Abstract
Colorectal cancer is subject to a high rate of mutations, with late stage tumors often containing many mutations. These tumors are difficult to treat, and even with the recently implemented methods of personalized medicine at modern hospitals aiming to narrow treatments, a gap still exists. Proper modeling of these tumors may help to recommend optimal treatments for individual patients, preferably utilizing a model that maintains proper signaling in respect to the derived parent tissue. In this study, we utilized an extracellular matrix-derived hydrogel to create a 3D micro-tumor construct platform capable of both supporting cells for long time durations and for high throughput drug screening. Experiments with cell lines demonstrated long-term viability with maintenance of cell proliferation. Furthermore, studies with several chemotherapeutics utilizing different mechanisms of action displayed differences in efficacy in comparing 3D and 2D cultures. Finally, patient colorectal tumor tissue was acquired and employed to reconstruct micro-tumor constructs, providing a system for the testing of novel chemotherapeutics against tumors in a patient-specific manner. Collectively, the results describe a system capable of high throughput testing while maintaining important characteristics of the parent tissue.
Keywords: Precision medicine, Personalized medicine, Organoids, Cancer models, In vitro, Ex vivo
INTRODUCTION
Colorectal cancers (CRC) are one of the top five most diagnosed cancers, consisting of 8% of all cancers with 4.4% of all people being diagnosed in their lifetime.30 Although early detection methods such as colonoscopy exist, at least 20% of newly detected colorectal cancer is in stage IV, often carrying a high mutational load, with the five-year survival rate at 12%.16 Common mutations include proto-oncogene mutations to the EGFR, PI3K/AKT, and TGFβRII pathways, tumor suppressor inactivating mutations to TP53, APC, and the SMAD family, along with other chromosome mutations, microsatellite instability, and CpG island methylation promotion affecting gene expression.3, 8, 28 The presence of these genetic abnormalities can alter the clinician’s regimen; it is important they are given the best information about the tumor and have the best treatments to treat them. Treatments may include surgery/radiotherapy followed by chemotherapy depending on the stage of the tumor.15
Development of chemotherapies for the treatment of various cancers have involved using immortalized cell lines derived from patient tumor samples. The cell lines chosen for the treatments should contain the targetable mechanism to confirm a novel therapy’s efficacy and other cell lines without the particular mutation or target of interest to determine if there is a difference in efficacy between the two. There have been numerous colorectal cancer cell lines established that encompass a wide range of genotypes and phenotypes.1 However, it is known that 2D culture can change the phenotype of a cell line over time, an observation that has been confirmed by observing a conversion of mesenchymal CRC cells into epithelial phenotypes and morphologies during traditional 2D culture.32 Furthermore, it has been determined that differences in signaling caused by such artificial conditions can cause drug resistance towards certain pathway targeting drugs.18, 22 This drift away from the original tissue creates motivation to develop models for drug compound screens that more closely mimic the tissue of origin.
At the current stage in treatment of CRC, oncologists prescribe cocktails of drugs to reduce the tumor’s size and ability to spread. Until recently, these cocktails consisted of several compounds designed to kill cells that reproducible quickly and non-specifically, such as FOLFOX or FOLFIRI regimens. However, with the advent of pathway specific inhibitors and monoclonal antibody-based drugs, the potential of personalized treatment has grown and allowed extended survival and improved quality of life. Today, precision medicine efforts include a biopsy of the tumor to determine the characteristics for the patient’s cancer, including staging and aggressiveness.10 Genomic screening can also be used to identify mutations to be targeted by pathway-specific drugs. However, determination of which drug will be most efficacious is still often a clinician’s “best guess”, with the chosen regimen not always working despite tumor genotype.
In cases of unresectable colorectal cancer, chemotherapy remains the best treatment option, yet the development of targeted therapies has lagged behind other cancers. The need for proper diagnosis and treatment has grown more important than ever, an area where gaps still exist between the doctor diagnosing the cancer genotype and determining treatment.6 However, despite identifying targets, there is no way to test these targets against a bank of drugs to confirm the response of the genotyping and determine the best treatment regimen in an empirical manner.14
Our laboratory has long held that accurate in vitro systems should preferably utilize a 3D architecture and use the cells that most closely resemble the composition of the tissue they are trying to mimic, both in numbers and with physiological function. By accomplishing these criteria, we believe drug testing performed will better mirror the response of the parent in vivo tissue, thus serving as more accurate models for preclinical drug and toxicity testing, as well as personalized medicine applications (Fig. 1). The hypothesis that inspired the studies herein is 3D tissue and tumor constructs and organoid models are able to more accurately mimic the physiological function of the human body because they properly maintain expression of tissue specific markers as well as specific cell-cell interactions, something 2D cultures are inferior in comparison.27 Notably, we have shown previously in such 3D systems that colorectal cancer cells of a mesenchymal origin retain this origin. Alternatively, when placed in 2D plastic tissue culture, they transitioned to an epithelial phenotype, no longer accurately representing the derived tissue source.32, 33 Because of this, cells drawn from patients and maintained as 3D constructs using ECM-derived hydrogels will better match the source from which they are drawn in comparison to 2D over time, a practice utilized in both patient derived xenografts along with organoid expansion models.24, 31, 34
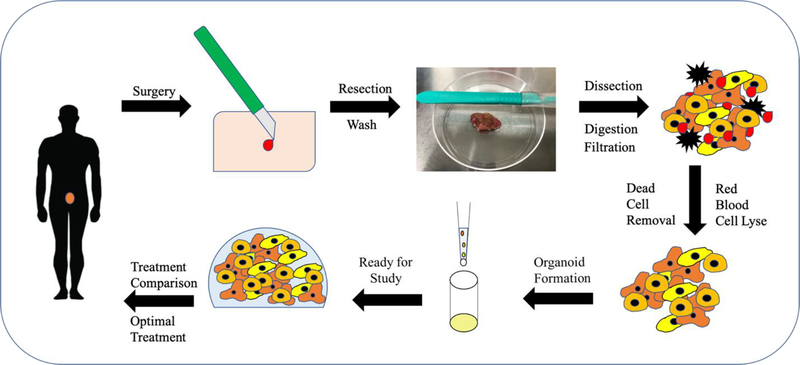
Figure 1. Proposed In Vitro treatment protocol for personalized medicine.
