Abstract
The interaction of the isolated EF-hand domain of phospholipase C δ1 with arachidonic acid (AA) was characterized using circular dichroism (CD) and fluorescence spectroscopy. The far-UV CD spectral changes indicate that AA binds to the EF domain. The near-UV CD spectra suggest that the orientations of aromatic residues in the peptide are affected when AA binds to the protein. The fluorescence of the single intrinsic tryptophan located in EF1 was enhanced by the addition of dodecylmaltoside (DDM) and AA suggesting that this region of the protein is involved in hydrophobic interactions. In the presence of a low concentration of DDM it was found that AA induced a change in fluorescence resonance energy transfer, which is indicative of a conformational change. The lipid induced conformational change may play a role in calcium binding because the isolated EF-hand domain did not bind Ca2+ in the absence of lipids, but Ca2+-dependent changes in the intrinsic tryptophan emission were observed when free fatty acids were present. These studies identify specific EF-hand domains as allosteric regulatory domains that require hydrophobic ligands such as lipids.
Keywords: Phospholipase C, EF-hand domain, Fatty acids, Fluorescence spectroscopy, Allosterism, FRET, Calcium binding, Circular dichroism, Arachidonic acid, Hydrophobic interaction
Phospholipase C plays an important role in calcium signaling in cells. The enzyme cleaves the polar head group from inositol phospholipids to generate second messengers, inositol 1,4,5-trisphosphate (IP3)1 and diacylglycerol (DAG). IP3 is a potent mobilizer of intracellular calcium from the endoplasmic reticulum and DAG is an activator of protein kinase C (PKC). The mammalian phospholipase C family consists of six subfamilies that differ in structure, mode of regulation, and function: beta, delta, gamma, epsilon, zeta, and eta [1–5].
Phospholipase C δ1 (PLC δ1) is widely expressed in various cell types. The enzyme exhibits a multidomain architecture that consists of the PH, EF-hand, catalytic TIM barrel, and C2 domains [6]. The PH domain-truncated enzyme and PH domain of this enzyme were crystallized and their structures were solved separately [6,7]. We have identified PIP2 and PS as allosteric lipid regulators for the PH domain and C2 domain of PLC δ1, respectively [8–10]. Recently, we also reported that free fatty acids (FFA) activate PLC δ1 via allosterism [11]. Our results suggested that the EF-hand domain may be involved in the FFA binding and mediate conformational changes.
EF-hand motifs appear in numerous proteins and are involved in the regulation of these proteins by calcium. The EF-hand motif is arranged in a helix-loop-helix conformation with a Ca2+-binding site in the loop [12]. A number of EF-hand motifs containing proteins that are involved in lipid binding have been reported. Among them there are neuronal calcium sensors such as recovering which is myristoylated [13]. There are also EF-hand containing proteins that are not myristoylated but can still bind to lipids. A hetero-oligomer of the S100 family, S100A8/S100A9 is able to bind free fatty acids [14,15]. Interaction of another S100 protein, S100D with a detergent has been explored using NMR [16]. A multidomain enzyme, diacylglycerol kinase-α contains the EF-hand motifs and is regulated by lipids and calcium [17,18]. However, little is known about the roles of the EF-hand domain of PLC δ1. In this report we characterized the interaction of the isolated EF-hand domain of PLC δ1 with amphiphiles using spectroscopy to elucidate the molecular mechanism of lipid-induced activation of the enzyme.
Experimental procedures
Materials
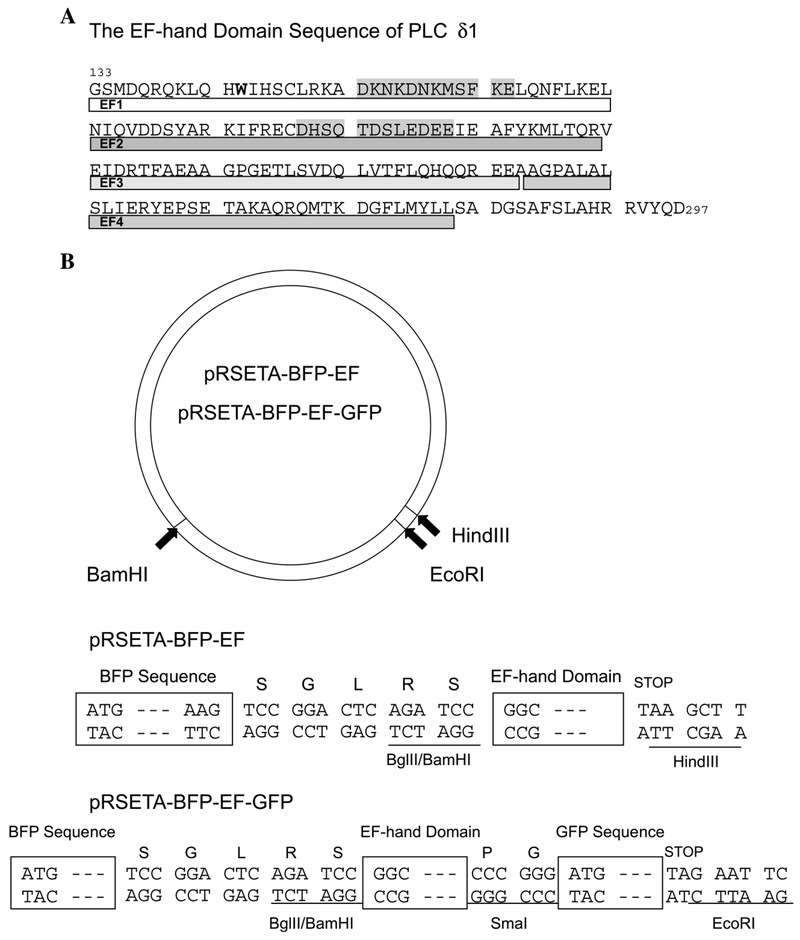
Arachidonic acid (AA) and DDM were obtained from Calbiochem; stearic acid (SA) was purchased from Sigma. The EF-hand sequence of human PLC δ1 (133–297) was inserted into a pGEX-2T vector between BamHI and SmaI cloning sites. The BFP-EF and BFP-EF-GFP constructs were made using the PCR method (Fig. 1). The templates for BFP and GFP, pEBFP-C1 and pEGFP-C1, respectively, were from Clontech. The PCR products were digested with the restriction enzymes indicated in Fig. 1 and inserted into the pRSETA vector.
Fig. 1.
Amino acid sequence of PLC δ1 EF (A) and construction of BFP-EF and BFP-EF-GFP expression plasmids (B). (A) The amino acid sequence (133–297) of PLC δ1 EF-hand domain is shown (PLC δ1 EF). Two putative Ca2+-binding sites are shaded in gray. The single intrinsic Trp reside is in bold-face. (B) BFP and GFP coding sequences were amplified by the PCR method using pEBFP-C1 and pEGFP-C1 as template. The BFP-GFP construct linked with the multiple cloning site was subcloned into pRSETA expression vector between BamHI and EcoRI. To construct BFP-EF the coding sequence of the EF-hand domain (133–297) preceded by BamHI site was inserted between BglII and HindIII in the BFP-GFP construct. For BFP-EF-GFP, the coding sequence of the EF-hand domain (133–296) sandwiched between BamHI and SmaI sites was inserted into BglII and SmaI sites in the BFP-GFP construct. The EF sequence for this construct was shortened to avoid repetitive Gs in the primer.
Expression and purification of proteins
The EF-hand fragment was expressed and purified according to the method previously described [11] with some modifications. Briefly, the fragment fused to GST was expressed in Escherichia coli. The lysate resulting from the cells was applied to a glutathione Sepharose 4B (Amersham-Pharmacia Biotech) column. The resin was washed several times with PBS containing 1% Triton X-100 (Sigma–Aldrich), then with 20 mM Tris-HCl, pH 8, containing 175 mM NaCl. The fusion protein was cleaved with thrombin (Roche) followed by several washes with the same buffer. For higher purity protein, an additional step using a Q-Sepharose (Amersham-Pharmacia Biotech) column was employed. The eluent from the first step, the thrombin-cleaved fragment, was applied to the Q-Sepharose column equilibrated with 30 mM Tris-HCl, pH 8.7. The flow-though fraction contained the desired protein, while impurities were retained on the resin.
