Abstract
Hepatocellular carcinoma (HCC) is the fifth most common malignancy worldwide and the second most lethal human cancer. A portion of patients with advanced HCC can significantly benefit from treatments with sorafenib, adriamycin, 5-fluorouracil and platinum drugs. However, most HCC patients eventually develop drug resistance, resulting in a poor prognosis. The mechanisms involved in HCC drug resistance are complex and inconclusive. Human transcripts without protein-coding potential are known as noncoding RNAs (ncRNAs), including microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), long noncoding RNAs (lncRNAs) and circular RNA (circRNA). Accumulated evidences demonstrate that several deregulated miRNAs and lncRNAs are important regulators in the development of HCC drug resistance which elucidates their potential clinical implications. In this review, we summarized the detailed mechanisms by which miRNAs and lncRNAs affect HCC drug resistance. Multiple tumor-specific miRNAs and lncRNAs may serve as novel therapeutic targets and prognostic biomarkers for HCC.
Keywords: Hepatocellular carcinoma, Drug resistance, Long non-coding RNA, microRNA
Background
Hepatocellular carcinoma (HCC) is the fifth most prevalent malignancy worldwide, accounting for approximately 90 % of primary liver cancer, characterized by high mortality, recurrence, metastasis and poor prognosis [1]. Due to the lack of symptoms in the early disease stages, most patients were diagnosed at the advanced disease stages with metastasis and poor hepatic reserve, thus losing the opportunity of curative resection. In the last decade, improved drug therapy agents have significantly prolonged the survival of HCC patients with advanced diseases. The commonly used therapeutic regimens include sorafenib, adriamycin (ADM), 5-fluorouracil (5-FU), platinum-containing anti-cancer drugs, camptothecin and gemcitabine [2–4]. However, the acquisition of multi-drug resistance (MDR) to these agents is the Achilles’ Heel in clinical oncology, and may result in a poor prognosis. Intrinsic or acquired drug resistance is defined as the resistance of malignant cells to different structurally and functionally unrelated anticancer drugs. The mechanisms involved in HCC drug resistance are complex and include the increased expression of drug efflux transporters that recognize and pump out anticancer drugs out of tumor cells, redistribution of intracellular accumulation of agents, inactivation of apoptosis signaling pathways, enhanced DNA damage repair capacity, accelerated drug metabolism and activation of cancer stem cells (CSCs) [5–7]. Up to now, however, the precise mechanisms underlying HCC drug resistance remain to be investigated.
Human transcripts without protein-coding potential are known as noncoding RNAs (ncRNAs). According to the length and shape, ncRNAs can be subdivided into the following types: tiny/short ncRNAs, long ncRNAs (lncRNAs) larger than 200 nucleotides (nt) and circular RNA (circRNAs). There are various small ncRNAs, such as microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) [8–16]. NcRNAs were once considered as the “dark matter” or “by-products” during gene transcription. It has been found that ncRNAs are involved in various cellular functions, including proliferation, apoptosis and the cell cycle progression [17, 18]. For instance, snoRNAs are essential for guidance of chemical modifications of other RNAs, whereas tRNAs and rRNAs are critical for protein translation. MiRNAs and lncRNAs play a vital role in regulating gene expression via their fine regulation at various levels, including transcription, translation and protein functions. Multiple miRNAs and lncRNAs not only can serve as biomarkers for diagnosis of multiple cancers, but also are involved in drug resistance [19–26]. Lots of ncRNAs have been found to be abnormally expressed in HCC and to be associated with the invasion, metastasis, drug resistance and radioresistance of HCC cells [19–26]. Among the abnormally expressed ncRNAs, multiple miRNAs and lncRNAs have been found to play essential roles in HCC drug resistance.
In this review, we systematically summarize the literatures on miRNAs and lncRNAs modulating HCC drug resistance as well as the underlying mechanisms, thereby providing insight into the role of ncRNAs as putative biomarkers and/or therapeutic targets of HCC in the future.
MiRNAs and therapy resistance
Currently, drug therapy for HCC includes targeted therapy, chemotherapy and immunotherapy. Molecular targeted drugs include sorafenib, regorafenib, lenvatinib and tivantinib. Chemotherapeutic drugs commonly used in clinics are adriamycin, 5-FU, cisplatin, and oxaliplatin. As for immunotherapy for HCC, it is a novel management choice and principally includes immune checkpoint blockers/monoclonal antibodies against the programmed cell death protein 1 (PD-1), PD-1 ligand (PD-L1) and receptor cytotoxic T lymphocyte antigen-4 (CTLA-4), such as nivolumab, pembrolizumab, MED14736, ipilimumab and tremelimumab.
As a class of endogenous, small, single-stranded ncRNAs of 19–24 nucleotides (nt) in length, miRNAs could bind to the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs) and suppress their expression and/or prohibit the translation. miRNAs are commonly involved in cellular differentiation, proliferation, death, angiogenesis and metabolic stress responses [27–29]. Dysregulated miRNAs act as oncogenes or tumor suppressors in the development and progression of many cancers, including HCC [30–32]. Interestingly, significantly altered miRNA expression have been found in a variety of drug-resistant HCC cells compared to those in drug-sensitive cells, suggesting that prediction of efficacy of different drugs using various miRNAs may promote individualized HCC therapy [25, 33–36]. miRNAs can regulate immune checkpoint molecules expression in tumor microenvironment [37]. However, it is still unclear how miRNAs are involved in resistance to immune checkpoint blockers currently. Moreover, emerging evidences indicate that multiple miRNAs involved in sorafenib resistance. By contrast, few miRNAs have been found to be participated in resistance to other novel targeted drugs.
MiRNAs and resistance to sorafenib
Sorafenib, an oral multikinase inhibitor, was initially demonstrated to suppress proliferation and angiogenesis by targeting BRAF, Raf-1, Flt3, VEGFR-2/3 and PDGFR-β. Subsequently, it was also indicated that sorafenib could target signaling pathways independent of Raf, especially pathways regulating apoptosis and cell cycle progression [38–40]. As an FDA-approved standard targeted therapy agent for HCC, sorafenib showed survival benefits in advanced HCC patients worldwide. However, most patients eventually progressed to drug resistance diseases. Currently, the mechanisms involved in sorafenib resistance include activation of EGFR, c-Jun and/or AKT, microenvironmental hypoxia, epithelial-mesenchymal transition (EMT), activation of CSCs, resistance to apoptosis, cell cycle dysregulation, autophagy, and the abnormal expression of miRNAs and lncRNAs [36, 41–48]. A variety of miRNAs have been reported to be involved in sorafenib resistant in HCC (Table 1).
Table 1.
MiRNAs and sorafenib resistance in HCC
| MiRNAs | Expression1 | Pathway | Reference |
|---|---|---|---|
| miR-21 | up-regulated | PTEN/Akt | [49] |
| miR-93 | up-regulated | PTEN; CDKN1A | [50] |
| miR-216a /217 | up-regulated | PTEN; SMAD7 | [51] |
| miR-181a | up-regulated | RASSF1 | [52] |
| miR-494 | up-regulated | PTEN | [53] |
| mTOR | [45] | ||
| miR-221 | up-regulated | caspase-3 | [46] |
| miR-122 | down-regulated | IGF-1R | [54] |
| IGF-1R; SRF; ADAM10 | [55] | ||
| PDK4 | [56] | ||
| SLC7a1 | [57] | ||
| GALNT10 | [58] | ||
| miR-34a | down-regulated | Bcl-2 | [59] |
| miR-27b | down-regulated | P53 | [60] |
| let-7 | down-regulated | BCL-XL | [61] |
| miR-193b | down-regulated | MCL1 | [62, 63] |
| miR-486 | down-regulated | CLDN10; CITRON | [64] |
| miR-367-3p | down-regulated | Androgen receptor | [65] |
| miR-338-3p | down-regulated | HIF-1α | [43] |
| miR-137 | down-regulated | ANT2 | [66] |
| miR-142-3p | down-regulated | ATG5/ATG16L1 | [36] |
| miR-7 | down-regulated | TYRO3 | [67] |
1miRNAs either up-regulated or down-regulated in sorafenib resistant hepatocellular carcinoma cells
Note: This table shows 18 miRNAs, their expression level and potential targets in sorafenib resistance of hepatocellular carcinoma
Several oncogenic miRNAs can promote sorafenib resistance [45, 49–53]. For example, miR-93, miR-216a and miR-217 could confer sorafenib resistance by targeting cell cycle protein-dependent kinase 1A (CDKN1A) and modulating apoptosis as well as the TGF-β signaling [50, 51]. Additionally, miR-181 can trigger sorafenib resistance by targeting and suppressing Ras association domain family member 1 (RASSF1) [52]. Exogenous expression of miR-494 has been found to increase resistance of HCC to sorafenib via targeting PTEN and activating the mTOR signaling. In animal models, sorafenib combined with anti-miR-494 potentiated the sensitivity of HCC to sorafenib, suggesting that miR-494 is a possible therapeutic target of advanced HCC [45, 53]. It was also found that miR-221 potentiates sorafenib resistance by suppressing caspase-3 mediated apoptosis in vivo. Moreover, its expression in serum has been found to be significantly lower in sorafenib responders compared to non-responders, suggesting that miR-221 may be a candidate biomarker to predict responders to sorafenib [46].
