Abstract
Magnetoencephelography (MEG) is a functional neuroimaging tool that records the magnetic fields induced by neuronal activity; however, signal from muscle activity often corrupts the data. Eye-blinks are one of the most common types of muscle artifact. They can be recorded by affixing eye proximal electrodes, as in electrooculography (EOG), however this complicates patient preparation and decreases comfort. Moreover, it can induce further muscular artifacts from facial twitching. We propose an EOG free, data driven approach. We begin with Independent Component Analysis (ICA), a well-known preprocessing approach that factors observed signal into statistically independent components. When applied to MEG, ICA can help separate neuronal components from non-neuronal ones, however, the components are randomly ordered. Thus, we develop a method to assign one of two labels, non-eye-blink or eye-blink, to each component.
Our contributions are two-fold. First, we develop a 10-layer Convolutional Neural Network (CNN), which directly labels eye-blink artifacts. Second, we visualize the learned spatial features using attention mapping, to reveal what it has learned and bolster confidence in the method’s ability to generalize to unseen data. We acquired 8-min, eyes open, resting state MEG from 44 subjects. We trained our method on the spatial maps from ICA of 14 subjects selected randomly with expertly labeled ground truth. We then tested on the remaining 30 subjects. Our approach achieves a test classification accuracy of 99.67%, sensitivity: 97.62%, specificity: 99.77%, and ROC AUC: 98.69%. We also show the learned spatial features correspond to those human experts typically use which corroborates our model’s validity. This work (1) facilitates creation of fully automated processing pipelines in MEG that need to remove motion artifacts related to eye blinks, and (2) potentially obviates the use of additional EOG electrodes for the recording of eye-blinks in MEG studies.
Keywords: MEG, Eye-Blink, Artifact, EOG, Automatic, Deep learning, CNN
1. Introduction
The functional neuroimaging method known as magnetoencephalography (MEG) offers better temporal resolution than fMRI [1, 2]. Moreover, MEG source space reconstruction is simpler than electroencephalography (EEG) as it is less dependent on intervening tissue’s characteristics [1, 3, 4]. However, any technique that attempts to measure electromagnetic radiation from the neurons must combat muscular artifacts. Unfortunately, there is significant overlap between the two sources of signal. For example, the spectral bandwidth of muscle activity is 20–300 Hz while some of the key neuronal frequency bands, such as gamma-frequency band (30–80 Hz), lie entirely in the muscle activity bandwidth [3, 5].
Independent Component Analysis (ICA) based artifact detection improves MEG signal to noise ratio by 35% compared to spectral approaches that improve ratio by 5–10% [6]. Even though ICA can transform the noise and the signal into individual components, the components are randomly ordered and must be manually identified by a trained expert [3, 7]. To automatically detect eye-blink artifacts, one of the most common type of muscle artifact in MEG data, some researchers opt for electrooculography (EOG) to simultaneously record the muscle activity originating near the eyes [8]. These artifacts are then flagged and later removed from the MEG data. However, the EOG electrodes lengthen the setup process and can be uncomfortable to wear for some patients [9] inducing additional artifacts from facial twitching and postural muscle movements [3].
Very little work has been done to automate eye-blink detection without using EOG. Duan et al. [2] train a support vector machine (SVM) using manually selected features from the temporal ICA. They train their model using data from 10 pediatric subjects and report cross-validation specificity of 99.65% and sensitivity of 92.01%. In comparison to this work, using a convolutional neural network (CNN), we report a slightly higher specificity (99.77%) and a 5.61% better sensitivity (97.62%). Furthermore, we have tested on a much larger set of subjects and, instead of cross-validation measures, use a more stringent form of accuracy reporting in which we hold out a test set until after all hyperparameter experimentation has concluded. Additionally, we gain an intuitive understanding of the automatically learned features as our visualizations show.
