Abstract
Schizophrenia (SCZ) is a neurodevelopmental disorder. Thus, studying pathogenetic mechanisms underlying SCZ requires studying the development of brain cells. Cortical interneurons (cINs) are consistently observed to be abnormal in SCZ postmortem brains. These abnormalities may explain altered gamma oscillation and cognitive function in patients with SCZ. Of note, currently used antipsychotic drugs ameliorate psychosis, but they are not very effective in reversing cognitive deficits. Characterizing mechanisms of SCZ pathogenesis, especially related to cognitive deficits, may lead to improved treatments. We generated homogeneous populations of developing cINs from 15 healthy control (HC) iPSC lines and 15 SCZ iPSC lines. SCZ cINs, but not SCZ glutamatergic neurons, show dysregulated Oxidative Phosphorylation (OxPhos) related gene expression, accompanied by compromised mitochondrial function. The OxPhos deficit in cINs could be reversed by Alpha Lipoic Acid/Acetyl-L-Carnitine (ALA/ALC) but not by other chemicals previously identified as increasing mitochondrial function. The restoration of mitochondrial function by ALA/ALC was accompanied by a reversal of arborization deficits in SCZ cINs. OxPhos abnormality, even in the absence of any circuit environment with other neuronal subtypes, appears to be an intrinsic deficit in SCZ cINs.
Introduction
Schizophrenia (SCZ) is a debilitating neurodevelopmental disorder with pathogenetic roots during development, much before the most prominent symptoms of psychosis appear in adolescence1, 2. Postmortem tissue studies3–5 and brain imaging6–9 provide a wealth of information on abnormalities in patient brains but are not suitable to study the molecular and cellular changes of early development. Peripheral cells have been used as a model for studying schizophrenia10, but it is not clear how informative these cells are, considering that a large portion of the pathogenesis mechanism could be specific to the nervous system. Lack of informative developmental tissues hampers understanding of the pathogenesis mechanism of SCZ. Based on the high heritability of SCZ11, iPSC technology12 can be used to generate disease-relevant developmental tissues and to study molecular and cellular abnormalities occurring during early development13–20, on the condition that well-defined and homogeneous populations of disease-relevant cell types can be consistently generated from iPSCs. Many iPSC-derived neurons used in SCZ studies have been mixed populations of different neuronal subtypes21, often with varying proportions of subtypes from batch to batch. Such heterogeneity can address some issues but could also have resulted in variability of culture composition, leading to missed or false differences, and an inability to in identify abnormalities of specific processes in specific cell types.
Numerous postmortem studies have shown that one of the most consistently affected neuronal types in SCZ patient brains are GABAergic cortical interneurons (cINs), especially parvalbumin (PV)+ or somatostatin (SST)+ expressing cINs22–24, derived from the medial ganglionic eminence (MGE). Functionally, it was suggested that altered GABA neurotransmission in SCZ results in abnormalities of gamma oscillation, thereby causing cognitive impairments in SCZ patients25. Accordingly, pharmacological activation of the GABAA receptor restored gamma band activity in SCZ patients, accompanied by some degree of improved cognitive function26. However, conventional studies using postmortem brains or brain imaging were not able to distinguish whether there are intrinsic abnormalities in cINs that cause cIN hypofunction in SCZ brains, or whether cIN hypofunction is secondary to impaired glutamatergic input onto cINs, as also observed post mortem. Understanding cIN-specific pathogenetic mechanisms will help in the design of therapeutics that better treat/prevent SCZ, especially its cognitive deficits, which are not adequately responsive to current anti-psychotic treatments. Thus, with the aim of modeling cIN-intrinsic SCZ pathogenetic mechanisms without the presence of other neuronal subtypes, we developed an efficient method of generating a homogeneous population of MGE-derived developmental cINs from human pluripotent stem cells (hPSCs), as described in our previous studies27, 28.
Mitochondria are critically involved in various aspects of neuronal functions, including synaptic transmission29–32, Ca2+ signaling33, 34, generation of action potentials35 and ion homeostasis36–38, enabling these energy demanding processes. Thus, it is conceivable that hypofunction of mitochondria could result in suboptimal neuronal function in circuit environments39–41. In line with the notion of OxPhox deficits being causal in SCZ pathogenesis35, mitochondrial abnormalities caused by haploinsufficiency of Mrpl40 (one of the 22q11DS locus genes) resulted in SCZ-relevant behavioral deficits in mice42. Moreover, Cox10 deletion in Parvalbumin+ cINs results in abnormal gamma oscillations in the Prefrontal cortex (PFC) and hippocampus, accompanied by deficits in sensory motor gating and sociability43.
In well-defined homogeneous developmental cIN populations, we observed deficits in the OxPhos pathway as one of the major abnormalities in developing SCZ cINs. Mild but significant decreases in OxPhos gene expression levels were functionally translated to mitochondria hypofunction in developing SCZ cINs. This abnormality is intrinsic to cINs, in the absence of any other neuronal subtypes that could affect their functionality. Hypofunction of mitochondria resulted in oxidative stress in the cells, which was successfully reversed by treating the cells with ALA/ALC44–47, but not by most other chemicals tested. Reversal of mitochondrial hypofunction in SCZ cINs was accompanied by the restoration of other SCZ cIN abnormalities, such as arborization48. Unlike SCZ cINs, the OxPhos deficit was not observed in SCZ glutamatergic neurons. Determining this was only possible through the use of homogeneous populations of iPSC-derived neuronal subtypes. This novel understanding of the intrinsic abnormalities of developing SCZ cINs may help in the development of new therapeutics to better treat/prevent SCZ, and especially cIN-associated symptoms.
Results
Homogeneous populations of developmental cINs were generated from HC vs. SCZ iPSCs
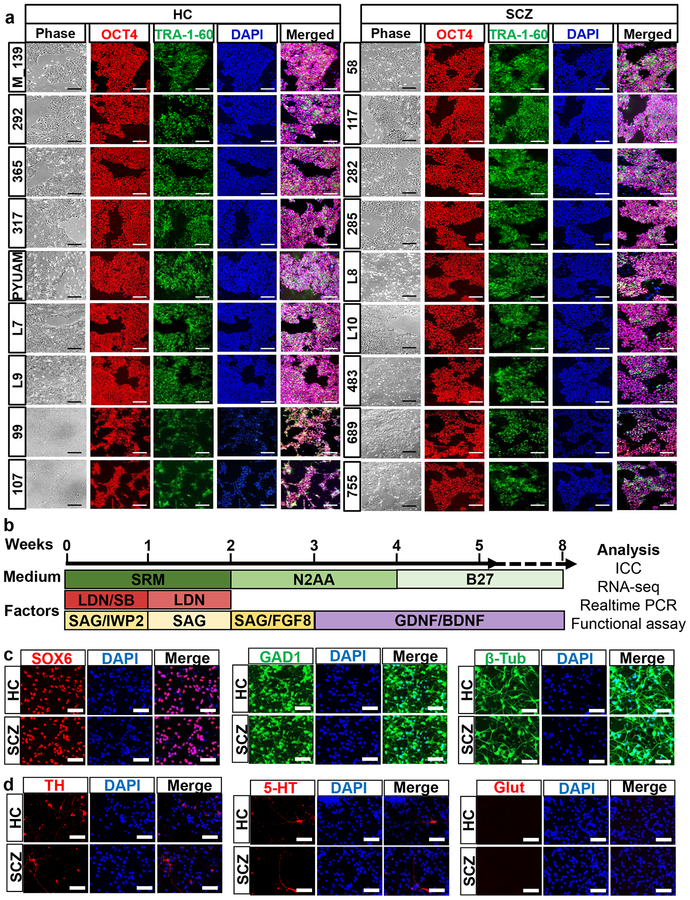
As the first step in studying SCZ pathogenetic mechanisms in disease-relevant developmental tissues, we generated iPSCs from 9 HC vs. 9 SCZ fibroblast lines using modified RNA methods49. It is critical to use an integration-free reprogramming method to study SCZ, since SCZ is a disease with complex genetics where hundreds of genetic loci with small effects culminate in illness. We narrowed down our patient selection criteria to male Caucasian patients to reduce variations caused by ethnicity and gender. In addition, we chose patients who required clozapine treatment to further narrow down the patient group to those with more severe, chronic cases of the disease (Supplementary Figure 1a). All reprogrammed iPSCs showed hPSC-like morphology and expressed hPSC markers (Fig. 1a).
Fig. 1. Generation of homogeneous population of developmental cINs from HC and SCZ iPSCs.
(a) Immunocytochemistry analysis of generated iPSCs for expression of human PSC markers, Oct4 and Tra-1–60. Scale bar=200μm.
(b) Differentiation scheme of cINs from hPSCs. SRM: serum replacement media, LDN: 100 nM LDN193189, SB: 10 μM SB431542, SAG: 0.1 μM Smoothened agonist, and IWP2: 5 μM Inhibitor of Wnt production-2. After 8 weeks’ differentiation, the cells were harvested for experiment.
(c) Immunocytochemistry analysis of generated cINs for expression of Sox6, GAD1 and β-Tubulin, analyzed after 8 weeks’ differentiation. Scale bar=50μm.
(d) Immunocytochemistry analysis for TH, 5-HT and Glutamate, analyzed after 8 weeks’ differentiation. Scale bar=50μm.
We differentiated generated iPSCs into homogeneous populations of developmental cINs, using the protocol we optimized previously27, 28 with a slight modification (Fig. 1b). This protocol efficiently generated a developmental cINs from multiple iPSC lines, as shown by immunocytochemistry where the majority of cells expressed the neuronal marker β-tubulin and the cIN markers SOX6 and GAD1 (Fig. 1c and Supplementary Figure 1b). There was only a minimal number of neurons with alternate neuronal subtypes, such as dopaminergic neurons (TH+), serotonergic neurons (5HT+) and glutamatergic neurons (glutamate+) (Fig. 1d and Supplementary Figure 1c).
