Abstract
The P16 (CDKN2Aink4a) gene is an endogenous CDK4/6 inhibitor. Palbociclib (PD0332991) is an anti-CDK4/6 chemical for cancer treatment. P16 is most frequently inactivated by copy number deletion and DNA methylation in cancers. It is well known that cancer cells with P16 deletion are more sensitive to palbociclib than those without. However, whether P16 methylation is related to palbociclib sensitivity is not known. By analyzing public pharmacogenomic datasets, we found that the IC50 of palbociclib in cancer cell lines (n = 522) was positively correlated with both the P16 expression level and P16 gene copy number. Our experimental results further showed that cancer cell lines with P16 methylation were more sensitive to palbociclib than those without. To determine whether P16 methylation directly increased the sensitivity of cancer cells to palbociclib, we induced P16 methylation in the lung cancer cell lines H661 and HCC827 and the gastric cancer cell line BGC823 via an engineered P16-specific DNA methyltransferase (P16-Dnmt) and found that the sensitivity of these cells to palbociclib was significantly increased. The survival rate of P16-Dnmt cells was significantly lower than that of vector control cells 48 hrs post treatment with palbociclib (10 μM). Notably, palbociclib treatment also selectively inhibited the proliferation of the P16-methylated subpopulation of P16-Dnmt cells, further indicating that P16 methylation can increase the sensitivity of cells to this CDK4/6 inhibitor. These results were confirmed in an animal experiment. In conclusion, inactivation of the P16 gene by DNA methylation can increase the sensitivity of cancer cells to palbociclib.
Introduction
The ability to sustain uncontrolled cell proliferation is one of the hallmarks of cancer cells [1]. The normal process of cell division depends on the cell cycle, a series of highly regulated steps manipulated by a set of specific cyclins that act in association with cyclin-dependent kinases (CDKs) [2–4]. The CDK4/6 complex plays a key role in cell cycle progression via monophosphorylation of retinoblastoma protein (RB) and subsequently promotes G1-S phase transition [5, 6].
The clinical implementation of first-generation nonselective CDK inhibitors was originally hampered by the high toxicity and low efficacy of these agents [7, 8]. Second-generation selective CDK4/6 inhibitors, including palbociclib, ribociclib, and abemaciclib, can induce G1 phase cell cycle arrest in RB-positive tumor models with improved effectiveness and reduced adverse effects [9–17]. On the basis of the significant improvements in progression-free survival (PFS) in the PALOMA-1, MONALEESA-2 and MONARCH-1 and 2 clinical trials, palbociclib, ribociclib, and abemaciclib received FDA approval for the treatment of hormone receptor (HR)-positive and human epidermal growth factor receptor 2 (HER2)-negative breast cancer [18–20]. However, not all of these patients could benefit from treatment with CDK4/6 inhibitors [21, 22]. Therefore, biomarkers for predicting the response to these drugs are needed.
The P16 protein, encoded by the CDKN2Aink4a gene, is an endogenous cellular CDK4/6 inhibitor that controls the G1-S phase transition of the cell cycle. This gene is one of the most frequently inactivated genes in cancer genomes; it is inactivated mainly by DNA methylation [23]. It has been reported that cancer cells with P16 copy number deletion are more sensitive to palbociclib than those without [24–28]. Hence, we sought to determine whether cancer cells with P16 methylation exhibit increased sensitivity to therapeutic CDK4/6 inhibitors. Building on these premises, we systematically investigated the relationship between P16 methylation and the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib using both public datasets and cell models of P16 methylation induced by an artificial P16-specific methyltransferase (P16-Dnmt) [29].
Materials and methods
Compliance with ethical standards
All animal experiments were approved by Peking University Cancer Hospital’s Institutional Animal Care and Use Committee and complied with the internationally recognized Animal Research: Reporting of In Vivo Experiments guidelines. This article does not contain any studies with human participants performed by any of the authors.
Dataset sources
P16 (CDKN2Aink4a) gene expression levels and copy numbers in cancer cell lines were downloaded from the Cancer Cell Line Encyclopedia (CCLE) [https://portals.broadinstitute.org/ccle] [30]. The half-maximal inhibitory concentration (IC50) value data for the CDK4/6 inhibitor palbociclib (PD0332991) in various cell lines were downloaded from the Genomics of Drug Sensitivity in Cancer (GDSC) database [https://www.cancerrxgene.org/translation/Drug/1054#t_IC50] [31].