2. Methods
2.1. Data Collection and Processing
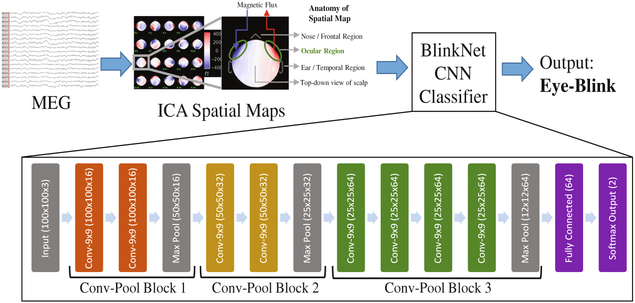
As part of the Imaging Telemetry And Kinematic modeLing in youth football (iTAKL) concussion study [10], 44 male high school football players underwent MEG scans for 8 min. Subjects had their eyes open and fixed on a target to minimize ocular saccadic movement. For preprocessing, we downsampled the signal to 250 Hz, set notch filter to remove harmonics of 60 Hz, and then, applied bandpass filter from 1 Hz to 100 Hz. The preprocessed MEG signal was then decomposed using ICA into 20 components with the InfoMax ICA implementation [11]. We empirically tested multiple component numbers and 20 yielded the most coherent, recognizable spatial maps for our expert MEG scientist. Each component consists of a spatial map indicating the areas of magnetic influx and outflux measured at the scalp and temporal time courses of these maps’ activation over the 8-minute acquisition. To build our classifier in this study, we use the spatial maps as the input. The spatial maps from all subjects were labeled by an expert from our radiology department with more than 5 years of experience in MEG image interpretation (ED). We then randomly selected 14 subjects for training, and 30 subjects were set aside for testing the classifier.
Our Brainstorm toolbox [12] preprocessing pipeline renders spatial topographic maps of ICA as colored RGB images for ease of human interpretation. The spatial maps are generated using the 2D Sensor cap option, which projects the 3D sensors onto a 2D plane and provides a realistic distribution of the sensors while minimizing distortion. Motivated by the success of CNNs in classifying RGB images for the Image-net Large Scale Visual Recognition Challenge (ILSVRC) [13], we decided to build our classifier using a CNN to label these images.
2.2. Recognized CNN Models/Model Selection
In 2012, AlexNet [14] reduced the top-5-error of ILSVRC general object classification from 26.2% to 15.3% using CNNs and established CNNs as the new state of the art methodology for classifying 2D RGB images. In 2014, VGG [15] reduced this error rate to 6.8% by using a deeper model. While these models (VGG and AlexNet) do not work well on our problem directly, our model (Fig. 1) takes inspiration from both of them. Our model required innovation in the appropriate integration and fusion of the best architectural aspects. Like these networks, our CNN architecture also increases in the number of convolution layers per pooling layer as we progress through the architecture. AlexNet uses various convolution filter sizes, some as large as 11 × 11, whereas, VGG uses only 3 × 3 filter size for all the convolution layers. Thus, we decided to use one filter size throughout our network and experimented with different sizes from 3 × 3 to 11 × 11. Similar to AlexNet and VGG, we add 0.5 dropout in the first fully connected layer for regularization [16]. Furthermore, while other researchers report using traditional input feature normalization, including zero-mean and unit-variance transformations, we found these to be insufficient to ensure model convergence. There-fore we applied batch-normalization [17] to the input layer and all the convolution layers, and this resulted in consistent convergence of our model.
Fig. 1.
Automatic eye-blink artifact detection using BlinkNet (CNN)
Our training parameters were as follows: batch size was 16; learning rate was 1e-5, which we reduced by factor of 10 when the training loss plateaus for 4 epochs; optimizer was Adam [18]; and, the number of epochs was 40.
2.3. Cross-Validation/Test Set
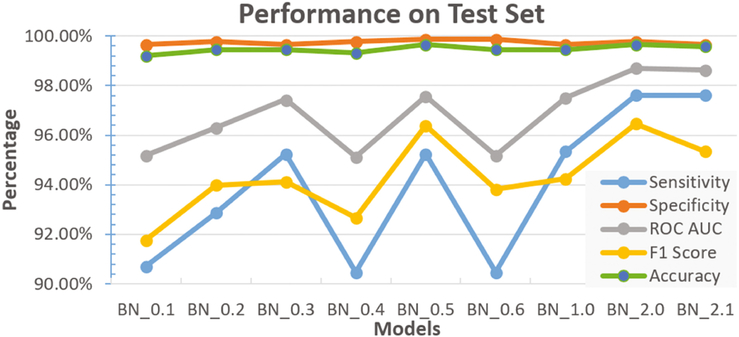
We employed a Leave-One-Subject-Out cross-validation strategy. Using this approach, we repeatedly trained on 13 subjects at a time and evaluated the performance on the 14th subject. Then we computed the mean performance across all folds. We did not use any of the 30 subjects from the test dataset for model selection. Table 1 lists the different models we tested. Our initial models, that we call BlinkNet 0.1 to 0.3, comprised of 6 convolution layers. We found that these models did not perfectly fit the training data set. We reasoned that our classification objective is likely more complex than the statistical complexity of these models, thus we experimented with deeper models consisting of 8 convolution layers. These models did manage to reach 100% training accuracy. Further-more, BlinkNet 1.0 and 2.0 had the best cross-validation scores while BlinkNet 2.1 had slightly worse performance. Our test set included 600 spatial maps from 30 never seen before subjects.