For production of the EF fluorescent fusion proteins, BL21(DE3) E. coli was transformed with the pRSETA-BFP-EF or pRSETA-BFP-EF-GFP constructs. Fusion protein production was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside and incubation of the culture at 20 °C overnight. The cells were collected and frozen at −80 °C. The cell pellet was resuspended with 50 mM sodium phosphate, pH 8.0, 0.1 M NaCl, 0.1% Tween 20 plus protease inhibitor cocktail. The suspension was sonicated and centrifuged at 13,000g at 4 °C for 45 min. The resulting supernatant was applied to an appropriate amount of Ni–NTA resin (Qiagen). The resin was washed several times with the suspension buffer and then several times more with 50 mM sodium phosphate, pH 8.0, containing 0.1 M NaCl and 15 mM imidazole. The protein was eluted with 50 mM sodium phosphate, pH 7.5, containing 0.1 M NaCl and 100 mM imidazole. The protein solutions were either dialyzed or applied to a PD10 (desalting column from Amersham-Pharmacia Biotech) column to exchange buffers for the different experiments.
CD spectroscopy
The folding of the EF fragment was assessed by CD spectroscopy using an AVIV 202 spectrophotometer. For far-UV CD spectra, the protein, 0.4 mg/ml in 10 mM sodium phosphate and 150 mM NaCl, pH 7.5, was placed in a 0.1 cm cell. The ellipticity was measured from 190 to 260 nm at 25 °C and converted, after subtraction of the buffer spectrum as the blank, to residue molar ellipticity using the molecular weight of the peptide (19,107 Da).
The breakdown into other secondary structure components was estimated using CDNN [19,20]. For near-UV CD spectra, protein solutions of 11 or 6 mg/ml were used; 10 scans were averaged and then smoothed to yield spectra in the absence and presence of AA.
Steady-state fluorescence
Protein solutions were in 0.1 M NaCl, 0.1 mM EGTA, 50 mM Hepes, pH 7.3. Stock solutions of fatty acids in ethanol were diluted to 20 mM for the titration experiments. The indicated amounts of dodecylmaltoside and fatty acids were added to the protein solution. A Model 2000-4 spectrofluorometer equipped with two model 814 PMT photon-counting detectors from Photon Technology International (PTI) was used for detection of fluorescence from the single tryptophan in the EF-hand fragment. The fluorescence spectrum was measured (from 310 to 450 nm) with excitation at 295 nm, 4 nm excitation and emission slit widths, and a constant temperature of 25 °C.
A calcium indicator, fluo-3 (Molecular Probes) was used to detect Ca2+ binding to the EF-hand fragment. The buffer was the same as in the experiments above except no EGTA was used. The indicator, 4.16 μM, was titrated with increasing concentrations of Ca2+ to determine the dissociation constant of this indicator under the conditions used here. The protein was dialyzed against the same buffer with Chelex resin (Bio-Rad) to remove any divalent cations. The same amount of indicator was added to 10.3 μM protein and the resulting solution was titrated with Ca2+ (0.0–51.4 μM). At the endpoint of the titration (51.4 μM Ca2+), EGTA was added to 200 μM, and the fluorescence intensity measured, F0, represented the dye in the absence of bound Ca2+. The dye was excited with 506 nm, and emission was monitored at 520 nm (1 nm excitation and emission slit widths). The temperature in the sample holder was kept constant at 25 °C. The maximum reading at 51.4 μM Ca2+, Fmax, was used to measure the change in fluorescence induced by saturating the dye with Ca2+. At each intermediate Ca2+ concentration, the fractional saturation of the dye by Ca2+ (α) was obtained from the equation, α = (F − F0)/(Fmax − F0). The dissociation constant of the indicator for Ca2+ was calculated using Prism (Graphpad). Eq. (2) was used to estimate both Kd1 [Ca]cont and defined below. The expected changes in the presence of a Ca2+-binding protein were simulated according to the model described below. In brief, at each Ca2+ concentration the value of β was varied so that the total Ca2+ concentration derived from Eq. (3) was equal to the value estimated by Eq. (1). This task was performed with Excel (Microsoft).
Fluorescence anisotropy was measured using a PC1 photon-counting fluorometer (ISS) with excitation at 492 nm, emission at 520 nm, and 8 nm excitation and emission slit widths. The buffer condition used was the same as in the tryptophan fluorescence experiment described above and the protein was added to 0.43 μM. Prior to the measurement the EF-hand fragment protein had been labeled as follows. An excess amount of a thiol-reactive 5-iodoacetamidofluorescein (Molecular Probe) dissolved in dimethyl sulfoxide was added to the protein solution and the mixture was incubated at room temperature. Then the unbound fluorescent dyes were removed by a PD-10 column equilibrated with 0.1 M NaCl, 0.1 mM EGTA, 50 mM Hepes, pH 7.3. The labeling efficiency was calculated by independently measuring protein concentration (Bradford assay) and absorption at 492 nm.
(A) Interaction between the Ca2+ indicator and Ca2+ C + Ca ↔ C-Ca C : Ca2+ indicator, Ca : Ca2+ ion
The buffer contains contaminating Ca2+([Ca]cont). Since the total Ca2+ concentration ([Ca]total) is the sum of the concentration of added Ca2+ ([Ca]added) and [Ca]cont, it is written as
| (1) |
To use total concentrations instead of free concentrations, the following equation was derived.
| (2) |
where
(B) Dye/Ca2+ interaction in the presence of a Ca2+-binding protein
When one Ca2+-binding site is assumed for a protein P, the dissociation constant Kd2 is written as
As in the above case, a fraction of the complex formation between P and Ca, β, is introduced to account for ion that is bound to the protein. The total Ca2+ concentration can be determined by
| (3) |
Time-resolved fluorescence measurements
Fluorescence lifetimes were measured using frequency-domain fluorometer equipped with a Hamamatsu 6 μm micro-channel plate detector (MCP-PMT) as previously described [21]. The system covers a wide frequency range allowing precise detection of lifetimes ranging from few picosecond to tens of nanoseconds. For excitation we used harmonic content of pulse train with a repetition rate of 3.75 MHz and a pulse with of 5 ps from synchronously pumped and cavity dumped DCS dye laser. The dye-laser was pumped with a mode-locked argon ion laser (Coherent, Innova 100) [21]. The dye laser output was frequency doubled to 355 nm for BFP excitation. For intensity decay measurements, magic angle conditions were used. The samples were placed in a 2 mm path cuvette. The emission was observed using 420 nm cut-off filter together with 460 nm interference filter.
The frequency-domain intensity decay data were fitted to the discreet sum of exponential components:
where αi is the pre-exponential factor (Σαi = 1.0) and τi the corresponding fluorescence lifetime.
Non-denaturing electrophoresis
Proteins were added to 400 μM DDM or 400 μM DDM plus 200 μM AA suspended in 40 mM sodium phosphate, 80 mM imidazole, pH 7.5, containing 80 mM NaCl. Agarose, 1% w/v, was dissolved in 50 mM MOPS, pH 7.0, and used to form the gel. The MOPS buffer was also used as the running buffer. Electrophoresis was performed at room temperature for 45 min at 110 V. The gel was visualized with UV light so that the emission from the fluorescent proteins was observed.