Dysregulation of OxPhos genes in SCZ cINs
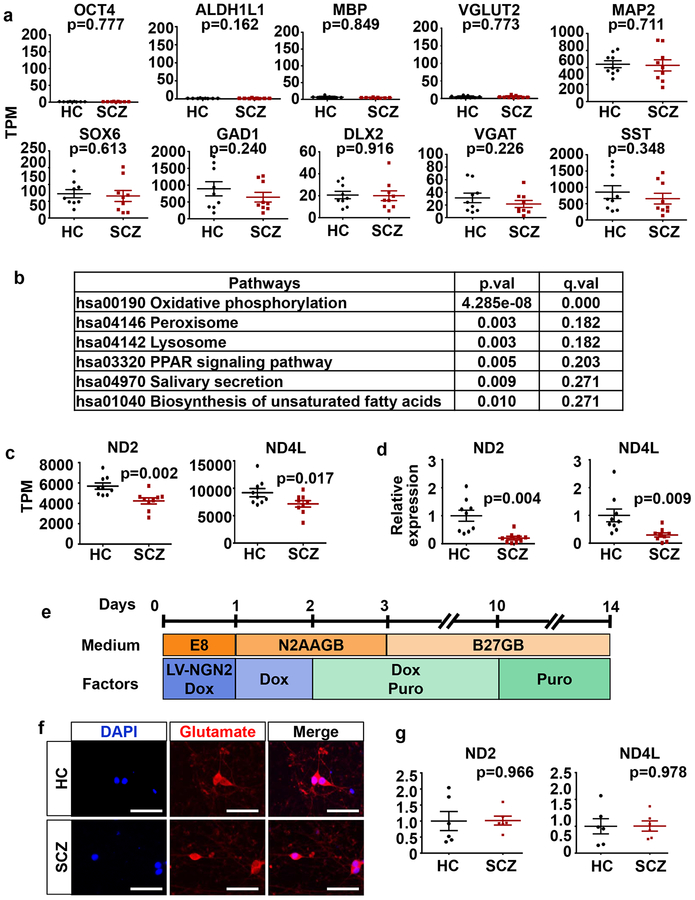
Having observed the induction of homogeneous populations of developmental cINs from multiple iPSCs, irrrespective of disease status, we began transcriptome analysis of these developmental cINs (Supplementary Figure 2a) to understand the intrinsic developmental abnormalities of SCZ cINs in the absence of other neuronal subtypes. In the initial pilot RNAseq we did, we used cINs from 9 HC and 9 SCZ subjects from a single differentiation per line (Supplementary Figure 1a). PCA analysis shows that generated cINs clustered with each other, clearly separated from the fibroblasts and iPSCs and from which they were sequentially derived (Supplementary Figure 2b). Whereas there was little expression of the pluripotent stem cell marker OCT4, astrocyte marker ALDH1L1, oligodendrocyte marker MBP or glutamatergic neuronal marker Vglut2, there was robust expression of the neuronal marker MAP2 and cIN marker genes, such as SOX6, GAD1, DLX2, VGAT and SST, regardless of disease status (Fig. 2a), confirming the efficient generation of developmental cINs, as in the immunocytochemistry result.
Fig. 2. Oxidative phosphorylation pathway is significantly altered in developing SCZ cINs, but not in SCZ glutamatergic neurons.
(a) RNAseq transcriptome profiling of cINs derived from 9 HC iPSCs vs. 9 SCZ iPSCs. Gene expression is shown as transcripts per kilobase million (TPM). Differential expression was analyzed by DESeq (n=9 RNAseq). Error bars are SEM.
(b) Pathway analysis of enriched genes with differential expression using GAGE and Pathview in R and KEGG database. q value: p value with multiple testing correction.
(c) ND2 and ND4L expression in 9 HC vs 9 SCZ cINs, analyzed by RNAseq. Gene expression is shown as transcripts per million (TPM). Differential expression was analyzed by DESeq2 (n=9 RNAseq). Error bars are SEM.
(d) Quantitative real-time PCR analysis of ND2 and ND4L mRNA expression in 9 HC vs 9 SCZ cINs. Data were normalized by GAPDH expression and are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n=9 lines).
(e) The differentiation scheme of glutaminergic neurons. iPSCs were infected with LV-TetO-Ngn2-puro, and Ngn2 expression was induced by doxycyclin for 10 days. Induced glutamatergic neurons were harvested after 14 days’ differentiation for analysis.
(f) Immunocytochemistry analysis of induced glutaminergic neurons with anti-Glutamate antibodies. Scale bar=20 μm.
(g) Quantitative real-time PCR analysis of ND2 and ND4L mRNA expression in HC and SCZ glutaminergic neurons. Data were normalized by GAPDH gene expression and are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n= 6 lines).
Though the generated cINs displayed similar gene expression patterns overall (Fig. 2a and Supplementary Figure 1), Pathview analysis using whole transcriptome data showed that OxPhos was one of the most significantly affected pathways (Fig. 2b. p=4.28e-8, q=7.03e-6). Among various mitochondrial complexes, the changes in mitochondrially encoded NADH dehydrogenases (complex I) were most pronounced (Supplementary Figure 2c). The most significant of these were ND2 and ND4L (Fig. 2c), whose decreased expression was also confirmed by real time PCR analysis (Fig. 2d). Next, we tested whether this abnormality in OxPhos gene expression was also observed in glutamatergic neurons. For this purpose, we differentiated iPSCs into induced postmitotic glutamatergic neurons by directly reprogramming them, using transient forced expression of NGN250, 51. We analyzed their gene expression patterns two weeks after differentiation, when they are fully postmitotic neurons50, 51 (Fig. 2e). Using this protocol, we could efficiently generate glutamatergic neurons from multiple iPSC lines, regardless of disease status (Fig. 2f). There were few other neuronal subtypes, such as dopaminergic (TH+), serotonergic (5HT+) or GABAergic (GAD+) neurons present, demonstrating the homogeneity of the generated glutamatergic neurons (Supplementary Figure 3a). Efficient generation of glutamatergic neurons was also confirmed by RNAseq analysis, which showed negligible expression levels of non-relevant markers (pluripotent stem cell markers and other neural lineage markers) and enriched expression of mature glutamatergic neuronal markers (Supplementary Figure 3b). Unlike SCZ cINs, neither the expression of ND2 nor ND4L was significantly changed in SCZ glutamatergic neurons compared to HC glutamatergic neurons (Fig. 2g), suggesting that homogeneous populations of neuronal subtypes are needed to tease apart cell specific SCZ pathogenetic mechanisms.
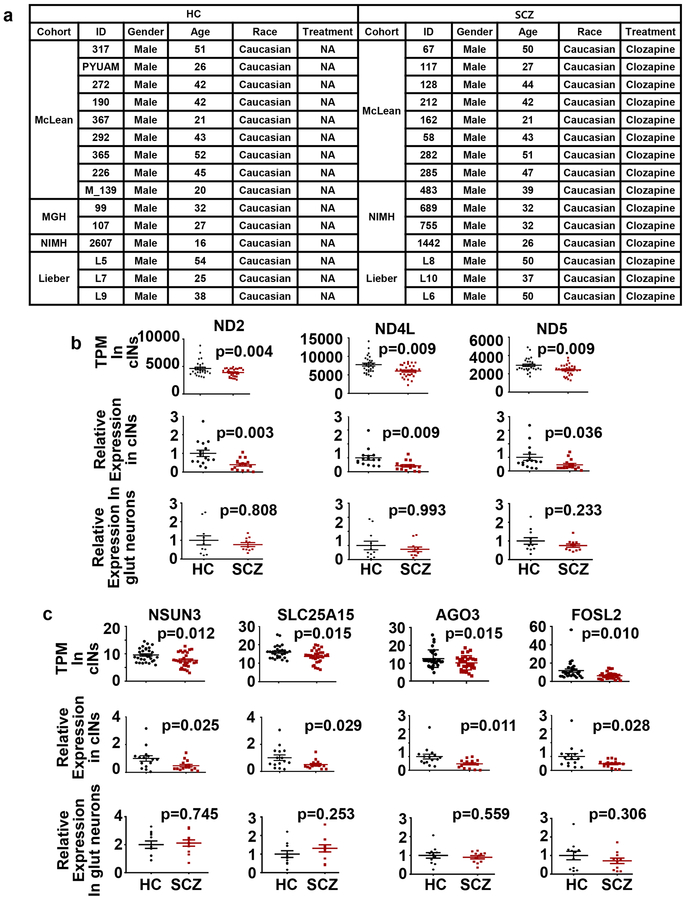
Considering the overall mild phenotypic changes in SCZ cINs, it is critical to test whether the initial results replicate in a larger sample set. Thus, we extended our RNAseq analysis to 15 HC and 15 SCZ lines, with additional independent differentiations (two independent differentiations per line, Fig. 3a). Additional iPSC lines’ phenotype and differentiation efficiency was confirmed in Supplementary Figure 4a–b and in our previous study48. As in the pilot RNAseq, PCA analysis showed that generated cINs clustered with each other, separated from the iPSCs and fibroblasts from which they were derived (Supplementary Figure 4c). The overall downregulation of mitochondrially encoded complex I gene expression was also observed in this expanded cohort (Supplementary Figure 4d and Supplementary Table 1). In addition to ND2 and ND4L, ND5 was significantly downregulated. These results were confirmed by real time PCR analysis (Fig. 3b). However, we did not observe downregulation of OxPhos genes in the larger cohort of SCZ glutamatergic neurons (Fig. 3b). We tested for over-representation of OxPhos genes among SCZ DE genes using Fisher’s Exact test and observed significant enrichment of the OxPhos genes’ among SCZ cIN DE genes (p=1.604e-11), but not in SCZ glutamatergic neurons (p>0.999).
Fig. 3. Multiple OxPhos genes are dysregulated in developing SCZ cINs in expanded cohort.
(a) Demographic information of subjects in the expanded cohort.
(b-c) RNAseq transcriptome profiling and quantitative real-time PCR analysis of cINs derived from 15 HC vs. 15 SCZ iPSCs and glutamatergic neurons derived from 11 HC vs. 11 SCZ iPSCs. For RNAseq, gene expression is shown as transcripts per million (TPM) and differentially expressed genes were analyzed by DESeq2. For real-tim PCR, data were normalized by GAPDH expression and analyzed using two-tailed unpaired t-test (n=15 lines per group, including two independent differentiations per line). Data are presented as mean±SEM.