Table 1.
Architectural aspects of the different versions of BlinkNet tested. Highlighted rows are models that achieved best performance during cross-validation.
| Model Name | Filter Size | Convolution Layers | Pool Layers | Total Parameters |
|---|---|---|---|---|
| BlinkNet_0.1 | 3×3 | 6 | 3 | 663,006 |
| BlinkNet_0.2 | 5×5 | 6 | 3 | 790,750 |
| BlinkNet_0.3 | 7×7 | 6 | 3 | 982,366 |
| BlinkNet_0.4 | 3×3 | 8 | 3 | 737,374 |
| BlinkNet_0.5 | 5×5 | 8 | 3 | 996,190 |
| BlinkNet_0.6 | 5×5 | 8 | 4 | 553,822 |
| BlinkNet_1.0 | 7×7 | 8 | 3 | 1,384,414 |
| BlinkNet_2.0 | 9×9 | 8 | 3 | 1,902,046 |
| BlinkNet_2.1 | 11×11 | 8 | 3 | 2,549,086 |
3. Results
We evaluated all of the candidate architectures summarized in Table 1 on the held out test data. The results of this comparison are shown in Fig. 3 and include five measures of performance: sensitivity , specificity , area under the receiver operating characteristic curve (ROC AUC), F1 score , and accuracy . Our proposed model, BlinkNet 2.0 (the model in which we used 8 convolution layers, 9 × 9 filters, and 3 pooling layers) achieved the best performance in all performance measures.
Fig. 3.
Evaluation of our models on previously unseen data from 30 subjects.
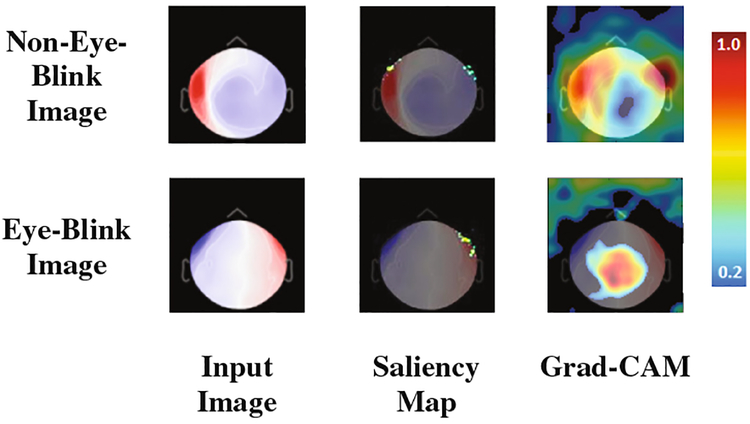
Neural networks (NN) can suffer from limited interpretability, which reduces user confidence and slows adoption of NN based solutions. Thus, we visualize the learned features using saliency maps [19] and gradient-weighted class activation maps (grad-CAM) [20]. Both of these methods give us insight into the image areas the network considers important for classification. Figure 4 shows the visualization results.
Fig. 4.
Attention maps from the CNN. Both Saliency maps and grad-CAM images are created using output from the softmax layer. On the right is the color scheme for the heatmaps.
Saliency maps [19] are created by computing gradients of the softmax output with respect to an input image. Pixels with higher values, shown in yellow-green in the middle column of Fig. 4, correspond to those with greater influence over the CNN output. This provides insight into the influence of each individual pixel on the output value of the model. The areas identified using the saliency map visualization technique highlights the ocular regions. This makes sense in that the presence or absence of magnetic influx or outflux from these areas should be of vital import to identify eye-blink and non-eye-blink spatial maps. While such insights are helpful, a limitation of saliency maps is that they tend to be less useful for determining class specific areas. For example, if a spatial feature is considered important for proper classification of two different classes, it is often observed in the saliency map of both classes. To overcome this limitation we apply an additional visualization method called gradient weighted class activation mapping (grad-CAM).