PLC δ1 activity assay
Both the wild-type and PLC δ1 (Δ1–210) deletion mutant were expressed using BL21(DE3) E. coli. The proteins were purified according to the method described previously [11]. The indicated amount of PI containing [3H]PI and fatty acid was dried under argon. The lipids were solubilized in a solution of 0.1 M NaCl, 50 mM Hepes, pH 7.3, and DDM. The mixtures were vortexed and sonicated briefly. Then acetylated BSA was added to 50 μg/ml and EGTA was added to 1 mM. After adding appropriate amount of the enzyme, reactions were initiated by adding CaCl2. Activity assays were performed at 30 °C. Reactions were stopped by adding chloroform/methanol/HCl (100:100:0.6), followed by 1 N HCl containing 5 mM EDTA. The aqueous and organic phases were separated by centrifugation. The aqueous phases were counted by liquid scintillation.
Results and discussion
Isolated EF-hand domain of PLC δ1 has secondary structure
PLC δ1 has four EF-hands that consist of four loop-helix-loop-helix motifs arrayed in two lobes. The overall structure of this region in PLC δ1 closely resembles that of calmodulin [6]. In the crystal structure of a truncated PLC δ1 [6], the three resolved EF-hands interact closely with the C2 domain. To show that the isolated EF-hand domain (PLC δ1 EF) adopts a similar structure when expressed independent of the rest of the protein, we examined secondary structure via circular dichroism (CD). As shown in Fig. 2, the protein is well-folded and predominantly helical as monitored by the magnitude of the residue molar ellipticity and double minima at 208 and 222 nm. Analysis of the entire spectra estimated 72.2% α-helix, very little β-sheet (5.3%), with a small amount of both β-turn (11.4%) and random coil (13.1%). This high helical content is consistent with the structure of the EF-hand observed in the crystal structure of the enzyme [6].
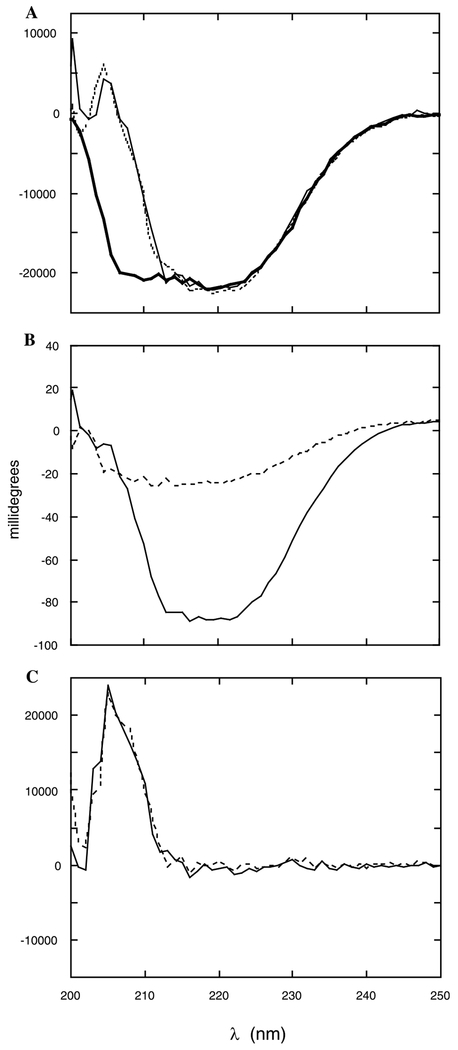
Fig. 2.
Far-UV CD spectra of PLC δ1 EF. (A) Far-UV spectra for 0.4 mg/ml PLC δ1 EF in the absence (thick solid line) and presence of 50 μM AA/100 μM DDM prior to subtraction of ligand alone spectrum (dotted line), and after the AA/DDM contribution has been subtracted (solid line). (B) Ellipticity, in millidegrees, for PLC δ1 EF in the presence of AA/DDM and the contribution to the spectrum for the same concentration of free AA/DDM: (dotted line) AA/DDM alone; (solid line) AA/DDM + PLC δ1 EF. (C) Difference spectra for the corrected PLC δ1 EF + AA spectrum minus PLC δ1 EF alone: (dotted line) AA added from an ethanol stock solution; (solid line) AA/DDM. Except for (B), the ordinate is expressed as residue molar ellipticity [θ] (deg Σ cm2/dmol).
The AA added to the EF peptide contributed positive ellipticity to the far-UV CD spectrum below 230 nm. Fig. 2B compares the spectrum for EF with 50 μM AA/100 μM DDM to the ‘blank’ of 50 μM AA/100 μM DDM, careful subtraction of an AA/DDM blank from the EF plus ligand spectrum generated far-UV CD spectra that showed distinct changes. (Fig. 2A, compare the bold line for EF alone with the dotted and dashed lines for EF plus AA). While the ellipticity at 222 nm was comparable to that in the absence of AA, there was a significant loss of negative ellipticity below 210 nm. Difference spectra emphasizing this change are shown in Fig. 2C. If one assumes that this represents changes in protein secondary structure, CDNN estimates the α-helix to have decreased (to 53.3%) with increased β-sheet (now 9.5%), β-turn (13.5%), and random coil (22.5%). Such large changes in secondary structure are unlikely for an EF-hand type motif. Contributors may include a large induced CD spectrum in this region observed when AA bound to the EF-hand. Regardless of the detailed interpretation, the far-UV CD spectral changes indicate that AA binds to the EF domain (Table 1).
Table 1.
Change in secondary structure (estimated by CDNN) upon AA binding to the EF peptide
| Sample | % α-Helix | % β-Sheet | % β-Turn | % Random |
|---|---|---|---|---|
| EF | 72.5 | 5.3 | 11.4 | 13.1 |
| +50 μM AA | 54.5 | 9.2 | 13.2 | 21.8 |
| +AA/DDM (50 μM/100 μM) | 53.3 | 9.5 | 13.5 | 22.5 |
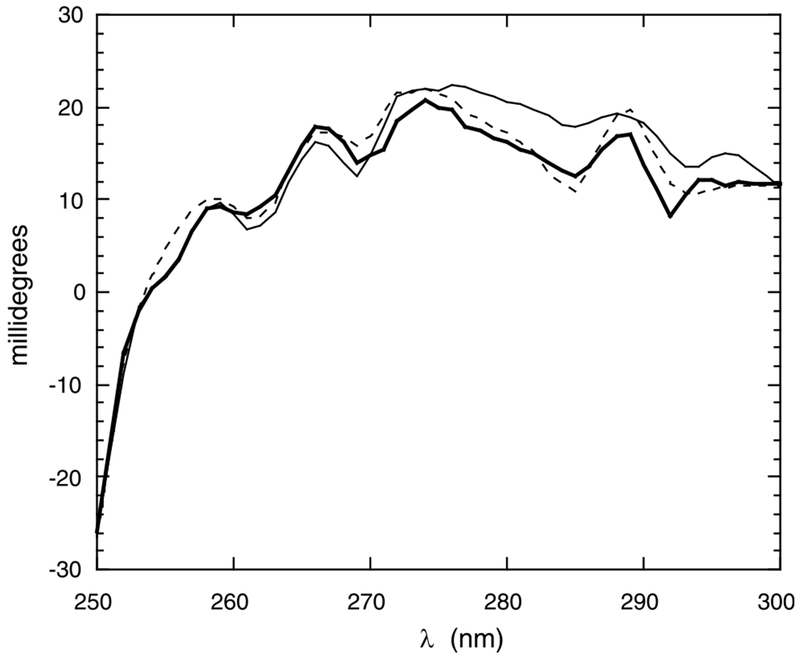
The near-UV region was also examined to see if AA binding effected changes in the tertiary structure as sensed by aromatic residues. Fig. 3 compares spectra for EF alone and with 50 and 200 μM AA/DMM (1:2). Ellipticity in millidegrees for the concentrated sample, rather than a molar ellipticity is shown. The changes are relatively small, with increased positive ellipticity in the 280–290 nm region. Since there are five Tyr and a Trp residue in the EF protein, it is not clear exactly which residues experience environmental changes that alter the ellipticity. While the exact interpretation of this change is unclear, the changes indicate that the orientations of aromatic residues in the peptide, structured in the EF domain by itself, are affected when AA binds to the protein.
Fig. 3.