In addition to OxPhos enzymes, several other OxPhos-related genes were also altered in SCZ cINs. These genes include NSUN3, SLC25A15, AGO3 and FOSL2 (Fig 3c). NSUN3, a mitochondrial tRNA methyltransferase, promotes mitochondrial activity52, 53. SLC25A15, a mitochondrial ornithine transporter, is critical for normal mitochondrial function54. AGO3, a core component of RNA induced silencing complexes (RISCs), regulates mitochondrial function by gene silencing55, 56. FOSL2, a component of the AP-1 transcription factor, participates in mitochondrial regulation of transcription57, 58. Dysregulation of these genes in SCZ cINs was confirmed by real time PCR analysis (Fig. 3c), but were not observed in SCZ glutamatergic neurons (Fig. 3c and Supplementary Figure 3c). Interestingly, some of the OxPhos DE genes were also significantly dysregulated in SCZ postmortem studies (Supplementary Table 2 and Table 3) suggesting that iPSC-derived developing cINs already harbor some of signatures of SCZ neurons in adult brains.
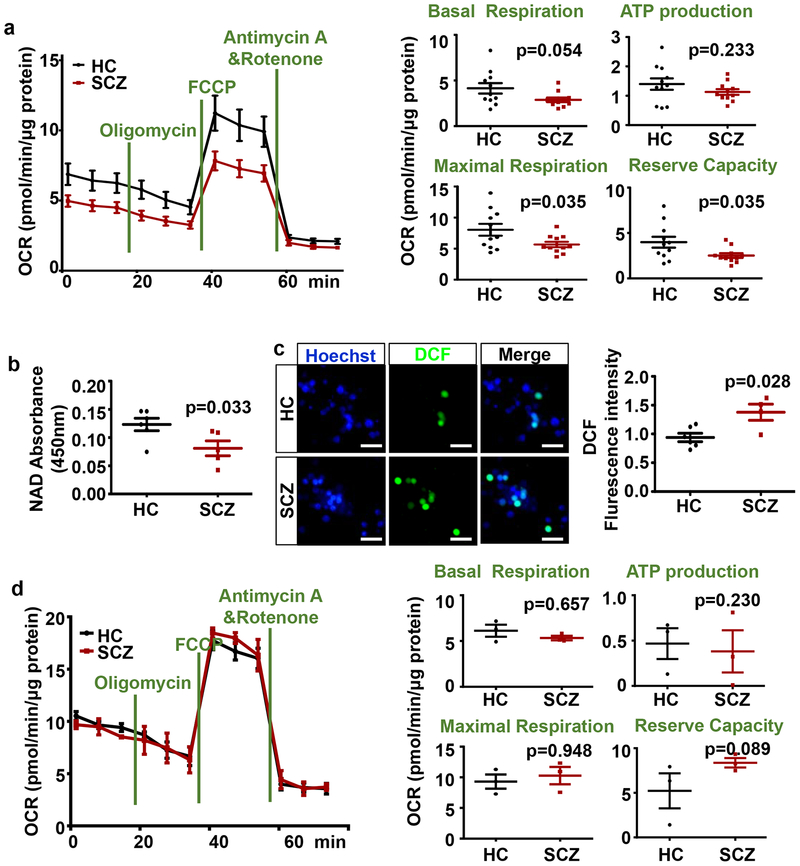
Dysregulations of OxPhos enzymes and regulators in SCZ cINs are associated with mitochondrial functional deficits in cINs
Having observed mild but significant dysregulation of multiple OxPhos genes, we sought to test whether observed mild dysregulation of multiple OxPhos genes did indeed result in a functional deficit in cIN mitochondria. Thus, we characterized the OxPhos functions of HC vs SCZ cINs using a Seahorse Analyzer. SZ derived cINs had significant decreases in maximal respiration (Fig. 4a) and reserve capacity (Fig. 4a), as defined in Materials and Methods section. Accordingly, when we assayed NAD level in HC vs SCZ cINs, a significant decrease of total NAD levels was observed (Fig. 4b), though there is a caveat that reduction in cellular NAD level is not necessarily a direct indicator of mitochondrial NAD level. Mild but significant decreases in mitochondrial function resulted in a significant increase in Oxidative stress in SCZ cINs as assayed by Dichlorofluorescein (DCF) (Fig. 4c), a membrane permeable dye that fluoresces upon oxidization by reactive oxygen species (ROS) in treated cells59. On the other hand, we did not observe any functional deficit of mitochondria in SCZ glutamatergic neurons (Fig. 4d), consistent with the unaffected OxPhos gene expression in these neurons.
Fig 4. Dysregulation of OxPhos genes in SCZ cINs resulted in defects in mitochondrial function.
(a) Analysis of oxidative phosphorylation using Seahorse Analyzer, showing significant decrease in mitochondrial function in SCZ cINs. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n=11 lines per group, with two independent differentiations per line).
(b) NAD assay in HC vs. SCZ cINs. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n=6 lines in HC group and 5 lines in SCZ group, with two independent differentiations per line).
(c) Decreased mitochondrial function in SCZ cINs resulted in increased oxidative stress, assayed by DCF assay. Scale bar=20 μm. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n=6 lines in HC group and 4 lines in SCZ group, with two independent differentiations per line).
(d) Analysis of oxidative phosphorylation using Seahorse Analyzer, showing no significant change in mitochondrial function in SCZ glutamatergic neurons. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (n=3 lines per group).
Reversal of SCZ cIN-specific deficits by chemical treatment
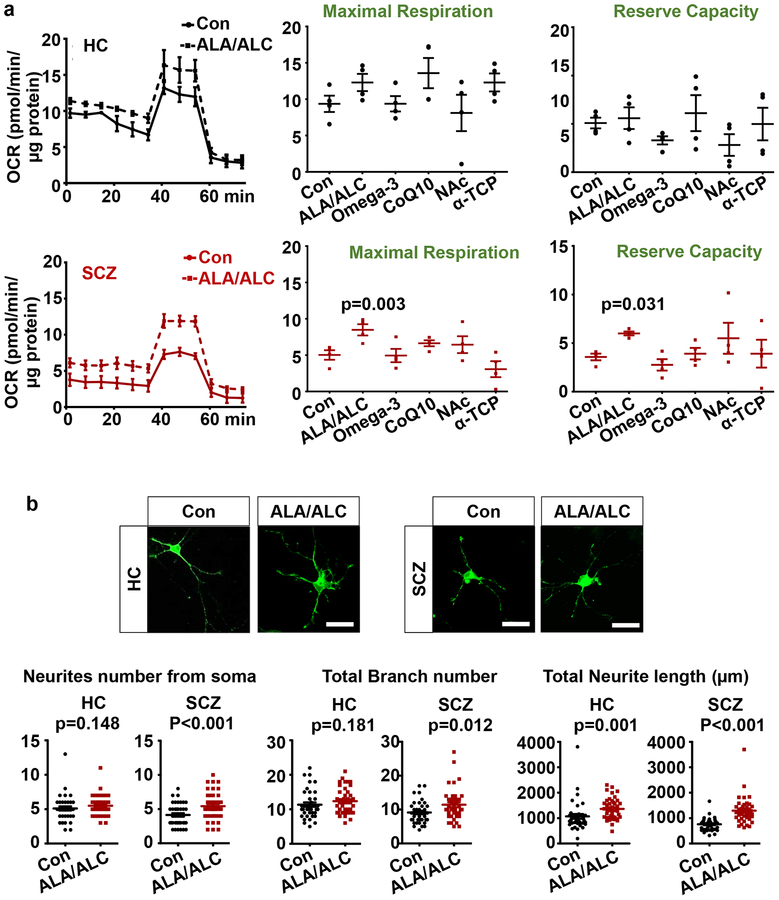
There have been reports of chemicals that boost mitochondrial function, and we tested whether these compounds could reverse the deficit observed in SCZ cINs. We treated cINs with chemicals previously linked to the enhancement of mitochondrial function, including ALA/ALC60–62, Omega-3 fatty acids63–66, CoQ10 (Coenzyme Q10)67, 68, NAc (N-Acetyl cysteine)69, 70, or α-tocopherol71 after 8 weeks’ differentiation and analyzed their mitochondrial function using the Seahorse analyzer after 24 hours’ treatment. No chemical significantly altered OxPhos in HC cINs (Fig. 5a). ALA/ALC significantly increased OxPhos in SCZ cINs, shown by significant increases in maximal respiration and reserve capacity (Fig. 5a), whereas all other chemicals tested did not affect OxPhos in SCZ cINs. Since we previously observed SCZ cIN-specific deficits in arborization48, a process that requires active energy consumption29, 31, we tested whether the reversal of OxPhos deficits by ALA/ALC treatment could help ameliorate arborization deficits. Thus, we treated 8 weeks-old HC vs. SCZ cINs infected with a limiting titer of LV-Ubc-GFP with ALA/ALC for 7 days and analyzed their arborization. Consistent with the previous study, we observed a mild but significant decrease in arborization in SCZ cINs compared to HC cINs (Supplementary Figure 5b). ALA/ALC treatment ameliorated arborization deficits in SCZ cINs, but there was no significant effect in HC cINs (Fig. 5b). This suggests that reversal of OxPhos deficits in SCZ cINs can ameliorate other energy consumption-related SCZ deficits, indicating that restoration of dysregulated mitochondrial function in cINs could be a valuable therapeutic target of SCZ.
Fig 5. Reversal of OxPhos deficits with chemical treatment.
(a) Screening of chemicals that reverse OxPhos deficits in SCZ cINs using Seahorse Analyzer. Top black graphs are analysis in HC lines and bottom red graphs are analysis SCZ lines. Con: no treatment control. ALA/ALC: Alpha Lipoic Acid/Acetyl-L-Carnitine; Omega-3: Omega-3 fatty acids (DHA+EPA+ALA); CoQ10: Coenzyme Q10; NAc: N-acetylcysteine; α-TCP: α-tocopherol. Data are presented as mean±SEM. Paired one-way ANOVA test was used for analysis (n=4 lines per group with two independent differentiations per line), followed by Dunnett post-hoc analysis (supplementary Table 4).
(b) ALA/ALC ameliorate arborization deficits in SCZ cINs. cINs infected with a limiting titer of GFP-expressing lentivirus were treated with ALA/ALC for 7 days and analyzed using ImageJ with the Neuron J plugin. Scale bar=20 μm. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (HC: n= 40 neurons).
