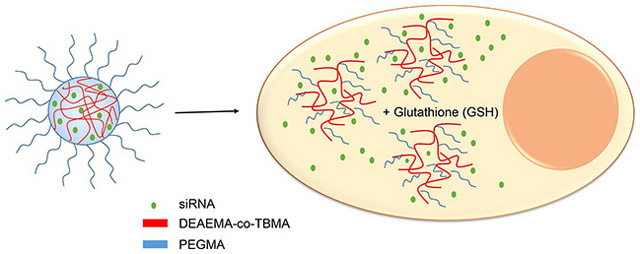
Abstract
The progress of short interfering RNA (siRNA) technologies has unlocked the development of novel alternatives for the treatment of a myriad of diseases, including viral infections, autoimmune disorders, or cancer. Nevertheless, the clinical use of these therapies faces significant challenges, mainly overcoming the charged and large nature of these molecules to effectively enter the cell. In this work, we developed a cationic polymer nanoparticle system that is able to load siRNA due to electrostatic interactions. The pH-responsiveness and membrane-disrupting ability of these carriers make them suitable intracellular delivery vehicles. In the work presented herein we synthesized, characterized, and evaluated the properties of nanoparticles based on 2-diethylaminoethyl methacrylate and tert-butyl methacrylate copolymers. A disulfide crosslinker was incorporated in the nanogels to enable the degradation of the nanoparticles in reductive environments, showing no significant changes on their physicochemical properties. The capability of the developed nanogels to be internalized, deliver siRNA, and induce gene knockdown were demonstrated using a human epithelial colorectal adenocarcinoma cell line. Overall, these findings suggest that this platform exhibits desirable characteristics as a potential siRNA-delivery platform.
Keywords: cationic, polymer, nanoparticles, drug delivery, intracellular, siRNA
Graphical Abstract
1. Introduction
The discovery of RNA interference (RNAi) has prompted an enormous research effort in all fields of biological science and our understanding of gene regulation mechanisms has increased.(Fire et al., 1998) In principle, RNAi mediated by siRNA could be used as a powerful and versatile treatment modality to treat nearly any disease resulting from aberrant gene expression. As with many biotherapeutics, efficient delivery has been implicated as the major hurdle to its widespread clinical application.(Whitehead et al., 2009)
Several materials-based strategies have been developed to circumvent the challenges associated with cytoplasmic delivery of siRNA. The most common classes of materials are lipids and lipidoids, polycationic polymers, and siRNA conjugates.(Whitehead et al., 2009) Lipids are perhaps the most widely studied class of materials for siRNA delivery and lipid-based formulations for RNAi have progressed to the clinic in greater number than other delivery systems.(Siegwart et al., 2011)
The first evidence of RNAi in humans was demonstrated using polycationic cyclodextrin nanoparticles.(Davis et al., 2010) However, alternative strategies have emerged to enable siRNA delivery to target cells and tissues. Khormaee et al.(Khormaee et al., 2012) reported the use of a membrane-disruptive anionic pseudo-peptide to deliver siRNA in vivo. Polysaccharides from yeast cell walls have been used to deliver inflammation-suppressing siRNA via oral administration.(Aouadi et al., 2009)
Amine-containing methacrylates, such as 2-(diethylaminoethyl) methacrylate (DEAMA) (Tamura et al., 2009) and 2-(dimethylaminoethyl) methacrylate (DMAEMA)(Convertine et al., 2009), are attractive foundations for polymeric nucleic-acid-delivery systems because of their ability to form cooperative electrostatic interactions with polyanionic siRNA. In our previous work, we explored the aqueous solution properties, membrane-disruptive properties, and internalization mechanisms of pH-responsive nanoscale hydrogels (nanogels).(Liechty et al., 2018)
As we have shown before (Fisher et al., 2008; Forbes and Peppas, 2012, 2014a, b; Hariharan and Peppas, 1996; Liechty et al., 2011), that we have found our most promising nanoparticle platform for siRNA delivery consists of a (1) ionizable core of 2-(diethylaminoethyl methacrylate) (DEAEMA), (2) the hydrophobic comonomer of tert-butyl methacrylate (TBMA), and (3) a grafted corona of poly(ethylene glycol) tethers. The polymer was synthesized using 30 mol of TBMA per 100 mol DEAEMA. The resulting nanogel (named PDETB30) undergoes a volume phase transition from collapsed hydrophobe to swollen hydrophile at approximately pH 6.5 and is highly disruptive to model membrane systems in this transition region.(Liechty et al., 2018) Additionally, PDETB30 displays excellent biocompatibility with Caco-2 and RAW 264.7 cells as evaluated using in vitro toxicity assays.(Liechty et al., 2013)
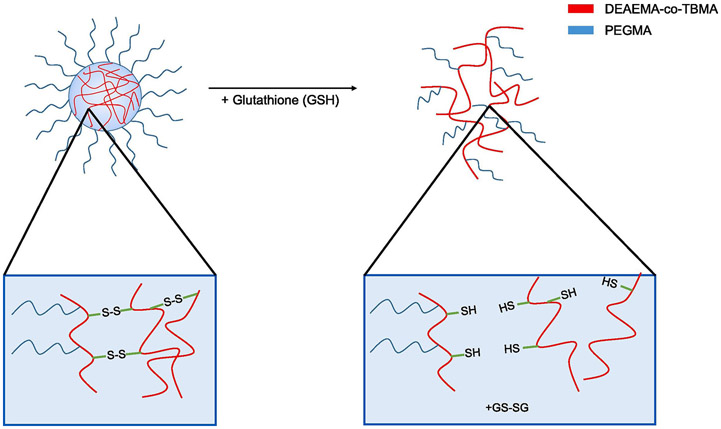
However, intracellular delivery systems require the incorporation of mechanisms that allow the release of the payload inside the cell under specific conditions.(Antimisiaris et al., 2017; Knipe et al., 2015; Knipe et al., 2016; Wagner, 2012) Disulfide linkers can be cleaved by the reductive tripeptide glutathione (GSH); present at intracellular concentrations of 1 - 11 mM.(Winter et al., 2006) By incorporating these linkers into polycationic nanogels, degradability can be imparted to the network while retaining their mechanical integrity and pH-responsive behavior. In this work, we concentrate on the synthesis and characterization of nanogels with reducible disulfide crosslinks and discuss studies showing their suitability as siRNA carriers in comparison to non-degradable PDETB30 nanogels.
2. Materials and Methods
2.1. Synthesis of disulfide crosslinker
Dichloromethane (>99.5%) was purchased from Fisher Scientific (Plainfield, NJ). Methacryloyl chloride (97%) and anhydrous pyridine (99.8%) were purchased from Sigma-Aldrich (St. Louis, MO). 2-Hydroxyethyldisulfide (90%) was purchased from Acros (Geel, Belgium).
The bifunctional disulfide crosslinker, bis(2-methacryloyloxyethyl) disulfide (SSXL), was synthesized according to the method reported by Gao, et al.(Gao et al., 2005) Briefly, organic solvents were dried over MgSO4 before use. Dichloromethane was purged with N2 for 15 min and placed in a dry nitrogen atmosphere (O2 <0.1 ppm, H2O < 0.1 ppm). Pyridine (15.8 mL, 0.195 mol) and bis(2-hydroxyethyl) disulfide (10.00 g, 0.065 mol) were added to cold (4°C) dichloromethane and agitated briefly. Methacryloyl chloride was added dropwise to the stirring organic mixture. The flask was then sealed, removed from the ice bath, and the reaction was allowed to proceed for 12 h in a nitrogen atmosphere.
