Abstract
Adipose tissue (AT) serves a crucial role in maintaining organismal metabolic homeostasis. Studies have demonstrated that AT is populated with a diverse array of immune cells that coordinate and regulate AT function. This adipo-immune system is highly dynamic, reflecting the physiologic state of the organism (e.g. obese, lean, aged, and young) as well as the constant physiologic remodeling of AT associated with the daily rhythms of fasting and feeding. Many of the adaptive and maladaptive functional changes of AT are regulated by changes in the quantity and quality of distinct sets of AT-resident immune cells. Here we developed a protocol to assess the dynamic state of the immune system within AT by constructing censuses of adipose-resident immune cells (macrophages, dendritic cells, neutrophils, eosinophils, NK cells, innate lymphocytes, T cells, and B cells, etc.) based on flow cytometry, which we term adipo-immune profiles (AIPs). Constructing AIPs can be an integral part of assessment for AT health and function. This unit describes the protocols to generate such AIPs.
Keywords: adipose tissue, immune cell profiling, macrophage, regulatory T cell, flow cytometry
INTRODUCTION
Adipose tissue (AT) function is intimately related with immune cells residing within AT (Hotamisligil, 2017; Y. S. Lee, Wollam, & Olefsky, 2018). Since the initial findings that macrophage number increases in obese AT in mice described in the 1960s (Hausberger, 1966) and again demonstrated in 2003 (Ferrante, 2013; Xu et al., 2003)), critical roles for several specific types of immune cells have been implicated in modulating distinct functions of AT, including insulin sensitivity (Lynch et al., 2012; Molofsky et al., 2013; Talukdar et al., 2012; Wu et al., 2011), adaptive thermogenesis (Brestoff et al., 2014; M.-W. Lee et al., 2014; Rao et al., 2014), and immune homeostasis (Han et al., 2017). While these studies have largely focused on the effect of one immune cell type at a time, the immune system usually carries out its function via coordination of an array of immune cell subsets, in AT as well as other tissue types. To gain more holistic insight about the composition of the immune system within AT, we found it helpful to assemble an immunological census within a defined AT depot, which we termed an adipo-immune profile (AIP). This method is reliant on conventional flow cytometry and allows for quantitation of numerous distinct immune cell types (macrophages, dendritic cells, neutrophils, eosinophils, NK cells, innate lymphocytes, T cells, and B cells, etc.), and thus allows investigators to understand how immune systems globally change within AT upon a given perturbation. We developed this method while characterizing immune changes in ATs from lean, young mice, obese mice, and lean, old mice and discovered a striking, previously undescribed increase between young and old mice in a unique type of regulatory T cell, we termed the fat-resident regulatory T cell (fTreg). This lynchpin finding was what facilitated our eventual discovery that fTregs drive age-associated insulin resistance (Bapat et al., 2015; Maillard & Saltiel, 2015). We have continued to refine this method and have found construction of AIPs useful in understanding the immune systems of AT from other important physiological contexts. The protocol described below describes in detail how to construct AIPs from murine AT depots.
BASIC PROTOCOL 1. Isolation of immune cell populations from adipose tissue
AT depots are located in multiple defined anatomic sites in the murine and human body. Distinct AT depots function and behave differently under different physiologic stresses, carry out distinct metabolic functions, and possess distinct adipo-immune cell compositions. In general, AT depots can be divided into visceral AT (VAT) and subcutaneous AT (SAT). In this protocol, we use epididymal AT, a visceral AT depot, as an example to construct an AIP. AIPs of other AT depots can be carried out using the same method.
Materials
Mice for Adipo-Immune Profiling (Wild-type C57BL/6 male mice at one year of age were used here, but this protocol can apply to mice of other genetic backgrounds and at different ages.)
Adipose tissue dissociation buffer (see recipe)
RPMI medium (Thermo-Fisher, Cat. No. 11875093)
1× PBS (Fisher Scientific, Cat. No. MT21040CM)
70 μm cell strainer (Fisher Scientific, Cat. No. 22363548)
Scissors and tweezers for mouse dissection
Petri Dish
Conical tubes
Centrifuge (Eppendorf 5810R)
Orbital Shaker
-
Euthanize mouse in a CO2 chamber. Open the thoracic cavity and cut a small incision in the right atrium of the heart as an outlet for blood. Perfuse the mouse by slowly injecting 10 mL of ice cold 1×PBS into the left ventricle to remove blood from tissues. See UNIT 15.1 (Miller, Karpus, & Davidson, 2010) for detailed instructions on mouse perfusion.