Meanwhile, multiple tumor suppressor miRNAs can reverse sorafenib resistance in HCC. MiR-122, for instance, has been found to be the most highly expressed miRNA in normal liver and remarkably reduced in sorafenib-resistant HCC cells. MiR-122 could promote sorafenib sensitivity of HCC cells through targeting and suppressing insulin-like growth factor 1 receptor (IGF-1R), serum response factor (SRF), depolymerization and metalloproteinase domain-containing protein 10 (ADAM10), pyruvate dehydrogenase kinase 4 (PDK4), solute carrier family 7 member 1 (SLC7A1) and peptide N-acetyl N-acetylgalactosaminyl transferase 10 (GALNT10) [54–58]. Similarly, miR-34a has been found to reverse the tolerance of HCC cells to sorafenib via silencing BCL-2 [59]. MiR-27b, let-7 and miR-193b which are curial regulators of apoptosis, also have been demonstrated to enhance sensitivity of HCC cells to sorafenib by silencing P53, Bcl-XL and/or myeloid leukemia cell differentiation protein (MCL1), respectively [60–63]. In addition, the tumor suppressor miR-486 and miR-367-3p have been shown to promote sorafenib sensitivity by inhibiting CITRON, claudin 10 (CLDN10) and androgen receptor (AR) [64, 65]. It has been reported that miR-338-3p can sensitize HCC cells to sorafenib by silencing hypoxia inducing factor 1α (HIF-1α) in vitro and in vivo [43]. MiR-137 has been found to be significantly down-regulated in sorafenib-resistant Huh7-R HCC cells. Exogenous miR-137 can promote sorafenib sensitivity and inhibit cancer initiation cell (CIC) phenotype through targeting adenine nucleotide transporter 2 (ANT2) [66]. Ectopic expression of miR-142-3p, a novel autophagy regulator, can sensitize HCC cells to sorafenib by silencing autophagy related 5 (ATG5) and autophagy related 16-like 1 (ATG16L1) and, thus, promote autophagy induced by sorafenib [36]. In addition, the tumor suppressor miR-7 has been found to promote sorafenib sensitivity in HCC cells through silencing expression of TYRO3, a member of TYRO3-AXL-MER family of receptor tyrosine kinases [67].
MiRNAs and resistance to adriamycin
Adriamycin, an anthracycline antibiotic and non-specific periodic drug, is a strong inhibitor of DNA and RNA synthesis in tumor cells during proliferation. Adriamycin could diffuse into the nucleus of HCC cells, interact with DNA and eventually induce apoptosis. There are some known mechanisms for development of adriamycin resistance of HCC [68–75]. Multiple miRNAs have been reported to be involved in adriamycin resistance in HCC (Table 2).
Table 2.
MiRNAs and adriamycin resistance in HCC
| MiRNAs | Expression1 | Pathway | Reference |
|---|---|---|---|
| Let-7a | up-regulated | Caspase–3 | [76] |
| miR-519d | up-regulated | CDKN1A/p21, PTEN, AKT3, TIMP2 | [77] |
| miR-26a/b | down-regulated | ULK1 | [78] |
| miR-26b | down-regulated | TAK1, TAB3 | [79] |
| miR-520b | down-regulated | ATG7 | [80] |
| miR-491-3p | down-regulated | Sp3/ABCB1 | [81] |
| miR-122 | down-regulated | MDR1 | [82] |
| ABCB1; ABCF2 | [83] | ||
| PKM2 | [84] | ||
| miR-31 | down-regulated | NDRG3 | [85] |
| miR-223 | down-regulated | ABCB1 | [86] |
| miR-133a,miR-326 | down-regulated | ABCC1 | [87] |
| miR-101 | down-regulated | EZH2 | [88] |
| Mcl-1 | [89] | ||
| miR-199a-3p | down-regulated | mTOR, c-Met | [90] |
| miR-215 | down-regulated | DHFR, TS | [91] |
| miR-145 | down-regulated | Smad3 | [92] |
| miR-503 | down-regulated | MDR1, MRP, ERCC1, Bcl-2 | [93] |
1miRNAs either up-regulated or down-regulated in adriamycin resistant hepatocellular carcinoma cells
Note: This table shows 17 miRNAs, their expression level and potential targets in adriamycin resistance of hepatocellular carcinoma
Several oncogenic miRNAs can promote adriamycin resistance. Let-7a has been found to increase resistance of HepG2 cells to adriamycin [76]. Exogenous expression of miR-519d has also been found to increase resistance of HCC cells to adriamycin by targeting multiple tumor suppressor genes, including p21 and PTEN [77].
By contrast, it has been found that many tumor suppressor miRNAs can reverse adriamycin resistance in HCC. MiR-26a/b, for instance, has been found to be significantly down-regulated in 30 HCC tissues compared to normal tissues. In addition, exogenous miR-26a/b expression has been found to promote adriamycin sensitivity of HCC cells by targeting ULK1 expression as well as the autophagy signaling pathway in vivo and in vitro [78]. Moreover, miR-26b can also sensitize HCC cells to adriamycin through silencing the TAK1 and TAB3 pathways [79]. Similarly, miR-520b has been found to increase the sensitivity of BEL-7402/ADM HCC cells to adriamycin via silencing expression of ATG7, a key autophagy regulator [80]. In HCC cells, expression of miR-491-3p are inversely associated with expression of ABCB1 or Sp3. Consistently, ectopic expression of miR-491-3p could sensitize HCC cells to adriamycin by silencing expression of ABCB1 or Sp3 [81]. MiR-122, a highly expressed liver-specific miRNA in normal liver tissue, was significantly down-regulated in HCC. The tumor suppressor miR-122 has been found to promote adriamycin sensitivity in HCC cells through inhibiting cell cycle, anti-apoptotic effector factors and ABC transporters [82–84]. Similarly, miR-31 has been found to increase the sensitivity of HCC cells to adriamycin via silencing the expression of NDRG3 [85]. In addition, down-regulated expression of tumor suppressor miRNAs, such as miR-223, miR-133a, miR-326, miR-101, miR-199a-3p, miR-215, miR-145 and miR-503, are significantly correlated with adriamycin resistance of HCC through inhibiting expression of the MDR-related genes. On the contrary, restoring the expression of these miRNAs could reverse adriamycin resistance of HCC cells [86–93].
MiRNAs and resistance to 5-FU
5-FU, a heterocyclic aromatic chemotherapeutic agent, is a broadly used chemotherapeutic drug for HCC treatment. Through inhibiting thymidylate synthase (TS), 5-FU can interfere DNA replication and, thus, result in cell cycle arrest and apoptosis in response to DNA damage [94, 95]. Multiple oncogenic or tumor suppressive miRNAs have been found to be involved in 5-FU resistance (Table 3).
Table 3.