The reaction product in dichloromethane was successively washed with 1 N HCl, 1 N NaOH, and DI water. The organic phase was retained and dried. The product was then dissolved in diethyl ether and passed through a column of sodium carbonate and basic alumina. Diethyl ether was removed through rotary evaporation.
Additional flash chromatography purification was performed using a Teledyne-Isco Companion Automated Flash Chromatography Instrument (Lincoln, NE) equipped with a 100 g silica column. The solvent gradient was established as follows: Solvent A – hexanes, Solvent B-ethyl acetate. The gradient was adjusted from 0 – 15% B over 40 minutes and then from 15 – 100% B over 10 minutes. Fractions of interest were determined by monitoring absorbance at 258 nm. Product fractions were pooled and solvent was removed by rotary evaporation.
2.2. Characterization of disulfide crosslinker
The composition of raw materials, purification fractions, and final product of the SSXL synthesis were confirmed using a Varian (Palo Alto, CA) DirectDrive 400 MHz nuclear magnetic resonance spectrometer equipped with an automatic sampler. Chloroform-d (CDCl3, 99.8%) was obtained from Acros Organics (Fairlawn, NJ). SSXL was dissolved at 1% (vol/vol) in CDC13 for 1H-NMR analysis. All NMR spectra were analyzed using SpinWorks 3™ software.
2.3. Synthesis and Characterization of Degradable Nanogels
The nomenclature of the copolymer nanogels presented in this work was established to reflect the copolymer composition. The primary monomer in this work was DEAEMA (DE), and the comonomers used were TBMA (TB), or TBAEMA (TBA). The number at the end of the polymer name represents the comonomer ratio (number of moles of the comonomer per 100 mol of the primary monomer). To impart a mechanism for biodegradation to the responsive nanogel PDETB30, SSXL was used as a replacement for tetra(ethylene glycol) dimethacrylate (TEGDMA) in the photoemulsion polymerization. SSXL was added to pre-polymer mixture at 2.5 mol% of total monomer and photoemulsion polymerization and nanogel purification proceeded as previously described.(Liechty et al., 2013; Liechty et al., 2018) The resulting nanogel, termed PDESSB30, was stored in a desiccator at −20°C until further use.
A fluorescent version of PDESSB30 was synthesized and purified as previously shown. The covalent conjugation of Oregon Green 488 (OG488) was enabled by the incorporation of primary amines in the PDESSB30 core. Briefly, 2-aminoethyl methacrylate hydrochloride (AEMA) was included in the pre-polymer feed mixture at 5 mol% of DEAEMA. The resulting copolymer was named PDESSB30f to describe the amine functionality. The primary amine of AEMA was verified with a fluorescamine assay after synthesis and purification.
Following the amine-modification of the PDESSB30f copolymer, Oregon Green 488 carboxylic acid, succinimidyl ester (OG488, Molecular Probes, Eugene, OR) was incorporated to the polymer. First, the fluorescent probe was dissolved in DMSO to yield a 10 mg ml−1 solution, and PDESSB30f was suspended at 10 mg ml−1 in 150 mM sodium bicarbonate buffer, pH 8.30. OG488 was then added to the PDESSB30f suspension to give a 1:1 mol ratio between AEMA and OG488. The reaction was stirred in the dark for 6 h. Following reaction completion, unreacted dye was separated from labeled PDESSB30-OG488 through dialysis against DI water. Dialysis proceeded for 3 days using 12,000 – 14,000 MWCO dialysis tubing (Spectrum Labs, Rancho Dominguez, CA). Labeled nanogels, PDESSB30-OG488, were lyophilized in the dark for 3 days.
Several characterization techniques were employed to study the physicochemical properties of the PDESSB30 nanogels in comparison to the TMBA analogue, PDETB30. Dynamic light scattering was used to determine the hydrodynamic diameter of PDESSB30 nanogels as a function of environmental pH. Measurements of the ζ-potential were also performed to evaluate the effective surface charge as a function of environmental pH. Additionally, transmission electron microscopy (TEM) was used to determine the diameter of the dry nanogels and was conducted as previously described. The metabolic activity of RAW 264.7 cells upon exposure to PDESSB30 and PDETB30 was measured using a commercially-available MTS Cell Proliferation assay kit as previously reported.
2.4. RNA Binding Studies
The RNA complexation buffer was prepared with 3.15 g sodium phosphate dibasic heptahydrate, 0.02 g potassium phosphate monobasic monohydrate, 0.20 g potassium chloride, and 8.01 g sodium chloride in DI water. Following salt dissolution, the solution pH was adjusted to pH 5.50 and the final solution volume was brought up to 100 mL. To remove nucleases from the buffer, diethylpyrocarbonate (DEPC) was added at 0.1% and incubated at room temperature overnight. The buffer solution was then autoclaved to inactivate the DEPC. Polymer-siRNA complexes were formed by combining aqueous solutions of nanogels, siRNA, 10x RNAse-free Phosphate Buffered Saline (PBS), and RNAse-free water to obtain the desired concentrations.
Silencer® GAPDH siRNA, Quant-iT™ Ribogreen® RNA Assay Kit, and RNAse Free H2O were purchased from Life Technologies (Carlsbad, CA). Free siRNA in solution was measured using the Ribogreen® assay according to manufacturer’s instructions. Nanogel suspensions were diluted in RNAse free complexation buffer (pH 5.50). Concentrated siRNA was added to yield 500 ng ml−1 RNA in a nanogel suspension at designated concentrations. Measurements of the free siRNA were taken after 60, 120, and 180 minute complexation periods.
2.5. Cell Culture Studies
Human colorectal adenocarcinoma cells (Caco-2) were cultured in Dulbecco’s Modified Eagles Medium (DMEM) supplemented with 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 0.25 μg mL−1 Amphotericin B, and 10% Fetal Bovine Serum. Caco-2 cells were used between passage 34 and 62. Caco-2 cells were passaged by incubation with 0.25% Trypsin-EDTA at 37°C. Then, trypsin was neutralized by addition of fresh DMEM and cells were separated by centrifugation. The cell suspension was resuspended in fresh media, diluted as necessary, and added to tissue-culture treated flasks or 6-well plates. Caco-2 cells were typically passaged at a 1:5 ratio with media replenished every 2–3 days. Cells were allowed to grow to 80% confluency before use.
2.6. siRNA Delivery
DyLight 647-labeled small interfering RNA (Sense: DY647-UAAGGCUAUGAAGAGAUACUU) was purchased from Thermo Scientific (Lafayette, CO). Cy3-labeled Silencer® Negative Control No. 1 siRNA was purchased from Life Technologies (Carlsbad, CA). Fluorescent nanogels, PDETB30-OG488 and PDESSB30-OG488 were synthesized and purified as previously described.