System for utilization of 3D in vitro system for personalized medicine as described above. Current personalized medicine regimens include doctor taking a biopsy from patient and sending it to be tested for certain mutations to determine drug regimen. Our proposal would include taking cells from the biopsy to create a 3D system to confirm patient cell response to certain chemotherapeutics.
Recently, we have applied these concepts to generate ex vivo micro-tumor constructs (μTCs) from mesothelioma and appendiceal tumor biospecimens and demonstrated their use in drug screening studies.23, 35 Herein we describe the development of analogous models of CRC. By utilizing a 3D ECM hydrogel-based model, we were able to create a reproducible database for the study of colorectal cancer and its response to chemotherapeutics. By comparing the results from the toxicity testing across multiple cell line-derived μTCs, our group was able to determine chemotherapeutic dosing to target a tumor with an unknown genetic profile. Patient-derived CRC tumor tissue was then procured and processed to form patient-specific CRC μTCs and tested to demonstrate the ability to rapidly treat patient cells against a range of treatments. We believe that the use of these 3D systems will lead towards utilization of a large-scale, reproducible and more accurate alternative to 2D cell culture and animal models for both the development of new drugs and the testing of patient tumor cells to determine the best treatment regimen in a patient-by-patient manner.
MATERIALS AND METHODS
Both cell lines and primary human tissues were employed in the described studies; all cell lines were purchased commercially, and all human tissue was acquired with consent under a protocol approved by the Wake Forest School of Medicine Institutional Review Board (IRB), entitled “Organoid Technology: a patient-specific tumor-on-a-chip platform for determining chemotherapy response prior to the initiation of treatment for abdominal malignancies“, overseen by Drs. Votanopoulos and Skardal. Additionally, all experiments involving biohazards, select agents, and toxins were performed in line with institutional safety guidelines of Wake Forest Baptist Medical Center.
Colorectal Cancer Cell Lines and Organoid Formation
Human colon carcinoma cells (HCT-116, HT-29, Caco-2, and SW480, originally sourced from ATCC, made available through the Cell and Viral Vector Core Shared Resource at Wake Forest) were expanded on tissue culture plastic using 15 cm tissue-treated dishes until 90% confluence with Dulbecco’s Minimum Essential Medium (DMEM, Sigma, St. Louis, MO), containing 10% fetal bovine serum (FBS, Hyclone, Logan, UT). Cells were detached from the substrate with Trypsin/EDTA (Hyclone) and resuspended for use in further studies.
Tumor Dissociation
Tumors were acquired on day of resection following IRB-approved guidelines, with dissociation and organoid formation performed within 24 hours. Tumors were washed twice in a solution of DPBS with 5 μg/mL Gentamicin (G1272, Sigma) 5 μg/mL Amphotericin B (A2942, Sigma) and 10 μL/mL Penicillin/Streptomycin for five minutes. Tumors were manually dissected into pieces measuring less than 2 mm, removing any fat or necrotic tissue. Tissue was placed into a 15 mL conical containing DMEM low glucose with no supplements, 100,000 CDA units/ mL Collagenase HA 200 Wunch Unit (001–1050, VitaCyte, Indianapolis, IN), and 22,000 NPA units/ mL BP Protease (003–1000, VitaCyte), with the total volume enough to cover the dissected tumor. The conical was then placed onto a mixing rack and kept at 37°C until tumor was dissociated, with a maximum time of two hours allowed. 5 mL of cold DMEM-10 was added to the tumor dissociation mixture to quench enzymatic activity and contents of tube were transferred to a sterile 50 mL conical tube. The dissociated tumor mixture was then placed into sterile filtration kit with 100 μM pore size (SCNY00100, Millipore) to remove undigested pieces and centrifuged. BD Pharm Lyse™ (555899, BD Biosciences) was then added to the cell pellet to perform a red blood cell lyse according to company protocol. Dead cell sorting was performed on the remaining cells (130–090-101, Miltenyi Biotec). Remaining cells were counted and ready for use in experiments.
Micro-Tissue Construct Formation
Constructs were formed using a thiolated hyaluronic acid, thiolated gelatin, and polyethylene glycol diacrylate (PEGDA)-based hydrogel system (ESI-BIO, Alameda, CA). Thiolated HA and gelatin components were dissolved at 1% w/v each in water containing 0.1% w/v photoinitiator (2-Hydroxy-4-(2-hydroxyethoxy)-2-methylpropiophenone, Sigma, St. Louis, MO), and mixed with a 1% w/v linear PEGDA crosslinker solution in a 2:2:1 volume ratio. For construct formation, the hydrogel-precursor solution was used to resuspend cells at a cell density of 10×106 cells/mL. Constructs were placed into the wells of a sterile 48 or 96 well plate previously coated with cured polydimethylsiloxane (PDMS, used as a hydrophobic coating) in 5 or 10 μL constructs per well, based on experiment. The constructs were then exposed to UV light from a DYMAX 75 V.2 UV spot lamp for one second each. The constructs were then covered with 200 μL DMEM media with media changes performed every three days. μTCs were allowed to incubate for 24 hours for cell lines or seven days for patient samples until treatment began.
