secreted biomolecular diffusion, generating an autocrine-dominated regime for HSC culture.[15, 56] Similarly, the significantly increased diffusivity of the Low and Med hydrogels supports a large radius of biomolecular diffusion that reduces autocrine feedback and enhances MSC-generated paracrine signaling. Dilute cell seeding densities were specifically chosen for this study to promote large cell to cell distances of 86–205μm of spacing between cells (assuming simple cubic unit cell). The diffusivity of the hydrogels and the dilute seeding conditions (2000 HSPCs/hydrogel) restricted cell-cell interactions to soluble signaling, highlighting the role the hydrogel variants play in determining long-range cell communication.
We observed HSC population maintenance was strongly influenced by initial hydrogel poroelastic properties, the presence of MSCs, and remodeling processes. When cultured in purely homotypic environments, a high diffusion, low stiffness hydrogel (Low) supported the largest HSC maintenance (Figure 5). To explore the influence of remodeling and paracrine signals from niche-associated MSCs in the Low, Med, and High hydrogels, we examined HSC lineage specification patterns in heterotypic cultures with MSCs at either 1×105 or 1×106 MSCs/mL (1:1, 1:10 HSPC:MSC). The addition of cells did not significantly alter the initial Young’s modulus (Figure 3), however, MSC matrix remodeling occurred as evidenced by downregulated matrix-deposition genes (Col1(a1), TIMP-1, TIMP-2, TIMP-3) and upregulated matrix-degradation genes (MMP-2, MMP-9) (Figure 4). While we observed significant matrix remodeling via compressive testing in High hydrogels, no clear trends were observed in the Low or Med conditions. Taken together, the gene expression, modulus testing, and differences in diffusive pathlengths and reaction times for MMPs and TIMPs[57] suggests that hydrogel remodeling was likely non-uniform. This makes it challenging to resolve local remodeling processes within the heterotypic HSC-MSC cultures, and is an area of ongoing effort that leverages new tools for local matrix analyses (e.g. microrheology[57]).[58] However, the observed remodeling and the significant changes in HSC activity suggest dynamic interaction between HSCs and MSCs is an exciting avenue for further exploration.
Oversaturation of the culture with MSCs (1:10 HSPC:MSC) reduced maintenance of early hematopoietic progenitors compared to HSC monocultures, perhaps due to increased soluble factors and significant remodeling given the 10x number of cells in the culture (Figure 5). However, a 1:1 (HSPC:MSC) culture shifted HSC population dynamics, with a significant increase in HSC maintenance overtime. This shift aligns with literature, confirming the ability of MSCs to influence HSC equilibrium through secreted factors within the in vivo niche.[17] Particularly, the highest maintenance was observed in the High hydrogel, with an increased fraction of quiescent HSCs. This is of clinical relevance for long-term homeostasis and repair,[59] and valuable for HSC lineage specification or expansion.[60] Notably, the High hydrogel had minimal initial biotransport characteristics but also displayed markers of MSC-mediated remodeling (MMP-9 expression). This suggests a time-dependent balance between initial autocrine-favored feedback and MSC paracrine signals for increasing HSC maintenance worthy of future exploration.
The development of a synthetic niche analog for HSC culture can take advantage of poroelastic properties to mediate the balance of heterotypic cell signaling via soluble factors. Examining HSC differentiation and cell-cycle patterns, as well as MSC-mediated remodeling suggests a path towards ex vivo expansion of HSCs without exhaustion. We show that combinations of initial hydrogel properties and MSCs increases the maintenance of HSCs in a culture system with initially hindered diffusion. These findings suggest that MSC-secreted paracrine signaling and initial autocrine feedback are essential tools to expand HSCs in vitro. Such findings offer exciting opportunities to explore the importance of dynamic reciprocity in the context of heterotypic cultures over timescales beyond 7-days. Future experiments with longer culture timescales can reveal long-term phenotypic changes of MSCs and HSCS within co-culture in response to extended matrix remodeling. Matrix remodeling as well as selective domains of autocrine feedback versus paracrine signaling can be leveraged for the intelligent design of an in vitro HSC culture systems.