Concentrated suspensions (20X) of fluorescent nanogels (PDETB30-OG488 or PDESSB30-OG488), fluorescent siRNA (DY647-siRNA or Cy3-siRNA), or fluorescent nanogels and fluorescent siRNA were prepared to contain 0.5 mg mL−1 nanogel, 26.5 μg mL−1 (~2000 nM) siRNA, 1X complexation buffer, and RNAse free H2O. Control samples (nanogel or siRNA) were prepared in a similar fashion, replacing the volume of the absent component(s) with RNAse free H2O.
To separate nanogel/siRNA complexes from complexation buffer, 4 volume equivalents of acetone were added to the suspension following complexation completion to induce a polyelectrolyte-ionomer transition.(Liechty et al., 2013) Suspensions were centrifuged at 15,000 rpm for 5 min, supernatant was discarded, and residual solvent was evaporated. Polymer/siRNA complexes were resuspended in the original complexation volume of RNAse free PBS at pH 7.40.
Following resuspension, 100 μL of nanogel/siRNA complexes at 500 μg ml−1 were diluted in PBS to yield a final concentration of 25 μg ml−1. Control wells received 100 μL PBS or 100 μL of the corresponding nanogel-only or siRNA-only solution. Nanogel exposure occurred for designated time points at 37°C or 4°C. Following the exposure period, cells were rinsed 3X with DPBS (with calcium and magnesium) and the media was replaced with 2 mL serum-free DMEM.
For imaging flow cytometry, Hoechst 33342 was added to each well for nuclear staining at a final concentration of 2.5 μg ml−1. The nuclear staining process was performed for 45 min for Caco-2 cells at 37°C, 5% CO2. Following Hoechst incubation, cells were rinsed 3X with DPBS (without calcium and magnesium). No nuclear stain was used in conventional flow cytometry experiments. Caco-2 cells were isolated by replacing the final DPBS wash with 500 μL 0.25% trypsin-EDTA and incubating at 37°C, 5% CO2 for 8 min. Trypsin was neutralized by adding 3 mL DMEM (without phenol red) supplemented with 10% FBS. Cell suspensions were centrifuged for 5 min at 500 g. The supernatant was discarded and the cell pellet re-suspended in flow cytometry buffer. All cell suspensions were kept on ice until analysis. Propidium iodide (PI) was used as a live/dead discriminator and was added to cell suspensions immediately before analysis at a final concentration of 1 μg mL−1.
2.6.1. Flow Cytometry
Efficiency of PDETB30- and PDESSB30-mediated Cy3-siRNA delivery was compared using a BD FACSCalibur (San Jose, CA) flow cytometer equipped with lasers at 488 nm and 635 nm. Fluorescence data were collected using FL-2 (570 – 600 nm, Cy3) and FL-3 (653 - 669 nm, PI). Dead or dying cells were identified with propidium iodide. Typically, 40,000 cells were collected per sample.
2.6.2. Multi-spectral Imaging Flow Cytometry
Analysis of uptake mechanisms and siRNA delivery was conducted using an Amnis Image Stream (Seattle, WA) imaging flow cytometer equipped with lasers at 405 nm, 488 nm, 658 nm, and 785 nm. Fluorescence data were collected using Channel 1 (430 – 505 nm, Hoechst), Channel 2 (505 – 595 nm, OG488), Channel 4 (595 – 660 nm, PI), Channel 5 (660 – 745 nm, DY647), and Channel 6 (745 – 800 nm, side scatter). Brightfield images were collected in Channel 3.
Cells were imaged with a 60X objective. Fluid velocity was set to a nominal value of 40 mm/sec. Fluorescence compensation matrices were constructed using Amnis IDEAS® software and verified manually for proper fit. At least 5,000 cells were collected for analysis. Dead cells (PI positive) were excluded from analysis. Out-of-focus cells were also excluded from further analysis by gating the Gradient RMS feature in IDEAS® software. Typically, cells with Gradient RMS value <40 were considered out of focus.
2.7. siRNA-Mediated Gene Silencing
GAPDH Positive Control siRNA, KD Alert Assay Kits, and 10X Phosphate Buffered Saline (RNAse free) were purchased from Life Technologies (Carlsbad, CA). Caco-2 cells were seeded in tissue-culture treated 96-well plates at 2,500 cells/well and allowed to equilibrate 24 hours before use. GAPDH siRNA-loaded PDETB30 or PDESSB30 nanogels were prepared as discussed before. Following 60 min incubation in complexation buffer, nanogel/siRNA complexes were precipitated through the addition of acetone and centrifuged at 15,000 rpm for 5 min. Supernatant was discarded and complexes were resuspended in RNAse-free PBS. Prior to use, Caco-2 cells were washed with PBS and media replaced with serum free DMEM. Concentrated (20X) nanogel/siRNA complexes or control suspensions were added to test wells and incubated at 37°C, 5% CO2 for 60 min. Following the exposure period, cells were washed 3X with pre-warmed PBS and media replaced with complete DMEM. Cells were incubated at 37°C, 5% CO2 prior to conducting the KD Alert gene silencing assay according to the manufacturer’s instructions. Care was taken to adjust the microplate reader sensitivity to remain within the GAPDH enzyme calibration curve established according to the manufacturer’s instructions.
2.8. Statistical Analysis
Statistical comparisons between experimental and control groups were made with two-tailed, unpaired, Student’s t-tests. Differences were accepted as statistically significant with p < 0.05.
3. Results and Discussion
3.1. Degradable Nanogel Synthesis and Characterization
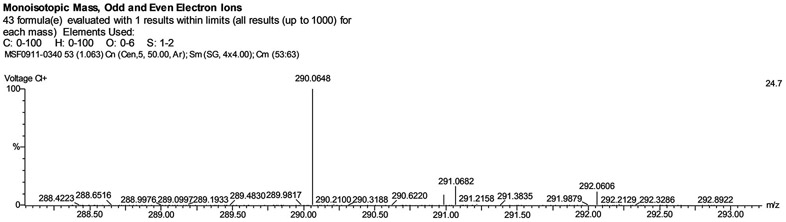
The homobifunctional crosslinker 2-bis-(2-methacryloyloxyethyl disulfide) was successfully synthesized to endow responsive DEAEMA-based nanogels with a mechanism for biodegradation, namely reductive cleavage of the disulfide bonds. The synthesis and purification of 2-bis-(2-methacryloyloxyethyl disulfide) (SSXL) resulted in a molar yield of approximately 50%. Analysis of the mass spectra (Figure 2) reveals a product of the expected molecular weight, 290 Da. The structure of SSXL was further confirmed with 1H-NMR. The spectra, shown in Supplementary Figure 1, show peaks at δ = 1.95 ppm (1H, H2C=C-), 2.96 ppm (1H, H2C=C-), 4.45 (2H, CH2-CH2-S-), 5.60 ppm (2H, -O-CH2-CH2-), and 6.14 ppm (3H, CH3-C=).
Figure 2. Mass spectrum of purified bis(2-methacryloyloxyethyl) disulfide.
Anticipated molecular weight of the bis(2-methacryloyloxyethyl) disulfide = 290.4 g/mol.