Discussion
In this study, by employing homogeneous populations of developing SCZ cINs, we could identify OxPhos dysfunction as a major intrinsic abnormality in these cells during early development. Interestingly, a significant deficit was observed in reserve capacity and maximal respiration without a significant change in basal respiration in SCZ cINs, suggesting that SCZ cINs may not respond to stressful environments as effectively as HC cINs. There have been postmortem studies that reported abnormal mitochondrial function in various regions in SCZ patient brains9, 72–76. However, due to side effects of antipsychotics on mitochondrial function77, it has not been clear whether the OxPhos deficits observed in postmortem SCZ brains were SCZ-innate abnormalities or the side effect of drug treatment. This point was successfully addressed in our study using developmental cINs in the absence of any antipsychotics.
We observed OxPhos deficits were associated with oxidative stress in developing cINs. Interestingly, oxidative stress in SCZ cINs has been consistently observed in post-mortem tissues78–80 which could have been resulted from intrinsic abnormalities or could be resulted from other circuit-based abnormalities such as lower input from glutamatergic neurons. Our results demonstrate that oxidative stress can result from cIN-intrinsic OxPhos abnormalities in the absence of any circuit environment. Furthermore, we observed that arborization deficits in developmental cINs48, like OxPhos deficits, was reversed by ALA/ALC, suggesting that regulation of cIN OxPhos deficits could serve as a new therapeutic intervention to reverse cIN-related SCZ symptoms. Considering that mitochondria function is critical for proper neuronal functions29–38, 81 (including redox regulation81 and proper arborization31, 32), it is conceivable that mitochondrial dysfunction in cINs will result in suboptimal neuronal function. For example, defects in regulating inhibition of local circuitry could cause compromised circuit function, such as abnormal gamma oscillation, in which cINs play a critical role, and which is abnormal in SCZ patients25, 43. Interestingly, a recent study observed restoration of mitochondrial function, accompanied by reversal of SCZ-like behavioral deficits, by transplanting healthy mitochondria in a rat model of SCZ82.
Unlike metabolic disorders where more severe systemic symptoms are observed83, 84, SCZ cINs displayed modest decreases in OxPhos gene expression accompanied by modest reductions in mitochondrial function, which is consistent with the overall modest pathophysiology of SCZ. We observed that expression of many complex I genes was decreased in SCZ cINs, as was that of other mitochondria-related genes, including a mitochondrial tRNA methyltransferase (NSUN3), a mitochondrial carrier (SLC25A15), a RISC component (AGO3), and a transcriptional regulator (FOSL2). Thus, it seems that modest decreases of expression of many genes, each of which might not have impacted overall function of mitochondria much, collectively contributed to the reduction of mitochondrial function, in line with the multigenic nature of SCZ pathogenesis.
We have screened a small number of chemicals reported to have enhanced mitochondrial function in various cellular contexts and observed that an ALC/ALA combination significantly restored OxPhos in SCZ cINs. ALC is present in the brain at high concentrations and enhances mitochondrial function and energy production47. ALA has been used together with ALC to improve mitochondrial function62 and reduce the possible oxidative stress of ALC85. For potential clinical applications, ALA/ALC can easily cross the blood-brain barrier60, 86 and shows low side effects87, thus having the potential to be used as a safe adjuvant therapy for SCZ. All chemicals tested are known to boost mitochondrial function and are potent anti-oxidants, but ALA/ALC has unique mechanisms such as its role as an essential cofactor of mitochondrial enzymes46, 62, 88, 89, chelating potentially toxic metal ions such as Cu2+, Zn2+, Pb2+, Hg2+ and Fe3+ 90, and transporting long chain fatty acid into mitochondria, facilitating fatty acid beta-oxidation91–93. Further studies will reveal which of the mechanisms of action is responsible for the specificity of ALA/ALC in modifying SCZ cIN abnormalities.
Though there were no previous studies that examined OxPhos changes in developing SCZ cINs, there have been studies that examined mitochondrial function of other neural subtypes derived from patient iPSCs. The results of these studies showed varying aspects and degrees of changes depending on the different neural subtypes and differentiation stages94–96. Considering these context-dependent differences in OxPhos abnormalities, it is likely that we would not have been able to determine tissue specific abnormalities without using a homogeneous population of progenies, especially for cIN populations which comprise a minority of generated neurons in the absence of specific induction procedures using developmentally relevant signaling pathways, as used in our current protocol.
Recent large-scale efforts by the second set of Psychiatric Genomics Consortium (PGC2) identified more than 100 SCZ risk loci with genome-wide significance (p<5×10−8)97. Interestingly, among >300 putative SCZ risk genes within these loci, there was significant enrichment of mitochondrial genes (total 22 genes, p<0.05 by hypergeometric test)76, 98, pointing to the potential role of mitochondrial abnormality in SCZ pathogenesis. We did not find any overlap between our OxPhos DE genes and these putative SCZ risk genes from PGC2, but it will be interesting to examine the much larger putative SCZ risk gene list that will be released from PGC3 for this purpose99.
Homogeneous populations of developmental cINs used in this study allowed us to observe cIN intrinsic transcriptome abnormalities in OxPhos gene expression, with corroborating mitochondria hypofunctions from SCZ cINs during early development. These observations were not a result of antipsychotic treatment and were confirmed in the absence of other neuronal subtypes that can affect the functionality of cINs, revealing what is inherent to these SCZ cINs. The OxPhox abnormalities in these cINs could be reversed by one of the chemical treatments reported to enhance mitochondrial function, accompanied by restoration of other downstream abnormalities, specifically, abnormal arborization. The results demonstrate the promise of studying homogeneous disease relevant cell populations to dissect aspects of SCZ pathogenetic mechanisms. Some of these abnormalities, which would not be detected in mixed cell populations, could provide novel potential target to treat or even prevent SCZ.
Materials and methods
Generation of induced pluripotent stem cells
These study protocols were approved by the McLean Hospital/Partners Healthcare Institutional Review Board and New York Medical College Institutional Review Board. All procedures were performed in accordance with the Institutional Review Board’s guidelines and all human samples were obtained with informed consent. Human fibroblasts were obtained from Dr. Bruce Cohen (McLean Hospital), Dr. Daniel Weinberger (Lieber Institute for Brain Development), and Dr. Judith Rapoport (National Institute of Mental Health). Skin biopsies were performed on healthy donors and SCZ patients. The skin tissue cut out one or two 4mm cylindrical pieces of skin. They were performed under local anesthesia (2% Lidocaine with or without epinephrine). The skin samples were washed with PBS 3 times and placed in 35mm culture dishes. One drop of DMEM with Glutamax with 10% fetal bovine serum (FBS), 2% L-Glutamine, 1% Penicillin/Streptomycin solution (PS), and 1% Amphotericin and Gentamicin solution was added on each piece of skin and put in an incubator (37°C/5% CO2). The skin samples in the culture dishes were changed with fresh media every other day to induce fibroblast cell growth as described in100.
Human fibroblasts were reprogrammed using modified RNA methods49 by Cellular Reprogramming, Inc. (San Diego, CA). Each fibroblast line was plated to 6-well plates without feeders at three different plating densities and subjected to messenger RNA reprogramming. Nascent colonies were bulk-passaged from the most productive well to establish passage 1 (P1) iPSC cultures on rLaminin-521 (BioLamina, Sweden) in Nutristem XF media (Biological Industries, Israel) and expanded in the same culture system until at least passage 3 before being characterized by 4’-6-diamidino-2-phenylindole (DAPI)/OCT4/TRA-1–60 immunostaining and frozen down for storage.
Differentiation of iPSCs into cINs
Thawed human iPSCs were maintained on Matrigel (BD, San Jose, CA) coated plates with Essential 8 (E8) media (Invitrogen, Carlsbad, CA). ROCK inhibitor, Y27632 (10uM, ApexBio, Boston, MA, USA) was added to the culture for 24 hours after passaging to prevent single cell-induced cell death of iPSCs.
cINs are generated from MGE progenitors during embryonic development through the action of relevant signaling molecules. Thus, first we induced the MGE phenotype by ventralizing iPSC-derived neuroectoderm by SHH activation and WNT inhibition. More specifically, iPSCs were trypsinized and grown as floating spheres in low adherent flasks in KSR media (DMEM, 15 % knockout serum replacement, 2 mM L-glutamine and 10 μM β-mercaptoethanol (all from Invitrogen) from day 0 to day 14 with the phenotype-inducing chemical combinations as described below. Y27632 (10uM) was added to the culture on the first day of differentiation again to prevent single cell-induced cell death of iPSCs. For neuroectoderm induction, cells were treated with LDN193189 (100 nM, Stemgent, Cambridge, MA) from day 0 to day 14 and SB431542 (10 μM, Tocris Cookson, Ellisville, MO) from day 0 to day 7. For MGE phenotype induction, media was supplemented with IWP2 (5 μM, EMD Millipore, Billerica, MA) from day 0 to day 7 and SAG (0.1 μM, EMD Millipore) from d0 to d21. From day 14, cells were grown in N2AA media (DMEM-F12 with N2-supplement (1:200, Invitrogen) and 200 μM ascorbic acid (AA, Sigma, St. Louis, MO)). FGF8 (100 ng/ml, Peprotech, Rocky Hill, NJ) was added from day 14 to day 21 to induce MGE phenotype at the expense of the CGE phenotype27. iPSCs passaged to low adherent flasks rapidly form spheres and MGE phenotype induction with the above-described chemicals was carried out in these sphere cultures. After three weeks’ induction as spheres, most of the cells take up the MGE phenotype, as we previously described in detail27. Induced MGE progenitors spontaneously differentiate and generate postmitotic cINs, as during their normal development, and thus once the MGE phenotype was established, we withdraw all morphogenic signaling molecules from the culture and then spontaneous differentiation proceeded as described below.