Supplementary Material
Acknowledgments:
The authors would like to acknowledge Dr. Barbara Pilas of the Roy J. Carver Biotechnology Center (Flow Cytometry Facility, UIUC) as well as Dr. Bhushan Mahadik and Dr. Ji-Sun Choi (ChBE, UIUC) for assistance with bone marrow cell isolation and flow cytometry. Preliminary Young’s modulus testing and mRNA extraction was aided by undergraduate researchers Kirsten Schroeder and Gabrielle Wolter respectively. Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Numbers R01 DK099528 (B.A.C.H) and F31 DK117514 (A.E.G.), as well as by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Numbers R21 EB018481 (B.A.C.H.) and T32 EB019944 (A.E.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Amino Acid Analysis was performed at the Molecular Structure Facility in the UC Davis Genome Center at the University of California, Davis. Material characterization was carried out in part in the Frederick Seitz Materials Research Laboratory Central Research Facilities, University of Illinois. The authors are also grateful for additional funding provided by the Department of Chemical & Biomolecular Engineering and the Institute for Genomic Biology at the University of Illinois at Urbana-Champaign.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References:
- [1].Doulatov S, Notta F, Laurenti E, Dick JE, Cell Stem Cell 2012, 10, 120; [DOI] [PubMed] [Google Scholar]; Mendelson A, Frenette PS, Nat Med 2014, 20, 833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ogawa M, Blood 1993, 81, 2844. [PubMed] [Google Scholar]
- [3].Pietras EM, Warr MR, Passegue E, J Cell Biol 2011, 195, 709; [DOI] [PMC free article] [PubMed] [Google Scholar]; Morrison SJ, Scadden DT, Nature 2014, 505, 327; [DOI] [PMC free article] [PubMed] [Google Scholar]; Adams GB, Scadden DT, Nat Immunol 2006, 7, 333; [DOI] [PubMed] [Google Scholar]; Purton LE, Scadden DT, in StemBook, Cambridge (MA) 2008; [Google Scholar]; Krause DS, Scadden DT, Preffer FI, Cytometry B Clin Cytom 2013, 84, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wilson A, Trumpp A, Nat Rev Immunol 2006, 6, 93. [DOI] [PubMed] [Google Scholar]
- [5].Copelan EA, N Engl J Med 2006, 354, 1813; [DOI] [PubMed] [Google Scholar]; D’Souza A, Zhu X, “Current Uses and Outcomes of Hematopoietic Cell Transplantation (HCT): CIBMTR Summary Slides”, 2016. [Google Scholar]
- [6].Zhang J, Li L, J Biol Chem 2008, 283, 9499. [DOI] [PubMed] [Google Scholar]
- [7].Choi JS, Mahadik BP, Harley BA, Biotechnol J 2015, 10, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, Park S-Y, Lu J, Protopopov A, Silberstein LE, Nat Cell Biol 2013, 15, 533; [DOI] [PMC free article] [PubMed] [Google Scholar]; Nilsson SK, Debatis ME, Dooner MS, Madri JA, Quesenberry PJ, Becker PS, J Histochem Cytochem 1998, 46, 371. [DOI] [PubMed] [Google Scholar]