Disulfide-crosslinked nanogels containing an ionizable core of DEAEMA-co-TBMA and PEG corona were successfully synthesized via photoemulsion polymerization. This synthesis was conducted as described in our previous reports (Fisher et al., 2008; Fisher and Peppas, 2009; Liechty et al., 2013) and resulted in a similar yield (~1.2 g of nanogel) to previous syntheses. Replacing the non-degradable linker TEGDMA (i.e. PDETB30) with SSXL had no identifiable change on physicochemical properties like pH-dependent swelling, ζ-potential, and size, which are consistent with previously reported results.(Forbes and Peppas, 2014a; Liechty et al., 2011; Liechty et al., 2013) After confirming that the physicochemical characteristics of the nanoscale hydrogels were unaffected by the change in crosslinker, studies were conducted to quantify the kinetics and extent of degradation in response to glutathione, a reductive tripeptide. A semi-quantitative, real-time measurement of degradation of SSXL-crosslinked nanoscale hydrogels was performed in a reductive aqueous suspension. The experiments were conducted using a Malvern ZetaSizer Nano ZS with the sample cell set to 37°C and measurements collected every 3 minutes. After a brief equilibration period, sample cuvettes were injected with PBS or aqueous glutathione to bring the final concentration to 1 mM or 10 mM glutathione in PBS.
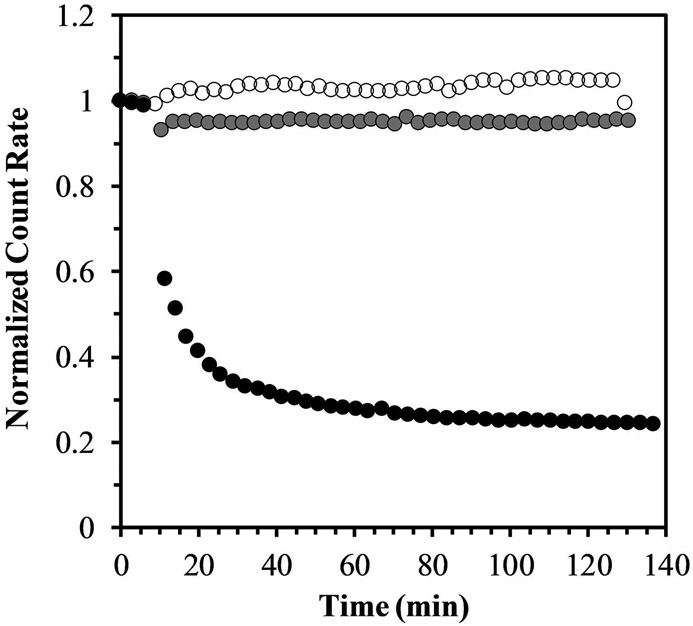
The glutathione-dependent polymer degradation analysis is shown in Figure 3. The presence of the degradable crosslinker SSXL in the PDESSB30 allows the synthesized nanogels to degrade in the presence of glutathione (10 mM solution) after 5 minutes of exposure, as demonstrated by the reduction on the particle count rate. This effect is not observed in the PDESSB30 when subjected to PBS-only. Furthermore, there is a glutathione-concentration dependence, since the polymer degradation does not occur in the presence of a 1 mM glutathione solution. This response confers the SSXL-crosslinked nanogels the ability to preserve their structural integrity in the extracellular milieu, and degrade upon entering the intracellular environment.
Figure 3. Light scattering analysis of glutathione-induced nanogel degradation.
PDESSB30 nanogels dissolved in PBS (at pH 7.4) and exposed to 1 mM (gray) and 10 mM (black) concentrations of glutathione (GSH) and incubated at 37°C. Normalized count rates are reduced following glutathione exposure as nanogel degradation occurs. As a control, nanogels suspended only in PBS maintained their count rate constant throughout the experiment (white).
3.2. Fluorescent Nanogel Synthesis and Characterization
To enable visualization of nanogel subcellular localization in siRNA delivery experiments, a fluorescent version of the PDESSB30 nanogel was necessary. A primary amine-containing analogue of PDESSB30, termed PDESSB30f, was successfully synthesized and purified.
Oregon Green 488 (OG488), an amine reactive dye, was conjugated to primary amines in the nanogel core. Prior to the conjugation reaction, the primary amine content of PDESSB30f was determined to be 17.0 ± 0.4 μmol g−1, which represents a 11.5% incorporation efficiency. OG488 was subsequently added to PDESSB30f at 1:1 mol ratio of dye to amine. Following dialysis and lyophilization, the Oregon Green 488 functionalization was verified using fluorescence spectroscopy and the percent functionalization was calculated with UV absorbance and comparison to an Oregon Green 488 standard curve.
The fluorescence emission (λex = 465 nm) spectra of the labeled nanogel (PDESSB30-OG488) was consistent to unbound dye. The fluorescent labeling was estimated at 16.9 ± 0.3 μmol g−1 using a standard curve of OG488 in PBS, and at 19.5 μmol g−1 calculated by the sample absorbance at 496 nm and the OG-488 extinction coefficient (ε) of 70,000 L mol−1 cm−1, suggesting near 100% conjugation efficiency.
3.3. siRNA Loading Efficiency of pH-responsive nanogels
The ability of the synthesized nanogels to encapsulate siRNA is a critical factor for this platform to efficaciously deliver therapeutically relevant dosages of RNA to disease sites and initiate gene silencing. Our previous work has shown that PDETB30 nanogels display the most desirable attributes as endosomolytic drug delivery vehicles, but also contain fewer ionizable DEAEMA groups than PDET or PDETBA30.(Liechty et al., 2011; Liechty et al., 2013) Previous work also demonstrated that cationic polymers were able to bind DNA more efficiently with increasing cationic density.(Dufresne et al., 2008) Thus, the RNA binding capacity was evaluated in a high-throughput fashion analogous to that described by Siegwart et al.(Siegwart et al., 2011) RNA binding was evaluated as a function of nanogel composition, RNA:nanogel mass ratio, and complexation time to determine the loading efficiency of each nanogel.
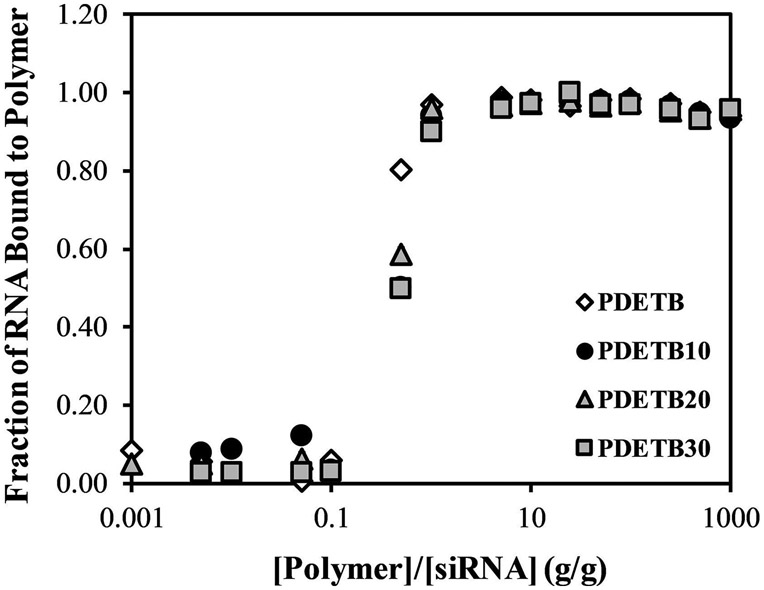
The fraction of free RNA (Ff) was calculated by taking the ratio of fluorescence intensity of sample solutions to fluorescence intensity of a polymer-free control RNA solution. Both measurements were corrected for background fluorescence and the fraction of bound RNA, Fb = 1 - Ff. These results indicate that after a 60 min complexation, all formulations can efficiently bind free siRNA up to a 1:1 mass ratio of polymer and siRNA and this binding is relatively independent of polymer composition as demonstrated in Figure 4. Nearly identical results were obtained for complexation periods of 120 and 180 min (data not shown).