MiRNAs and 5-FU resistance in HCC
| MiRNAs | Expression1 | Pathway | Reference |
|---|---|---|---|
| miR-200a-3p | up-regulated | DUSP6 | [96] |
| miR-183 | up-regulated | IDH2/SOCS6-HIF-1α | [97] |
| miR-141 | up-regulated | Keap1 | [98] |
| miR-193a-3p | up-regulated | SRSF2 | [99] |
| miR-195 | down-regulated | Bcl-w | [100] |
| miR-125b | down-regulated | Hexokinase II | [101] |
| Let-7 g | down-regulated | HMGA2 | [102] |
| miR-133a, miR-326 | down-regulated | Bcl-xl | [103] |
| miR-503 | down-regulated | EIF4E | [104] |
1miRNAs either up-regulated or down-regulated in resistant hepatocellular carcinoma cells
Note: This table shows 10 miRNAs, their expression level and potential targets in 5-FU resistance of hepatocellular carcinoma
It has been found that several oncogenic miRNAs can promote 5-FU resistance in HCC cells, including miR-200a-3p, miR-183, miR-141 and miR-193a-3p. For example, exogenous expression of miR-200a-3p enhanced 5-FU resistance of HCC cells by silencing dual specificity phosphatase 6 (DUSP6) [96]. There was significantly higher miR-183 expression in 5-FU resistant HCC cells compared to 5-FU sensitive HCC cells. Knockdown of oncogenic miR-183 might significantly reverse the 5-FU tolerance [97]. It has been reported that miR-141 can promote the resistance to 5-FU in HCC cells by inhibiting kelche-like ECH 1-related protein 1 (Keap1) and activating the antioxidant pathway [98]. Moreover, miR-193a-3p has been shown to enhance 5-FU resistance of HCC cells via suppressing serine/arginine-rich splicing factor 2 (SRSF2) [99].
On the contrary, several tumor suppressor miRNAs may be associated with 5-FU resistance. Accumulated evidences indicate that upregulated BCL-2 expression confers 5-FU resistance in HCC. It has been found that miR-195 could inhibit expression of BCL-2, and an increased sensitivity of drug-resistant HCC cells to 5-FU has been observed after exogenous overexpression of miR-195 [100]. Ectopic expression of miR-125b has been found to lead to reduced hexokinase 2 (HK2) protein expression and to sensitize HCC cells to 5-FU by inhibiting glycolysis [101]. 5-FU resistant HCC cells have been found to exhibit reduced expression levels of let-7 g and exogenous expression of let-7 g could significantly sensitize HCC cells to 5-FU [102]. Similarly, miR-133a and miR-326 may restore chemosensitivity of HCC to 5-FU by targeting Bcl-xl [103]. MiR-503, which has been found to be significantly down-regulated in HCC tissues, could lead to significantly increased 5-FU toxicity of HCC cells by suppressing eukaryotic translation initiation factor 4E (EIF4E) [104].
MiRNAs and resistance to cisplatin
Cisplatin, the first-generation of the platinum chemotherapeutic drugs, can inhibit DNA replication and transcription by forming crosslinks between DNA double strands and exhibits broad-spectrum antitumor activity. Cisplatin is one of the most commonly used chemotherapeutic agents to treat advanced HCC. Several miRNAs have been reported to be involved in cisplatin resistance in HCC (Table 4).
Table 4.
MiRNAs and cisplatin resistance in HCC
| MiRNA | Expression1 | Pathway | Reference |
|---|---|---|---|
| miR-130a | up-regulated | RUNX3 | [105] |
| miR-182 | up-regulated | TP53INP1 | [106] |
| miR-340 | down-regulated | Nrf2 | [107] |
| miR-363 | down-regulated | Mcl-1 | [108] |
1miRNAs either up-regulated or down-regulated in cisplatin resistant hepatocellular carcinoma cells
Note: This table shows 4 miRNAs, their expression level and potential targets in cisplatin resistance of hepatocellular carcinoma
Multiple oncogenic miRNAs can promote cisplatin resistance, such as miR-130a and miR-182. Significantly elevated expression of miR-130a and miR-182 has been observed in tumor tissues from HCC patients and cisplatin-resistant Huh7 and HepG2 cells [105, 106]. Exogenous expression of miR-130a led to cisplatin tolerance of Huh7 cells by targeting the tumor suppressor RUNX3 and activating the Wnt/β-catenin pathway [105]. Similarly, inhibition of miR-182 may partially conquer cisplatin resistance in cisplatin-resistant HepG2 cells by inhibiting tumor suppressor tumor protein 53-induced nucleoprotein 1 (TP53INP1) [106].
By contrast, multiple tumor suppressor miRNAs can reverse cisplatin resistance of HCC. MiR-340, for instance, has been found to be significantly down-regulated in cisplatin-resistant HCC cells. In addition, exogenous miR-340 expression has been found to sensitize chemo-resistant HepG2/CDDP cells to cisplatin through silencing Nrf2 expression, as well as the antioxidant pathway [107]. Additionally, miR-363 has been found to be significantly down-regulated in cisplatin resistant HepG2 cells compared to parental cells, and exogenous miR-363 could significantly reverse cisplatin tolerance in HepG2 cells by targeting Mcl-1 [108].
MiRNAs and resistance to other drugs
Paclitaxel, cetuximab and etoposide are also commonly used to treat HCC [109–111]. As shown in Table 5, several miRNAs are involved in resistance to these drugs. The tumor suppressor miR-16 has been found to be down-regulated in HCC tissues and to sensitize HCC cells to paclitaxel by suppressing the expression of IKBKB as well as the NF-κB signaling in vitro and in vivo [35]. Similarly, tumor suppressor miR-9 and miR-23a could sensitize HCC to cetuximab and etoposide by inhibiting expression of eukaryotic translation initiation factor 5A2 (eIF-5A-2) and topoisomerase 1 (TOP1), respectively [112, 113]. As a potential antitumor protein, tumor necrosis factor-related apoptosis inducing ligand (TRAIL) could selectively eliminate various types of HCC cells without exerting toxic effects in normal tissues. MiR-26b and miR-138 have been found to be involved in TRAIL-induced apoptosis and anti-malignancy in HCC. MiR-26b could promote the cytotoxicity of TRAIL in HCC cells by inhibiting Mcl-1 [114]. Similarly, miR-138 was significantly down-regulated in TRAIL resistant HCC cells compared to TRAIL sensitive HCC cells. Ectopic expression of miR-138 has been found to improve the sensitivity of HCC cells to TRAIL [115].
Table 5.
MiRNAs and other drugs resistance in HCC
| MiRNAs | Expression1 | Pathway | Drug | Reference |
|---|---|---|---|---|
| miR-16 | down-regulated | IKBKB | paclitaxel | [35] |
| miR-9 | down-regulated | eIF-5A-2 | cetuximab | [112] |
| miR-23a | down-regulated | Topoisomerase 1(TOP1) | etoposide | [113] |
| miR-26b | down-regulated | Mcl-1 | TRAIL | [114] |
| miR-138 | down-regulated | interferon stimulating gene 15 | TRAIL | [115] |
| miR-93 | up-regulated | PTEN; CDKN1A | tivantinib | [50] |
1miRNAs either up-regulated or down-regulated in other chemo-drugs resistant hepatocellular carcinoma cells
Note: This table shows 6 miRNAs, their expression level and potential targets in chemoresistance of hepatocellular carcinoma
In addition, miRNAs are also involved in resistance to tivantinib and regorafenib, two novel targeted drugs. Tivantinib, a small, selective oral inhibitor of c-Met receptor tyrosine kinase, provides an option as a second-line treatment for advanced HCC patients who have failed or are intolerant to sorafenib. It has been found that oncogenic miR-93 could confer tivantinib resistance by targeting PTEN and CDKN1A [50]. Interestingly, it has been reported that nine plasma miRNAs, including upregulated plasma expression of miR-30a, miR-122, miR-125b, miR-200a, and miR-374b, diminished levels of miR-15b, miR-107, and miR-320b, as well as the absence of miR-645, can predict regorafenib survival of advanced HCC patients [116].
LncRNAs and therapy resistance
LncRNAs are a new class of ncRNAs longer than 200 nt and have no protein coding potential. Based on the positions and characteristics in human genome, lncRNAs can be divided into five categories: sense, antisense, bidirectional, intronic and intergenic lncRNAs [117–120]. Multiple lncRNAs are aberrantly expressed in HCC and are significantly associated with metastasis, recurrence, prognosis and chemoresistance through various mechanisms, including interactions with DNA, RNA or proteins to form complexes that regulate the expression of target genes [118, 121–130]. Several lncRNAs have been found to be involved in drug resistance in HCC (Table 6).