Preparation of drug stock solutions
Nine treatment regimens, eight single agents and one combinational treatment, were used in colorectal cancer cell line screening. Novel chemotherapeutics F10 and CF10 were gifted from the Gmeiner research group at the Wake Forest School of Medicine. 5FU (F6627), cisplatin (479036 Aldrich), irinotecan (I1406) oxaliplatin (O9512) were purchased from Sigma-Aldrich. Regorafenib (S1178), sorafenib (S7397), dabrafenib (S2807) and trametinib (S2673) were purchased from SelleckChem© (Houston, Texas). Stocks of 10 mM were created for 5FU, F10, CF10, irinotecan, regorafenib, sorafenib, dabrafenib and trametinib by dissolving in DMSO; stocks of 10 mM were created for cisplatin, carboplatin, and oxaliplatin, respectively by dissolving in H2O. Doses were selected based on literature findings, and ranges for F10, CF10, 5FU, cisplatin, carboplatin, oxaliplatin, regorafenib and sorafenib were selected as 1–100 μM, with additional doses added if no LD50 was determined. Treatments of dabrafenib/trametinib had doses selected after a literature review; a ratio of 20:1 for dabrafenib: trametinib.2, 7 The range for treatments was 20, 40, 80 μM dabrafenib, scaling trametinib accordingly.
LIVE/DEAD Analysis
CRC cells in 2D and μTCs were incubated for four days under normal conditions, with additional seven, and fourteen days for μTCs. Media was removed viability was assessed by LIVE/DEAD® Viability/Cytotoxicity Kit assays (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions and allowed to incubate for 1 hour. Imaging was then performed by macro-confocal microscopy (Leica TCS LSI, Leica, Wetzlar, Germany) and composite images were created with ethidium bromide red fluorescence representing dead nuclei and calcein AM green fluorescence representing live cells.
MTS Assay
CRC cells in 2D and μTCs were treated for 72 hours at 37°C with 5% CO2 with n=5 or higher in 96 well plates. After treatments, media was aspirated and 100 μL of DMEM with additional 20 μL of Cell Titer® 96AQueous One Solution Cell Proliferation (Promega) was added to wells. Plates were then incubated at 37°C and 5% CO2 for a total of 30 minutes in 2D and 90 minutes for 3D, respectively. Upon completion, 100 μL from each well was placed into a sterile 96 well plate and read on a mass spectrometer at 490 nM wavelength. Readings were recorded and processed according to manufacturer’s instructions and data was entered into GraphPad Prism © to determine IC50 and to analyze differences in viability between 2D and 3D.
ATP activity assay
Treatments were added in 200 μL solution to wells of 96 well plates containing μTCs and allowed to incubate for 72 hours at 37°C with 5% CO2 with n=4 or higher. Media was then removed, with 200 μL of 1:1 mixture of DMEM and 3D Cell-Titer Glo Luminescent Cell Viability Assay solution, prepared according to manufacturer’s instructions (G968B, Promega, Madison, WI), was then added The entire liquid contents of the wells containing μTCs were then added to Costar White Polystrene 96 well Assay Plate (3912, Corning, NY) wells and the contents were read on a Veritas Microplate Luminometer (Promega) using default settings. Values were then averaged for experimental groups and analyzed using Graph Pad Prism© (Graphpad, La Jolla, CA) software.
Immunohistochemistry Staining and Imaging
3D μTCs were maintained for 96 hours, seven days and fourteen days in 96 well plates. Afterwards, μTCs were placed into 4% paraformaldehyde for four hours. Fixed 3D μTCs were then histologically processed to produce slides for analysis. A hematoxylin and eosin stain to visualize living cells and their positions was performed. Ki67(1:100) (ab16667) and CK18 (1:100) (ab82254) antibodies were utilized for chromogenic stains to determine cell division and identity of colorectal cancer origin, respectively. Slides were imaged on a Leica DM4000B.
Statistical Analysis
The data are generally presented as the means of number of replicates ± the standard deviation. All data are graphed and analyzed for significance using a Student’s T-test. For MTS and ATP assay results, p-values were considered significant under 0.05. Data samples were eliminated from the experimental groups if they fell outside two standard deviations from experimental group averages. Sample sizes (n=4 or n=5, depending on experimental parameters) were determined based on preliminary experiments. These sample sizes, with typically observed standard deviations, allowed statistical significance at p=0.05 with statistical power greater than 80%.
RESULTS
LIVE/DEAD imaging displays long term viability of cell line μTCs
Four colorectal cancer cell lines were utilized for experiments (Supplemental Table 1). Each line has been well studied and observations about their phenotype and genotypes can help to create a more complete and comprehensive data set when planning studies, particularly when utilizing targeted chemotherapeutics to determine treatments.4
μTCs and 2D cell lines were imaged after four days to confirm viability (Figure 2A), with μTCs imaged at additional time points of seven and fourteen days (Supplemental Figure 1) to determine if treatment time was acceptable to maximize proliferation assay results and to confirm health and growth of μTCs for potential longer-term studies. 2D wells were found to be 75% confluent at 24 hours, which leaves the cells in a large quantity but also in a proliferative state as advised by manufacturer. Overall μTCs displayed viability over the course of the fourteen day study and between time points, with cellular growth at fourteen days including cells growing around the perimeter and migrating from μTCs, potentially making viability readings beyond this point unreliable.