Figure 4. Cationic nanogels were successfully loaded with siRNA.
RNA Loading capacity of poly(DEAEMA-g-PEGMA) (PDET), and poly(DEAEMA-co-BMA-g-PEGMA) (PDETB10), (PDETB20), and (PDETB30) in PBS at pH 5.50. Data represent mean ± S.D. (n = 3).
The positive surface-charge of the evaluated polymer formulations at pH 5.5 may induce the adsorption of the anionic RNA onto the particle surface through electrostatic interactions, instead of the encapsulation of the cargo within the polymer matrix (Forbes and Peppas, 2014b). Therefore, to separate the effects of surface adsorption and electrostatic encapsulation, nanogels, polymer, and siRNA were allowed to complex in the acidic complexation buffer (PBS, pH 5.50) for 60 minutes and were subsequently transferred to 3X volume of serum-free DMEM. By immersing the polymer/siRNA complexes in a more basic solution, the effective surface charge can be reduced from approximately 30 mV to nearly neutral (0 ± 5 mV). This step change in surface charge serves to reduce the electrostatic interactions between the nanogel surface and siRNA, allowing desorption of RNA from the particle surface.
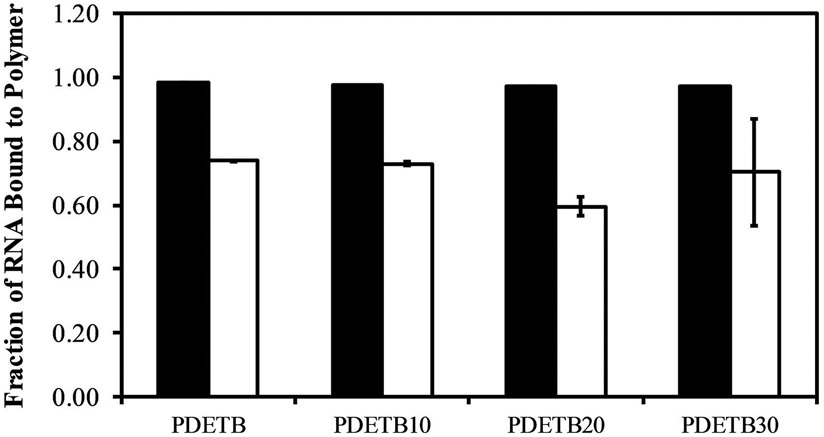
According to the light scattering data presented in previous studies (Liechty et al., 2013; Liechty et al., 2018), this pH-level will completely or partially (depending on nanogel composition) collapse the network structure, serving to entrap RNA in the network core and limit the diffusion out of the particle. As seen in Figure 5 for a 10:1 mass ratio of polymer to siRNA, approximately 70% of the RNA is retained in the bound state following immersion in DMEM in all evaluated formulations.
Figure 5. RNA loading of poly(DEAEMA-g-PEGMA) (PDET), and poly(DEAEMA-co-BMA-g-PEGMA) (PDETB10), (PDETB20), and (PDETB30) in PBS (at pH 5.50, filled) and in serum-free DMEM (at pH 7.40, white).
Polymer and siRNA were combined at a ratio of 10:1 [polymer]:[siRNA]. Fraction of bound RNA determined by Ribogreen assay following 180 min incubation in PBS and 30 min incubation in DMEM. Bars represent the mean ± S.D. (n = 3).
Further confirmation of siRNA and nanogel binding can be seen in Supplementary Figure 2. Complexes of PDETB30-OG488 and DY647-siRNA were visualized using Image Stream cytometry. As expected, the siRNA-loaded nanogels are too small to visualize with brightfield microscopy, but the fluorescence signal from PDETB30-OG488 and DY647-siRNA was visible. As seen in the representative figure showing DY647 vs. OG488 intensity plot presented in Supplementary Figure 2, nearly all PDETB30-OG488 nanogels contain DY647-siRNA. Analogous observations were made for complexes of PDESSB30-OG488 and DY647-siRNA (data not shown).
3.4. Nanogel-mediated delivery of siRNA
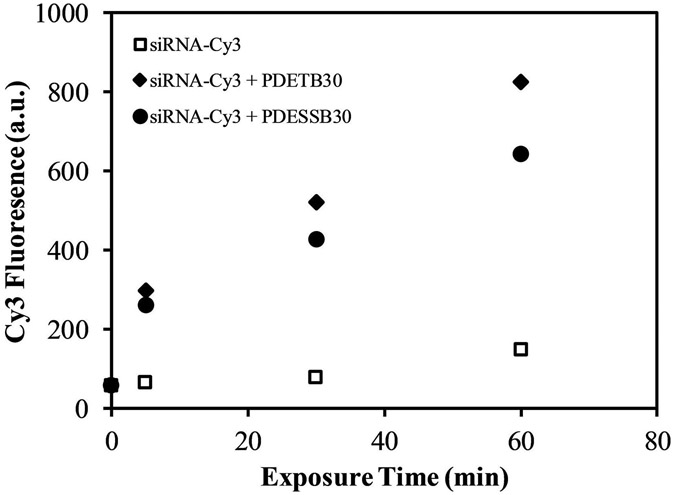
The ability of PDETB30, and its degradable analogue PDESSB30, to deliver fluorescent siRNA to Caco-2 cells was evaluated using flow cytometry. Figure 6 shows the influence of exposure time on uptake of Cy3-siRNA. As expected, naked siRNA is not able to efficiently enter Caco-2 cells. However, by complexation with PDETB30 or PDESSB30, the median fluorescence intensity is increased 5X after 5 min of exposure. Notably, siRNA delivery via PDETB30 and PDESSB30 results in a rapid increase in siRNA fluorescence from 0 – 5 min, followed by an approximately linear increase in median fluorescence from 5 min – 60 min.
Figure 6. siRNA delivery to Caco-2 cells as a function of incubation time.
Nanogel/Cy3-siRNA complexes were prepared at a 20:1 nanogel/siRNA ratio (g/g) and incubated with cells for designated time points. Data points represent the median fluorescence of live cells as determined by flow cytometry. Dead cells were excluded via propidium iodide.