Table 6.
LncRNAs and drug resistance in HCC
| LncRNAs | Expression1 | Pathway | Drugs | Reference |
|---|---|---|---|---|
| NR2F1-AS1 | up-regulated | miR-363-ABCC1 | oxaliplatin | [131] |
| HANR | up-regulated | GSKIP/P-GSK3β | adriamycin | [132] |
| lncARSR | up-regulated | PTEN-PI3K/Akt | adriamycin | [134] |
| HULC | up-regulated | USP22/Sirt1/ autophagy | oxaliplatin;5-FU; pirarubicin | [135] |
| MALAT1 | up-regulated | miR-216b/ autophagy | 5-FU; adriamycin;mitomycin | [136] |
| TUC338 | up-regulated | RASAL1 | sorafenib | [137] |
| VLDLR | up-regulated | ABCG2 | sorafenib | [138] |
1lncRNAs either up-regulated or down-regulated in chemo-resistant hepatocellular carcinoma cells
Note: This table shows 7 lncRNAs, their expression level and underlying pathways in the chemoresistance of hepatocellular carcinoma
It has been reported that lncRNA NR2F1-AS1 is significantly up-regulated in oxaliplatin-resistant HCC tissues and cell lines and confer HCC resistance to oxaliplatin. NR2F1-AS1 can promote tumor progression in vitro and in vivo. ABCC1 protein is a member of the superfamily of ATP-binding cassette (ABC) transporters which is involved in multi-drug resistance. In HCC cells, lncRNA NR2F1-AS1 could induce ABCC1 expression via endogenous sponging miR-363 and, thus, antagonize chemosensitivity to oxaliplatin [131].
LncRNA HANR (HCC associated long non-coding RNA) shows evidently increased expression in HCC tissues and is associated with poor prognosis of HCC patients. Silencing of lncRNA HANR suppresses HCC proliferation in vitro and in vivo, enhances apoptosis and promotes sensitivity to doxorubicin. It has been found lncRNA HANR could bind to GSKIP and suppress the phosphorylation of GSK3β. In hepatocarcinogenesis, suppression of GSK3β phosphorylation and enhanced GSK3β total protein expression regulate glycogen metabolism and cell growth. As a result, alteration of endogenous cellular HANR expression influenced the sensitivity of HCC to doxorubicin-mediated chemotherapy [132].
LncARSR (lncRNA activated in renal cell carcinoma with sunitinib resistance) was firstly reported to be correlated with clinically poor sunitinib response in renal cell carcinoma. It has been found that lncARSR promotes sunitinib resistance via competitively binding miR-34/miR-449 to facilitate AXL and c-MET expression in renal cell carcinoma cells [133]. Recently, LncARSR is also found to play a part in doxorubicin resistance of HCC. LncARSR is significantly upregulated in HCC, associates with large tumor size and advanced disease stage. Overexpression of lncARSR promotes doxorubicin resistance of HCC cells in vitro and in vivo. In HCC cells, lncARSR physically associates with PTEN mRNA, enhances PTEN mRNA degradation, reduces PTEN expression, and activates the PI3K/Akt signaling pathway [134].
It has been found that lncRNA HULC (highly upregulated lncRNA in HCC) could induce autophagy through silencing expression of silent information regulator 1 (Sirt1) protein and weaken the chemosensitivity of oxaliplatin, 5-FU and pirarubicin (THP) in HCC cells. It was also indicated that lncRNA HULC could up-regulate expression of ubiquitin-specific peptidase 22 (USP22) and reduce the ubiquitin-mediated degradation of Sirt1 protein by removing the conjugated polyubiquitin chain of Sirt1 [135]. In addition, lncRNA MALAT1 has been found to be dramatically increased in 5-FU resistant HCC cells of BEL-7402/5-FU and can modulate MDR through impacting autophagy. Knockdown of MALAT1 can reverse 5-FU, adriamycin and mitomycin resistance, diminish LC3-II level and potentiate 5-FU induced apoptosis, which are similar with the effects of autophagy inhibitor of 3-Methyladenine (3-MA) [136].
Therapy resistance to targeted therapy for HCC, i.e. sorafenib, is one of major hurdles in clinics. LncRNA TUC338, for instance, has been found to be involved in the HCC sorafenib resistance. Elevated levels of lncRNA TUC338 were found both in HCC tissues and cell lines. Knockdown of lncRNA TUC338 sensitized HCC cells to sorafenib, which was accompanied by increased expression of RASAL1 [137]. Similarly, lncRNA VLDLR is also involved in sorafenib resistance of HCC cells. VLDLR is one of lncRNAs contained within extracellular vesicles (EV) during chemotherapeutic stress in human HCC. Silencing VLDLR led to the decreased cell viability and altered cell cycle distribution, accompanied with the reduced expression of drug-resistant protein ABCG2 (ATP-binding cassette, subfamily G member 2). Overexpression of ABCG2 protein reversed the effects of VLDLR knockdown on sorafenib-induced cell death [138].
There are several novel lncRNAs have been indicated to play crucial functions in HCC oxaliplatin resistance. Yin et al. evaluated the difference of genome-wide lncRNA expression profiling between oxaliplatin-sensitive and oxaliplatin-resistant HCC cells. A total of 120 differentially expressed lncRNAs were identified, among which 61 lncRNAs were up-regulated and 59 lncRNAs were down-regulated (fold changes > 2, P < 0.05). It was found that ENST00000502804, NR_073453 and ENST00000438347 were also up-regulated in drug-resistant HCC tissues and high expression of ENST00000438347 and ENST00000518376 was associated with bad outcomes of HCC patients [139].
Conclusions and perspectives
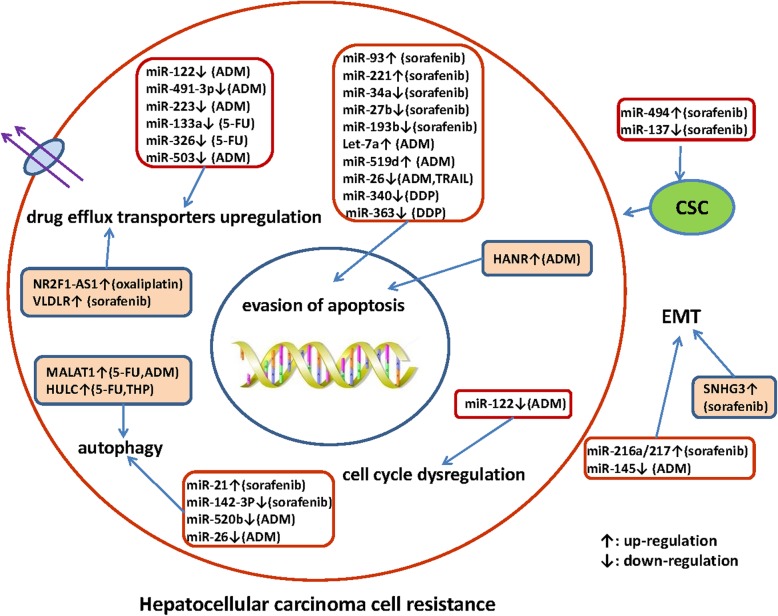
More and more ncRNAs have been identified to be associated with chemoresistance in HCC. The mechanisms underlying the role of ncRNAs in HCC drug resistance are summarized in Fig. 1. Targeting these dysregulated endogenous miRNAs and/or lncRNAs may be a promising way to reverse drug resistance. For example, nanoparticle delivery of synthetic oligonucleotides targeting oncogenic miRNAs or synthetic tumor suppressor miRNAs or administration with natural agents for the regulation of miRNAs have been investigated as a proof-of-concept for HCC treatment. For lncRNAs, it is applicable that the direct delivery of tumor suppressive lncRNAs molecules to the target cells or the knockdown of oncogenic lncRNAs through specific siRNAs or shRNAs against them. Targeting ncRNAs in combination with conventional chemotherapeutics against HCC may be a promising alternative for reversing drug resistance and contributing to a better outcome in advanced HCC patients. However, it remains challenging to select critical target ncRNAs from numerous candidates. In order to promote ncRNA-based therapeutic interventions that benefit HCC patients, further investigations on additional translation research and clinical trials are urgently needed, which may ultimately open up potential approaches for overcoming HCC drug resistance.