For optimal result, perfuse shortly after euthanization.
-
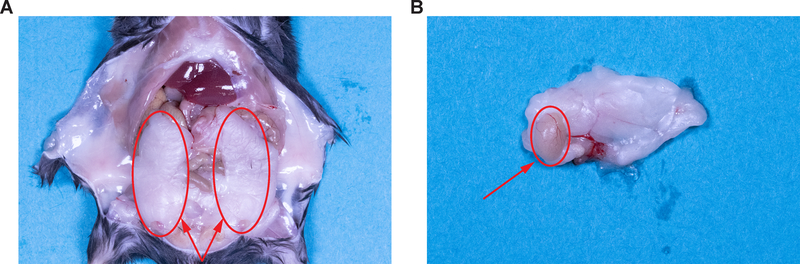
Use scissors and forceps to open the peritoneal cavity and remove the epididymal AT (Figure 1).
Be careful to dissect the testes away from the epididymal ATs.
Measure the weight of the AT (The epididymal AT from a one-year-old mouse is typically 0.4 g to 0.8 g.).
In a 10 cm dish, use scissors to mince individual AT depots into fine pieces (2–5 mm3).
Transfer the minced ATs to a 15 mL conical tube. Add 10 mL adipose tissue dissociation buffer, and incubate at 37°C with intermittent shaking for 1.5 hrs.
Pass the suspension through an 80 μm mesh to remove undigested clumps and debris.
Let the flow-through stand for 10 min to separate the floating adipocyte fraction and the infranatant containing the stromal vascular fraction.
Carefully transfer the infranatant to a new 15 ml conical tube without disturbing the floating adipocyte fraction.
Centrifuge at 400g for 10 min.
Wash the pellet once with 10 ml RPMI.
The resultant isolated cells can be immediately used for FACS staining and analysis (see basic Protocol 2 for details), or stored on ice for up to 4 hours before FACS staining.
Figure 1. Isolation of the epididymal fat pad.
(A) Location of the two epididymal fat pads marked by the red circles. (B) Location of the testis imbedded in the epididymal fat pad marked by the red circle.
BASIC PROTOCOL 2. Staining and flow cytometric analysis of AT immune cells
Here we describe how to perform staining of the immune cells isolated from AT for FACS analysis. Additionally, we show how FACS data is analyzed to identify each immune cell population.
Materials
Adipose tissue immune cell samples isolated from the Basic Protocol 1
Cell staining buffer (see recipe)
Cell fixation/permeabilization buffer (see recipe)
1× cell permeabilization buffer (see recipe)
Fc block: anti-mouse CD16/CD32 clone 2.4G2 (BioXcell, Cat. No. BE0307) in cell staining buffer
Fluorescently labeled antibodies (Table 3)
Table 3.
Information on antibodies commonly used in AT immune cell profiling by flow cytometry.
| Antibody | Clone | Vendor |
|---|---|---|
| CD3ε | 145–2C11 | Tonbo |
| CD4 | RM4–5 | Biolegend |
| CD5 | 53–7.3 | Biolegend |
| CD8a | 53–6.7 | Biolegend |
| CD11b | M1/70 | Biolegend |
| CD11c | N418 | Tonbo |
| CD19 | 1D3 | Tonbo |
| CD25 | PC61.5 | Biolegend |
| CD45.2 | 104 | Biolegend |
| CD62L | MEL-14 | Biolegend |
| CD90.2 | 53–2.1 | Thermo-Fisher |
| CD206 | C068C2 | Biolegend |
| B220 | RA3–6B2 | Biolegend |
| F4/80 | BM8.1 | Biolegend |
| FcεR1 | MAR-1 | Tonbo |
| Foxp3 | FJK-16s | Thermo-Fisher |
| GATA3 | TWAJ | Thermo-Fisher |
| LY6G(Gr-1) | RB6–8CS | Tonbo |
| NK1.1 | PK136 | Biolegend |
| RORγ | Q31–378 | BD Biosciences |
| Siglec-F | E50–2440 | BD Biosciences |
| TCRβ | H57–597 | Biolegend |
| TCRγ/δ | GL3 | Biolegend |
Cell fixation buffer (see recipe)
LSR II flow cytometer (Becton Dickinson) or equivalent
FlowJo analysis software
Surface and intracellular stain of the cells
-
1
Resuspend 0.5–1×105 AT immune cells obtained from Basic Protocol 1 in 200 μl Fc block solution. Incubate on ice for 10 min.