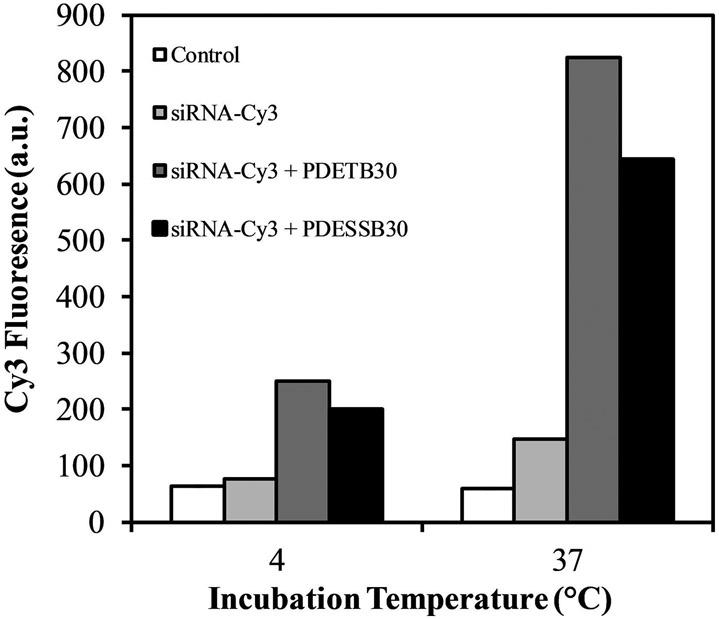
As discussed in our previous work, the mechanism of cationic nanogel internalization in Caco-2 cells is primarily macropinocytosis. This is an energy dependent internalization pathway and the uptake of nanogel/Cy-3siRNA complexes is predictably inhibited at 4°C (Figure 7). Moreover, these data corroborate the observations of previously reported results, whereby PDETB30 can be internalized through an energy-independent pathway (Liechty et al., 2018). Following 60 min uptake at 4°C, cells exposed to PDETB30/Cy3-siRNA and PDESSB30/Cy3-siRNA exhibit greater median fluorescence than cells exposed to Cy3-siRNA alone. These data suggest that PDETB30 and PDESSB30 are capable of delivery of siRNA to Caco-2 cells. This work served as the basis for further study of the intracellular distribution of fluorescent nanogel/siRNA complexes and the evaluation of the gene silencing activity of encapsulated siRNA.
Figure 7. siRNA delivery to Caco-2 cells as a function of nanogel composition and incubation temperature.
Nanogel/Cy3-siRNA complexes were prepared at a 20:1 nanogel/siRNA ratio (g/g) and incubated with Caco-2 cells for 60 min. Data points represent the median fluorescence of live cells as determined by flow cytometry. Dead cells excluded via propidium iodide.
3.5. Internalization analysis of nanogel/siRNA complexes
Multi-spectral imaging flow cytometry was used to simultaneously acquire statistical flow cytometry data and high-resolution fluorescence micrographs. Additionally, this analysis allowed the sorting and gating of events with particular image features, specifically cellular internalization (vs. surface adsorption) or probe colocalization.
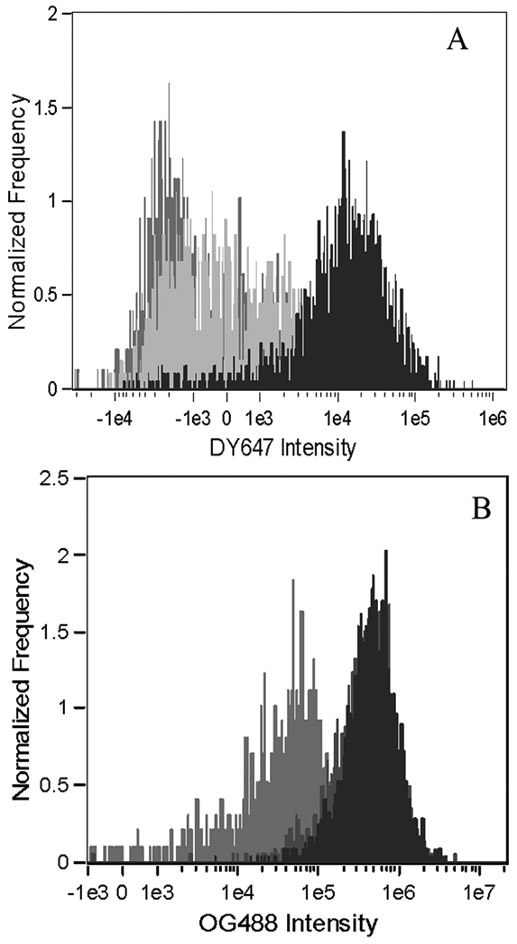
As shown in Figure 8, panel A, PDETB30-OG488 is an efficient delivery vehicle for DY647-siRNA in Caco-2 cells. Both PDETB30-OG488 and PDESSB30-OG488 (data not shown) enhance the cytoplasmic fluorescence of DY647-siRNA relative to the siRNA only (light gray histogram) and untreated control (gray histogram) cells. Figure 8, panel B shows the OG488 fluorescence intensity histogram in untreated (gray), PDETB30-OG488 (dark grey), and PDETB30-OG488/DY647-siRNA (black) treated samples. Notably, the fluorescence signal in the cells treated with nanogels only is greater than that of cells treated with nanogels/siRNA. This suggests that the internalization of nanogel/siRNA complexes is less efficient than nanogels alone.
Figure 8. Fluorescence intensity of Caco-2 cells in siRNA delivery experiments.
Fluorescence intensity histograms of DY647-siRNA and PDETB30-OG488 are shown in Panels A and B, respectively. Fluorescence histograms generated from live, focused, single cells exposed to PBS (untreated, gray), DY647-siRNA alone (light gray), PDETB30-OG488 alone (dark gray), or PDETB30-OG488/DY647-siRNA (black). Data represent the results of two pooled experiments.