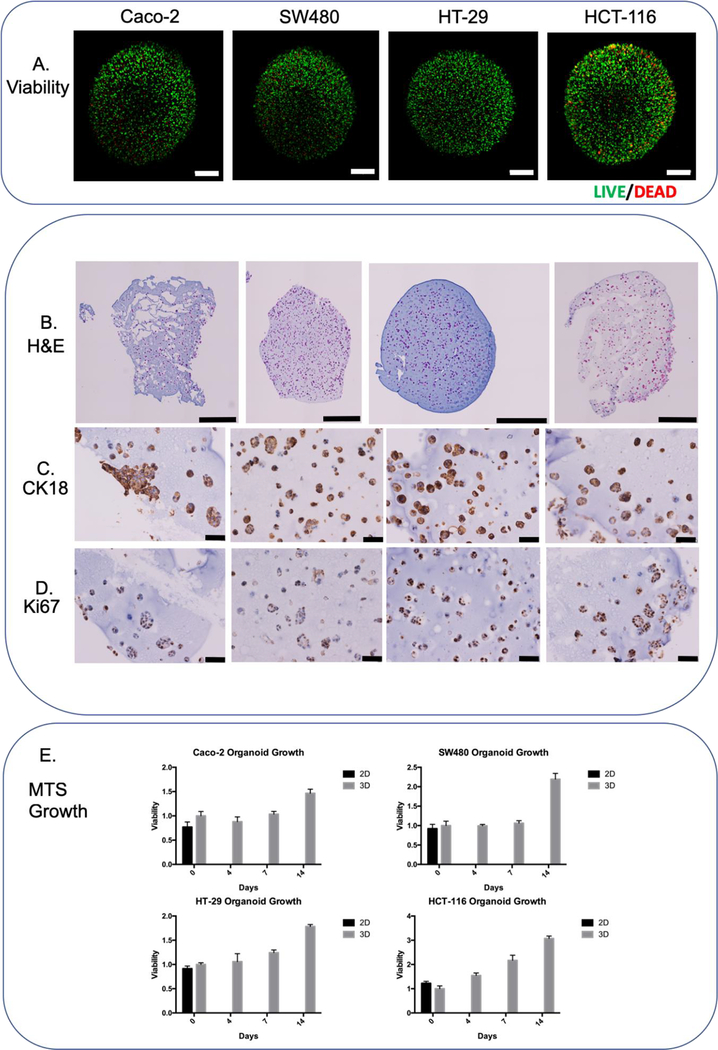
Figure 2-. Colorectal cancer cell line panel.
A) LIVE/DEAD images of colorectal cell lines (96 hours) B) H&E whole μTCs images (96 hours) C) CK18 chromogenic staining (96 hours) D) Ki67 chromogenic staining (96 hours) E) MTS growth chart over 2 weeks. Scale bars are equivalent to A) and B) 500 μM; C) and D) 50 μM.
Immunohistochemistry and Chromogenic staining display healthy μTCs with markers of proliferation and colorectal tumor identity
In order to determine cell distribution inside the μTCs, H&E staining was performed to assess the location of the cells inside the colorectal μTCs at time points of 4, (Figure 2B) 7 and 14 days. (Supplemental Figure 2). Ki67 is a marker that becomes localized to the nucleus only during active mitotic cell cycle; observation of this marker can be interpreted cells undergoing active growth in preparation for division29. Cells with active cell cycle, confirmed by positive Ki67 staining (Figure 2D), were observed in the μTCs with large growth noticed around the edges, presenting differently between the cell lines at two weeks (Figure 2 and Supplemental Figures 1 and 2). For example, HT-29 cells formed “spheroids” along the edge and eventually migrated from the μTC, whereas other cell lines grew very thick groups around the edge before eventually spreading off the main μTC. In addition, all cells for colorectal cell lines stained positive for CK18, a histopathology marker often found in highly expressed in colorectal tumors13 (Figure 2C).
Colorectal cancer cell line μTCs display metabolic growth over time display differences in drug sensitivity between 2D and 3D
Colorectal cancer cell line μTCs displayed steady growth over time, displaying a twofold increase in metabolic rate over two weeks as determined by MTS assay (Figure 2E). HCT-116, a highly proliferative and invasive CRC cell line, displayed the largest increase compared to the day zero group, increasing around three-fold.
Calculations were made to determine doses at which significance p=0.05 was reached using a student’s t-test between cells exposed to chemotherapeutics compared to control. Additionally, significance of p=0.05 was calculated between doses for 2D cell line cultures and 3D μTCs systems to determine if one system displayed more sensitivity in treatment efficacy. By using both significance against control and LD50 calculated by GraphPad Prism, it becomes clearer as to where true drug efficacy exists.
MTS assays were performed on colorectal μTCs exposed to numerous chemotherapeutics (Supplemental Table 2) over ranges of doses. Dose ranges were chosen based on previous literature which examined the concentrations required to cause cell death in 2D cultures. In comparing 2D vs 3D populations, a noticeable trend emerges: classes of chemotherapeutics utilizing DNA replication and repair mechanisms, including platinum-based compounds, fluoropyrimidines, and irinotecan, a topoisomerase I inhibitor, are in most cases more effective in 2D than 3D in determining LD50 (Figures 3, 4 and Table 1). Some groups have analyzed this trend in 2D and 3D cultures; one possible explanation for this disparity could be due to the distribution of growth media; in 2D cultures, the growth media is uniformly distributed across all cells present compared to a gradient like distribution for 3D cultures, creating an unequal distribution of chemotherapeutics.9, 17 Another proposed explanation involves the increased time 2D cells spend in active cell cycle and DNA synthesis compared to 3D cultures, allowing for more opportunity for these compounds to act22. Other groups have hypothesized differences in cultures are due to increased expression of genes associated with resistance, particularly those involved with drug metabolism.5, 17 In particular, a group analyzing the differences in treatment efficacy for paclitaxel described enhanced efficacy for 2D cultures compared to 3D cultures in ovarian cancer cell-based models, potentially due to increased drug resistance developed in 3D cultures compared to 2D.21 Similarly, groups analyzing differences in therapies such as 5FU, irinotecan and oxaliplatin for colorectal cancer have observed increased efficacy of these drugs in 2D over 3D.18, 33 It is likely that multiple mechanisms are responsible for this observation.