Cell numbers should be counted at this time using a hemocytometer after trypan blue staining. It is important to be able to quantitate the actual number of specific immune cell populations identified by FACS, which generally provides frequencies. Typically, we can harvest 2–5×105 cells per gram of fat at this stage.
-
2
Prepare antibody cocktails in cell staining buffer at 100 μL volume per sample. The four commonly used antibody cocktails for AT immune cells are listed in Table 1.
Cocktail #1 is used to identify myeloid cell subsets. Cocktails #2 and #3 are used to identify T, B, and NK subsets. Cocktail #4 is used to identify innate lymphocyte subsets. All antibody concentrations should be titrated for optimal results. As a common practice, we use 1:400 dilution of each antibody (~1 ng/ml) as a starting point. Design of proper compensation matrices is essential for multiparameter FACS analysis. Fluorescence-minus-one (FMO) controls should be conducted when setting up this analysis for the first time to allow for proper gating. A more detailed discussion on multiparameter FACS analysis is described in UNIT 5.8 (Roederer, 2002).
-
3
Aliquot immune cells in Fc block solution into three tubes at 50μl each.
-
4
Add 50 μl premixed antibody cocktail 1, 2, 3, or 4 (Table 1) into each tube with immune cell samples in Fc block solution to reach final volume at 100 μl.
-
5
Incubate cells in antibody cocktail for 30 min on ice in the dark.
-
6
Centrifuge 5 min at 500 g at 4°C.
-
7
Remove supernatant and wash pellet with 200 μl cell staining buffer.
-
8
Centrifuge 5 min at 500 g at 4°C.
-
9
For samples that don’t need intracellular staining (Stain 1 and 3), resuspend cell pellet in 100 μl cell fixation buffer. Store at 4°C in the dark until ready for FACS analysis (Basic Protocol 2 Step 22. For samples that need intracellular staining (Stain 2 and 4), proceed to the next step (Basic Protocol 2, Step 10–21).
-
10
Resuspend cells in 200 μl cell fixation/permeabilization buffer.
-
11
Incubate on ice for 1 hr in the dark.
-
12
Centrifuge 5 min at 725 g at 4°C.
-
13
Remove supernatant and wash pellet with 200 μl 1× cell permeabilization buffer.
-
14
Centrifuge 5 min at 725 g at 4°C.
-
15
Prepare antibody cocktail for intracellular staining in 1× cell permeabilization buffer (Table 1).
-
16
Remove supernatant and resuspend cell pellet in 100 μl antibody cocktail.
-
17
Incubate cells in antibody cocktail for 30 min on ice in the dark.
-
18
Centrifuge 5 min at 725 g at 4°C.
-
19
Remove supernatant and wash pellet with 200 μl 1× cell permeabilization buffer.
-
20
Centrifuge 5 min at 725 g at 4°C.
-
21
Remove supernatant and resuspend cell pellet in 100 μl cell fixation buffer. Store at 4°C in the dark until ready for FACS analysis.
-
22
Stained cells should be analyzed by FACS within 24 hours to achieve optimal result.
Table 1.
Antibody cocktails for AT immune profiling.
| Antibody Cocktail #1 | Antibody Cocktail #2 | Antibody Cocktail #3 | Antibody Cocktail #4 | ||||
|---|---|---|---|---|---|---|---|
| Antibody | Color | Antibody | Color | Antibody | Color | Antibody | Color |
| CD11b | APC-Cy7 | CD4 | APC | CD19 | Alexa 700 | CD45.2 | Alexa 700 |
| CD11c | PerCP-Cy5.5 | CD8 | Brilliant Violet 510 | CD45.2 | PerCP-Cy5.5 | CD90.2 | FITC |
| CD45.2 | Alexa 700 | CD25 | FITC | B220 | eFluor 450 | GATA3 (IC)* | APC |
| CD206 | PE-Cy7 | CD45.2 | PerCP-Cy5.5 | NK1.1 | PE | NK1.1 | Brilliant Violet 605 |
| F4/80 | APC | CD62L | Brilliant Violet 605 | TCRβ | FITC | RORγ (IC)* | eFluor 450 |
| Ly6G | FITC | Foxp3 (IC)* | eFluor 450 | CD3ε** | PE | ||
| Siglec-F | PE | TCRβ | PE-Cy7 | CD5** | PE | ||
| TCRγ/δ | PE | CD8** | PE | ||||
| CD11b** | PE | ||||||
| CD19** | PE | ||||||
| TCRβ** | PE | ||||||
| TCRγ/δ** | PE | ||||||
| FcεR1** | PE | ||||||
| F4/80** | PE | ||||||
IC: intracellular staining. These antibodies need to be used after permeabilization of the cells.