Fig. 1.
A summary diagram of miRNAs and lncRNAs involved in the drug resistance of hepatocellular carcinoma (HCC). Various miRNAs and lncRNAs could participate in drug resistance of HCC by affecting cell apoptosis, proliferation, cell cycle, autophagy, epithelial-mesenchymal transition, and cancer stem cell via modulating the expression of downstream target genes
Acknowledgements
Not applicable.
Abbreviations
- 3′-UTR
3′-untranslated region
- 3-MA
3-Methyladenine
- 5-FU
5-fluorouracil
- ABCG2
ATP-binding cassette, subfamily G member 2
- ADAM10
Depolymerization and metalloproteinase domain-containing protein 10
- ANT2
Adenine nucleotide transporter 2
- AR
Androgen receptor
- ATG16L1
Autophagy related 16-like 1
- ATG5
Autophagy related 5
- CDKN1A
Cell cycle protein-dependent kinase 1A
- CIC
Cancer initiation cell
- circRNA
circular RNA
- CSC
Cancer stem cells
- DUSP6
Dual specificity phosphatase 6
- EIF4E
Eukaryotic translation initiation factor 4E
- eIF-5A-2
Eukaryotic translation initiation factor 5A2
- EMT
Epithelial-mesenchymal transition
- GALNT10
Peptide N-acetyl N-acetylgalactosaminyl transferase 10
- HCC
Hepatocellular carcinoma
- HIF-1α
Hypoxia inducing factor 1α
- HK2
Hexokinase 2
- IGF-1R
Insulin-like growth factor 1 receptor
- Keap1
Kelche-like ECH 1-related protein 1
- lncRNA
Long noncoding RNA
- MDR
Multi-drug resistance
- miRNA/miR
microRNA
- mRNA
messenger RNA
- ncRNA
noncoding RNA
- nt
nucleotide
- PDK4
Pyruvate dehydrogenase kinase 4
- piRNA
PIWI-interacting RNA
- RASSF1
Ras association domain family member 1
- rRNA
Ribosomal RNA
- shRNA
Short hairpin RNA
- siRNA
Small interfering RNA
- Sirt1
Silent information regulator 1
- SLC7A1
Solute carrier family 7 member 1
- snoRNA
Small nucleolar RNA
- SRF
Serum response factor
- SRSF2
Serine/arginine-rich splicing factor 2
- THP
Pirarubicin
- TOP1
Topoisomerase 1
- TP53INP1
Tumor protein 53-induced nucleoprotein 1
- TRAIL
Tumor necrosis factor-related apoptosis inducing ligand
- tRNA
Transfer RNA
- TS
Thymidylate synthase
- USP22
Ubiquitin-specific peptidase 22
Author’s contribution
LW and MY conceived the review, acquired data, provided project funding and drafted the manuscript. XW, LL, JL, HX, YS, MX and TL undertook the initial research. NZ was involved in writing and reviewing the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by Science and Technology Development Project of Shandong Province (2016GSF201211) and Shandong Health Department (2016WS0554); National Natural Science Foundation of China (31671300, 31871306); Taishan Scholars Program of Shandong Province (tsqn20161060). The funding bodies did not play any role in writing the manuscript.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nasha Zhang, Email: wownseva@126.com.
Ming Yang, Phone: 86531-67626536, Email: aaryoung@yeah.net.
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Shen YC, Lin ZZ, Hsu CH, Hsu C, Shao YY, Cheng AL. Clinical trials in hepatocellular carcinoma: an update. Liver Cancer. 2013;2(3–4):345–364. doi: 10.1159/000343850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grem JL. 5-fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Investig New Drugs. 2000;18(4):299–313. doi: 10.1023/A:1006416410198. [DOI] [PubMed] [Google Scholar]
- 4.Kalyan A, Nimeiri H, Kulik L. Systemic therapy of hepatocellular carcinoma: current and promising. Clin Liver Dis. 2015;19(2):421–432. doi: 10.1016/j.cld.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12(7):408–424. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 6.Butler EB, Zhao Y, Munoz-Pinedo C, Lu J, Tan M. Stalling the engine of resistance: targeting cancer metabolism to overcome therapeutic resistance. Cancer Res. 2013;73(9):2709–2717. doi: 10.1158/0008-5472.CAN-12-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salehan MR, Morse HR. DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br J Biomed Sci. 2013;70(1):31–40. doi: 10.1080/09674845.2013.11669927. [DOI] [PubMed] [Google Scholar]
- 8.Xing F, Wang S, Zhou J. The expression of MicroRNA-598 inhibits ovarian Cancer cell proliferation and metastasis by targeting URI. Mol Ther Oncolytics. 2019;12:9–15. doi: 10.1016/j.omto.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wang WJ, Li HT, Yu JP, Han XP, Xu ZP, Li YM, et al. A competing endogenous RNA network reveals novel potential lncRNA, miRNA, and mRNA biomarkers in the prognosis of human Colon adenocarcinoma. J Surg Res. 2019;235:22–33. doi: 10.1016/j.jss.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 10.Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8(19):5213–5230. doi: 10.7150/thno.28001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of Colon Cancer. Cancer Epidemiol Biomark Prev. 2018;27(9):1019–1028. doi: 10.1158/1055-9965.EPI-18-0318. [DOI] [PubMed] [Google Scholar]
- 12.Yi C, Wan X, Zhang Y, Fu F, Zhao C, Qin R, et al. SNORA42 enhances prostate cancer cell viability, migration and EMT and is correlated with prostate cancer poor prognosis. Int J Biochem Cell Biol. 2018;102:138–150. doi: 10.1016/j.biocel.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Ma P, Liu P, Chen B, Liu Z. Small nucleolar RNA U2_19 promotes hepatocellular carcinoma progression by regulating Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;500(2):351–356. doi: 10.1016/j.bbrc.2018.04.074. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Zheng J, Chen P, Liu Q, Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;90:705–712. doi: 10.1016/j.biopha.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66(1):107–117. doi: 10.1136/gutjnl-2015-309359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin XG, Zeng JH, Lin P, Mo WJ, Li Q, Feng ZB, et al. Prognostic value of small nuclear RNAs (snRNAs) for digestive tract pan- adenocarcinomas identified by RNA sequencing data. Pathol Res Pract. 2019;215(3):414–426. doi: 10.1016/j.prp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration. EMT and metastasis Int J Cancer. 2016;139(2):269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Jiang Y, Lv LZ. Long non-coding RNA XLOC_010235 correlates with poor prognosis and promotes tumorigenesis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(21):4867–4874. [PubMed] [Google Scholar]
- 20.Tan HY, Zheng YB, Liu J. Serum miR-199a as a potential diagnostic biomarker for detection of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8657–8663. doi: 10.26355/eurrev_201812_16630. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Yang K, Yuan W, Gao Z. Determination of serum Exosomal H19 as a noninvasive biomarker for bladder Cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi: 10.12659/MSM.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu D, Zhan Y, Zhu K, Bai M, Han J, Si Y, et al. Plasma Exosomal Long non-coding RNAs serve as biomarkers for early detection of colorectal Cancer. Cell Physiol Biochem. 2018;51(6):2704–2715. doi: 10.1159/000495961. [DOI] [PubMed] [Google Scholar]
- 23.Gao H, Hao G, Sun Y, Li L, Wang Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001–8012. doi: 10.2147/OTT.S172379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L, Liao Q, Zou Z, Wen Y, Wang J, Liu C, et al. Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric Cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell Physiol Biochem. 2018;51(3):1221–1236. doi: 10.1159/000495499. [DOI] [PubMed] [Google Scholar]