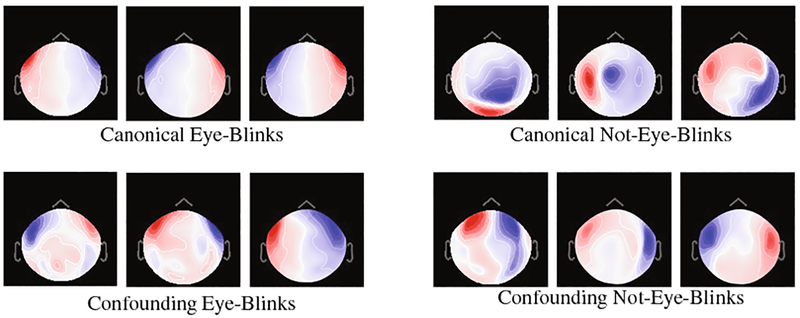
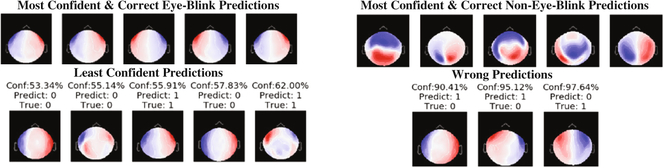
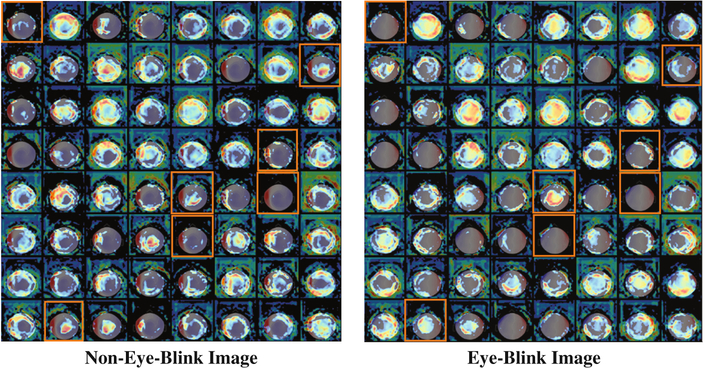
Grad-CAM [20] heatmaps, illustrated in Fig. 4 (third column), are created by visualizing the gradients from the feature maps of the last convolution layer for a given image. Unlike saliency maps, grad-CAM visualizations are class-discriminative, i.e., a feature region is associated with the class with strongest reliance on that region. Applying the approach to the eye-blink and non-eye-blink images reveals the intuitive class discriminative regions that our CNN has learned. For example, the heatmap in the top row shows the bilateral ocular regions while the bottom row shows the center of the scalp region. These are the same regions that human experts tend to rely upon to discriminate these two categories. This information can be combined with the images for which the CNN classification is most confident. Figures 2 and 5 show us the canonical and most confident correct predictions of non-eye-blink images have some signal (prominent red/blue colors) in the central scalp region, and minimal bilateral signal in the ocular regions. Meanwhile, confident and correct predictions of eye-blink images have the inverse: they lack strong signals in the central area and have significant signal in the bilateral ocular regions. These two facts are used together by the two neurons in the softmax layer to make a decision. In Fig. 6, we use grad-CAM to visualize the 64 filters learned in an earlier layer (the last convolution layer). Here we observe that some of this layer’s grad-CAM heatmaps highlight either the left or right ocular area or the central scalp region. This suggests that the CNN is learning individual areas in earlier layers and then combining them in subsequent layers to form discriminative heatmaps for labeling the MEG component images. Finally, we point out that visualization of learned features allows us to appreciate the failure modes of our CNN. If we analyze the three wrong predictions shown in the lower right of Fig. 5, we observe that false positive eye-blink classifications (first two in lower right of Fig. 5) have some bilateral signal in the ocular regions, while the false negative eye-blink (third image in lower right) contains strong signal in central scalp region.
Fig. 2.
Examples of Eye-Blink and Non-Eye-Blink spatial maps. Top row represents canonical examples. Bottom row represents confounding examples.
Fig. 5.
Analyzing the confidence of our model. Confidence level ranges from 50% to 100%. Images in the bottom row state the confidence of the model in the prediction, the prediction, and the true label (0 = Non-Eye-Blink; 1 = Eye-Blink Artifact).
Fig. 6.
Applying grad-CAM to last convolution layer. This illustrates the important spatial areas from the second to last convolution layer that are responsible for activating each of the 64 filters in the last convolution layer. Activations corresponding to the Fig. 4 grad-CAM features are highlighted by orange boxes.