These PE-conjugated antibodies are used to gate out lineage positive cells for innate lymphocyte stain.
FACS data acquisition and analysis
-
23
Stained cell samples can be acquired on a BD LSRII or equivalent FACS machine with more than 8 color capacity.
During data acquisition, record all events instead of events in selected gates so that there is more flexibility in gating choices during the data analysis step.
-
24
Typically, 20,000 events are acquired. For analysis of more rare immune cell populations, it is advisable to increase acquisition events to 200,000 or more.
-
25
For FACS data analysis, use FlowJo software to process FCS sample file. Detailed gating strategy for each immune cell subset is listed in Table 2. A representative FACS analysis of AT immune cells from a one-year old male mouse’s epididymal fat is shown in Figure 2.
-
26
Use total cell number from the hemocytometer cell count and percentages generated from FACS analysis to calculate the number for each immune cell subset. Normalize cell numbers using the weight of the fat pad to generate quantitation in the form of cell numbers per gram of fat.
Table 2.
Cell surface and intracellular markers used to identify immune cell subsets in AT.
| Immune cell subsets | Markers for identification of immune cell subsets |
|---|---|
| M1 macrophage | CD45.2+ F4/80+ CD11b+ CD11c+ CD206− |
| M2 macrophage | CD45.2+ F4/80+ CD11b+ CD11c+ CD206+ |
| Double negative (DN) macrophage | CD45.2+ F4/80+ CD11b+ CD11c− CD206– |
| Eosinophil | CD45.2+ CD11b+ Siglec-F+ |
| Neutrophil | CD45.2+ CD11c− F4/80 CD11b+ Ly6Ghigh |
| NK cell | CD45.2+ TCRβ− NK1.1+ |
| NK T cell | CD45.2+ TCRβ+ NK1.1+ |
| B cell | CD45.2+ B220+ CD19+ |
| γ/δ T cell | CD45.2+ TCRβ− TCRγδ+ |
| CD4+ T cell | CD45.2+ TCRβ+ CD4+ CD8– |
| CD8+ T cell | CD45.2+ TCRβ+ CD4− CD8+ |
| Regulatory T cell | CD45.2+ TCRβ+ CD4+ CD8− Foxp3+ |
| Naïve CD4+ T cell | CD45.2+ TCRβ+ CD4+ CD8− Foxp3− CD62Lhigh |
| Activated CD4+ T cell | CD45.2+ TCRβ+ CD4+ CD8− Foxp3− CD62Llow |
| Innate lymphocyte group 1 (ILC1) | CD45.2+ Lineage− CD90.2+ NK1.1+ RORγ− |
| Innate lymphocyte group 2 (ILC2) | CD45.2+ Lineage− CD90.2+ NK1.1− RORγ− |
| Innate lymphocyte group 3 (ILC3) | CD45.2+ Lineage− CD90.2+ NK1.1− RORγ+ |
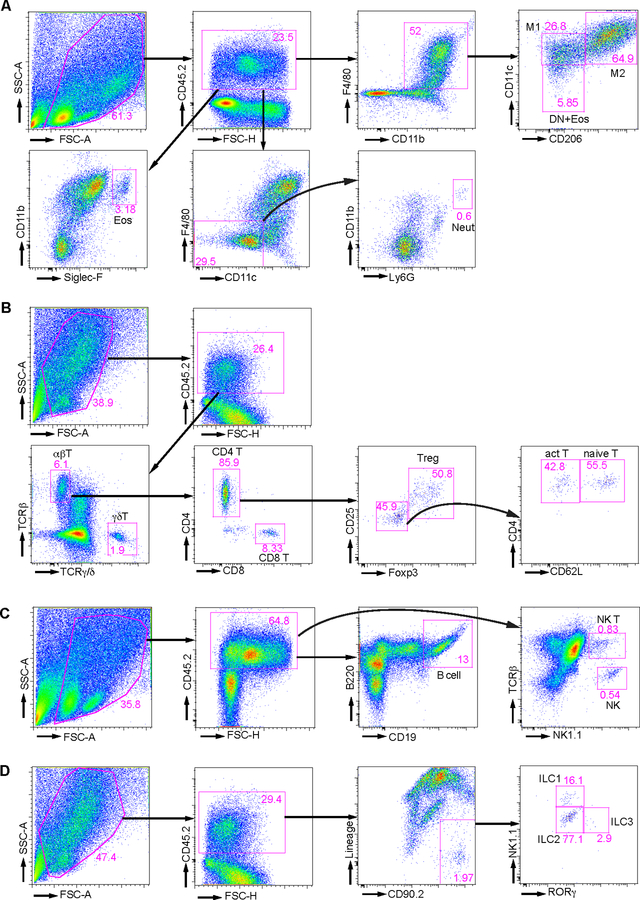
Figure 2. Gating strategy for FACS analysis of AT immune cell subsets.