- 25.Que KT, Zhou Y, You Y, Zhang Z, Zhao XP, Gong JP, et al. MicroRNA-31-5p regulates chemosensitivity by preventing the nuclear location of PARP1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37(1):268. doi: 10.1186/s13046-018-0930-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Chen M, Wu L, Tu J, Zhao Z, Fan X, Mao J, et al. miR-590-5p suppresses hepatocellular carcinoma chemoresistance by targeting YAP1 expression. EBioMedicine. 2018;35:142–154. doi: 10.1016/j.ebiom.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan G, Wu H, Du Y, He F. Tumor suppressor role of microRNA-545 in oral squamous cell carcinoma. Oncol Lett. 2019;17(2):2063–2068. doi: 10.3892/ol.2018.9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Gao H, Liang J, Qiao J, Duan J, Shi H, et al. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. 2018;8(12):2387–2401. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu B, Lu X, Zhao Y, Liu C, Huang X, Chen S, et al. MicroRNA-135a induces prostate cancer cell apoptosis via inhibition of STAT6. Oncol Lett. 2019;17(2):1889–1895. doi: 10.3892/ol.2018.9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni JS, Zheng H, Huang ZP, Hong YG, Ou YL, Tao YP, et al. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17(2):2317–2327. doi: 10.3892/ol.2018.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37(1):324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Chang W, Chang W, Chang X, Zhai S, Pan G, et al. MicroRNA-376c-3p facilitates human hepatocellular carcinoma progression via repressing AT-rich interaction domain 2. J Cancer. 2018;9(22):4187–4196. doi: 10.7150/jca.27939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang XM, Yu XN, Liu TT, Zhu HR, Shi X, Bilegsaikhan E, et al. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;105:1147–1154. doi: 10.1016/j.biopha.2018.06.097. [DOI] [PubMed] [Google Scholar]
- 34.Tian T, Fu X, Lu J, Ruan Z, Nan K, Yao Y, et al. MicroRNA-760 inhibits doxorubicin resistance in hepatocellular carcinoma through regulating Notch1/Hes1-PTEN/Akt signaling pathway. J Biochem Mol Toxicol. 2018;32(8):e22167. doi: 10.1002/jbt.22167. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Chen G, Wang Y, He R, Du J, Jiao X, et al. Inhibition of microRNA-16 facilitates the paclitaxel resistance by targeting IKBKB via NF-kappaB signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;503(2):1035–1041. doi: 10.1016/j.bbrc.2018.06.113. [DOI] [PubMed] [Google Scholar]
- 36.Zhang K, Chen J, Zhou H, Chen Y, Zhi Y, Zhang B, et al. PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018;9(3):312. doi: 10.1038/s41419-018-0344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, Li N, Li Z, Zhu Q, Li F, Yang C, et al. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6(22):18933–18944. doi: 10.18632/oncotarget.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 39.Yu C, Bruzek LM, Meng XW, Gores GJ, Carter CA, Kaufmann SH, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene. 2005;24(46):6861–6869. doi: 10.1038/sj.onc.1208841. [DOI] [PubMed] [Google Scholar]
- 40.Sonntag R, Gassler N, Bangen JM, Trautwein C, Liedtke C. Pro-apoptotic Sorafenib signaling in murine hepatocytes depends on malignancy and is associated with PUMA expression in vitro and in vivo. Cell Death Dis. 2014;5:e1030. doi: 10.1038/cddis.2013.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Hu P, Tang F, Xie C. HDAC6-mediated EGFR stabilization and activation restrict cell response to sorafenib in non-small cell lung cancer cells. Med Oncol. 2016;33(5):50. doi: 10.1007/s12032-016-0765-5. [DOI] [PubMed] [Google Scholar]
- 42.Xiang QF, Zhan MX, Li Y, Liang H, Hu C, Huang YM, et al. Activation of MET promotes resistance to sorafenib in hepatocellular carcinoma cells via the AKT/ERK1/2-EGR1 pathway. Artif Cells Nanomed Biotechnol. 2019;47(1):83–89. doi: 10.1080/21691401.2018.1543195. [DOI] [PubMed] [Google Scholar]
- 43.Xu H, Zhao L, Fang Q, Sun J, Zhang S, Zhan C, et al. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1alpha. PLoS One. 2014;9(12):e115565. doi: 10.1371/journal.pone.0115565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Wang M, Liang N, Guan L. PDCD2 sensitizes HepG2 cells to sorafenib by suppressing epithelialmesenchymal transition. Mol Med Rep. 2019;19(3):2173–2179. doi: 10.3892/mmr.2019.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, et al. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 2018;9(1):4. doi: 10.1038/s41419-017-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fornari F, Pollutri D, Patrizi C, La Bella T, Marinelli S, Casadei Gardini A, et al. In hepatocellular carcinoma miR-221 modulates Sorafenib resistance through inhibition of Caspase-3-mediated apoptosis. Clin Cancer Res. 2017;23(14):3953–3965. doi: 10.1158/1078-0432.CCR-16-1464. [DOI] [PubMed] [Google Scholar]
- 47.Liu K, Liu S, Zhang W, Ji B, Wang Y, Liu Y. miR222 regulates sorafenib resistance and enhance tumorigenicity in hepatocellular carcinoma. Int J Oncol. 2014;45(4):1537–1546. doi: 10.3892/ijo.2014.2577. [DOI] [PubMed] [Google Scholar]
- 48.Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, et al. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. 2019;234(3):2788–2794. doi: 10.1002/jcp.27095. [DOI] [PubMed] [Google Scholar]
- 49.He C, Dong X, Zhai B, Jiang X, Dong D, Li B, et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6(30):28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, et al. MicroRNA-93 activates c-met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6(5):3211–3224. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58(2):629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 52.Azumi J, Tsubota T, Sakabe T, Shiota G. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci. 2016;107(9):1256–1262. doi: 10.1111/cas.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, et al. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34(2):1003–1010. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Huang J, Ma L, Shan J, Shen J, Yang Z, et al. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016;371(2):171–181. doi: 10.1016/j.canlet.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 55.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song K, Kwon H, Han C, Zhang J, Dash S, Lim K, et al. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: regulation by MIR-122. Oncotarget. 2015;6(38):40822–40835. doi: 10.18632/oncotarget.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kishikawa T, Otsuka M, Tan PS, Ohno M, Sun X, Yoshikawa T, et al. Decreased miR122 in hepatocellular carcinoma leads to chemoresistance with increased arginine. Oncotarget. 2015;6(10):8339–8352. doi: 10.18632/oncotarget.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, et al. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290(2):1170–1185. doi: 10.1074/jbc.M114.601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13(1):77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 60.Mu W, Hu C, Zhang H, Qu Z, Cen J, Qiu Z, et al. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Res. 2015;25(4):477–495. doi: 10.1038/cr.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 62.Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, et al. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16(3):957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao K, Zhang J, He C, Xu K, Liu J, Sun J, et al. Restoration of miR-193b sensitizes hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352(2):245–252. doi: 10.1016/j.canlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X, et al. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatol Res. 2015;45(13):1312–1322. doi: 10.1111/hepr.12500. [DOI] [PubMed] [Google Scholar]
- 65.Xu J, Lin H, Li G, Sun Y, Chen J, Shi L, et al. The miR-367-3p increases Sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55–67. doi: 10.1016/j.ebiom.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu AQ, Lv B, Qiu F, Wang XY, Cao XH. Upregulation of miR-137 reverses sorafenib resistance and cancer-initiating cell phenotypes by degrading ANT2 in hepatocellular carcinoma. Oncol Rep. 2017;37(4):2071–2078. doi: 10.3892/or.2017.5498. [DOI] [PubMed] [Google Scholar]
- 67.Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V, et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67(1):216–231. doi: 10.1002/hep.29478. [DOI] [PubMed] [Google Scholar]
- 68.Liu W, Meng Q, Sun Y, Wang C, Huo X, Liu Z, et al. Targeting P-glycoprotein: Nelfinavir reverses Adriamycin resistance in K562/ADR cells. Cell Physiol Biochem. 2018;51(4):1616–1631. doi: 10.1159/000495650. [DOI] [PubMed] [Google Scholar]