- [9].Jansen LE, Birch NP, Schiffman JD, Crosby AJ, Peyton SR, J Mech Behav Biomed Mater 2015, 50, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi JS, Harley BA, Sci Adv 2017, 3, e1600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Muller E, Wang W, Qiao W, Bornhauser M, Zandstra PW, Werner C, Pompe T, Sci Rep 2016, 6, 31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Janowska-Wieczorek A, Majka M, Ratajczak J, Ratajczak MZ, Stem Cells 2001, 19, 99; [DOI] [PubMed] [Google Scholar]; Majka M, Janowska-Wieczorek A, Ratajczak J, Ehrenman K, Pietrzkowski Z, Kowalska MA, Gewirtz AM, Emerson SG, Ratajczak MZ, Blood 2001, 97, 3075. [DOI] [PubMed] [Google Scholar]
- [13].Stylianopoulos T, Poh MZ, Insin N, Bawendi MG, Fukumura D, Munn LL, Jain RK, Biophys J 2010, 99, 1342; [DOI] [PMC free article] [PubMed] [Google Scholar]; Kamali-Zare P, Nicholson C, Basic Clin Neurosci 2013, 4, 282; [PMC free article] [PubMed] [Google Scholar]; Pluen A, Netti PA, Jain RK, Berk DA, Biophysical Journal 1999, 77, 542; [DOI] [PMC free article] [PubMed] [Google Scholar]; van Donkelaar CC, Chao G, Bader DL, Oomens CW, Comput Methods Biomech Biomed Engin 2011, 14, 425. [DOI] [PubMed] [Google Scholar]
- [14].Taqvi S, Dixit L, Roy K, J Biomed Mater Res A 2006, 79, 689; [DOI] [PubMed] [Google Scholar]; Csaszar E, Kirouac DC, Yu M, Wang W, Qiao W, Cooke MP, Boitano AE, Ito C, Zandstra PW, Cell Stem Cell 2012, 10, 218. [DOI] [PubMed] [Google Scholar]
- [15].Mahadik BP, Bharadwaj NA, Ewoldt RH, Harley BA, Biomaterials 2017, 125, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ehninger A, Trumpp A, J Exp Med 2011, 208, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS, Nature 2010, 466, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Anthony BA, Link DC, Trends Immunol 2014, 35, 32; [DOI] [PMC free article] [PubMed] [Google Scholar]; Li T, Wu Y, Bone Marrow Res 2011, 2011, 353878; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wagner W, Roderburg C, Wein F, Diehlmann A, Frankhauser M, Schubert R, Eckstein V, Ho AD, Stem Cells 2007, 25, 2638; [DOI] [PubMed] [Google Scholar]; El Marsafy S, Journal of Stem Cell Research & Therapy 2014, 04. [Google Scholar]
- [19].Sugiyama T, Kohara H, Noda M, Nagasawa T, Immunity 2006, 25, 977. [DOI] [PubMed] [Google Scholar]
- [20].Mirantes C, Passegue E, Pietras EM, Exp Cell Res 2014, 329, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tocci A, Forte L, Hematol J 2003, 4, 92. [DOI] [PubMed] [Google Scholar]
- [22].Page-McCaw A, Ewald AJ, Werb Z, Nat Rev Mol Cell Biol 2007, 8, 221; [DOI] [PMC free article] [PubMed] [Google Scholar]; Sassoli C, Nosi D, Tani A, Chellini F, Mazzanti B, Quercioli F, Zecchi-Orlandini S, Formigli L, Exp Cell Res 2014, 323, 297; [DOI] [PubMed] [Google Scholar]; Daley WP, Peters SB, Larsen M, J Cell Sci 2008, 121, 255. [DOI] [PubMed] [Google Scholar]
- [23].Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM, Wound Repair Regen 2011, 19, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alexander J, Cukierman E, Curr Opin Cell Biol 2016, 42, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benton JA, DeForest CA, Vivekanandan V, Anseth KS, Tissue Eng Part A 2009, 15, 3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pedron S, Harley BA, J Biomed Mater Res A 2013, 101, 3404. [DOI] [PubMed] [Google Scholar]