Typical FACS plots generated from (A) Antibody cocktail #1, myeloid cells; (B) Antibody cocktail #2, T cell subsets; (C) Antibody cocktail #3, B cell, NK cell, and NK T cell; and (D) Antibody cocktail #4, innate lymphocytes. M1: M1 adipose tissue associated macrophage. M2: M2 adipose tissue associated macrophage. DN: double negative adipose tissue associated macrophage. Eos: eosinophil. Neut: neutrophil. Treg: regulatory T cell. Naïve T: naïve CD4+ T cell. Act T: activated CD4+ T cell. NK: natural killer cell. NK T: natural killer T cell. ILC1: group 1 innate lymphocyte. ILC2: group 2 innate lymphocyte. ILC3: group 3 innate lymphocyte.
REAGENTS AND SOLUTIONS
Adipose tissue dissociation buffer
100mM HEPES buffer pH7.4, 120mM NaCl, 50mM KCl, 5mM glucose, 1mM CaCl2, 1.5% BSA, and 1mg/ml collagenase.
Make 1M stock solutions of NaCl, KCl, glucose, and CaCl2•2H2O. To make 50 ml stock solutions, the following amount of materials is needed.
2.922g NaCl
3.727g KCl
9.0g glucose
7.35g CaCl2·2H2O (Be certain to use the dihydrate form of CaCl2 as opposed to the anhydrous form. The anhydrous form has less purity, and the stock solution will form a foggy precipitate in a few days.)
Once the stock solutions are made, make the buffer fresh each time. For 25 ml of adipose tissue dissociation buffer, combine
3 ml of 1M NaCl
1.25 ml of 1M KCl
125 μl of 1M glucose
25 μl of 1M CaCl2
2.5 ml of 1M HEPES
0.375 g BSA
Adjust pH of the solution to 7.4. Add collagenase at 1mg/ml (Worthington Biochemical, Collagenase Type 1, Cat no LS004196). Dissolve in 37°C water bath. Sterilize the solution through a 0.2μm filtration unit, and put it on ice until use.
FACS staining buffer
0.5% BSA in 1× PBS.
Cell fixation/permeabilization buffer
1 to 4 dilution from Foxp3/Transcription Factor Fixation/Permeabilization Cencentrate and Diluent. (Thermo Fisher, Cat. No. 00–5521-00)
1× Cell permeabilization buffer
1 to 10 dilution in distilled water from 10x Cell Permeabilization Buffer. (Thermo Fisher, Cat no 00–8333-56)
Cell fixation buffer
1% paraformaldehyde in 1× PBS.
COMMENTARY
Background Information
Since the reports in 2003 of an increase in macrophage content associated with obesity-associated insulin resistance, there has been a steady increase in interest of various immune cell types found in AT, at first focusing on innate immune cells but eventually focusing on adaptive immune cells as well. These studies have largely studied the given immune cell of interest in isolation from other AT-resident immune cells. Yet it is clear that the immune system functions in an interdependent manner with extensive crosstalk between different immune cell subsets to collectively preserve or dysregulate immune-metabolic homeostasis in AT. As such, we developed a method to globally assess immune state within AT at single-cell resolution.