- 69.Han J, Jun Y, Kim SH, Hoang HH, Jung Y, Kim S, et al. Rapid emergence and mechanisms of resistance by U87 glioblastoma cells to doxorubicin in an in vitro tumor microfluidic ecology. Proc Natl Acad Sci U S A. 2016;113(50):14283–14288. doi: 10.1073/pnas.1614898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao Zhuanglei, Li Zhaoxia, Liu Yuelin, Liu Zhonghao. Forkhead box O3 promotes colon cancer proliferation and drug resistance by activating MDR1 expression. Molecular Genetics & Genomic Medicine. 2019;7(3):e554. doi: 10.1002/mgg3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen JM, Bai JY, Yang KX. Effect of resveratrol on doxorubicin resistance in breast neoplasm cells by modulating PI3K/Akt signaling pathway. IUBMB Life. 2018;70(6):491–500. doi: 10.1002/iub.1749. [DOI] [PubMed] [Google Scholar]
- 72.Fan Y, Li M, Ma K, Hu Y, Jing J, Shi Y, et al. Dual-target MDM2/MDMX inhibitor increases the sensitization of doxorubicin and inhibits migration and invasion abilities of triple-negative breast cancer cells through activation of TAB1/TAK1/p38 MAPK pathway. Cancer Biol Ther. 2018:1–16. [DOI] [PMC free article] [PubMed]
- 73.Shin DH, Choi YJ, Park JW. SIRT1 and AMPK mediate hypoxia-induced resistance of non-small cell lung cancers to cisplatin and doxorubicin. Cancer Res. 2014;74(1):298–308. doi: 10.1158/0008-5472.CAN-13-2620. [DOI] [PubMed] [Google Scholar]
- 74.Mi H, Wang X, Wang F, Li L, Zhu M, Wang N, et al. miR-381 induces sensitivity of breast cancer cells to doxorubicin by inactivation of MAPK signaling via FYN. Eur J Pharmacol. 2018;839:66–75. doi: 10.1016/j.ejphar.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 75.Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. 2018;106:850–857. doi: 10.1016/j.biopha.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13(10):1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- 77.Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227(3):275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 78.Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8(1):e2540. doi: 10.1038/cddis.2016.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-kappaB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. doi: 10.1186/1476-4598-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao AM, Zhang XY, Hu JN, Ke ZP. Apigenin sensitizes hepatocellular carcinoma cells to doxorubic through regulating miR-520b/ATG7 axis. Chem Biol Interact. 2018;280:45–50. doi: 10.1016/j.cbi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Y, Qi X, Chen J, Wei W, Yu C, Yan H, et al. The miR-491-3p/Sp3/ABCB1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett. 2017;408:102–111. doi: 10.1016/j.canlet.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 82.Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310(2):160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 83.Yahya SMM, Fathy SA, El-Khayat ZA, El-Toukhy SE, Hamed AR, Hegazy MGA, et al. Possible role of microRNA-122 in modulating multidrug resistance of hepatocellular carcinoma. Indian J Clin Biochem. 2018;33(1):21–30. doi: 10.1007/s12291-017-0651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan C, Wang X, Shi K, Zheng Y, Li J, Chen Y, et al. MiR-122 reverses the doxorubicin-resistance in hepatocellular carcinoma cells through regulating the tumor metabolism. PLoS One. 2016;11(5):e0152090. doi: 10.1371/journal.pone.0152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du Z, Niu S, Xu X, Xu Q. MicroRNA31-NDRG3 regulation axes are essential for hepatocellular carcinoma survival and drug resistance. Cancer Biomark. 2017;19(2):221–230. doi: 10.3233/CBM-170568. [DOI] [PubMed] [Google Scholar]
- 86.Yang T, Zheng ZM, Li XN, Li ZF, Wang Y, Geng YF, et al. MiR-223 modulates multidrug resistance via downregulation of ABCB1 in hepatocellular carcinoma cells. Exp Biol Med (Maywood) 2013;238(9):1024–1032. doi: 10.1177/1535370213497321. [DOI] [PubMed] [Google Scholar]
- 87.Ma J, Wang T, Guo R, Yang X, Yin J, Yu J, et al. Involvement of miR-133a and miR-326 in ADM resistance of HepG2 through modulating expression of ABCC1. J Drug Target. 2015;23(6):519–524. doi: 10.3109/1061186X.2015.1015536. [DOI] [PubMed] [Google Scholar]
- 88.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60(3):590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 89.He H, Tian W, Chen H, Deng Y. MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep. 2016;13(2):1923–1929. doi: 10.3892/mmr.2015.4727. [DOI] [PubMed] [Google Scholar]
- 90.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70(12):5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 91.Wang L, Wang YM, Xu S, Wang WG, Chen Y, Mao JY, et al. MicroRNA-215 is upregulated by treatment with Adriamycin and leads to the chemoresistance of hepatocellular carcinoma cells and tissues. Mol Med Rep. 2015;12(4):5274–5280. doi: 10.3892/mmr.2015.4012. [DOI] [PubMed] [Google Scholar]
- 92.Ju BL, Chen YB, Zhang WY, Yu CH, Zhu DQ, Jin J. miR-145 regulates chemoresistance in hepatocellular carcinoma via epithelial mesenchymal transition. Cell Mol Biol (Noisy-le-grand) 2015;61(3):12–16. [PubMed] [Google Scholar]
- 93.Wang D, Zhang N, Ye Y, Qian J, Zhu Y, Wang C. Role and mechanisms of microRNA503 in drug resistance reversal in HepG2/ADM human hepatocellular carcinoma cells. Mol Med Rep. 2014;10(6):3268–3274. doi: 10.3892/mmr.2014.2591. [DOI] [PubMed] [Google Scholar]
- 94.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 95.Noordhuis P, Holwerda U, Van der Wilt CL, Van Groeningen CJ, Smid K, Meijer S, et al. 5-fluorouracil incorporation into RNA and DNA in relation to thymidylate synthase inhibition of human colorectal cancers. Ann Oncol. 2004;15(7):1025–1032. doi: 10.1093/annonc/mdh264. [DOI] [PubMed] [Google Scholar]
- 96.Lee H, Kim C, Kang H, Tak H, Ahn S, Yoon SK, et al. microRNA-200a-3p increases 5-fluorouracil resistance by regulating dual specificity phosphatase 6 expression. Exp Mol Med. 2017;49(5):e327. doi: 10.1038/emm.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang XJ, Zhang DL, Fu C, Wei BZ, Li GJ. MiR-183 modulates multi-drug resistance in hepatocellular cancer (HCC) cells via miR-183-IDH2/SOCS6-HIF-1alpha feedback loop. Eur Rev Med Pharmacol Sci. 2016;20(10):2020–2027. [PubMed] [Google Scholar]
- 98.Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, et al. MiR-141 activates Nrf2-dependent antioxidant pathway via Down-regulating the expression of Keap1 conferring the resistance of hepatocellular carcinoma cells to 5-fluorouracil. Cell Physiol Biochem. 2015;35(6):2333–2348. doi: 10.1159/000374036. [DOI] [PubMed] [Google Scholar]
- 99.Ma K, He Y, Zhang H, Fei Q, Niu D, Wang D, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287(8):5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang X, Yin J, Yu J, Xiang Q, Liu Y, Tang S, et al. miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol Rep. 2012;27(1):250–257. doi: 10.3892/or.2011.1472. [DOI] [PubMed] [Google Scholar]
- 101.Jiang JX, Gao S, Pan YZ, Yu C, Sun CY. Overexpression of microRNA-125b sensitizes human hepatocellular carcinoma cells to 5-fluorouracil through inhibition of glycolysis by targeting hexokinase II. Mol Med Rep. 2014;10(2):995–1002. doi: 10.3892/mmr.2014.2271. [DOI] [PubMed] [Google Scholar]
- 102.Tang H, Zhang P, Xiang Q, Yin J, Yu J, Yang X, et al. Let-7 g microRNA sensitizes fluorouracil-resistant human hepatoma cells. Pharmazie. 2014;69(4):287–292. [PubMed] [Google Scholar]
- 103.Ma J, Wang T, Guo R, Yang X, Yin J, Yu J, et al. MicroRNA133a and microRNA326 cocontribute to hepatocellular carcinoma 5fluorouracil and cisplatin sensitivity by directly targeting Bcell lymphomaextra large. Mol Med Rep. 2015;12(4):6235–6240. doi: 10.3892/mmr.2015.4134. [DOI] [PubMed] [Google Scholar]
- 104.Yang X, Zang J, Pan X, Yin J, Xiang Q, Yu J, et al. miR-503 inhibits proliferation making human hepatocellular carcinoma cells susceptible to 5fluorouracil by targeting EIF4E. Oncol Rep. 2017;37(1):563–570. doi: 10.3892/or.2016.5220. [DOI] [PubMed] [Google Scholar]