For differentiation into postmitotic cINs, we supplemented N2AA media with 10 ng/ml glial cell derived neurotrophic factor (GDNF, Peprotech) and 10 ng/ml brain derived neurotrophic factor (BDNF, Peprotech) from day 21 to provide trophic support for postmitotic cINs. At day 45 of differentiation, cIN spheres were trypsinized in the presence of 0.1M trehalose (Sigma) and the resuspended cells were filtered through a cell strainer cap (35μm nylon mesh, Corning, NY, USA) to exclude the dead cell cluster, and then plated on polyornithine (PLO; 15 mg/ml; Sigma) and fibronectin (FN; 1 mg/ml; Sigma)-coated plates in B27GB media (DMEM-F12 media with B27 supplement (1:100, Invitrogen), 10 ng/ml GDNF and 10 ng/ml BDNF) supplemented with 10uM Y27632 on the first day of passaging only. Cells were fixed for immunocytochemistry at 6w of differentiation and harvested for RNA preparation at 8w of differentiation. All cell lines are routinely tested for mycoplasma contamination using a Mycoplasma Detection Kit (InvivoGen, San Diego, CA, USA). Cell lines used in this study were verified to be mycoplasma free before undertaking any experiment with them. Cell lines used for each experiment are summarized in Supplementary Table 5.
Differentiation of iPSCs into Glutamatergic neurons
Human iPSC cells were seeded at a density of 106 cells/well on Matrigel-coated 6-well plates with E8 media and 5 μM Rock inhibitor Y-27632 (Selleck Chemicals, Houston, TX), a lentivirus that inducibly expresses Ngn2 and constitutively expresses the Puromycin resistance gene (packaged using pLV_TRET_hNgn2_UBC_Puro plasmid from Addgene, Plasmid #61474 (Addgene, Cambridge, MA)), 2 μg/ml of Doxycycline and 2 μg/ml of polybrene. After overnight incubation (day 1), the media was replaced with N2AAGB media (DMEMF12 with N2 supplement (1:200), 200 μM ascorbic acid, 10 ng/ml GDNF and 10 ng/ml BDNF) with 2 μg/ml of Doxycycline. At day 2, 1 μg/ml of Puromycin was added to the culture. At day 3, the media was changed with B27GB media (DMEMF12 with B27 Supplement (1:100), 10 ng/ml GDNF and 10 ng/ml BDNF) and 2 μg/ml of Doxycycline and changed every other day until day 14. At day 14, the glutaminergic neurons were harvested for RNA preparation using Trizol or fixed with 4% paraformaldehyde (PFA).
Immunocytochemistry, Cell counting and Arborization analysis
Cells were fixed in 4% PFA for 10 min. The fixed cells were blocked in PBS with 10% normal serum and 0.1% Triton X-100 for 10 min at room temperature (RT). Cells were incubated with the primary antibody in PBS with 2% normal serum overnight at 4°C. After washing in PBS, the cells were incubated with the fluorescently-labeled secondary antibody in PBS with 2% normal serum for 1 hour at RT. Cell nuclei were also counterstained with DAPI (Invitrogen, 1:10,000). The antibodies used are summarized in Supplementary Table 6. After washing in PBS, coverslips were mounted onto slide glass using Fluoromount-G (Southern Biotech, Birmingham, AL). Fluorescent images were taken using the EVOS FL Auto microscope (Life Technologies, Carlsbad, CA) or Olympus DSU Spinning Disc Confocal on an IX81 inverted microscope (Olympus, Center Valley, PA).
For cell counting, Image J software (Version 1.51p, NIH, Bethesda, MD) was used to count the cell number using the multi point function. Percentages of cells positive for each marker were quantified in relation to DAPI-stained nuclei from three independent differentiations. At least 500 cells were counted for each line.
For arborization analysis, cells were infected with a limiting concentration of LV-UbiC-GFP virus (MOI=0.001) to induce scarce infections, treated with different chemicals in B27GB media for 7 days and fixed for analysis. GFP+ cells were traced using ImageJ software version 1.51 (NIH) with the NeuronJ plug in to obtain parameters of arborization such as neurite length, neurite number from soma, and branch number.
RNA preparation and Real time PCR
RNA samples were isolated using a TRIzol-reagent (Invitrogen) and 300ng of the total RNA was used for cDNA synthesis using the Oligo(dT)12–18 primer (Gene Link, Hawthorne, NY), 0.5 mM dNTP mix (Thermo Scientific, Waltham, MA), 5 mM DTT (Invitrogen), RevertAid H minus RT (Thermo Scientific), and 5× Reaction Buffer (Thermo Scientific) in SimpliAmp™ Thermal Cycler (Applied Biosystems, Waltham, MA). The real time PCRs were performed using the CFX Connect™ Real-Time PCR Detection System (BioRad, Hercules, CA), with 40 cycles of denaturation (95°C for 15sec), annealing (55°C for 30sec), and extension (72°C for 30sec). The primer sequences used for real time PCR are summarized in Supplementary Table 7.
RNA-seq Analysis
For the RNAseq analysis, RNA quality was examined by the 4200 TapeStation (Agilent Technologies, Santa Clara, CA) and RNA concentration was determined by the Qubit Fluorometric Quantitation (Life Technologies, Carlsbad, CA). For each sample, 100–200ng of RNA was used to construct a cDNA sequencing library using the TruSeq Stranded mRNA Library Preparation Kit (Illumina, San Diego, CA), in accordance with the protocol using the poly-adenylated RNA isolation. Paired-end sequencing (75 bp × 2) was performed in the Illumina NextSeq 550 system. Raw sequence reads were de-multiplexed and trimmed for adapters by using the Illumina bcl2fastq conversion software (v2.19). Sequence reads of each sample were pseudo-aligned to human hg38 reference transcriptome and the gene transcript abundance was quantified by using Kallisto101. Pathway analysis was performed by GAGE and Pathview packages102 using whole transcriptome data. The differential expression of genes and transcripts were achieved in paired groups by using tximport and DESeq2 packages103 in R platform. The RNA-Seq data are available at the GEO website (https://www.ncbi.nlm.nih.gov/geo/) under accession GSE125805 and GSE GSE125999.
Mitochondria-related gene list (Supplementary Table 8) were generated by combining mitochondria-related gene sets from the KEGG OxPhos pathway gene set104, the MitoCarta mitochondrially expressed gene set105, nucleus-mitochondria crosstalk gene sets106–109 as well as genes with high PubMed hit numbers with OxPhos search terms (>10 hits). After removing duplicate genes among these gene sets, a total of 1308 mitochondria-related genes were used as a reference gene set for Fisher’s exact test and for OxPhos annotation of DE genes. For the enrichment analyses of OxPhos genes for cINs and glutamatergic neurons, a one-sided Fisher’s exact test was used to test the over-representation (enrichment) using R statistical software (version 3.5.1, http://www.R-project.org/).
Chemical preparation and storage
All chemicals were obtained from Sigma (St. Louis, MO). α-lipoic acid (ALA) was dissolved in pure ethanol to make a 50mM stock. acetyl-L-carnitine (ALC) was dissolved in phosphate buffer saline (PBS) to make a 10mM stock. cis-5,8,11,14,17-Eicosapentaenoic acid (EPA) and cis-4,7,10,13,16,19-Docosahexaenoic acid (DHA) were dissolved in pure ethanol to make 50mM stocks. Coenzyme Q10 (CoQ10) was dissolved in DMSO to make a 30mM stock. N-Acetyl-L-cysteine (NAC) was dissolved in pure ethanol to make a 100mM stock. α-tocopherol (α-TCP) was dissolved in pure ethanol to make a 50mM stock. All stocks were aliquoted and stored at −20°C to avoid repeated freezing and thawing. All chemicals were diluted by 1:1000 in the culture media and used for cell culture.
OxPhos analysis using Seahorse analyzer
Mitochondrial activity of cINs was measured using the Seahorse XFp8 analyzer (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s instructions. Briefly, 2e5 cells were plated in the XF cell culture miniplate and incubated at 37°C with 5% CO2. One day before the test, the cartridge with XF calibrant was incubated in a non-CO2 incubator overnight to equilibrate. Before the assay, the media was changed to the XF assay medium supplemented with 5 mM sodium pyruvate (Thermo Scientific, Waltham, MA), 10 mM glucose (Thermo Scientific, Waltham, MA), and 2 mM glutamine, and equilibrated in a non-CO2 incubator for 1 hour. Oxygen consumption rate (OCR) were monitored through sequential injections of 1 μM oligomycin, 0.3 μM FCCP and 1 μM rotenone/antimycin A (Seahorse XF Cell Mito Stress Test Kit, Agilent, Santa Clara, CA). The various parameters of the mitochondrial activity were calculated as illustrated in Supplementary Figure 5: Basal Respiration=baseline OCR-Rotenone/antimycin A OCR, ATP production=Baseline OCR-Oligomycin OCR, Maximum Respiration=FCCP OCR-Rotenone/antimycin A OCR and Space Capacity=Maximum Respiration-Basal Respiration. Following analysis, plates were washed with PBS a couple of times to remove dead cells or cell debris before lysing cells. The result was normalized to total protein levels quantified using a BCA protein assay (Thermo Scientific, Cambridge, MA).
Cellular total NAD content assay
Cellular total NAD content of cINs was measured using an NAD/NADH cell-based assay kit (Cayman chemical, Ann Arbor, MI) according to the manufacturer’s instructions. Briefly, cells were plated in a clear 96-well plate and incubated at 37°C with 5% CO2. The cells were washed with assay buffer, followed by the addition of the prepared permeabilization buffer and incubation with gentle shaking for 30min at room temperature. The permeabilization buffer from each well was transferred to a new well in a new 96-well plate, along with prepared standard samples for the standard curve. Reaction buffer was added to each well and the cells were incubated with gentle shaking for 90min at room temperature. The absorbance of each sample was then calculated using a microplate reader at a wavelength of 450nm, and the total NAD content was calculated using the standard curve.
Cellular Oxidative stress assay using DCF
Cells were plated in a clear 96-well plate and incubated at 37°C with 5% CO2. DCFDA Cellular Reactive Oxygen Species Detection Assay Kit was used to test the cellular oxidative stress level. Stock DCF reagent was diluted using PBS (final working concentration is 1 μM) and added to each well. After 30min incubation in 37 °C, the fluorescence signal was read using a microplate reader of 485nm/529nm.