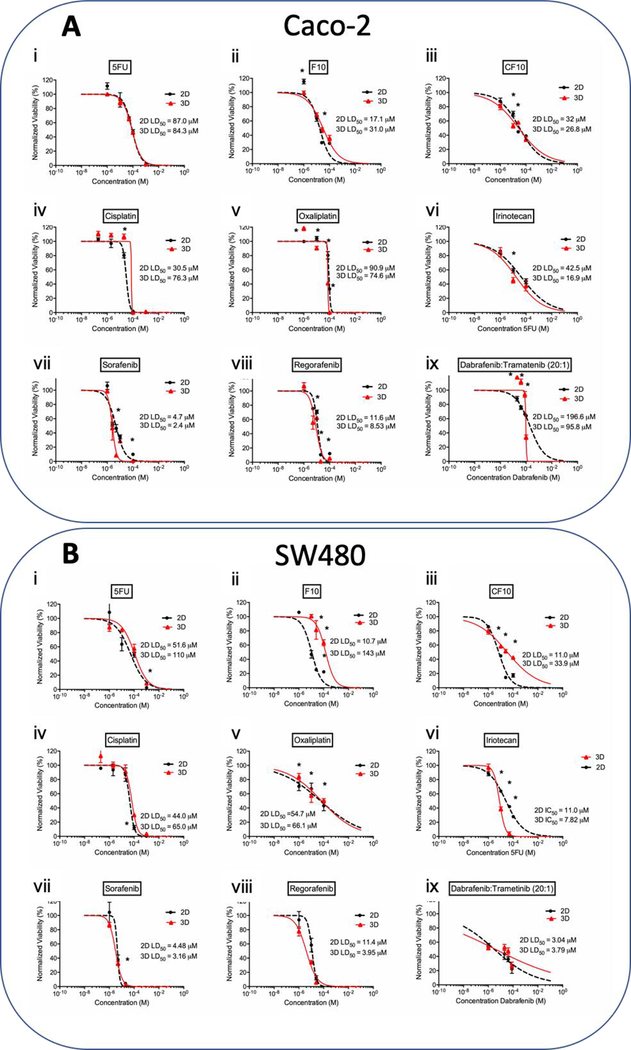
Figure 3. LD50 charts of chemotherapeutic treatments on A) Caco-2 and B) SW480 cell lines.
The effects of fluoropyrimidine based drugs i) 5FU, ii) F10, iii) CF10, platinum-based drugs iv) cisplatin, v) oxaliplatin, vi) topoisomerase I inhibitor irinotecan, small molecule inhibitors vii) sorafenib, viii) regorafenib, and ix) dabrafenib/trametinib combination treatment, on the proliferation and viability of the cell line in 2D and 3D organoid. Statistical significance: * is equivalent to p-value under 0.05 between 2D and 3D at the same dose.
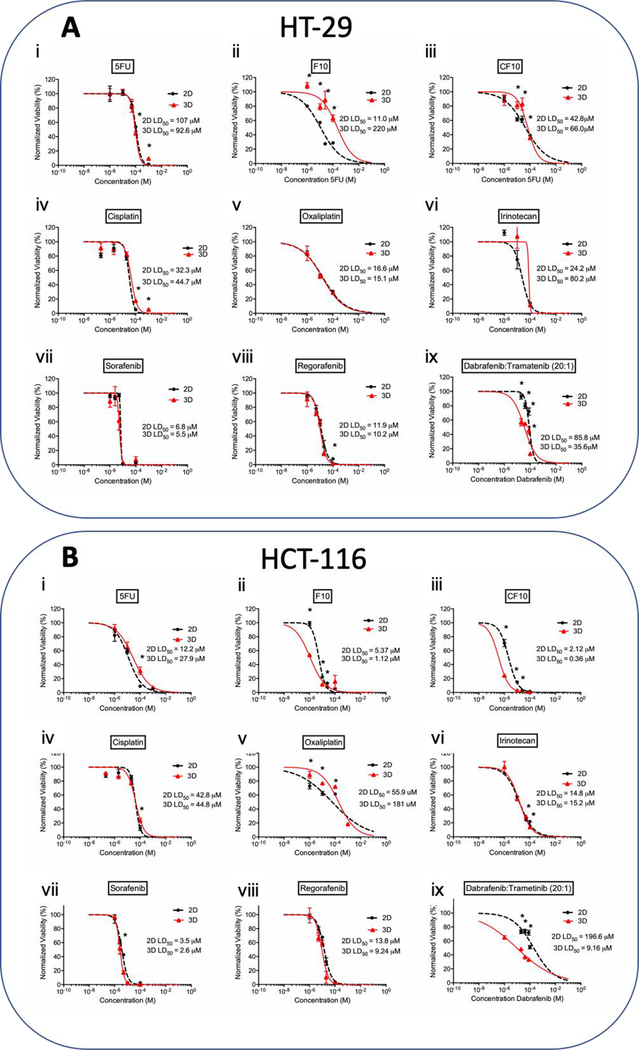
Figure 4. LD50 charts of chemotherapeutic treatments on A) HT-29 and B) HCT-116 cell lines.
The effects of fluoropyrimidine based drugs i) 5FU, ii) F10, iii) CF10, platinum-based drugs iv) cisplatin, v) oxaliplatin, vi) topoisomerase I inhibitor irinotecan, small molecule inhibitors vii) sorafenib, viii) regorafenib, and ix) Dabrafenib/trametinib combination treatment, on the proliferation and viability of the cell line in 2D and 3D organoid. Statistical significance: * is equivalent to p-value under 0.05 between 2D and 3D at the same dose.
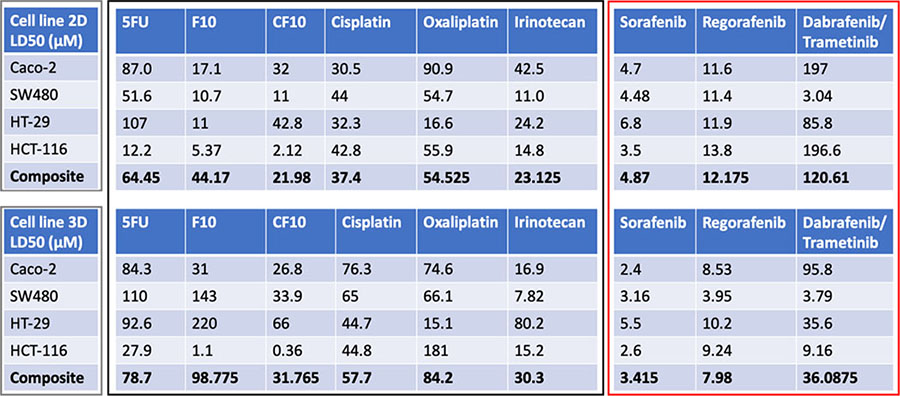
Table 1. Comparison of the LD50 doses for chemotherapeutics on all cell lines.