Near-UV CD spectra of PLC δ1 EF. Near-UV CD spectra for 6 mg/ml PLC δ1 EF in the absence (thick solid line) and presence of 50 μM (dotted line) and 200 μM (solid line) AA (with 1:2 AA/DDM).
Intrinsic tryptophan emission monitors amphiphile binding
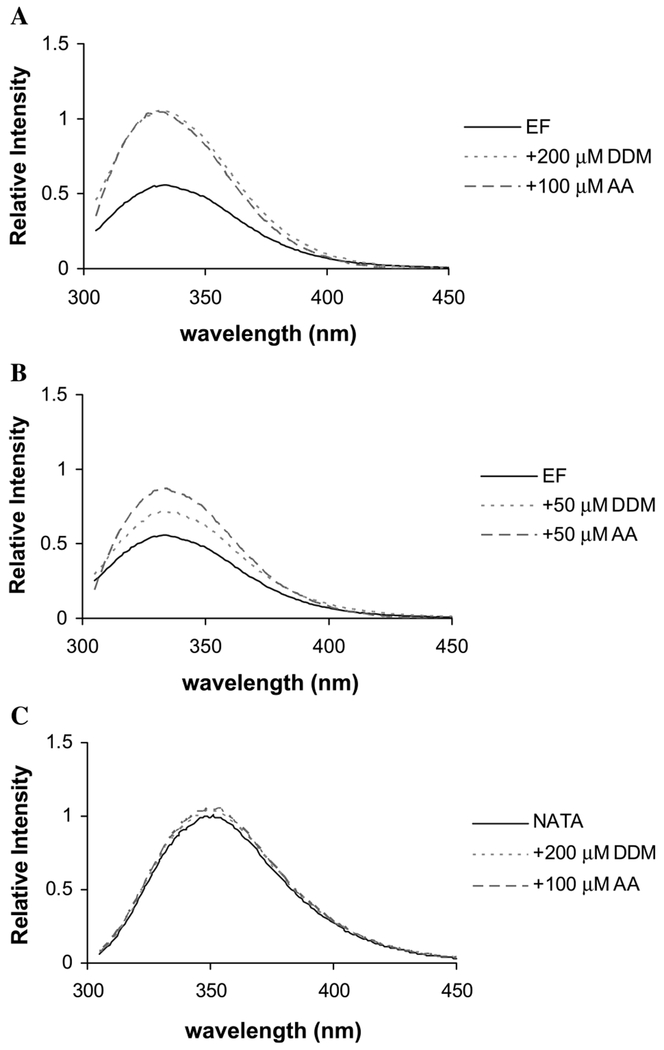
Since there is only one tryptophan residue in the PLC δ1 EF, we used this residue as a probe of lipid/amphiphile binding. Tryptophan fluorescence is widely used for detection of changes in the local environment of the fluorophore upon ligand binding. Tryptophan emission spectra were compared with and without DDM or AA. As seen in Fig. 4C, addition of the detergent or further addition of AA to a tryptophan analog, NATA, had no effect on its emission spectrum indicating that NATA does not partition into the lipids. On the other hand, addition of the 200 μM detergent (above the DDM CMC of 176 μM [22]) to PLC δ1 EF caused a roughly twofold increase in intensity suggesting that the Trp may be part of the binding site or that the Trp fluorescence is monitoring a conformational change that occurs when the detergent binds to the protein. Next, we examined the effect of AA on the tryptophan fluorescence intensity. Since it is difficult to add AA at high concentrations without detergent present, we compared spectra taken first with 200 or 50 μM DDM, then with both DDM and AA present (Fig. 4). The addition of 50 μM DDM caused a 25% increase in fluorescence intensity; 50 μM AA plus 50 μM DDM increased the fluorescence 70% over the protein alone. The presence of 100 μM AA along with 200 μM DDM produced no additional increase in EF-hand fluorescence over what was observed with the DDM alone. This suggests that detergent can interact with PLC δ1 EF and compete for AA binding. At the lower DDM concentration, the detergent should be mostly monomeric, although the protein interaction could lower its CMC. In the AA/DDM mixture, a mixed micelle interface must be available because the AA is solubilized (at least initially). Thus, the enhanced fluorescence in this sample could reflect interactions of the protein with the DDM interface (a non-specific interfacial binding) as well as specific binding of AA to the protein.
Fig. 4.
Intrinsic tryptophan emission spectra of PLC δ1 EF. (A) The protein concentration was 4.1 μM (solid line). DDM was added to 200 μM (dotted line) and then AA was added to 100 μM (dashed line). (B) DDM was added to 50 μM (dotted line) and then AA was added to 50 μM (dashed line). (C) NATA (1 μM in the same buffer) was used as control. The same experiment as in (A) was performed. All the intensities are expressed relative to the maximum of the NATA emission in the absence of DDM or AA.
Many enzymes are regulated by interactions with interfaces, and while AA may have a higher affinity or be more potent than DDM, DDM appears to be able to interact with the EF-hand domain of PLC δ1. The results of these studies show that this domain of the enzyme is involved in hydrophobic interactions, and that the amphiphiles DDM or AA can provide such interactions. In fact the hydrolysis toward soluble substrates including cIP catalyzed by full-length PLC δ1 was enhanced by the formation of DDM micelles [11]. It has also been shown that deletion of EF1 of PLC δ1 significantly reduced its enzymatic activity [23]. Is the EF-hand domain responsible for the interfacial effect? To determine if PLC δ1 EF associates with the aggregated or monomeric amphiphiles, we attempted to measure fluorescence anisotropy. We were unable to utilize the Trp fluorescence for this purpose because the residue is highly flexible regardless of the presence or absence of the lipid ligands. Therefore, the protein was labeled with a thiol-reactive 5-iodoacetamidofluorescein (5-IAF), then fluorescence anisotropy was measured (Table 2). The fluorescent anisotropy obtained is likely to be a mixture of two components since there are two cysteines in the protein (the labeling efficiency was estimated to be 61% per site provided each Cys residue is equally reactive). Addition of DDM did not cause a significant change in the fluorescence anisotropy. However, it increased from 0.193 to 0.234 by further addition of AA. A preliminary NMR result showed that addition of DDM/AA mixture to PLC δ1 EF caused a line broadening to the spectrum (M. Roberts, unpublished data). These results indicate that in the presence of AA the protein associates with the aggregated state of amphiphiles rather than monomeric state. An alternative interpretation is that the protein aggregates when AA is present. Although anisotropy of the IAF-labeled protein did not suggest its association to the DDM micelles, the Trp fluorescence intensity increase showed hydrophobic interaction with the detergent. At this point it is difficult to determine how the protein interacts with the detergent. An NMR study on S100D suggested that a few isolated detergent molecules interact with a hydrophobic patch of the protein when it is loaded with Ca2+ [16]. A similar binding process may occur to PLC δ1 EF in the presence of DDM. Nonetheless, it appears that two distinct types of interactions, i.e., one with DDM and another with AA, are involved at the EF-hand domain of PLC δ1.
Table 2.
Fluorescence anisotropy of IAF-labeled PLC δ1 EF
| Anisotropy | Intensity | |
|---|---|---|
| No DDM/No AA | 0.190 | 38,529 |
| 200 μM DDM | 0.193 | 40,100 |
| 200 μM DDM/100 μM AA | 0.234 | 40,287 |
The two Cys residues were labeled with 5-IAF. The fluorescence anisotropy was measured according to Experimental procedures.
It is noteworthy that the emission spectrum of PLC δ1 EF without any lipid exhibited its maximum at 333 nm while that of NATA fluorescence was 351 nm. This blue-shifted spectrum of PLC δ1 EF indicates that the single tryptophan is in a hydrophobic environment and is shielded from the solvent. One can assume that the segment around this tryptophan is likely to be folded. Other evidence that supports this hypothesis is that the emission of Trp in the protein was more quenched than that of NATA in the same solution, which implies that Trp is in the vicinity of fluorescence-quenching residues or amide bonds. In fact, when the secondary structure of PLC δ1 EF was predicted with GOR4 [24], the segment around Trp was suggested to be helical. In the crystal structure of PLC δ1 which lacks the PH domain, this region of the EF-hand domain is not visible, suggesting significant flexibility of this region [6]. Taken together with our results, it points to the EF1 region having a dynamic structure with perhaps multiple conformations possible that shield the tryptophan from solvent.