The protocol described herein to assemble an immunological census within AT, which we call an adipo-immune profile or AIP, utilizes conventional flow cytometry. In our own practice, we have found this approach to be affordable, adaptable, and requiring a relatively short amount of time from initial recovery of AT-resident immune cells to data acquisition and analysis (one full working day). Recently, additional methodologies beyond conventional flow cytometry to globally assess immune cells at a single cell resolution have emerged including mass cytometry and single cell RNA-sequencing (scRNA-Seq). In principle, these approaches also will be able to quantitatively define distinct immune cell types within AT. However, caution should be taken to validate that these emerging methodologies accurately preserve relative cell abundance across different immune cell types, perhaps via cross-comparison utilizing conventional flow cytometry. Regardless of methodology, the overarching principle of assembling a holistic immunological census within AT is crucial when assessing AT-immune relationships. In our assessment, for most initial investigations, flow cytometry-based AIPs provide a reliable, reproducible, and affordable snapshot of immune state within a given AT.
Critical Parameters
The immune cell composition in ATs is highly dynamic, and varies depending on the condition of the mouse. Major factors include age, sex, diet, and anatomic site of the AT depot being interrogated. For example, fTreg cell frequency increases as the C57BL/6J mouse gets older, from about 10–15% of total CD4+ T cells at 6 weeks of age to 60–70% in one-year old mice. Therefore, it is important to always have an appropriately matched control group for equitable comparison.
One of the critical factors in constructing AIPs involves the process of AT digestion. AT needs to be minced very well into small pieces for digestion to work well. If the AT is under-digested, not enough immune cells will be liberated into the infranatant for downstream analysis. Oppositely, if AT is over-digested, a large portion of the cells will die and detrimentally affect FACS staining and analysis. It is helpful to evaluate the status of the cells by cell counting after the digestion step before FACS staining. Most of the cells at this stage should be alive as assessed by Trypan Blue stain; live cells will not take up the stain. Additionally, we noticed that certain cell surface markers, such as CD4, are downregulated during digestion, perhaps due to regulated endocytosis and other mechanisms. One way to mitigate this issue is to incubate immune cells in RPMI medium at 37°C for one hour right after they are isolated from AT, and then proceed to FACS staining. Cell surface CD4 expression increases significantly after this recovery step.
Troubleshooting
It can be challenging to identify multiple AT immune cell subsets from FACS data initially. We always stain the splenic cells from the same mouse to help set up gates for our AT-resident immune cell populations. AIP construction might require a new compensation matrix setup on the flow cytometer due to the unique properties of these AT-resident immune cells as well as the different isolation protocols required to analyze the AT-resident immune cells, in contrast to isolation of cells from lymphoid tissues. When it is not clear if a particular antibody is staining correctly, we perform a FMO, “X-1” control stain, a stain with all other antibodies in the cocktail except the one antibody in question. We can validate the antibody by comparing the FACS signal of the experimental stain with the “X-1” stain.
Understanding Results
In general, immune cells are more abundant in visceral ATs than subcutaneous ATs. Obese mice, induced by high-fat diet or genetic mutations, usually have AT depots that are several times larger than lean mice. For meaningful comparisons, we calculate the density of AT infiltrating immune cells based on cell number per gram of fat. The most abundant immune cell population in AT is macrophages. They can be further divided into M1 and M2 subsets, which are shown to be pro-inflammatory or anti-inflammatory, respectively. Although the frequencies of other immune cells in ATs are not as abundant as macrophages, numerous studies have shown the importance of nearly every immune cell subset present in AT, including CD4+ T cells, CD8+ T cells, Tregs, B cells, dendritic cells, NK cells, Eosinophils, and ILCs, etc. Therefore, if we are wanting to investigate non-macrophage populations more carefully, we find it helpful to gate out the macrophage populations early in our gating schema to better understand how the non-macrophage populations behave within AT.
Time Considerations
Harvesting the AT depots from mouse and processing them into single cell suspension takes about 3 to 4 hours. FACS staining of the immune cells takes about 3 hours. Acquisition of FACS data takes about 3 to 5 minutes per sample. For a typical experiment analyzing AT immune cells from 5–6 mice, the total experimental time is about 8 hours.
Significance Statement.