Table summarizes the LD50 doses for all chemotherapeutics on all four cell lines tested. Composite LD50 doses for all compounds affecting DNA replication and repair mechanisms were lower for all 2D cultures compared to 3D cultures, and all small molecule inhibitors displayed higher composite efficacy in 3D cultures compared to 2D cultures.
In contrast, compounds that target downstream EGFR pathway mutations appear to cause cell death more effectively in 3D than 2D. Small molecule inhibitors including those that target the EGFR pathway and its’ downstream proteins are used for patients with tumors containing targets that these molecules can interact and inhibit. These patient’s tumor cells often contain mutated or upregulated pathways which, while providing enhanced growth due to constant expression, can also lead to increased sensitivity if targeted. Observations in literature debate which therapies utilizing pathway targeting inhibitors show more efficacy in either 2D or 3D. Therapies targeting AKT-mTOR pathway components have shown increased efficacy in 3D cultures compared to 2D; it was theorized to be related to the increased rate of phosphorylation for AKT in 2D cultures27. Similarly, increased rates of phosphorylated and total p42/ p44 MAPK have been described in 3D for both HT-29 and SW480 as compared to 2D cultures. In these cultures, testing of IC50 for drugs utilizing inhibition of EGFR tyrosine kinase displayed resistance for 3D cultures of WT EGFR pathway cell line Caco-2 in comparison to 2D.22 Very few studies have been performed to measure activity of K-Ras, B-Raf and MEK½ or measuring the efficacy of available drugs targeting B-Raf or MEK½ in comparing 2D and 3D cultures. It is our belief these pathways are favored in 3D and expressed at a higher rate in our μTCs and by using small molecule inhibitors to target these pathways will display increased efficacy compared to their 2D counterparts. Our group has previously observed upregulations in pathways related to epithelial to mesenchymal transition such as WNT- beta catenin signaling from transitioning cell lines from 2D to 3D culture over a period of time32, 33. It is perhaps due to differences in signaling pathways that may account for the increased efficacy of small molecule inhibitors in our 3D cultures.
Patient derived μTCs show viability and characteristics of colorectal tumors
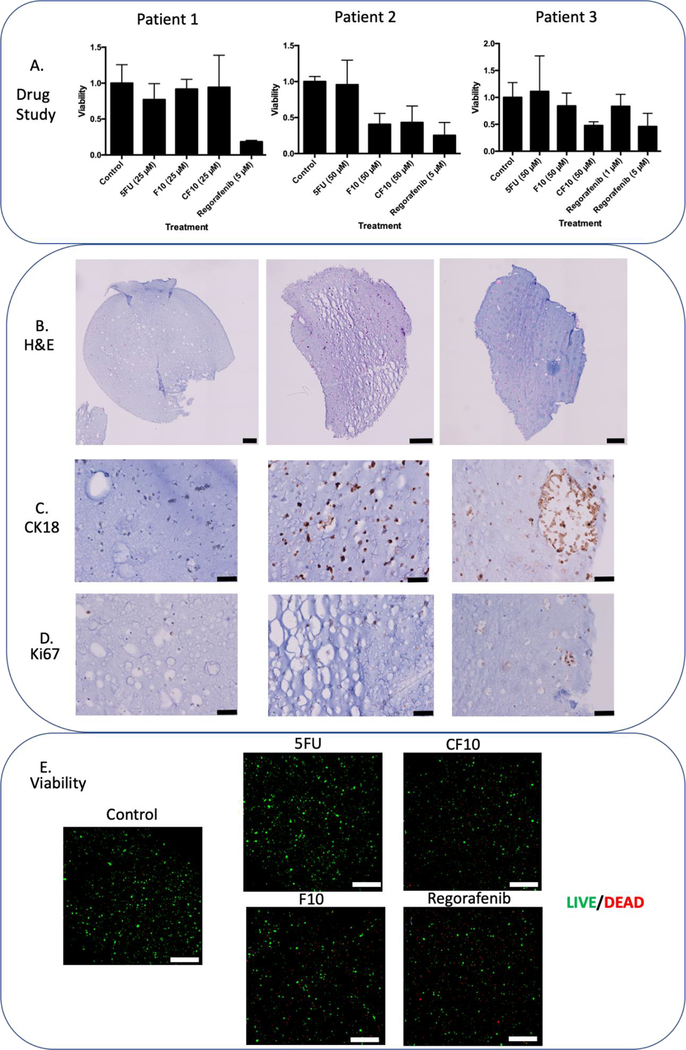
Three patient tumors were acquired according to IRB protocols and dissociated to create μTCs for study. All μTCs were created at 10 μl size while maintaining cellular density to increase potential signal for viability assays. μTCs displayed uniform cell distribution similar to cell line organoids of four and seven days (Figure 5A). Patient-derived μTCs displayed positive Ki67 and CK18 stain, displaying both proliferation and colorectal tumor origin (Figure 5B and 5C). LIVE/DEAD image analysis of patient two displayed high viability (Figure 5E, control).
Figure 5. Panel of Patient Tumor μTCs.
Summary information of three patient tumors that have been processed and placed into organoids for one week, followed histology and immunohistochemistry, specifically B) H&E chromogenic stain, C) CK18 chromogenic stain, and D) Ki67 chromogenic stain. Following three-day exposure drug studies and A) ATP activity quantification and E) LIVE/DEAD imaging for patient two-derived μTCs reveal drug treatment efficacy. Scale bars are equivalent to A) and E) 200 μM; B) and C) 50 μM.