Cell lines and culture
Human lung cancer cell lines H1975, H1395, H661, H596, H460, H358, H292 and HCC827, as well as human breast cancer cell lines HCC1937, MDA-MD-468 and MDA-MD-157, were purchased from the National Infrastructure of Cell Line Resource (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences). Human lung cancer cell lines H1299 and A549; human breast cancer cell lines MDA-MB-231 and MCF7; human gastric cancer cell lines BGC823, MGC803 and AGS; human colon cancer cell lines HCT116, RKO and SW480; and human liver cancer cell line HepG2 were obtained from laboratories at Peking University Cancer Hospital and Institute. The H596 and MCF7 cell lines were cultured in DMEM (Gibco, USA) containing 10% FBS (Gibco) and 1% penicillin-streptomycin (Gibco), the AGS cell line was cultured in F12 medium (Gibco), and the remaining cell lines were cultured in RPMI 1640 medium (Gibco). All cells were maintained at 37°C in humidified air with 5% CO2.
Cell viability assay
The CDK4/6 inhibitor palbociclib (PD0332991) was purchased from Selleck Chemicals (#S1116; USA) and dissolved in double-distilled H2O as a stock solution at concentration of 10 mM. Cells were seeded in 96-well plates at 4,000 cells per well (4 wells/group). Forty-eight hrs post treatment with various concentrations of palbociclib, cell confluence was evaluated and analyzed using the IncuCyte ZOOM system (Essen BioSci, USA). The half-maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 6 software.
Transfection of P16-Dnmt and empty control vector
The established P16-Dnmt expression vector and pTRIPZ empty control vector were prepared and used to transfect cells as previously described [29]. Briefly, to induce methylation of CpG islands around the P16 transcription start site, an engineered P16 promoter-specific seven zinc finger protein (7ZFP) was fused with the catalytic domain of mouse Dnmt3a (approximately 608–908 aa) and integrated into the pTRIPZ vector, which contained a “Tet-On” switch (Open Biosystem, USA). The lung cancer cell lines H661 and HCC827 and the gastric cancer cell line BGC823 were infected with lentiviral particles containing the P16-Dnmt or control vector and incubated for 48 hrs. Then, puromycin (Sigma, USA) was added to the medium (final concentration, 1 μg/mL) to kill nontransfected cells. The pooled cells treated with puromycin for two weeks were considered stably transfected cells. Then, these cells were treated with 0.25 μg/mL doxycycline (Sigma, USA) for 14 days to induce P16-Dnmt expression.
DNA extraction and bisulfite modification
Genomic DNA was extracted from cells or tumor tissues and subjected to bisulfite treatment using an EZ DNA Methylation-Gold Kit (ZYMO RESEARCH, USA) according to the manufacturer’s instructions. The modified DNA was stored at -20°C until use.
Methylation-specific PCR (MSP) and MethyLight assay
The methylation status of P16 CpG islands was assessed by a 150/151-bp methylation-specific PCR (MSP) or 115-bp quantitative MethyLight assay as previously described [32–34]. Briefly, bisulfite-modified genomic DNA was amplified using the methylated/unmethylated P16-specific primer sets in the MSP assay or using the P16-specific primers and probe in the 115-bp MethyLight assay (Table 1). All PCR products were amplified with HotStart Taq DNA polymerase (QIAGEN, Germany). In the MethyLight assay, the level of P16 methylation was determined using an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, USA) and normalized to that of the internal control gene, COL2A1 (collagen type II alpha 1). The relative copy number (RCN) of the methylated P16 CpG islands was calculated according to the formula [2−deltaCt, (deltaCt = Ctmethylated-P16 − CtCOL2A1)].