Adipose tissue serves an essential function in maintaining organismal metabolic homeostasis. Dysfunction of adipose tissue can lead to metabolic diseases such as type-2 diabetes and atherosclerosis. Studies have demonstrated that adipose tissue is populated with a diverse array of immune cells that coordinate and regulate adipose tissue function. This adipo-immune system is highly dynamic, reflecting the physiologic state of the organism as well as the constant physiologic remodeling of adipose tissue. Here we describe a protocol to assess the dynamic state of the immune system within adipose tissue by constructing censuses of adipose-resident immune cells based on flow cytometry, which we term adipo-immune profiles (AIPs). Constructing AIPs can be an integral part of assessment for adipose tissue health and function.
ACKNOWLEDGEMENT
This work is partly supported by NOMIS Foundation, Rita Allen Foundation, and National Institutes of Health (R01-AI107027, P30-CA014195, S10-OD023689).
LITERATURE CITED
- Bapat SP, Suh JM, Fang S, Liu S, Zhang Y, Cheng A, et al. (2015). Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature, 528, 137–141. 10.1038/nature16151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, Kim BS, Saenz SA, Stine RR, Monticelli LA, Sonnenberg GF, et al. (2014). Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature, 1–17. 10.1038/nature14115 [DOI] [PMC free article] [PubMed]
- Ferrante AW Jr. (2013). Macrophages, fat, and the emergence of immunometabolism. The Journal of Clinical Investigation, 123(12), 4992–4993. 10.1172/JCI73658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-J, Zaretsky AG, Andrade-Oliveira V, Collins N, Dzutsev A, Shaik J, et al. (2017). White Adipose Tissue Is a Reservoir for Memory T Cells and Promotes Protective Memory Responses to Infection. Immunity, 47(6), 1154–1168.e6 10.1016/j.immuni.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausberger FX (1966). Pathological changes in adipose tissue of obese mice. Anat. Rec 154, 651–660. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS (2017). Foundations of Immunometabolism and Implications for Metabolic Health and Disease. Immunity, 47(3), 406–420. 10.1016/j.immuni.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-W, Odegaard JI, Mukundan L, Qiu Y, Molofsky AB, Nussbaum JC, et al. (2014). Activated Type 2 Innate Lymphoid Cells Regulate Beige Fat Biogenesis, 1–14. 10.1016/j.cell.2014.12.011 [DOI] [PMC free article] [PubMed]
- Lee YS, Wollam J, & Olefsky JM (2018). An Integrated View of Immunometabolism. Cell, 172(1–2), 22–40. 10.1016/j.cell.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch L, Nowak M, Varghese B, Clark J, Hogan AE, Toxavidis V, et al. (2012). Adipose Tissue Invariant NKT Cells Protect against Diet-Induced Obesity and Metabolic Disorder through Regulatory Cytokine Production. Immunity, 37(3), 574–587. 10.1016/j.immuni.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, & Saltiel AR (2015). Metabolism: Inflammation keeps old mice healthy. Nature, 486, 549–2. 10.1038/nature15648 [DOI] [PubMed] [Google Scholar]
- Miller SD, Karpus WJ, Davidson TS (2010). Experimental Autoimmune Encephalomyelitis in the Mouse. Current Protocols in Immunology, 88: 15.1.1–15.1.20. 10.1002/0471142735.im1501s88 [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A, et al. (2013). Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. The Journal of Experimental Medicine, 210(3), 535–549. 10.1084/jem.20020656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, et al. (2014). Meteorin-like Is a Hormone that Regulates Immune-Adipose Interactions to Increase Beige Fat Thermogenesis, 157(6), 1279–1291. 10.1016/j.cell.2014.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M (2002). Multiparameter FACS analysis. Current Protocols in Immunology, 5: 5.8.1–5.8.10. 10.1002/0471142735.im0508s49 [DOI] [PubMed] [Google Scholar]
- Talukdar S, Da Young Oh, Bandyopadhyay G, Li D, Xu J, McNelis J, et al. (2012). Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nature Medicine, 18(9), 1407–1412. 10.1038/nm.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Molofsky AB, Liang H-E, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, et al. (2011). Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science (New York, NY), 332(6026), 243–247. 10.1126/science.1201475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. (2003). Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. The Journal of Clinical Investigation, 112(12), 1821–1830. 10.1172/JCI19451DS1 [DOI] [PMC free article] [PubMed] [Google Scholar]