Patient tumors displays drug sensitivity in similar regimens to cell lines
Drug studies were performed on the patient tumor samples to identify potential therapy candidates for the tumor specimens and to compare novel fluoropyrimidine chemotherapeutics F10 and CF10 to 5FU. μTCs were exposed to treatments for 72 hours. Doses were chosen based on composite LD50 doses from the cell line data as follows: 5FU (78.7 μM), CF10 (31 μM) and regorafenib 7.98 μM). A treatment dose was decided from CF10 at 25 μM and at 5 μM for regorafenib, with the intention comparing fluoropyrimidine treatments and regorafenib. F10 and CF10 was developed to incorporate selectively towards FdUMP over FUMP, effectively working to eliminate RNA incorporation causing GI tract toxicity.12 Patient one displayed no significant sensitivity to any of the fluoropyrimidine treatments compared to the control, only regorafenib displayed significant efficacy (5D). After an increase in dose for fluoropyrimidines to 50 μM, patient two displayed significant sensitivity towards F10 and CF10 as compared to both 5FU and the control, with 5 μM regorafenib continuing to display high sensitivity. LIVE/DEAD imaging would further reveal both lower calcein AM cytoplasmic staining in addition to higher ethidium bromide binding in both modified fluoropyrimidines and regorafenib, signifying a lower number of viable cells in these treatments (Figure 5E) A higher yield in cells for patient three allowed for an additional treatment of 1 μM for regorafenib. No significant efficacy was determined from using 1 μM regorafenib, as opposed to a significant effect with 5 μM regorafenib, displaying dose sensitivity. Overall patient μTCs displayed variable sensitivity to several chemotherapeutics utilizing different mechanisms of action.
Discussion
Bioengineered 3D cell line and tissue constructs are an effective model in the study of the human body, in both healthy and diseased state. By using human cells, many of the shortcomings of animal models are averted and by using them in 3D models, cellular interactions are preserved as opposed to 2D models, where genes and RNA expression are skewed. Our studies demonstrated the cells present in these constructs displayed high viability and growth over the course of two weeks, with continued expression of colorectal cancer marker CK18 as well as displaying division marker Ki67 antigen. Due to extensive growth at two weeks, experimental timelines were kept below this time length for all studies. Further studies could continue past two weeks to observe cellular migration and further analyze differences in gene and RNA expression that could analyze the changes from 2D to 3D culture, and even for cells that have left the construct back to 2D.
To choose effective doses for patient derived tumors, we performed drug studies using cell lines in both 2D and our 3D hydrogel culture. Many studies have been performed to determine proper concentrations for cell line viability to determine effective concentrations; however, there is little consensus on dosing. This is due to the myriad differences in experimental design, along with the expected measurable outcomes of experiments ranging from measuring cell death, measuring changes in expression of genes or RNA, and whole organism studies to measure rates of curative and stable disease progression.19, 20, 26 Of note are the differences in determining IC50 dosing in comparison of 2D monolayer and 3D cultures in cell lines. In comparing the classes and functions of chemotherapeutics, treatments utilized to target DNA replication and repair machinery display enhanced efficacy in the 2D cultures compared to the 3D cultures, while small molecule inhibitors displayed higher efficacy in 3D over 2D cultures. Further experimentation may focus on differences in effective treatment between 2D and 3D cultures; these changes may be due to signaling changes or even differences in genomic expression over time. By performing these experiments, we demonstrated the potential for this system to emphasize differences in treatment due to cell culture methods and furthermore provide the system for more advanced analysis into these differences.
Difficulty in culturing patient derived cells has been an issue noted in many scientific journals, with primary focus on difficulty in maintaining self-renewing cancer cell populations25 Loss of stem cell like markers can cause cancer cells to lose their ability to continuously reproduce or cause terminal differentiation in culture, resulting in senescence.11 Although this paper did not investigate the activity of these genes, future studies can be designed to detect their activity. Utilizing immunohistochemistry to identify markers Ki67 and CK18 can preliminarily determine the identity of the cells and their ability to reproduce; combined with H&E staining, this can give a view into cellular health and viability. Further studies may include tracking cellular growth and potential genetic drift over time to confirm maintenance of the cancer cell population.
The enhanced efficacy of modulated fluoropyrimidines F10 and CF10 in comparison to 5FU the patient tumors displays a secondary function of our system, the rapid testing of novel chemotherapeutics and the ability to compare treatment regimens. Observations of variable drug efficacies between patients suggest the patient tumors are of differing genetic origin, thereby creating different optimal treatment regimens. Further drug studies could involve utilizing complete drug regimens, such as FOLFOX and FOLFIRI. With the need for more accurate personalized medicine for cancer treatments, we have demonstrated the utility of a 3D in vitro system, incorporating both cell lines and primary tumor tissue, to demonstrate treatment differences between cell line 2D and 3D culture and to utilize these results to prepare dosing for primary samples for patients.
Supplementary Material
Acknowledgements:
We wish to thank Libby McWilliams (Procurement Manager), Kathleen Cummings (Protocol and Data Manager) and the Wake Forest Advanced Tumor Bank Shared Resource.
AS acknowledges funding through the Wake Forest Clinical and Translational Science Institute Open Pilot Program, supported by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through Grant Award Number UL1TR001420. AS and KV acknowledge funding through the Comprehensive Cancer Center at Wake Forest Baptist Medical Center’s Clinical Research Associate Director Pilot Funds, and services from the Tumor Tissue and Pathology Shared Resource supported by the Comprehensive Cancer Center at Wake Forest Baptist Medical Center’s NCI Cancer Center Support Grant P30CA012197.