Table 1. Sequences of oligonucleotides used as primers and probes in various PCR assays.
| Gene | Assay | Oligo name | Primer sequence (5’→3’) | Product size | PCR Tm |
|---|---|---|---|---|---|
| P16 | MSP-M | P16M-F | ttattagagggtggggcggatcgc | 150 bp | 62°C |
| P16M-R | gaccccgaaccgcgaccgtaa | ||||
| P16 | MSP-U | P16U-F | ttattagagggtggggtggattgt | 151 bp | 62°C |
| P16U-R | caaccccaaaccacaaccataa | ||||
| P16 | MethyLight | P16ML-F | cgcggtcgtggttagttagt | 115 bp | 62°C |
| P16ML-R | tacgctcgacgactacgaaa | ||||
| P16ML-Probe | FAM-gttgtttttcgtcgtcggtt-TAMRA | ||||
| COL2A1 | MethyLight | COL2A1-F | tctaacaattataaactccaaccaccaa | 91 bp | 62°C |
| COL2A1-R | gggaagatgggatagaagggaatat | ||||
| COL2A1-Probe | FAM-ccttcattctaacccaatac- ctatcccacctctaaa-BHQ | ||||
| P16 | (q)RT-PCR | P16-qF | gctgcccaacgcaccgaata | 180 bp | 60°C |
| P16-qR | accaccagcgtgtccaggaa | ||||
| GAPDH | (q)RT-PCR | GAPDH-qF | gagatggtgatgggatttc | 224 bp | 58°C |
| GAPDH-qR | gaaggtgaaggtcggagt |
RNA extraction and quantitative RT-PCR (qRT-PCR)
Cells were harvested at a confluence of approximately 70%. Total RNA was extracted from transfected cells or tumor tissues by TRIzol reagent (Thermo Fisher, USA) and reverse transcribed using TransScript First-Strand cDNA Synthesis SuperMix (TransGen, China). The qRT-PCR for P16 mRNA was performed using primer sets shown in Table 1. GAPDH was used as the reference gene. The amplification was performed with FastStart Universal SYBR Green Master (ROX) (Roche, Switzerland) in the Applied Biosystems 7500 Real-Time PCR System, as previously described [35].
Immunofluorescence and confocal microscopy analysis
Cells were fixed in 4% polyformaldehyde for 10 min at room temperature, treated with 1% Triton X-100 in PBS for 10 min, blocked with 5% bovine serum albumin (BSA) for 1 hr, and hybridized to a mouse monoclonal antibody against the P16 protein (Ventana Roche Diagnostics, E6H4, Switzerland) overnight at 4°C. Samples were incubated with FITC-labeled secondary antibody (KPL, 172–1806, USA) for 1 hr at room temperature, followed by nuclear staining with DAPI. Fluorescence images were acquired and analyzed with an ImageXpress Micro High Content Screening System (Molecular Devices, USA).
Western blot analysis
Primary monoclonal antibodies against RB (Abcam, ab181616, UK), Ser780-phosphorylated RB (p-RB) (Cell Signaling Technology, #9307, USA), P16 (Abcam, ab108349), and GAPDH (Proteintech Group, 60004-1-Ig, USA) were diluted at 1:1000, 1:1000, 1:1000 and 1:15000, respectively. Signals were visualized using Immobilon Western HRP Substrate (Millipore, USA).
BALB/c nude mouse xenograft model and treatment
HCC827 cells (2 × 106 cells in 200 μL of PBS) stably transfected with the P16-Dnmt vector or control vector and induced with 0.25 μg/mL doxycycline for 7 days were injected subcutaneously into the lower limbs of BALB/c nude mice (female, 5 weeks old, weighing 18–22 g, purchased from Beijing Huafukang Biotech). Mice were provided access to distilled, sterile water containing 2 μg/mL doxycycline ad libitum. When the tumor size reached approximately 200 mm3, on the 30th day post transplantation, mice were randomized into the control group and the palbociclib group (6 mice/group) and treated with control buffer or palbociclib [100 mg/kg in 50 mM sodium lactate buffer (pH 4.0)] via oral gavage (i.g.) daily for 3 weeks. Then, mice were sacrificed by spine dislocation, and the tumors were weighed. Tumor samples were processed for formalin-fixed and paraffin-embedded (FFPE) sections or Western blot analysis.
Immunohistochemistry (IHC)
For IHC, after dewaxing, rehydration, endogenous peroxidase quenching and blocking with 5% BSA according to standard procedures, 4-μm thick formalin-fixed, paraffin-embedded (FFPE) sections were incubated with primary antibody against Ki-67 (1:300) overnight at 4°C. The remaining IHC steps were the same as previously described [36].