- [27].Mahadik BP, Pedron Haba S, Skertich LJ, Harley BA, Biomaterials 2015, 67, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen JE, Pedron S, Harley BAC, Macromol Biosci 2017, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].A. International, ASTM International, 2017;; Pedron S, Becka E, Harley BA, Biomaterials 2013, 34, 7408. [DOI] [PubMed] [Google Scholar]
- [30].Kalcioglu ZI, Mahmoodian R, Hu Y, Suo Z, Van Vliet KJ, Soft Matter 2012, 8; [Google Scholar]; Hu Y, Zhao X, Vlassak JJ, Suo Z, Applied Physics Letters 2010, 96. [Google Scholar]
- [31].Hu YH, Chen X, Whitesides GM, Vlassak JJ, Suo ZG, Journal of Materials Research 2011, 26, 785; [Google Scholar]; Lai Y, Hu Y, Soft Matter 2017, 13, 852. [DOI] [PubMed] [Google Scholar]
- [32].Peppas NA, Merrill EW, Journal of Applied Polymer Science 1977, 21, 1763. [Google Scholar]
- [33].Bray JC, Merrill EW, Journal of Applied Polymer Science 1973, 17, 3779. [Google Scholar]
- [34].Canal T, Peppas NA, J Biomed Mater Res 1989, 23, 1183. [DOI] [PubMed] [Google Scholar]
- [35].Schiel JE, Hage DS, Talanta 2005, 65, 495. [DOI] [PubMed] [Google Scholar]
- [36].Fels IG, Journal of Applied Polymer Science 1964, 8, 1813; [Google Scholar]; Fessler JH, Hodge AJ, Journal of Molecular Biology 1962, 5, 446. [DOI] [PubMed] [Google Scholar]
- [37].Anjum F, Lienemann PS, Metzger S, Biernaskie J, Kallos MS, Ehrbar M, Biomaterials 2016, 87, 104. [DOI] [PubMed] [Google Scholar]
- [38].Flory PJ, Rehner J, The Journal of Chemical Physics 1943, 11, 521. [Google Scholar]
- [39].Burchard W, in Physical Techniques for the Study of Food Biopolymers, 1994, 151; [Google Scholar]; Bryngelson JD, E. M. Billings, in Physics of Biological Systems, Springer, 1997, 80. [Google Scholar]
- [40].Challen GA, Boles N, Lin KKY, Goodell MA, Cytom Part A 2009, 75a, 14; [DOI] [PMC free article] [PubMed] [Google Scholar]; Okada S, Nakauchi H, Nagayoshi K, Nishikawa SI, Miura Y, Suda T, Blood 1992, 80, 3044. [PubMed] [Google Scholar]
- [41].Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE, Blood 2005, 105, 2717. [DOI] [PubMed] [Google Scholar]
- [42].Yang LP, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SEW, Blood 2005, 105, 2717; [DOI] [PubMed] [Google Scholar]; Beaudin AE, Boyer SW, Forsberg EC, Exp Hematol 2014, 42, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tian C, Zhang Y, Ann Hematol 2016, 95, 543. [DOI] [PubMed] [Google Scholar]
- [44].Zhang Y, Yan X, Sashida G, Zhao X, Rao Y, Goyama S, Whitman SP, Zorko N, Bernot K, Conway RM, Witte D, Wang QF, Tenen DG, Xiao Z, Marcucci G, Mulloy JC, Grimes HL, Caligiuri MA, Huang G, Blood 2012, 120, 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Challen GA, Boles N, Lin KK, Goodell MA, Cytometry A 2009, 75, 14; [DOI] [PMC free article] [PubMed] [Google Scholar]; A. Mayle, M. Luo, M. Jeong, M. A. Goodell, Cytometry A 2013, 83, 27;22736515 [Google Scholar]; Spangrude GJ, Heimfeld S, Weissman IL, Science 1988, 241, 58. [DOI] [PubMed] [Google Scholar]
- [46].in Part Number 4371095, Rev. B, Applied Biosystems, 2008.