Statistical analysis
All statistical analyses were performed using GraphPad Prism7 (GraphPad Software, La Jolla CA) and SPSS (version 16; SPSS Inc., Chicago, IL). The normal distribution of data was tested using the Shapiro-Wilks test and when significant, log-transformed data were used for comparison or alternatively, a nonparametric Kolmogorov-Smirnov test was used for comparison. We tested equal variance by the F-test or Levine’s test and if there was a significant difference, we used a t-test with Welch’s correction. A two-tailed unpaired t-test was used to compare the means between two groups when the assumptions of normal distribution and equal variance were met. When comparing more than 2 groups, a one-way ANOVA was used. For comparison of OCR after chemical treatment, mixed effect model were used to handle clustering and covariance among correlated samples110, 111. Dunnett’s test was used to compare each of groups with a control group as a post hoc analysis with an adjustment for multiple comparisons. P-values <0.05 were considered to be statistically significant.
No statistical method was used to pre-determine the sample sizes. However, the sample size we used in this study is similar to the largest sample among previous publications. This sample size was adequate to identify the dysfunction of Oxidative Phosphorylation in schizophrenia interneurons. No data were excluded. Experimental cohorts were chosen based on our selection criteria (Caucasian male patients treated with Clozapine vs. age- and gender-matched Caucasian male healthy controls) without randomization to reduce variation caused by age, ethnicity and gender. Blinding was used during cell counting and arborization analysis. For each figure, the statistical test was justified as appropriate, meeting the assumption of the tests as summarized in Supplementary Table 9.
Supplementary Material
Supplementary Figure 1. Generation of homogeneous populations of cINs from HC and SCZ iPSCs. Related to Fig. 1.
(a) Table of subjects analyzed in pilot study. HC refers to healthy control subjects and SCZ refers to schizophrenia patients.
(b) Cell counting analysis of generated cINs for expression of Sox6, GAD1 and β-Tubulin, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by one-way ANOVA (SOX6, p=0.531; GAD1, p=0.123; β-Tubulin, p=0.318).
(c) Cell counting analysis of generated cINs for expression of TH, 5-HT, Glutamate and GAD1, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by two-tailed unpaired t-test (TH, p=0.967; 5-HT, p=0.781; Glutamate, p=1.000; GAD1, p=0.089).
Supplementary Figure 2. RNAseq Analysis of HC vs SCZ cINs. Related to Fig. 2.
(a) Number of reads generated. Rate (%) = Read number of Pseudoaligned/Read number of Input×100.
(b) Principal component analysis of HC and SCZ cINs. RNA-seq data of iPSC and fibroblasts were from Choi et al. (Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nature biotechnology 2015; 33(11): 1173–1181).
(c) Heat map depicting expression changes of mitochondrial Complex I in SCZ cINs. The color in each box corresponds to the value of Log2Fold Changes (Log2FC) for each gene. Blue color indicates down-regulation in SCZ cINs and red color indicates up-regulation in SCZ cINs. The scale bar on the right shows the color of designated Log2Fold Changes values.
Supplementary Figure 3. Generation of homogeneous population of glutamatergic neurons from HC vs. SCZ iPSCs. Related to Fig. 2.
(a) Immunocytochemistry analysis of induced glutaminergic neurons for Brn2, TH, 5-HT and GAD1 expression. Scale bar=100 μm.
(b-c) RNAseq transcriptome profiling of NGN2-induced glutaminergic neurons derived from 5 HC iPSCs vs. 5 SCZ iPSCs. Gene expression is shown as transcripts per million (TPM). Differential expression was analyzed by DESeq2. Error bars are SEM.
Supplementary Figure 4. RNAseq Analysis of HC vs SCZ cINs in a larger cohort. Related to Fig. 3.
(a) Immunocytochemistry analysis of iPSCs for expression of human PSC markers, Oct4 and Tra-1–60. Scale bar=200μm.
(b) Cell counting analysis of generated cINs for expression of Sox6, GAD1 and β-Tubulin, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by one-way ANOVA (SOX6, p=0.714; GAD1, p=0.479; β-Tubulin, p=0.490).
(c) Principal component analysis of HC and SCZ cINs. RNA-seq data of iPSC and fibroblasts were from Choi et al. (Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nature biotechnology 2015; 33(11): 1173–1181).
(d) Heat map depicting expression changes of mitochondrial Complex I in SCZ cINs. The color in each box corresponds to the value of Log2Fold Changes (Log2FC) for each gene. Blue color indicates down-regulation in SCZ cINs and red color indicates up-regulation in SCZ cINs. The scale bar on the right shows the color of designated Log2Fold Changes value.
Supplementary Figure 5. Arborization analysis of HC and SCZ cINs with and without ALA/ALC treatment. Related to Fig. 5.
(a) Tracing of ALA/ALC-treated HC or SCZ cINs (Scale bar=50 μm).
(b) Arborization deficits in SCZ cINs compared with HC cINs. cINs infected with a limiting titer of GFP-expressing lentivirus were analyzed using ImageJ with the Neuron J plugin. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (HC: n= 40 neurons per group).
Acknowledgement
This study was supported by MH107884 (S.C.) and NYSTEM C32607GG (S.C.).
Footnotes
Conflict of interest
We do not have anything to disclose.
Supplementary information is available at MP’s website.
References
- 1.Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry 2012; 17(12): 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry 1987; 44(7): 660–669. [DOI] [PubMed] [Google Scholar]
- 3.Harrison PJ. Postmortem studies in schizophrenia. Dialogues in clinical neuroscience 2000; 2(4): 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffe AE. Postmortem human brain genomics in neuropsychiatric disorders--how far can we go? Current opinion in neurobiology 2016; 36: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Burgos G, Cho RY, Lewis DA. Alterations in cortical network oscillations and parvalbumin neurons in schizophrenia. Biological psychiatry 2015; 77(12): 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arslan A Imaging genetics of schizophrenia in the post-GWAS era. Progress in neuro-psychopharmacology & biological psychiatry 2018; 80(Pt B): 155–165. [DOI] [PubMed] [Google Scholar]
- 7.Kotrla KJ, Weinberger DR. Brain imaging in schizophrenia. Annual review of medicine 1995; 46: 113–122. [DOI] [PubMed] [Google Scholar]
- 8.Kim SY, Cohen BM, Chen X, Lukas SE, Shinn AK, Yuksel AC et al. Redox Dysregulation in Schizophrenia Revealed by in vivo NAD+/NADH Measurement. Schizophrenia bulletin 2017; 43(1): 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du F, Cooper AJ, Thida T, Sehovic S, Lukas SE, Cohen BM et al. In vivo evidence for cerebral bioenergetic abnormalities in schizophrenia measured using 31P magnetization transfer spectroscopy. JAMA psychiatry 2014; 71(1): 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matigian NA, McCurdy RD, Feron F, Perry C, Smith H, Filippich C et al. Fibroblast and lymphoblast gene expression profiles in schizophrenia: are non-neural cells informative? PLoS One 2008; 3(6): e2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM et al. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biological psychiatry 2018; 83(6): 492–498. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131(5): 861–872. [DOI] [PubMed] [Google Scholar]
- 13.Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE et al. Open Chromatin Profiling in hiPSC-Derived Neurons Prioritizes Functional Noncoding Psychiatric Risk Variants and Highlights Neurodevelopmental Loci. Cell stem cell 2017; 21(3): 305–318 e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nature medicine 2018; 24(10): 1579–1589. [DOI] [PubMed] [Google Scholar]
- 15.Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA et al. Genomic DISC1 Disruption in hiPSCs Alters Wnt Signaling and Neural Cell Fate. Cell reports 2015; 12(9): 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windrem MS, Osipovitch M, Liu Z, Bates J, Chandler-Militello D, Zou L et al. Human iPSC Glial Mouse Chimeras Reveal Glial Contributions to Schizophrenia. Cell stem cell 2017; 21(2): 195–208 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes J, Salas F, Mokhtari R, Dolstra H, Pedrosa E, Lachman HM. Modeling the neuropsychiatric manifestations of Lowe syndrome using induced pluripotent stem cells: defective F-actin polymerization and WAVE-1 expression in neuronal cells. Molecular autism 2018; 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell stem cell 2013; 12(4): 487–496. [DOI] [PubMed] [Google Scholar]
- 19.Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S et al. Human Neuropsychiatric Disease Modeling using Conditional Deletion Reveals Synaptic Transmission Defects Caused by Heterozygous Mutations in NRXN1. Cell stem cell 2015; 17(3): 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl Psychiat 2017; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noh H, Shao ZC, Coyle JT, Chung SM. Modeling schizophrenia pathogenesis using patient-derived induced pluripotent stem cells (iPSCs). Bba-Mol Basis Dis 2017; 1863(9): 2382–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benes FM. The GABA system in schizophrenia: cells, molecules and microcircuitry. Schizophrenia research 2015; 167(1–3): 1–3. [DOI] [PubMed] [Google Scholar]
- 23.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci 2005; 6(4): 312–324. [DOI] [PubMed] [Google Scholar]
- 24.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends in neurosciences 2012; 35(1): 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volk DW, Lewis DA. Early developmental disturbances of cortical inhibitory neurons: contribution to cognitive deficits in schizophrenia. Schizophrenia bulletin 2014; 40(5): 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis DA, Cho RY, Carter CS, Eklund K, Forster S, Kelly MA et al. Subunit-selective modulation of GABA type A receptor neurotransmission and cognition in schizophrenia. Am J Psychiatry 2008; 165(12): 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TG, Yao R, Monnell T, Cho JH, Vasudevan A, Koh A et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem cells 2014; 32(7): 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn S, Kim TG, Kim KS, Chung S. Differentiation of human pluripotent stem cells into Medial Ganglionic Eminence vs. Caudal Ganglionic Eminence cells. Methods 2016; 101: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Domenech G, Higgs NF, Vaccaro V, Ros H, Arancibia-Carcamo IL, MacAskill AF et al. Loss of Dendritic Complexity Precedes Neurodegeneration in a Mouse Model with Disrupted Mitochondrial Distribution in Mature Dendrites. Cell reports 2016; 17(2): 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza E, Miranda-Barrientos JA, Vazquez-Roque RA, Morales-Herrera E, Ruelas A, De la Rosa G et al. In vivo mitochondrial inhibition alters corticostriatal synaptic function and the modulatory effects of neurotrophins. Neuroscience 2014; 280: 156–170. [DOI] [PubMed] [Google Scholar]