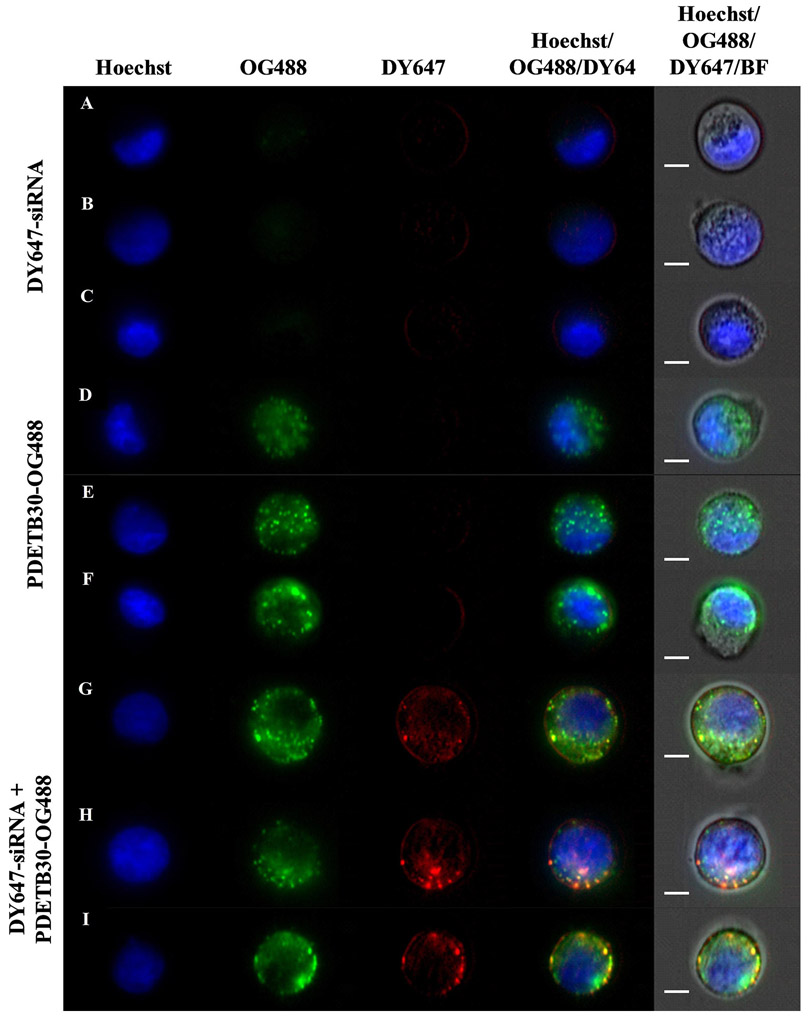
Figure 9 shows representative fluorescence micrographs of Caco-2 cells. Cell nuclei are shown in blue (Hoechst), PDETB30-OG488 in green, and siRNA in red (DY647). Areas of nanogel/siRNA colocalization appear yellow on the fluorescence overlay. Panels A-C show representative images of cells exposed only to 100 nM DY647-siRNA for 60 min, panels D-F show representative images of cells exposed only to 25 μg mL−1 PDETB30-OG488 for 60 min, and panels G – I show representative images of cells exposed 25 μg mL−1 PDETB30-OG488 and 100 nM DY647-siRNA for 60 minutes. As expected due to its high MW (~13 kDa) and negative charge, little to no internalization was observed by the naked siRNA. In panels G – I (PDETB30-OG488/DY647-siRNA), the siRNA staining pattern appears mostly diffuse and distributed near the cell membrane. Several bright, colocalized spots appear in panels H and I, suggesting vesicular entrapment of polymer/siRNA complexes. Vesicular entrapment, and/or lysosomal accumulation, is expected in a portion of the nanogel/siRNA complexes due to the reliance on macropinocytosis by Caco-2 cells to internalize PDETB30-OG488.
Figure 9. Representative fluorescent micrographs of DY647-siRNA delivery to Caco-2 cells.
Nuclear stain (Hoechst 33342, blue), PDETB30-OG488 (Oregon Green 488, green), and DY647-siRNA (DyLight 647, red) are shown. Three representative samples of Caco-2 cells exposed to DY647-siRNA alone (A-C), PDETB30-OG488 alone (D – F), or PDETB30-OG488/DY647-siRNA (G – I) are shown. Images sampled from median region of DY647 histogram. Scale bar represents 7 μm.
3.6. siRNA-Mediated Gene Silencing
The efficacious delivery of functional siRNA to model cells lines is an important step in the assessment of polymeric delivery systems. In these studies, GAPDH was chosen as the target gene for siRNA knockdown. GAPDH is a well-known housekeeping gene, ubiquitously expressed in nearly all cell types, and is involved in the reduction of NAD+ to NADH in the glycolysis pathway (Tristan et al., 2011; Zainuddin et al., 2010). Knockdown was assessed by using a KDAlert™ GAPDH Assay Kit and monitoring the increase in fluorescence (em:520/ex:590) over a 4 minute period.
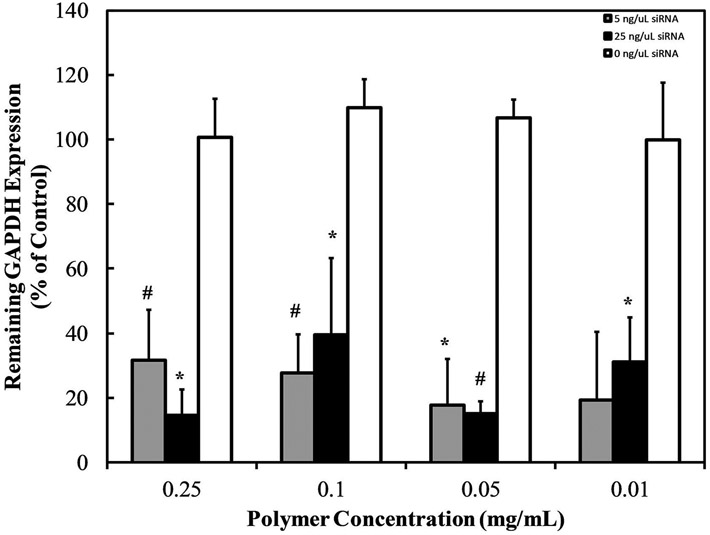
GAPDH knockdown, shown in Figure 10, reveals that GAPDH siRNA delivered via PDETB30 induces a robust gene silencing effect, reducing GAPDH expression by 60 – 85%. This knockdown effect occurred at multiple polymer:siRNA ratios ranging from 8:1 – 1000:1. These effects were not observed upon exposure to the non-specific siRNA negative control.
Figure 10. GAPDH knockdown in Caco-2 cells following exposure to PDETB30/siRNA.
Cells exposed to siRNA-loaded nanogels at designated concentrations of PDETB30 and 250 ng mL−1 (~20 nM, gray) or 1250 ng mL−1 (~100 nM, black) GAPDH siRNA, or siRNA negative control (white). Expression levels measured 48 hrs after transfection. Bars represent the mean of % remaining GAPDH expression ± S.D. (n = 3). * p < 0.05, # p < 0.01.
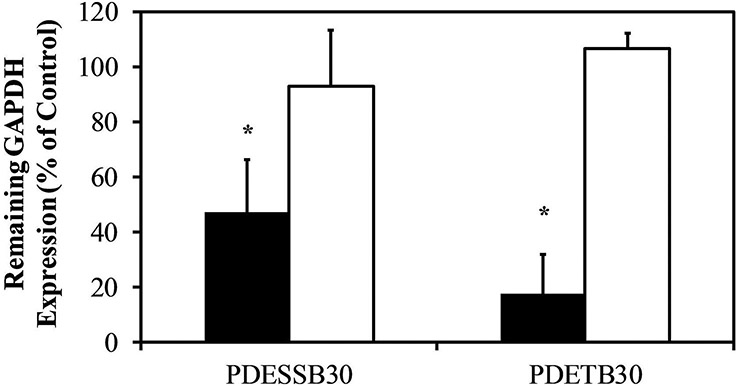
Furthermore, evaluation between the efficacy of siRNA-mediated gene silencing between Caco-2 cells treated with PDETB30/siRNA or PDESSB30/siRNA at a 200:1 nanogel:siRNA ratio was performed as shown in Figure 11. Both nanogel formulations were capable of delivering functional siRNA. Knockdown efficiency was 53% for cells treated with PDESSB30/siRNA and 83% for cells treated with PDETB30/siRNA. Knockdown was not observed upon exposure to the non-specific siRNA negative control. Based on flow cytometry data in Figure 5, Caco-2 cells treated with PDETB30/siRNA exhibited characteristically higher siRNA fluorescence than did cells treated with PDESSB30/siRNA. Therefore, the superior knockdown efficiency of PDETB30/siRNA relative to PDESSB30/siRNA is likely due to increased siRNA delivery efficiency by the former combination.
Figure 11. GAPDH knockdown in Caco-2 cells following exposure to PDETB30/siRNA or PDESSB30/siRNA.