EF-hand domain sandwiched between BFP and GFP exhibits FRET changes with AA binding
Previously, we reported that free fatty acids (FFAs) activate PLC δ1 through allosterism and that the EF-hand domain appeared to represent a site for binding of these lipids. Conformational changes upon FFA binding are thought to occur within the enzyme. Fluorescence resonance energy transfer (FRET) was used to distinguish whether conformational changes occur in the EF-hand domain itself upon binding to FFAs vs. changes in relative orientations of helices or loops vs. overall changes in secondary structure. Fluorescence resonance energy transfer (FRET) is one of the widely used techniques to detect conformational changes in proteins. We engineered BFP-EF (donor only) and BFP-EF-GFP (donor-acceptor) (Fig. 1) constructs to tell us if any global conformational changes occur when amphiphiles bind the EF-hand protein. The original method utilizing GFP variants was developed by Miyawaki et al. [25] for a calcium indicator using calmodulin and MLCK peptide linked together. Although the EF-hand domain of PLC δ1 has not been studied as well as calmodulin and the MLCK peptide, this method appears to be useful for our purpose.
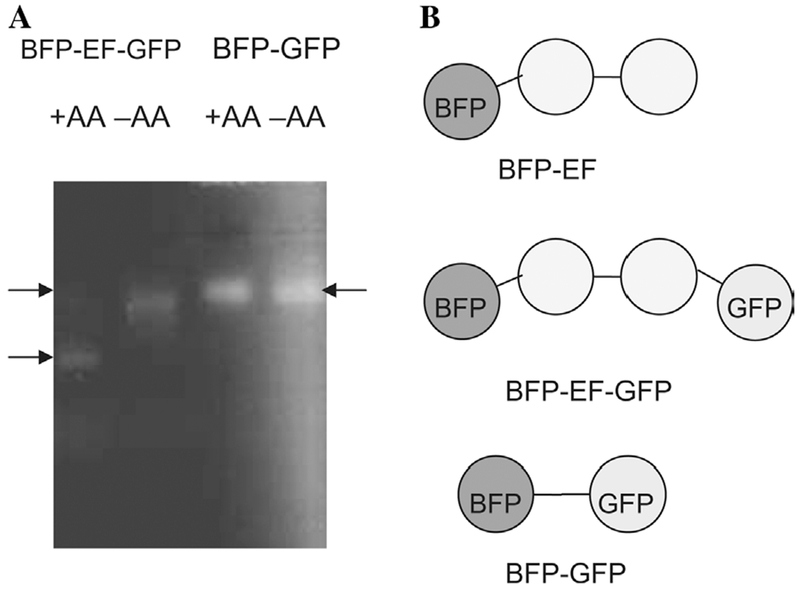
Previously, we showed that the mobility of the isolated EF-hand domain shifted in non-denaturing PAGE when AA was added to the protein [11]. To determine if the EF-hand domain with BFP/GFP moieties are able to bind AA, non-denaturing gel electrophoresis was performed (Fig. 5A). The BFP-GFP construct that has 12 residues (SGLRSRAQASRA) between the two domains served as a control. The mobility of BFP-EF-GFP shifted when AA was included, while BFP-GFP did not show a detectable shift in mobility under the same conditions. Therefore, the gel shift is specific to the inserted protein, i.e., the EF-hand domain, and our new constructs with BFP/GFP did not lose the ability to bind fatty acids (Fig. 6).
Fig. 5.
AA causes a mobility shift in non-denaturing electrophoresis. (A) To assess if the EF-hand domain with additional BFP/GFP moieties are able to bind AA, non-denaturing electrophoresis was performed. BFP-EF-GFP was added to 400 μM DDM (−AA) or 400 μM DDM containing 200 μM AA (+AA). BFP-GFP was used as control. (B) Schematic representation of the constructs is shown. The EF-hand domain is drawn as a dumbbell.
Fig. 6.
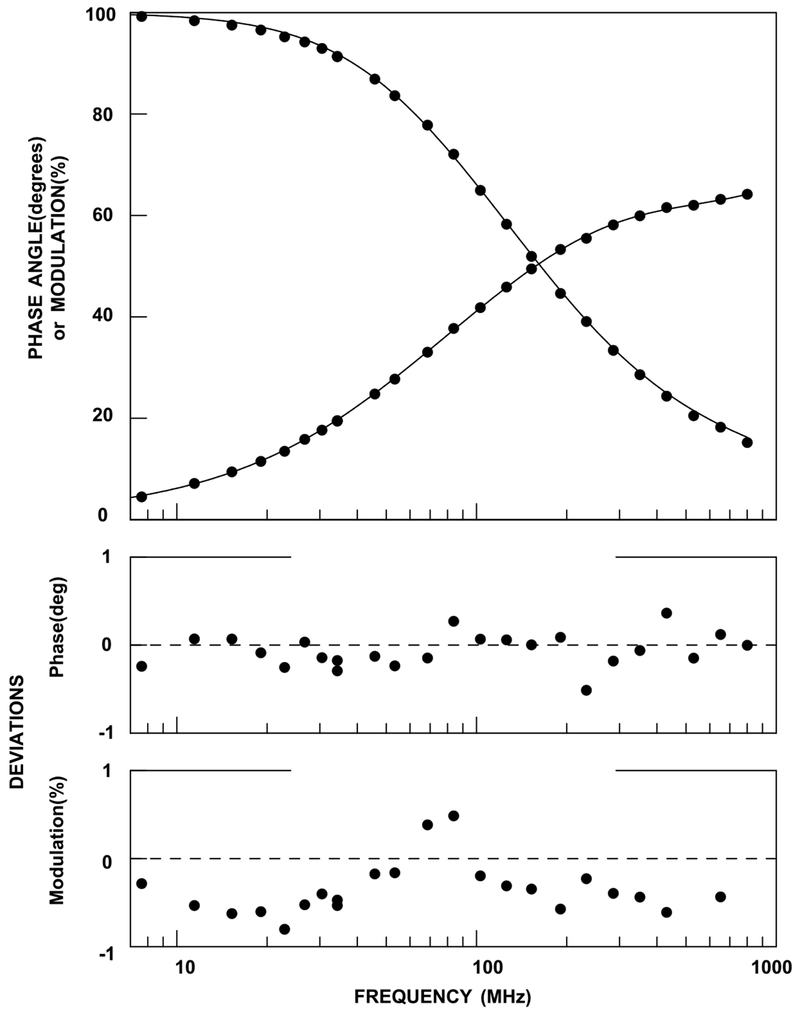
Fluorescence lifetime measurement of the donor in BFP-EF. Measurements were done for >20 modulation frequencies and lifetimes of the donor (BFP) were calculated by fitting the values of phase angle and modulation. The upper panel shows a frequency-domain decay for BFP-EF in the presence of 50 μM AA and 77 μM DDM. The lower two panels are deviations for a fit to the decay.
The fluorescence lifetimes of BFP for BFP-EF (donor only) or BFP-EF-GFP (donor-acceptor) were measured and transfer efficiencies were calculated according to the equation, E =1 − τda/ τd. Here, τda is the lifetime in a donor–acceptor pair, and τd is the lifetime in a donor only construct [26]. When AA was dissolved with 77 μM DDM (below the CMC of the pure detergent), there was a significant difference in transfer efficiency (14–32%) in samples with 0 or 150 μM AA (Table 3). However, there was no apparent difference in their transfer efficiency (~20% for each concentration) when AA was introduced with 200 μM DDM. Since PLC δ1 EF binds DDM (Fig. 4), high concentrations of DDM masks the effect of AA, presumably due to a competitive mechanism. When 50 μM SA was added, the transfer efficiency was similar to that of 50 μM AA (Table 3) indicating no specificity in the type of fatty acid bound. It is possible that our observation using this technique reflects tight binding of an anionic lipid rather than differences between saturated and unsaturated FFAs. Nonetheless, these findings with low DDM concentrations suggest that there is a global conformational change when the EF-hand domain binding to FFAs, although whether it is due to a change in distance of the probes or orientation or both, is not clear. The conformational change within the isolated domain could at least partly account for the activation of the intact enzyme by AA. It would be of interest to study how this conformational change within the domain propagates through the enzyme, which then leads to activation of the enzyme. As previously stated, the EF-hands are in close apposition to the C2 domain in the crystal structure, and the C2 domain is an allosteric regulator of PLC δ1 [10].