- 105.Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q, et al. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem Biophys Res Commun. 2012;425(2):468–472. doi: 10.1016/j.bbrc.2012.07.127. [DOI] [PubMed] [Google Scholar]
- 106.Qin J, Luo M, Qian H, Chen W. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene. 2014;538(2):342–347. doi: 10.1016/j.gene.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 107.Shi L, Chen ZG, Wu LL, Zheng JJ, Yang JR, Chen XF, et al. miR-340 reverses cisplatin resistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Asian Pac J Cancer Prev. 2014;15(23):10439–10444. doi: 10.7314/APJCP.2014.15.23.10439. [DOI] [PubMed] [Google Scholar]
- 108.Ou Y, Zhai D, Wu N, Li X. Downregulation of miR-363 increases drug resistance in cisplatin-treated HepG2 by dysregulating Mcl-1. Gene. 2015;572(1):116–122. doi: 10.1016/j.gene.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 109.Chen L, Liu Y, Wang W, Liu K. Effect of integrin receptor-targeted liposomal paclitaxel for hepatocellular carcinoma targeting and therapy. Oncol Lett. 2015;10(1):77–84. doi: 10.3892/ol.2015.3242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Poggi G, Montagna B, Melchiorre F, Quaretti P, Delmonte A, Riccardi A, et al. Hepatic intra-arterial cetuximab in combination with 5-fluorouracil and cisplatin as salvage treatment for sorafenib-refractory hepatocellular carcinoma. Anticancer Res. 2011;31(11):3927–3933. [PubMed] [Google Scholar]
- 111.Dostal Z, Kosina P, Mlejnek P, Kikalova K, Modriansky M. Mifepristone potentiates etoposide toxicity in Hep G2 cells by modulating drug transport. Toxicol in Vitro. 2019;54:33–40. doi: 10.1016/j.tiv.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 112.Xue F, Liang Y, Li Z, Liu Y, Zhang H, Wen Y, et al. MicroRNA-9 enhances sensitivity to cetuximab in epithelial phenotype hepatocellular carcinoma cells through regulation of the eukaryotic translation initiation factor 5A-2. Oncol Lett. 2018;15(1):813–820. doi: 10.3892/ol.2017.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang N, Zhu M, Tsao SW, Man K, Zhang Z, Feng Y. MiR-23a-mediated inhibition of topoisomerase 1 expression potentiates cell response to etoposide in human hepatocellular carcinoma. Mol Cancer. 2013;12(1):119. doi: 10.1186/1476-4598-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang C, Long J, Liu B, Xie X, Kuang M. Mcl-1 is a novel target of miR-26b that is associated with the apoptosis induced by TRAIL in HCC cells. Biomed Res Int. 2015;2015:572738. doi: 10.1155/2015/572738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuo C, Sheng X, Liu Z, Ma M, Xiong S, Deng H, et al. MicroRNA-138 enhances TRAIL-induced apoptosis through interferon-stimulated gene 15 downregulation in hepatocellular carcinoma cells. Tumour Biol. 2017;39(6):1010428317710410. doi: 10.1177/1010428317710410. [DOI] [PubMed] [Google Scholar]
- 116.Teufel M, Seidel H, Kochert K, Meinhardt G, Finn RS, Llovet JM, et al. Biomarkers associated with response to Regorafenib in patients with hepatocellular carcinoma. Gastroenterology. 2019;156(6):1731–1741. doi: 10.1053/j.gastro.2019.01.261. [DOI] [PubMed] [Google Scholar]
- 117.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, et al. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory Axis. Cancer Cell. 2018;33(3):417–434. doi: 10.1016/j.ccell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 118.Zhang P, Dong Q, Zhu H, Li S, Shi L, Chen X. Long non-coding antisense RNA GAS6-AS1 supports gastric cancer progression via increasing GAS6 expression. Gene. 2019;696:1–9. doi: 10.1016/j.gene.2018.12.079. [DOI] [PubMed] [Google Scholar]
- 119.Tzadok S, Caspin Y, Hachmo Y, Canaani D, Dotan I. Directionality of noncoding human RNAs: how to avoid artifacts. Anal Biochem. 2013;439(1):23–29. doi: 10.1016/j.ab.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 120.Hamazaki N, Nakashima K, Hayashi K, Imamura T. Detection of bidirectional promoter-derived lncRNAs from small-scale samples using pre-amplification-free directional RNA-seq method. Methods Mol Biol. 2017;1605:83–103. [DOI] [PubMed]
- 121.Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q, et al. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting AS a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10(3):150. doi: 10.1038/s41419-019-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lou Y, Yu Y, Xu X, Zhou S, Shen H, Fan T, et al. Long non-coding RNA LUCAT1 promotes tumourigenesis by inhibiting ANXA2 phosphorylation in hepatocellular carcinoma. J Cell Mol Med. 2019;23(3):1873–1884. doi: 10.1111/jcmm.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu D, Gao M, Wu K, Zhu D, Yang Y, Zhao S. LINC00152 facilitates tumorigenesis in esophageal squamous cell carcinoma via miR-153-3p/FYN axis. Biomed Pharmacother. 2019;112:108654. doi: 10.1016/j.biopha.2019.108654. [DOI] [PubMed] [Google Scholar]
- 124.Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18(1):28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen LL, He J, Qiu XT, Yu J, Wang ZM. The prognostic roles of long non-coding RNA SNHG17 in the patients with gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23(3):1063–1068. doi: 10.26355/eurrev_201902_16994. [DOI] [PubMed] [Google Scholar]
- 126.Gao R, Fang C, Xu J, Tan H, Li P, Ma L. LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145. Arch Biochem Biophys. 2019;663:183–191. doi: 10.1016/j.abb.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 127.Shi C, Wang M. LINC01118 modulates paclitaxel resistance of epithelial ovarian Cancer by regulating miR-134/ABCC1. Med Sci Monit. 2018;24:8831–8839. doi: 10.12659/MSM.910932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang L, Shang X, Feng Q. LncRNA TATDN1 contributes to the cisplatin resistance of non-small cell lung cancer through TATDN1/miR-451/TRIM66 axis. Cancer Biol Ther. 2019;20(3):261–271. doi: 10.1080/15384047.2018.1529091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X, et al. Knockdown of Long noncoding RNA HOXA-AS2 suppresses Chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 Axis. Cell Physiol Biochem. 2018;51(2):886–896. doi: 10.1159/000495387. [DOI] [PubMed] [Google Scholar]
- 130.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(12):1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol Med. 2018;22(6):3238–3245. doi: 10.1111/jcmm.13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y, et al. LncRNA HANR promotes tumorigenesis and increase of Chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem. 2017;43(5):1926–1938. doi: 10.1159/000484116. [DOI] [PubMed] [Google Scholar]
- 133.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes Sunitinib resistance in renal Cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 134.Li Y, Ye Y, Feng B, Qi Y. Long noncoding RNA lncARSR promotes doxorubicin resistance in hepatocellular carcinoma via modulating PTEN-PI3K/Akt pathway. J Cell Biochem. 2017;118(12):4498–4507. doi: 10.1002/jcb.26107. [DOI] [PubMed] [Google Scholar]
- 135.Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 136.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478(3):1067–1073. doi: 10.1016/j.bbrc.2016.08.065. [DOI] [PubMed] [Google Scholar]
- 137.Jin W, Chen L, Cai X, Zhang Y, Zhang J, Ma D, et al. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol Rep. 2017;37(1):273–280. doi: 10.3892/or.2016.5248. [DOI] [PubMed] [Google Scholar]
- 138.Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12(10):1377–1387. doi: 10.1158/1541-7786.MCR-13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yin X, Zheng SS, Zhang L, Xie XY, Wang Y, Zhang BH, et al. Identification of long noncoding RNA expression profile in oxaliplatin-resistant hepatocellular carcinoma cells. Gene. 2017;596:53–88. doi: 10.1016/j.gene.2016.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.