4. Conclusion
In this paper, we have proposed a CNN that accurately detects eye-blink artifacts in MEG and obviates the need for problematic EOG electrodes and wires. Our solution is fully automated; it does not require any manual input at test time. Our end-to-end CNN learns the important features from data derived spatial maps. Through advanced visualization, we reveal the learned features, which largely match those features used by the human experts. We achieve this success without making use of the temporal time courses of the ICA components. We suspect this information is complementary and should improve our model further. In the future, we aim to build on this work by automatically identifying other anomalies such as cardiac artifacts.
Acknowledgements.
The authors would like to thank Jillian Urban, Mireille Kelley, Derek Jones, and Joel Stitzel for their assistance in providing recruitment and study oversight. Support for this research was provided by NIH grant R01NS082453 (JAM, JDS), R03NS088125 (JAM), and R01NS091602 (CW, JAM, JDS).
References
- 1.Fatima Z, Quraan MA, Kovacevic N, et al. : ICA-based artifact correction improves spatial localization of adaptive spatial filters in MEG. NeuroImage 78, 284–294 (2013) [DOI] [PubMed] [Google Scholar]
- 2.Duan F, Phothisonothai M, Kikuchi M, et al. : Boosting specificity of MEG artifact removal by weighted support vector machine. In: Conference proceedings: Annual International Conference of the IEEE EMBS Annual Conference 2013, pp. 6039–6042 (2013) [DOI] [PubMed] [Google Scholar]
- 3.Muthukumaraswamy SD: High-frequency brain activity and muscle artifacts in MEG/EEG: a review and recommendations. Front Hum. Neurosci 7, 138 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buzsáki G, Anastassiou Ca, Koch C.: The origin of extracellular fields and currents-EEG, ECoG: LFP and spikes. Nat. Rev. Neurosci 13(6), 407–420 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Criswell E: Cram’s Introduction to Surface Electromyography (2011) [Google Scholar]
- 6.Gonzalez-Moreno A, Aurtenetxe S, Lopez-Garcia ME, et al. : Signal-to-noise ratio of the MEG signal after preprocessing. J. Neurosci. Methods 222, 56–61 (2014) [DOI] [PubMed] [Google Scholar]
- 7.Gross J, Baillet S, Barnes GR, et al. : Good practice for conducting and reporting MEG research. NeuroImage 65, 349–363 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breuer L, Dammers J, Roberts TPL, et al. : Ocular and cardiac artifact rejection for real-time analysis in MEG. J. Neurosci. Methods 233, 105–114 (2014) [DOI] [PubMed] [Google Scholar]
- 9.Roy RN, Charbonnier S, Bonnet S: Eye blink characterization from frontal EEG electrodes using source separation and pattern recognition algorithms. Biomed. Sig. Process. Control 14, 256–264 (2014) [Google Scholar]
- 10.Davenport EM, Whitlow CT, Urban JE, et al. : Abnormal white matter integrity related to head impact exposure in a season of high school varsity football. J. Neurotrauma 31(19), 1617–1624 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell AJ, Sejnowski TJ: An information-maximization approach to blind separation and blind deconvolution. Neural Comput 7(6), 1129–1159 (1995) [DOI] [PubMed] [Google Scholar]
- 12.Tadel F, Baillet S, Mosher JC, et al. : Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci 2011, 879716 (2011). doi: 10.1155/2011/879716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russakovsky O, Deng J, Su H, et al. : ImageNet large scale visual recognition challenge. Int. J. Comput. Vis 115(3), 211–252 (2015) [Google Scholar]
- 14.Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1–9 (2012) [Google Scholar]
- 15.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. In: International Conference on Learning Representations (ICRL), pp. 1–14 (2015) [Google Scholar]
- 16.Srivastava N, Hinton G, Krizhevsky A, et al. : Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res 15, 1929–1958 (2014) [Google Scholar]
- 17.Ioffe S, Szegedy C: Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Arxiv, pp. 1–11 (2015) [Google Scholar]
- 18.Kingma DP, Adam BJ: A Method for Stochastic Optimization CoRR abs/1412.6980 [Google Scholar]
- 19.Simonyan K, Vedaldi A, Zisserman A: Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps CoRR abs/1312.6034 (2013) [Google Scholar]
- 20.Selvaraju RR, Das A, Vedantam R, et al. : Grad-CAM: Why did you say that? Visual Explanations from Deep Networks via Gradient-based Localization(Nips), pp. 1–5 (2016) [Google Scholar]