Table 3.
The fluorescence lifetimes of BFP in BFP-EF (donor only) and BFP-EF-GFP (donor-acceptor)
| BFP-EF τd (ns) |
BFP-EF-GFP τda (ns) |
Transfer efficiency E =1 − τda/τd |
||||
|---|---|---|---|---|---|---|
| 77 μM DDM | 200 μM DDM | 77 μM DDM | 200 μM DDM | 77 μM DDM | 200 μM DDM | |
| 0 μM AA | 1.67 | 1.56 | 1.42 | 1.26 | 0.14 | 0.19 |
| 50 μM AA | 1.73 | 1.58 | 1.37 | 1.26 | 0.21 | 0.20 |
| 150 μM AA | 1.87 | 1.55 | 1.28 | 1.28 | 0.32 | 0.18 |
| 50 μM SA | 1.69 | 1.32 | 0.22 | |||
Transfer efficiencies were calculated according to the equation, E =1 − τda/τd, where τda is the lifetime in a donor-acceptor pair and τd is the lifetime in a donor only construct. An error for each lifetime is small (less than a few percent). Even in the transfer efficiencies calculated based on the values, they are significantly less than 10%.
Trp in the EF-hand domain detects Ca2+ -dependent changes only in the presence of fatty acids
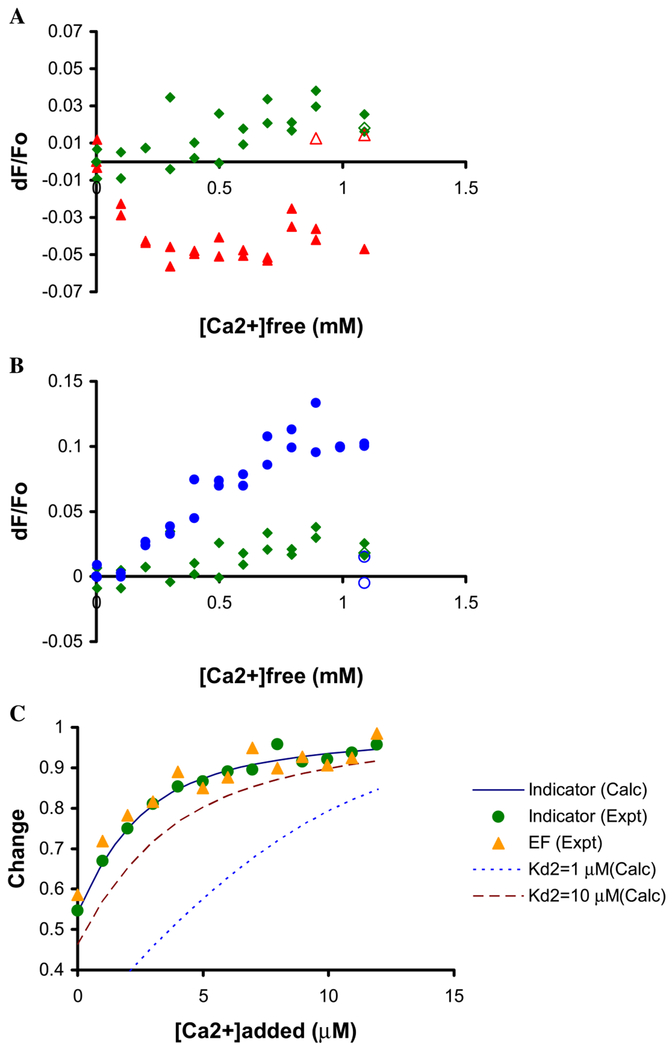
There are two putative Ca2+-binding sites in the EF-hand domain of PLC δ1, EF1, and EF2 [6]. Until this report, there has been no direct evidence showing that this domain is able to bind Ca2+, although it has been shown that the PH-domain linked with the EF-hand domain is able to bind PIP2 in a Ca2+-dependent manner [27]. We hypothesized that PLC δ1 EF may be able to bind Ca2+ when lipids such as FFAs are present. Ca2+ binding was monitored using the intrinsic tryptophan fluorescence. There were no Ca2+-dependent changes in the tryptophan emission in the absence of any lipid (data not shown). However, as shown in Fig. 7A, there was a small (up to 5%) but reproducible decrease in fluorescence intensity when PLC δ1 EF was titrated with Ca2+ in the presence of 100 μM AA plus 200 μM DDM (the reduction in fluorescence was observed for two preparations of PLC δ1 EF). When 2mM EGTA was added at the final point of the titration (0.9 or 1.1 mM free Ca2+), the original fluorescence intensity was fully recovered (indicating that the observed change in the presence of AA was not due to artifacts such as drifts in the spectrum). Addition of Ca2+ with only DDM present did not cause any change in the tryptophan fluorescence. This suggests that the tryptophan residue is sensitive to reversible Ca2+ binding by PLC δ1 EF or the interface and that AA is critical to this effect. Next, we performed the same experiment with SA present in DDM micelles to see if similar changes were caused by Ca2+ when a saturated chain FFA was present. Interestingly, with SA the EF-hand fluorescence exhibited an increase in intensity up to 10% of the original value (Fig. 7B). The changes in the Trp fluorescence intensity by addition of Ca2+ were in the opposite direction to those of AA, which suggests that a different environment is created around the residue depending on types of lipids used. The Ca2+ concentration necessary to achieve the half-maximal change, EC50, was 96 μM in the presence of AA and 348 μM in the presence of SA. Again, these changes were observed only when fatty acids were present. Although there was no apparent Ca2+-dependent change in intensity when fluorescently labeled PC was added to the micelles in the absence of PLC δ1 EF (data not shown), it is possible that the carboxyl group of FFA can coordinate a divalent cation such as Ca2+ [28]. Such an interaction may cause changes in the micelle structure. However, because the Trp residue in PLC δ1 EF was able to detect the Ca2+-dependent changes, the protein is likely to be involved in Ca2+ binding.
Fig. 7.
The intrinsic tryptophan fluorescence exhibits Ca2+-dependent changes in the presence of FFAs. (A) PLC δ1 EF (4.0 μM) was titrated with Ca2+ in the absence (diamond) or presence of 100 μM AA (triangle). DDM was added up to 200 μM prior to titration. 2 mM EGTA was added at the endpoint of titration (open symbols). Data points generated from two separate experiments were superimposed. (B) PLC δ1 EF (4.0 μM) was titrated with Ca2+ in the presence of 100 μM SA (circle). DDM was added up to 200 μM prior to titration. A measure of 2 mM EGTA was added at the endpoint of titration (open symbols). Data points generated from two separate experiments were superimposed. The same control experiments with DDM are shown as in (A). (C) A calcium indicator, fluo-3 was titrated with Ca2+ in the absence (circle) or presence of 10.3 μM PLC δ1 EF (triangle). Change, α was plotted as a function of added Ca2+ concentration (see Experimental procedures). Expected changes for a calcium binding protein with Kd2 = 1 μM or Kd2 = 10 μM were simulated.