- 31.Tsuyama T, Tsubouchi A, Usui T, Imamura H, Uemura T. Mitochondrial dysfunction induces dendritic loss via eIF2alpha phosphorylation. The Journal of cell biology 2017; 216(3): 815–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 2004; 119(6): 873–887. [DOI] [PubMed] [Google Scholar]
- 33.Haak LL, Grimaldi M, Smaili SS, Russell JT. Mitochondria regulate Ca2+ wave initiation and inositol trisphosphate signal transduction in oligodendrocyte progenitors. Journal of neurochemistry 2002; 80(3): 405–415. [DOI] [PubMed] [Google Scholar]
- 34.Montalvo GB, Artalejo AR, Gilabert JA. ATP from subplasmalemmal mitochondria controls Ca2+-dependent inactivation of CRAC channels. The Journal of biological chemistry 2006; 281(47): 35616–35623. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Berghe P, Kenyon JL, Smith TK. Mitochondrial Ca2+ uptake regulates the excitability of myenteric neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 2002; 22(16): 6962–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demaurex N, Poburko D, Frieden M. Regulation of plasma membrane calcium fluxes by mitochondria. Biochimica et biophysica acta 2009; 1787(11): 1383–1394. [DOI] [PubMed] [Google Scholar]
- 37.Katsura K, Kristian T, Siesjo BK. Energy metabolism, ion homeostasis, and cell damage in the brain. Biochemical Society transactions 1994; 22(4): 991–996. [DOI] [PubMed] [Google Scholar]
- 38.Hiraoka M. Metabolic pathways for ion homeostasis and persistent Na(+) current. Journal of cardiovascular electrophysiology 2006; 17 Suppl 1: S124–S126. [DOI] [PubMed] [Google Scholar]
- 39.Bergman O, Ben-Shachar D. Mitochondrial Oxidative Phosphorylation System (OXPHOS) Deficits in Schizophrenia: Possible Interactions with Cellular Processes. Canadian journal of psychiatry Revue canadienne de psychiatrie 2016; 61(8): 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace DC. A Mitochondrial Etiology of Neuropsychiatric Disorders. JAMA psychiatry 2017; 74(9): 863–864. [DOI] [PubMed] [Google Scholar]
- 41.Cuperfain AB, Zhang ZL, Kennedy JL, Goncalves VF. The Complex Interaction of Mitochondrial Genetics and Mitochondrial Pathways in Psychiatric Disease. Molecular neuropsychiatry 2018; 4(1): 52–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devaraju P, Yu J, Eddins D, Mellado-Lagarde MM, Earls LR, Westmoreland JJ et al. Haploinsufficiency of the 22q11.2 microdeletion gene Mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Molecular Psychiatry 2016; 22: 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inan M, Zhao M, Manuszak M, Karakaya C, Rajadhyaksha AM, Pickel VM et al. Energy deficit in parvalbumin neurons leads to circuit dysfunction, impaired sensory gating and social disability. Neurobiology of disease 2016; 93: 35–46. [DOI] [PubMed] [Google Scholar]
- 44.Kim H, Kim HJ, Lee K, Kim JM, Kim HS, Kim JR et al. alpha-Lipoic acid attenuates vascular calcification via reversal of mitochondrial function and restoration of Gas6/Axl/Akt survival pathway. Journal of cellular and molecular medicine 2012; 16(2): 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiller S, DeKroon R, Xu LQ, Robinette J, Winnik W, Alzate O et al. alpha-Lipoic acid protects mitochondrial enzymes and attenuates lipopolysaccharide-induced hypothermia in mice. Free Radical Bio Med 2014; 71: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel SP, Sullivan PG, Lyttle TS, Rabchevsky AG. Acetyl-L-carnitine ameliorates mitochondrial dysfunction following contusion spinal cord injury. Journal of neurochemistry 2010; 114(1): 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Zhang C, Zhang X, Wang S, Meng Q, Wu S et al. An acetyl-L-carnitine switch on mitochondrial dysfunction and rescue in the metabolomics study on aluminum oxide nanoparticles. Particle and fibre toxicology 2016; 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao Z, Noh H, Bin Kim W, Ni P, Nguyen C, Cote SE et al. Dysregulated protocadherin-pathway activity as an intrinsic defect in induced pluripotent stem cell-derived cortical interneurons from subjects with schizophrenia. Nature neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L, Ni Y, Wang J, Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Scientific reports 2012; 2: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013; 78(5): 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell reports 2018; 23(8): 2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Haute L, Powell CA, Minczuk M. Dealing with an Unconventional Genetic Code in Mitochondria: The Biogenesis and Pathogenic Defects of the 5-Formylcytosine Modification in Mitochondrial tRNA(Met). Biomolecules 2017; 7(1). [Google Scholar]
- 53.Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C et al. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cellular and molecular life sciences : CMLS 2018; 75(8): 1483–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korman SH, Kanazawa N, Abu-Libdeh B, Gutman A, Tsujino S. Hyperornithinemia, hyperammonemia, and homocitrullinuria syndrome with evidence of mitochondrial dysfunction due to a novel SLC25A15 (ORNT1) gene mutation in a Palestinian family. Journal of the neurological sciences 2004; 218(1–2): 53–58. [DOI] [PubMed] [Google Scholar]
- 55.Geiger J, Dalgaard LT. Interplay of mitochondrial metabolism and microRNAs. Cellular and molecular life sciences : CMLS 2017; 74(4): 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meister G Argonaute proteins: functional insights and emerging roles. Nature reviews Genetics 2013; 14(7): 447–459. [DOI] [PubMed] [Google Scholar]
- 57.Ogita K, Okuda H, Kitano M, Fujinami Y, Ozaki K, Yoneda Y. Localization of activator protein-1 complex with DNA binding activity in mitochondria of murine brain after in vivo treatment with kainate. The Journal of neuroscience : the official journal of the Society for Neuroscience 2002; 22(7): 2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogita K, Fujinami Y, Kitano M, Yoneda Y. Transcription factor activator protein-1 expressed by kainate treatment can bind to the non-coding region of mitochondrial genome in murine hippocampus. Journal of neuroscience research 2003; 73(6): 794–802. [DOI] [PubMed] [Google Scholar]
- 59.Marrocco I, Altieri F, Peluso I. Measurement and Clinical Significance of Biomarkers of Oxidative Stress in Humans. Oxidative medicine and cellular longevity 2017; 2017: 6501046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagen TM, Liu J, Lykkesfeldt J, Wehr CM, Ingersoll RT, Vinarsky V et al. Feeding acetyl-L-carnitine and lipoic acid to old rats significantly improves metabolic function while decreasing oxidative stress. Proceedings of the National Academy of Sciences of the United States of America 2002; 99(4): 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J. The effects and mechanisms of mitochondrial nutrient alpha-lipoic acid on improving age-associated mitochondrial and cognitive dysfunction: an overview. Neurochemical research 2008; 33(1): 194–203. [DOI] [PubMed] [Google Scholar]
- 62.Shay KP, Moreau RF, Smith EJ, Smith AR, Hagen TM. Alpha-lipoic acid as a dietary supplement: molecular mechanisms and therapeutic potential. Biochimica et biophysica acta 2009; 1790(10): 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afshordel S, Hagl S, Werner D, Rohner N, Kogel D, Bazan NG et al. Omega-3 polyunsaturated fatty acids improve mitochondrial dysfunction in brain aging--impact of Bcl-2 and NPD-1 like metabolites. Prostaglandins, leukotrienes, and essential fatty acids 2015; 92: 23–31. [DOI] [PubMed] [Google Scholar]
- 64.Kitajka K, Puskas LG, Zvara A, Hackler L Jr., Barcelo-Coblijn G, Yeo YK et al. The role of n-3 polyunsaturated fatty acids in brain: modulation of rat brain gene expression by dietary n-3 fatty acids. Proceedings of the National Academy of Sciences of the United States of America 2002; 99(5): 2619–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kitajka K, Sinclair AJ, Weisinger RS, Weisinger HS, Mathai M, Jayasooriya AP et al. Effects of dietary omega-3 polyunsaturated fatty acids on brain gene expression. Proceedings of the National Academy of Sciences of the United States of America 2004; 101(30): 10931–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harbeby E, Jouin M, Alessandri JM, Lallemand MS, Linard A, Lavialle M et al. n-3 PUFA status affects expression of genes involved in neuroenergetics differently in the fronto-parietal cortex compared to the CA1 area of the hippocampus: effect of rest and neuronal activation in the rat. Prostaglandins, leukotrienes, and essential fatty acids 2012; 86(6): 211–220. [DOI] [PubMed] [Google Scholar]
- 67.Schniertshauer D, Gebhard D, Bergemann J. Age-Dependent Loss of Mitochondrial Function in Epithelial Tissue Can Be Reversed by Coenzyme Q10. Journal of aging research 2018; 2018: 6354680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chokchaiwong S, Kuo YT, Lin SH, Hsu YC, Hsu SP, Liu YT et al. Coenzyme Q10 serves to couple mitochondrial oxidative phosphorylation and fatty acid beta-oxidation, and attenuates NLRP3 inflammasome activation. Free radical research 2018: 1–11. [DOI] [PubMed] [Google Scholar]
- 69.Martinez M, Ferrandiz ML, De Juan E, Miquel J. Age-related changes in glutathione and lipid peroxide content in mouse synaptic mitochondria: relationship to cytochrome c oxidase decline. Neuroscience letters 1994; 170(1): 121–124. [DOI] [PubMed] [Google Scholar]
- 70.Martinez Banaclocha M N-acetylcysteine elicited increase in complex I activity in synaptic mitochondria from aged mice: implications for treatment of Parkinson’s disease. Brain research 2000; 859(1): 173–175. [DOI] [PubMed] [Google Scholar]
- 71.Navarro A, Bandez MJ, Lopez-Cepero JM, Gomez C, Boveris A. High doses of vitamin E improve mitochondrial dysfunction in rat hippocampus and frontal cortex upon aging. American journal of physiology Regulatory, integrative and comparative physiology 2011; 300(4): R827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maurer I, Zierz S, Moller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophrenia research 2001; 48(1): 125–136. [DOI] [PubMed] [Google Scholar]