Cells exposed to siRNA-loaded nanogels at 200:1 nanogel:siRNA ratio (g/g, 50 μg mL−1 nanogel and 0.25 μg ml−1 siRNA). Expression levels measured 48 hrs after transfection. Non-specific siRNA was used as a negative control (white). Bars represent the mean of % remaining GAPDH expression ± S.D. (n = 3). * p < 0.05.
In this work we have shown that GAPDH siRNA and KD Alert assays confirmed the ability of the synthesized nanogels to induce gene silencing in Caco-2 cells. Based on these results, further assessment of the gene silencing capabilities of this platform are necessary (i.e. qPCR), before the in vivo evaluation of these systems.
4. Conclusions
A disulfide crosslinker was synthesized to allow degradation of pH-responsive nanogels in reductive environments. This crosslinker was incorporated into responsive nanogels, termed PDESSB30, without significant changes on their physicochemical properties. These nanogels degrade within minutes upon exposure to physiological levels of glutathione as determined by light scattering. It was shown that both, degradable (PDESSB30) and non-degradable (PDETB30) nanogels, are capable of delivering functional siRNA to Caco-2 cells, achieving gene silencing of 47% and 83%, respectively. The combination of attractive physicochemical properties and siRNA delivery efficiency make PDETB30 and PDESSB30 attractive candidates for further development as therapeutic siRNA delivery systems.
Supplementary Material
Figure 1. Cationic nanogel degradation in response to the presence of glutathione.
Disulfide crosslinks within the polymer core are sensitive to reductive conditions.
5. Acknowledgments
This work was supported in part by the National Institutes of Health Grants R01-EB022025-2, R01-EB00246-18, and National Science Foundation Grant 1033746, the William “Tex” Moncrief, Jr. Foundation, the Pratt Foundation, and the Cockrell Family Regents Chair. W.B.L. acknowledges the U.S. National Science Foundation for a Graduate Research Fellowship. Mass spectrometry studies were performed courtesy of the Mass Spectrometry Facility in the Department of Chemistry and Biochemistry, The University of Texas at Austin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
7. REFERENCES
- Antimisiaris S, Mourtas S, Papadia K, 2017. Targeted si-RNA with liposomes and exosomes (extracellular vesicles): How to unlock the potential. International Journal of Pharmaceutics 525, 293–312. [DOI] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, Ostroff GR, Czech MP, 2009. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature 458, 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS, 2009. Development of a novel endosomolytic diblock copolymer for siRNA delivery. Journal of Controlled Release 133, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A, 2010. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne M-H, Elsabahy M, Leroux J-C, 2008. Characterization of Polyion Complex Micelles Designed to Address the Challenges of Oligonucleotide Delivery. Pharmaceut Res 25, 2083–2093. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC, 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806. [DOI] [PubMed] [Google Scholar]
- Fisher OZ, Kim T, Dietz SR, Peppas NA, 2008. Enhanced Core Hydrophobicity, Functionalization and Cell Penetration of Polybasic Nanomatrices. Pharmaceut Res 26, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher OZ, Peppas NA, 2009. Polybasic Nanomatrices Prepared by UV-Initiated Photopolymerization. Macromolecules 42, 3391–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DC, Peppas NA, 2012. Oral delivery of small RNA and DNA. Journal of Controlled Release 162, 438–445. [DOI] [PubMed] [Google Scholar]
- Forbes DC, Peppas NA, 2014a. Polycationic Nanoparticles for siRNA Delivery: Comparing ARGET ATRP and UV-Initiated Formulations. ACS Nano 8, 2908–2917. [DOI] [PubMed] [Google Scholar]
- Forbes DC, Peppas NA, 2014b. Polymeric Nanocarriers for siRNA Delivery to Murine Macrophages. Macromolecular Bioscience 14, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Gao H, Tsarevsky NV, Matyjaszewski K, 2005. Synthesis of Degradable Miktoarm Star Copolymers via Atom Transfer Radical Polymerization. Macromolecules 38, 5995–6004. [Google Scholar]
- Hariharan D, Peppas NA, 1996. Characterization, dynamic swelling behaviour and solute transport in cationic networks with applications to the development of swelling-controlled release systems. Polymer 37, 149–161. [Google Scholar]
- Khormaee S, Choi Y, Shen MJ, Xu B, Wu H, Griffiths GL, Chen R, Slater NKH, Park JK, 2012. Endosomolytic Anionic Polymer for the Cytoplasmic Delivery of siRNAs in Localized In Vivo Applications. Advanced Functional Materials 23, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe JM, Chen F, Peppas NA, 2015. Enzymatic Biodegradation of Hydrogels for Protein Delivery Targeted to the Small Intestine. Biomacromolecules 16, 962–972. [DOI] [PubMed] [Google Scholar]
- Knipe JM, Strong LE, Peppas NA, 2016. Enzyme- and pH-Responsive Microencapsulated Nanogels for Oral Delivery of siRNA to Induce TNF-α Knockdown in the Intestine. Biomacromolecules 17, 788–797. [DOI] [PubMed] [Google Scholar]
- Liechty WB, Caldorera-Moore M, Phillips MA, Schoener C, Peppas NA, 2011. Advanced molecular design of biopolymers for transmucosal and intracellular delivery of chemotherapeutic agents and biological therapeutics. Journal of Controlled Release 155, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechty WB, Scheuerle RL, Peppas NA, 2013. Tunable, responsive nanogels containing t-butyl methacrylate and 2-(t-butylamino)ethyl methacrylate. Polymer 54, 3784–3795. [Google Scholar]
- Liechty WB, Scheuerle RL, Vela Ramirez JE, Peppas NA, 2018. Uptake and Function of Membrane-destabilizing Cationic Nanogels for Intracellular Drug Delivery. Bioengineering & Translational Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, Levins CG, Love KT, Lee H, Cortez C, Collins SP, Li YF, Jang J, Querbes W, Zurenko C, Novobrantseva T, Langer R, Anderson DG, 2011. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proceedings of the National Academy of Sciences 108, 12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Oishi M, Nagasaki Y, 2009. Enhanced Cytoplasmic Delivery of siRNA Using a Stabilized Polyion Complex Based on PEGylated Nanogels with a Cross-Linked Polyamine Structure. Biomacromolecules 10, 1818–1827. [DOI] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A, 2011. The diverse functions of GAPDH: Views from different subcellular compartments. Cellular Signalling 23, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, 2012. Polymers for siRNA Delivery: Inspired by Viruses to be Targeted, Dynamic, and Precise. Accounts of Chemical Research 45, 1005–1013. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG, 2009. Knocking down barriers: advances in siRNA delivery. Nature Reviews Drug Discovery 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter PM, Cai K, Chen J, Adair CR, Kiefer GE, Athey PS, Gaffney PJ, Buff CE, Robertson JD, Caruthers SD, Wickline SA, Lanza GM, 2006. Targeted PARACEST nanoparticle contrast agent for the detection of fibrin. Magnetic Resonance in Medicine 56, 1384–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainuddin A, Chua KH, Rahim NA, Makpol S, 2010. Effect of experimental treatment on GAPDH mRNA expression as a housekeeping gene in human diploid fibroblasts. BMC Molecular Biology 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.