The intrinsic fluorescence of the single tryptophan may not detect Ca2+ binding to all of the possible sites in the EF-hand domain. For this reason, we used the Ca2+ indicator fluo-3 to further explore Ca2+ binding to the EF-hands in the absence of amphiphiles. As shown in Fig. 7C, the fluorescence changes of the dye with and without PLC δ1 EF were plotted according to the method described in Experimental procedures. Under the condition used here, the dissociation constant of fluo-3 for Ca2+ was calculated to be 0.61 μM. Simulation of the curve for protein binding Ca2+ with a Kd of 1 or 10 μM puts limits on the binding constant of Ca2+ to the protein: it must be greater than 10 μM. A value of 100 μM, roughly what we see in the intrinsic fluorescence experiments, would be indistinguishable from what was observed. Therefore, we concluded that in the absence of any lipid PLC δ1 EF does not have detectable Ca2+-binding affinity (that is, higher than 10 μM). This is corroborated by the fact that we did not observe significant binding in a 45Ca2+ assay (M. Kobayashi, unpublished results) which was performed in a similar fashion to the experiments described earlier [10,29]. In that assay the radioactivity on the membrane was counted after a mixture containing PLC δ1 EF and 120 μM 45CaCl2 was passed through a membrane. These dye fluorescence experiments coupled with the intrinsic protein fluorescence indicate that the EF-hand domain has weak affinity for Ca2+ unless AA is present.
The EF-hand domain of PLC δ1 did not bind Ca2+ or Ca2+ analogs when the crystals were soaked with cations [6]. Isothermal titration calorimetry performed for Ca2+ binding to the PH-domain truncation mutants of PLC δ1, demonstrated a stoichiometry of four [30]. This is consistent with a single Ca2+ binding to the catalytic core, three to the C2 domain and none to the EF-hand motifs [30]. Using Dictyostelium discoideum PLC, Drayer et al. [31] demonstrated that mutation of the putative Ca2+-binding site in the EF-hand domain did not alter Ca2+ sensitivity of this enzyme. However, the maximum activity at saturating concentrations of Ca2+ for these mutants was markedly decreased suggesting that the mutated residues in the EF-hand are important for the enzymatic activity. The authors did not show whether the decrease in the enzymatic activity was due to a loss of Ca2+ binding to the EF-hand. Another group showed that mutations in which acidic residues in the putative Ca2+-binding site of PLC δ1 were replaced with alanine did not affect the enzymatic activity [23]. Our results showed that Ca2+-dependent changes in the EF-hand fragment were observed only when FFAs were present. The Ca2+ binding could be the result of conformational change induced by FFA binding to the protein. However, it is known that the carboxyl group of FFAs is able to interact with Ca2+ [28]. Hence and alternate explanation is that these observed changes are due to a direct interaction between Ca2+ and FFA (although one might have expected similar EC50 for AA and SA rather than the threefold difference observed). Regardless of which is correct, the fact that the tryptophan residue in EF1 responds to Ca2+ when FFA is present indicates that the protein participates in Ca2+ binding. A similar mechanism of Ca2+ binding was proposed for Ca2+-dependent interaction between the C2 domain of PLC δ1 and PS where the head group of PS and C2 domain coordinate Ca2+ [10]. Our results suggest that the interaction between the EF-hand domain and the lipids (anionic ones such as FFA in particular) may be a prerequisite for this domain to interact with Ca2+, which is possibly attracted to the head group of anionic lipids.
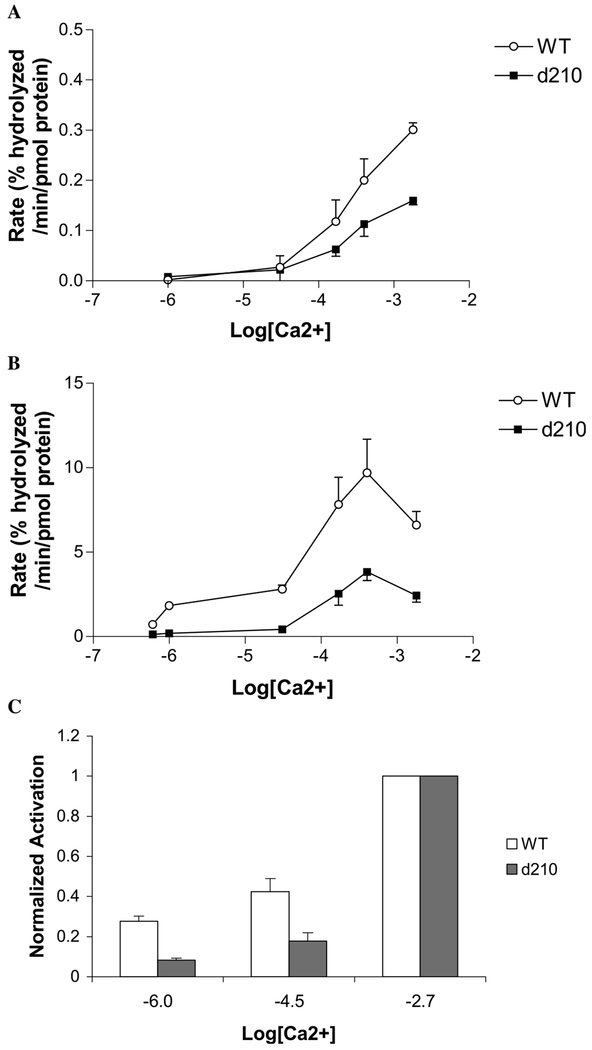
The affinities of PLC δ1 EF for Ca2+ in the presence of FFAs were weak (EC50 100–350 μM) indicating that under physiological conditions (intracellular Ca2+ concentration at resting level is in the order of 100 nm [32]) the EF-hand domain is not likely to be saturated with Ca2+. While high Ca2+ affinity is usually associated with EF-hand proteins, there are examples of other EF-hand domains with weak affinity for Ca2+ ions. For α-spectrin EF-hands, the Kd is estimated to be 50–100 μM [33], and the authors speculate that a Ca2+ binding event occurs where Ca2+ concentration significantly increases at least transiently. It has been suggested that such locations exist in cells [34,35]. A transient increase in Ca2+ and EF-hand binding may also occur with PLC δ1. However, an alternative explanation is that the presence of another domain such as the PH domain might enhance the Ca2+ binding affinity by the EF-hand domain. In fact our results from activity assays using the wild-type and deletion mutant support this hypothesis. The deletion mutant which lacks the PH and half of the EF-hand domains was less activated by AA between 1 and 10 μM free Ca2+, which corresponds to intracellular Ca2+ concentration in activated cells (Fig. 8). It has been shown that the PH domain linked with the EF-hand domain (1–175) is able to bind PIP2 in a Ca2+-dependent manner below 10 μM Ca2+ [27]. Recently, it was reported that Ca2+ sensitivity of the zeta isozymes is regulated thorough the EF-hand domain [36]. Taken all together, it is not unreasonable to conclude that the EF-hand domain is necessary for Ca2+ activation of the whole enzyme via anionic lipid binding to the domain.
Fig. 8.
The N-terminal region of PLC δ1 modulates Ca2+-sensitivity of the enzymatic activity in the presence of AA. (A) The rates of PI hydrolysis in the absence of AA were compared between the wild-type (open circle) and PLC δ1 (Δ1–210) deletion mutant (filled square). The substrate (3 μM) was solubilized with 250 μM DDM. The rates were determined from 2 to 5 experiments and plotted as a function of free Ca2+ concentration in logarithm. (B) The rates of PI hydrolysis in the presence of 100 μM AA were compared between the wild-type (open circle) and PLC δ1 (Δ1–210) deletion mutant (filled square). The substrate (3 μM) and 100 μM were solubilized with 200 μM DDM. (C) The rates in the presence of AA (B) were normalized so that the rate at log[Ca2+] = −2.7 for each enzyme, i.e., wild-type (open bar) or deletion mutant (gray bar) becomes unity.