- 73.Karry R, Klein E, Ben Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biological psychiatry 2004; 55(7): 676–684. [DOI] [PubMed] [Google Scholar]
- 74.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Human molecular genetics 2005; 14(2): 241–253. [DOI] [PubMed] [Google Scholar]
- 75.Roberts RC. Postmortem studies on mitochondria in schizophrenia. Schizophrenia research 2017; 187: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duncan LE, Holmans PA, Lee PH, O’Dushlaine CT, Kirby AW, Smoller JW et al. Pathway analyses implicate glial cells in schizophrenia. PLoS One 2014; 9(2): e89441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scaini G, Quevedo J, Velligan D, Roberts DL, Raventos H, Walss-Bass C. Second generation antipsychotic-induced mitochondrial alterations: Implications for increased risk of metabolic syndrome in patients with schizophrenia. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology 2018; 28(3): 369–380. [DOI] [PubMed] [Google Scholar]
- 78.Toker L, Mancarci BO, Tripathy S, Pavlidis P. Transcriptomic Evidence for Alterations in Astrocytes and Parvalbumin Interneurons in Subjects With Bipolar Disorder and Schizophrenia. Biological psychiatry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bitanihirwe BK, Woo TU. Oxidative stress in schizophrenia: an integrated approach. Neuroscience and biobehavioral reviews 2011; 35(3): 878–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Do KQ, Cuenod M, Hensch TK. Targeting Oxidative Stress and Aberrant Critical Period Plasticity in the Developmental Trajectory to Schizophrenia. Schizophrenia bulletin 2015; 41(4): 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Handy DE, Loscalzo J. Redox regulation of mitochondrial function. Antioxidants & redox signaling 2012; 16(11): 1323–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robicsek O, Ene HM, Karry R, Ytzhaki O, Asor E, McPhie D et al. Isolated Mitochondria Transfer Improves Neuronal Differentiation of Schizophrenia-Derived Induced Pluripotent Stem Cells and Rescues Deficits in a Rat Model of the Disorder. Schizophrenia bulletin 2018; 44(2): 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moraes CT. Mitochondrial disorders. Current opinion in neurology 1996; 9(5): 369–374. [DOI] [PubMed] [Google Scholar]
- 84.Suomalainen A. Mitochondrial roles in disease: a box full of surprises. EMBO molecular medicine 2015; 7(10): 1245–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tarnopolsky MA. The mitochondrial cocktail: rationale for combined nutraceutical therapy in mitochondrial cytopathies. Advanced drug delivery reviews 2008; 60(13–14): 1561–1567. [DOI] [PubMed] [Google Scholar]
- 86.Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM et al. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proceedings of the National Academy of Sciences of the United States of America 2002; 99(4): 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arnold LE, Amato A, Bozzolo H, Hollway J, Cook A, Ramadan Y et al. Acetyl-L-carnitine (ALC) in attention-deficit/hyperactivity disorder: a multi-site, placebo-controlled pilot trial. Journal of child and adolescent psychopharmacology 2007; 17(6): 791–802. [DOI] [PubMed] [Google Scholar]
- 88.Applegate MA, Humphries KM, Szweda LI. Reversible inhibition of alpha-ketoglutarate dehydrogenase by hydrogen peroxide: glutathionylation and protection of lipoic acid. Biochemistry 2008; 47(1): 473–478. [DOI] [PubMed] [Google Scholar]
- 89.Packer L, Witt EH, Tritschler HJ. alpha-Lipoic acid as a biological antioxidant. Free radical biology & medicine 1995; 19(2): 227–250. [DOI] [PubMed] [Google Scholar]
- 90.Ou P, Tritschler HJ, Wolff SP. Thioctic (lipoic) acid: a therapeutic metal-chelating antioxidant? Biochemical pharmacology 1995; 50(1): 123–126. [DOI] [PubMed] [Google Scholar]
- 91.Rebouche CJ. Carnitine function and requirements during the life cycle. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 1992; 6(15): 3379–3386. [PubMed] [Google Scholar]
- 92.Costell M, Grisolia S. Effect of carnitine feeding on the levels of heart and skeletal muscle carnitine of elderly mice. FEBS letters 1993; 315(1): 43–46. [DOI] [PubMed] [Google Scholar]
- 93.Gulcin I. Antioxidant and antiradical activities of L-carnitine. Life sciences 2006; 78(8): 803–811. [DOI] [PubMed] [Google Scholar]
- 94.Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Molecular psychiatry 2013; 18(10): 1067–1076. [DOI] [PubMed] [Google Scholar]
- 95.Paulsen Bda S, de Moraes Maciel R, Galina A, Souza da Silveira M, dos Santos Souza C, Drummond H et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell transplantation 2012; 21(7): 1547–1559. [DOI] [PubMed] [Google Scholar]
- 96.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular psychiatry 2015; 20(3): 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G et al. Evidence of Mitochondrial Dysfunction within the Complex Genetic Etiology of Schizophrenia. Molecular Neuropsychiatry 2015; 1(4): 201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corvin A, Sullivan PF. What Next in Schizophrenia Genetics for the Psychiatric Genomics Consortium? Schizophrenia bulletin 2016; 42(3): 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bliss LA, Sams MR, Deep-Soboslay A, Ren-Patterson R, Jaffe AE, Chenoweth JG et al. Use of postmortem human dura mater and scalp for deriving human fibroblast cultures. PloS one 2012; 7(9): e45282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 2016; 34(5): 525–527. [DOI] [PubMed] [Google Scholar]
- 102.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013; 29(14): 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014; 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research 2017; 45(D1): D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK et al. Gene map of the extended human MHC. Nature reviews Genetics 2004; 5(12): 889–899. [DOI] [PubMed] [Google Scholar]
- 106.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nature reviews Molecular cell biology 2016; 17(4): 213–226. [DOI] [PubMed] [Google Scholar]
- 107.Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene 2005; 354: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochimica et biophysica acta 2013; 1833(2): 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cagin U, Enriquez JA. The complex crosstalk between mitochondria and the nucleus: What goes in between? The international journal of biochemistry & cell biology 2015; 63: 10–15. [DOI] [PubMed] [Google Scholar]
- 110.Herring AH. Applied Longitudinal Analysis, 2nd Edition, by Garrett M. Fitzmaurice, Nan M. Laird, and James H. Ware, John Wiley & Sons, 2011. Journal of Biopharmaceutical Statistics 2013; 23(4): 940–941. [Google Scholar]
- 111.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics 1982; 38(4): 963–974. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Generation of homogeneous populations of cINs from HC and SCZ iPSCs. Related to Fig. 1.
(a) Table of subjects analyzed in pilot study. HC refers to healthy control subjects and SCZ refers to schizophrenia patients.
(b) Cell counting analysis of generated cINs for expression of Sox6, GAD1 and β-Tubulin, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by one-way ANOVA (SOX6, p=0.531; GAD1, p=0.123; β-Tubulin, p=0.318).
(c) Cell counting analysis of generated cINs for expression of TH, 5-HT, Glutamate and GAD1, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by two-tailed unpaired t-test (TH, p=0.967; 5-HT, p=0.781; Glutamate, p=1.000; GAD1, p=0.089).
Supplementary Figure 2. RNAseq Analysis of HC vs SCZ cINs. Related to Fig. 2.
(a) Number of reads generated. Rate (%) = Read number of Pseudoaligned/Read number of Input×100.
(b) Principal component analysis of HC and SCZ cINs. RNA-seq data of iPSC and fibroblasts were from Choi et al. (Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nature biotechnology 2015; 33(11): 1173–1181).
(c) Heat map depicting expression changes of mitochondrial Complex I in SCZ cINs. The color in each box corresponds to the value of Log2Fold Changes (Log2FC) for each gene. Blue color indicates down-regulation in SCZ cINs and red color indicates up-regulation in SCZ cINs. The scale bar on the right shows the color of designated Log2Fold Changes values.
Supplementary Figure 3. Generation of homogeneous population of glutamatergic neurons from HC vs. SCZ iPSCs. Related to Fig. 2.
(a) Immunocytochemistry analysis of induced glutaminergic neurons for Brn2, TH, 5-HT and GAD1 expression. Scale bar=100 μm.
(b-c) RNAseq transcriptome profiling of NGN2-induced glutaminergic neurons derived from 5 HC iPSCs vs. 5 SCZ iPSCs. Gene expression is shown as transcripts per million (TPM). Differential expression was analyzed by DESeq2. Error bars are SEM.
Supplementary Figure 4. RNAseq Analysis of HC vs SCZ cINs in a larger cohort. Related to Fig. 3.
(a) Immunocytochemistry analysis of iPSCs for expression of human PSC markers, Oct4 and Tra-1–60. Scale bar=200μm.
(b) Cell counting analysis of generated cINs for expression of Sox6, GAD1 and β-Tubulin, analyzed after 8 weeks’ differentiation. Data is presented as mean±SEM from three independent differentiations. There was no significant difference among lines by one-way ANOVA (SOX6, p=0.714; GAD1, p=0.479; β-Tubulin, p=0.490).
(c) Principal component analysis of HC and SCZ cINs. RNA-seq data of iPSC and fibroblasts were from Choi et al. (Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nature biotechnology 2015; 33(11): 1173–1181).
(d) Heat map depicting expression changes of mitochondrial Complex I in SCZ cINs. The color in each box corresponds to the value of Log2Fold Changes (Log2FC) for each gene. Blue color indicates down-regulation in SCZ cINs and red color indicates up-regulation in SCZ cINs. The scale bar on the right shows the color of designated Log2Fold Changes value.
Supplementary Figure 5. Arborization analysis of HC and SCZ cINs with and without ALA/ALC treatment. Related to Fig. 5.
(a) Tracing of ALA/ALC-treated HC or SCZ cINs (Scale bar=50 μm).
(b) Arborization deficits in SCZ cINs compared with HC cINs. cINs infected with a limiting titer of GFP-expressing lentivirus were analyzed using ImageJ with the Neuron J plugin. Data are presented as mean±SEM. Two-tailed unpaired t-test was used for analysis (HC: n= 40 neurons per group).