- [47].Abraham LC, Dice JF, Lee K, Kaplan DL, Exp Cell Res 2007, 313, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cohen J, Statistical power analysis for the behavioral sciences, L. Erlbaum Associates, Hillsdale, N.J. : 1988; [Google Scholar]; Cohen J, Psychol Bull 1992, 112, 155. [DOI] [PubMed] [Google Scholar]
- [49].Ghasemi A, Zahediasl S, Int J Endocrinol Metab 2012, 10, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Carroll RJ, Schneider H, Statistics & Probability Letters 1985, 3, 191. [Google Scholar]
- [51].Dean RB, Dixon WJ, Analytical Chemistry 1951, 23, 636. [Google Scholar]
- [52].Choi JS, Harley BAC, Science Advances 2017, 3, e1600455; [DOI] [PMC free article] [PubMed] [Google Scholar]; Altrock E, Muth CA Klein G, Spatz JP, Lee-Thedieck C, Biomaterials 2012, 33, 3107; [DOI] [PubMed] [Google Scholar]; Cuchiara ML, Coşkun S, Banda OA, Horter KL, Hirschi KK, West JL, Biotechnol Bioeng 2016, 113, 870; [DOI] [PubMed] [Google Scholar]; Cuchiara ML, Horter KL, Banda OA, West JL, Acta Biomater 2013, 9, 9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Csaszar E, Daniel C Kirouac M Yu W Wang W Qiao Michael Cooke P, Boitano Anthony E., Ito C, Zandstra Peter W., Cell Stem Cell 2012, 10, 218. [DOI] [PubMed] [Google Scholar]
- [54].Müller E, Wang W, Qiao W, Bornhäuser M, Zandstra PW, Werner C, Pompe T, Sci Rep 2016, 6, 31951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Alexander J, Cukierman E, Current Opinion in Cell Biology 2016, 42, 80; [DOI] [PMC free article] [PubMed] [Google Scholar]; Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM, Wound Repair and Regeneration 2011, 19, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pedron S, Pritchard AM, Vincil GA, Andrade B, Zimmerman SC, Harley BA, Biomacromolecules 2017, 18, 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schultz KM, Kyburz KA, Anseth KS, Proc Natl Acad Sci U S A 2015, 112, E3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Daviran M, Longwill SM, Casella JF, Schultz KM, Soft Matter 2018, 14, 3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lio P, Macdonald HR, Trumpp A, Cell 2008, 135, 1118. [DOI] [PubMed] [Google Scholar]
- [60].Li J, Exp Hematol 2011, 39, 511; [DOI] [PubMed] [Google Scholar]; Aggarwal R, Lu J, Pompili VJ, Das H, Current Molecular Medicine 2012, 12, 34; [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakamura-Ishizu A, Takizawa H, Suda T, Development 2014, 141, 4656. [DOI] [PubMed] [Google Scholar]
- [61].ThermoFisher.
- [62].Ofner IIICM, Bubnis WA, Pharmaceutical Research 1996, 13, 1821. [DOI] [PubMed] [Google Scholar]
- [63].Kasapis S, Al-Marhoobi IM, Mitchell JR, Biopolymers 2003, 70, 169. [DOI] [PubMed] [Google Scholar]
- [64].Berg JM TJ, Stryer L, in Biochemistry. 5th edition, W H Freeman, New York: 2002. [Google Scholar]
- [65].Pezron I, Djabourov M, Leblond J, Polymer 1991, 32, 3201; [Google Scholar]; Gupta A, Mohanty B, Bohidar HB, Biomacromolecules 2005, 6, 1623. [DOI] [PubMed] [Google Scholar]
- [66].Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL, BMC Bioinformatics 2012, 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B, BMC Genomics 2008, 9, 633; [DOI] [PMC free article] [PubMed] [Google Scholar]; Spandidos A, Wang X, Wang H, Seed B, Nucleic Acids Res 2010, 38, D792; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang X, Seed B, Nucleic Acids Res 2003, 31, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.