In summary, this is the first report of involvement of the EF-hand domain of PLC δ1 in Ca2+ binding. FFA binding is necessary for this domain to interact with Ca2+, although it is not known whether a high local Ca2+ concentration is necessary or whether the affinity for Ca2+ becomes higher in the context of the whole protein under physiological conditions. Lipid binding to the EF-hand domain induced conformational changes as assessed with both CD and fluorescence spectroscopy. This physical evidence supports our recent kinetic studies suggesting that the EF-hand acts as a lipid mediated allosteric regulatory domain. Future studies will investigate how Ca2+ binds with this domain.
Acknowledgments
This work was supported by National Institutes of Health Grants HL55591 and HL03961 to J.W.L, and GM60418 to M.F.R. We thank Dr. Carl Waltenbaugh for letting us use a fluorescence plate reader for preliminary results and Dr. John Solaro and Dr. Tomoyoshi Kobayashi at UIC for letting us use their instrument. We also thank Ms. Li Lin for helping us make some of the constructs. For anisotropy measurement we thankfully acknowledge the use of instruments in the Keck Biophysics Facility at Northwestern University [http://www.biochem.northwestern.edu/Keck/keckmain.html].
Footnotes
Abbreviations used: PLC δ1, phospholipase C δ1; PI, phosphatidylinositol; PIP2, phosphatidylinositol 4,5-bisphosphate; IP3, inositol 1,4,5-trisphosphate; DAG, diacylglycerol; PS, phosphatidylserine; PH domain, pleckstrin homology domain; DDM, dodecyl maltoside; AA, arachidonic acid or arachidonate; SA, stearic acid or stearate; FFA, free fatty acid; Hepes, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid; EGTA, ethylene glycol-bis(2-aminoethyl)-N,N,N′,N′-tetraacetic acid; MOPS, 3-(N-Morpholino)propanesulfonic acid; GST, glutathione S-transferase; NATA, N-acetyl-tryptophan-amide; CMC, critical micelle concentration; 5-IAF, 5-iodoacetamidofluorescein.
References
- [1].Rebecchi MJ, Pentyala SN, Physiol. Rev 80 (2000) 1291–1335. [DOI] [PubMed] [Google Scholar]
- [2].Rhee SG, Annu. Rev. Biochem 70 (2001) 281–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Parrington J, Jones ML, Tunwell R, Devader C, Katan M, Swann K, Reproduction 123 (2002) 31–39. [DOI] [PubMed] [Google Scholar]
- [4].Hwang JI, Oh YS, Shin KJ, Kim H, Ryu S, Suh PG, Biochem. J (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nakahara M, Shimozawa M, Nakamura Y, Irino Y, Morita M, Kudo Y, Fukami K, J. Biol. Chem (2005). [DOI] [PubMed] [Google Scholar]
- [6].Essen LO, Perisic O, Cheung R, Katan M, Williams RL, Nature 380 (1996) 595–602. [DOI] [PubMed] [Google Scholar]
- [7].Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB, Cell 79 (2) (1994) 199–209. [DOI] [PubMed] [Google Scholar]
- [8].Lomasney JW, Cheng HF, Wang LP, Kuan YS, Liu SM, Fesik SW, King K, J. Biol. Chem 271 (41) (1996) 25316–25326. [DOI] [PubMed] [Google Scholar]
- [9].Bromann PA, Boetticher EE, Lomasney JW, J. Biol. Chem 272 (1997) 16240–16246. [DOI] [PubMed] [Google Scholar]
- [10].Lomasney JW, Cheng HF, Roffler SR, King K, J. Biol. Chem 274 (1999) 21995–22001. [DOI] [PubMed] [Google Scholar]
- [11].Kobayashi M, Mutharasan RK, Feng J, Roberts MF, Lomasney JW, Biochemistry 43 (2004) 7522–7533. [DOI] [PubMed] [Google Scholar]
- [12].Kretsinger RH, Nockolds CE, J. Biol. Chem 248 (1973) 3313–3326. [PubMed] [Google Scholar]
- [13].Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M, Nature 389 (1997) 198–202. [DOI] [PubMed] [Google Scholar]
- [14].Kerkhoff C, Klempt M, Kaever V, Sorg C, J. Biol. Chem 274 (1999) 32672–32679. [DOI] [PubMed] [Google Scholar]
- [15].Kerkhoff C, Vogl T, Nacken W, Sopalla C, Sorg C, FEBS Lett. 460 (1999) 134–138. [DOI] [PubMed] [Google Scholar]
- [16].Malmendal A, Vander Kooi CW, Nielsen NC, Chazin WJ, Biochemistry 44 (2005) 6502–6512. [DOI] [PubMed] [Google Scholar]
- [17].Jiang Y, Qian W, Hawes JW, Walsh JP, J. Biol. Chem 275 (2000) 34092–34099. [DOI] [PubMed] [Google Scholar]
- [18].Sakane F, Yamada K, Kanoh H, Yokoyama C, Tanabe T, Nature 344 (1990) 345–348. [DOI] [PubMed] [Google Scholar]
- [19].Andrade MA, Chacon P, Merelo JJ, Moran F, Protein Eng. 6 (1993) 383–390. [DOI] [PubMed] [Google Scholar]
- [20].Bohm G, Muhr R, Jaenicke R, Protein Eng. 5 (1992) 191–195. [DOI] [PubMed] [Google Scholar]
- [21].Laczko G, Gryczynski I, Gryczynski Z, Wiczk W, Malak H, Lakowicz JR, Rev. Sci. Instrum 61 (1990) 2331–2337. [Google Scholar]
- [22].Banerjee P, Dasgupta A, Chromy BA, Dawson G, Arch. Biochem. Biophys 305 (1993) 68–77. [DOI] [PubMed] [Google Scholar]
- [23].Nakashima S, Banno Y, Watanabe T, Nakamura Y, Mizutani T, Sakai H, Zhao YT, Sugimoto Y, Nozawa Y, Biochem. Biophys. Res. Commun 211 (2) (1995) 364–369. [DOI] [PubMed] [Google Scholar]
- [24].Combet C, Blanchet C, Geourjon C, Deleage G, Trends Biochem. Sci 25 (2000) 147–150. [DOI] [PubMed] [Google Scholar]
- [25].Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY, Nature 388 (1997) 882–887. [DOI] [PubMed] [Google Scholar]
- [26].Lakowicz JR, Principles of Fluorescence Spectroscopy, second ed., Kluwer Academic, New York, 1999. [Google Scholar]
- [27].Yamamoto T, Takeuchi H, Kanematsu T, Allen V, Yagisawa H, Kikkawa U, Watanabe Y, Nakasima A, Katan M, Hirata M, Eur. J. Biochem 265 (1999) 481–490. [DOI] [PubMed] [Google Scholar]
- [28].Yamaguchi T, Kaneda M, Kakinuma K, Biochim. Biophys. Acta 861 (1986) 440–446. [DOI] [PubMed] [Google Scholar]
- [29].Nakamura J, Biochim. Biophys. Acta 870 (1986) 495–501. [DOI] [PubMed] [Google Scholar]
- [30].Grobler JA, Essen LO, Williams RL, Hurley JH, Nat. Struct. Biol 3 (9) (1996) 788–795. [DOI] [PubMed] [Google Scholar]
- [31].Drayer AL, Meima ME, Derks MW, Tuik R, van Haastert PJ, Biochem. J 311 (1995) 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miyazaki S, Jpn. J. Physiol 43 (1993) 409–434. [DOI] [PubMed] [Google Scholar]
- [33].Buevich AV, Lundberg S, Sethson I, Edlund U, Backman L, Cell. Mol. Biol. Lett 9 (2004) 167–186. [PubMed] [Google Scholar]
- [34].Macrez N, Mironneau J, Curr. Mol. Med 4 (2004) 263–275. [DOI] [PubMed] [Google Scholar]
- [35].Zucker RS, J. Physiol. Paris 87 (1993) 25–36. [DOI] [PubMed] [Google Scholar]
- [36].Kouchi Z, Fukami K, Shikano T, Oda S, Nakamura Y, Takenawa T, Miyazaki S, J. Biol. Chem 279 (2004) 10408–10412. [DOI] [PubMed] [Google Scholar]