Abbreviations:
- 2D
two dimensional
- 3D
three dimensional
- 5-FU
5-fluorouracil
- CK18
cytokeratin 18
- CRC
colorectal cancer
- DMEM
Dulbecco’s Minimum Essential Medium
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- HA
hyaluronic acid
- IHC
immunohistochemistry
- IRB
Institutional Review Board
- Ki67
Antigen Ki-67, a biomarker for proliferation
- μTCs
micro-tumor constructs
- PDMS
polydimethylsiloxane
- PEGDA
polyethylene glycol diacrylate
- RNA
ribonucleic acid
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: Dr. Skardal is an inventor on several patents associated with this work related to the generation of patient-derived tumor models for drug screening and personalized medicine
References:
- 1.Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, Lind GE and Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2: e71, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, Nishimura K, Lynch KD, Mermel CH, Lockerman EL, Kalsy A, Gurski JM Jr., Bahl S, Anderka K, Green LM, Lennon NJ, Huynh TG, Mino-Kenudson M, Getz G, Dias-Santagata D, Iafrate AJ, Engelman JA, Garraway LA and Corcoran RB. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 5: 358–367, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armaghany T, Wilson JD, Chu Q and Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res 5: 19–27, 2012. [PMC free article] [PubMed] [Google Scholar]
- 4.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, Bjornslett M, Meza-Zepeda LA, Eknaes M, Lind GE, Myklebost O, Skotheim RI, Sveen A and Lothe RA. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer 16: 116, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breslin S and O’Driscoll L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget 7: 45745–45756, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin L, Andersen JN and Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med 17: 297–303, 2011. [DOI] [PubMed] [Google Scholar]
- 7.de Gramont A FA, Seymour, Homerin, Hmissi, Cassidy, Boni, Cortes-Funes, Cervantes, Freyer, Papamichael, Le Bail, Louvet, Hendler, de Braud, Wilson, Morvan, Bonetti. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol 18: 2938–2947, 2000. [DOI] [PubMed] [Google Scholar]
- 8.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P and Delrio P. Genetics, diagnosis and management of colorectal cancer (Review). Oncol Rep 34: 1087–1096, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmondson R, Broglie JJ, Adcock AF and Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 12: 207–218, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming M, Ravula S, Tatishchev SF and Wang HL. Colorectal carcinoma: Pathologic aspects. J Gastrointest Oncol 3: 153–173, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert CA and Ross AH. Cancer stem cells: cell culture, markers, and targets for new therapies. J Cell Biochem 108: 1031–1038, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gmeiner WH, Debinski W, Milligan C, Caudell D and Pardee TS. The applications of the novel polymeric fluoropyrimidine F10 in cancer treatment: current evidence. Future Oncol 12: 2009–2020, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greystoke A, Ayub M, Rothwell DG, Morris D, Burt D, Hodgkinson CL, Morrow CJ, Smith N, Aung K, Valle J, Carter L, Blackhall F, Dive C and Brady G. Development of a circulating miRNA assay to monitor tumor burden: From mouse to man. Mol Oncol 10: 282–291, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamburg MA and Collins FS. The path to personalized medicine. N Engl J Med 363: 301–304, 2010. [DOI] [PubMed] [Google Scholar]
- 15.Hu T, Li Z, Gao CY and Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol 22: 6876–6889, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Institute N. S. Cancer Stat Facts: Colon and Rectum Cancer. NIH, 2017. [Google Scholar]
- 17.Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V, Ibbs M, Blizniak R, Luczewski L and Lamperska K. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci 14: 910–919, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson H, Fryknas M, Larsson R and Nygren P. Loss of cancer drug activity in colon cancer HCT-116 cells during spheroid formation in a new 3-D spheroid cell culture system. Exp Cell Res 318: 1577–1585, 2012. [DOI] [PubMed] [Google Scholar]
- 19.Katt ME, Placone AL, Wong AD, Xu ZS and Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol 4: 12, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langhans SA Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol 9: 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA and Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31: 8494–8506, 2010. [DOI] [PubMed] [Google Scholar]
- 22.Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schafer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, Krieg A and Stoecklein NH. Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One 8: e59689, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzocchi AR, Rajan SAP, Votanopoulos KI, Hall AR and Skardal A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci Rep 8: 2886, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzocchi AR, Soker S and Skardal A. Biofabrication Technologies for Developing In Vitro Tumor Models In: Tumor Organoids, edited by Soker S and Skardal A. Berlin, Germany: Springer Nature, 2017, pp. 51–70. [Google Scholar]
- 25.Miserocchi G, Mercatali L, Liverani C, De Vita A, Spadazzi C, Pieri F, Bongiovanni A, Recine F, Amadori D and Ibrahim T. Management and potentialities of primary cancer cultures in preclinical and translational studies. J Transl Med 15: 229, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parasuraman S Toxicological screening. J Pharmacol Pharmacother 2: 74–79, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschlager M, Kenner L, Pfeiffer D, Krupitza G and Dolznig H. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci 130: 203–218, 2017. [DOI] [PubMed] [Google Scholar]
- 28.Sameer AS Colorectal cancer: molecular mutations and polymorphisms. Front Oncol 3: 114, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholzen T and Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol 182: 311–322, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A and Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin 67: 177–193, 2017. [DOI] [PubMed] [Google Scholar]
- 31.Skardal A Biopolymers for In Vitro Tissue Model Biofabrication In: Biopolymers for Medical Applications, edited by Ruso JM and Messina PV. Boca Raton, FL: CRC Press, 2016. [Google Scholar]
- 32.Skardal A, Devarasetty M, Forsythe S, Atala A and Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng 113: 2020–2032, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skardal A, Devarasetty M, Rodman C, Atala A and Soker S. Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann Biomed Eng 43: 2361–2373, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skardal A, Smith L, Bharadwaj S, Atala A, Soker S and Zhang Y. Tissue specific synthetic ECM hydrogels for 3-D in vitro maintenance of hepatocyte function. Biomaterials 33: 4565–4575, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Votanopoulos KI, Mazzocchi A, Sivakumar H, Forsythe S, Aleman J, Levine EA and Skardal A. Appendiceal Cancer Patient-Specific Tumor Organoid Model for Predicting Chemotherapy Efficacy Prior to Initiation of Treatment: A Feasibility Study. Ann Surg Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.