Statistical analysis
Statistical analyses were performed using SPSS 23 software or GraphPad Prism 6 software. The Kolmogorov–Smirnov test and Shapiro–Wilk test were used to estimate the normality of distributions. The relationship between the IC50 value of palbociclib and the relative P16 (CDKN2Aink4a) mRNA level or copy number in various cancer cell lines was assessed using the nonparametric Spearman correlation test and a linear regression model. The Mann–Whitney test was used for nonnormally distributed data. Student’s t-test was used for normally distributed data. Statistical significance was accepted at p<0.05.
Results
P16 methylation is positively correlated with high sensitivity of cancer cells to palbociclib
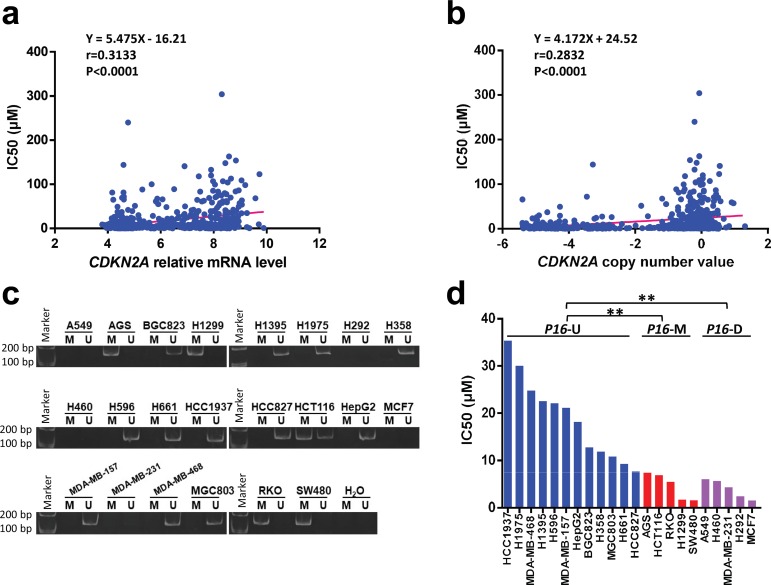
To explore the relationship between P16 methylation and the sensitivity of cancer cells to palbociclib, we first analyzed the correlation between the IC50 levels of palbociclib in 522 cell lines and the transcription level and copy number of the CDKN2A/P16 gene using public pharmacogenomics datasets [30, 31]. The analysis results revealed that the palbociclib IC50 levels in these cell lines were positively correlated with the CDKN2A mRNA level (r = 0.3133, p<0.0001; Fig 1A) and CDKN2A copy number (r = 0.2832, p<0.0001; Fig 1B). This result suggests that cancer cell lines with low or no P16 expression are more sensitive to palbociclib than those with high P16 expression.
Fig 1. Association between P16 expression and methylation and palbociclib sensitivity in cancer cell lines.
(a and b) Correlations between the palbociclib IC50 levels and CDKN2A/P16 mRNA levels and copy number in cancer cell lines (n = 522). The p-values were measured by a nonparametric Spearman correlation test and linear regression model. (c) The methylation status of the P16 gene in different cell lines as assessed by MSP. (d) The palbociclib IC50 values in 22 cancer cell lines with different P16 alterations, including P16 copy number deletion (P16-D), P16 methylation (P16-M), and P16 unmethylated and undeleted (16-U). Student’s t-test, **p<0.01.
Then, we validated the correlation in 22 cancer cell lines, which were divided into P16-unmethylated (P16-U), P16-methylated (P16-M), and P16-deleted (P16-D) groups according to the results of MSP (Fig 1C). We found that the IC50 values in the P16-U cell lines (n = 12) were consistently higher than those in the P16-M cell lines (n = 5) and P16-D cell lines (n = 5) [18.88 vs. 4.61 and 3.99 (μM), p<0.01; Fig 1D]. There was no difference in the IC50 values between the P16-M and P16-D cell lines. These results indicate that similar to copy number deletion, P16 methylation is positively associated with high sensitivity of cancer cells to palbociclib.
P16-specific methylation directly increases the sensitivity of cancer cells to palbociclib
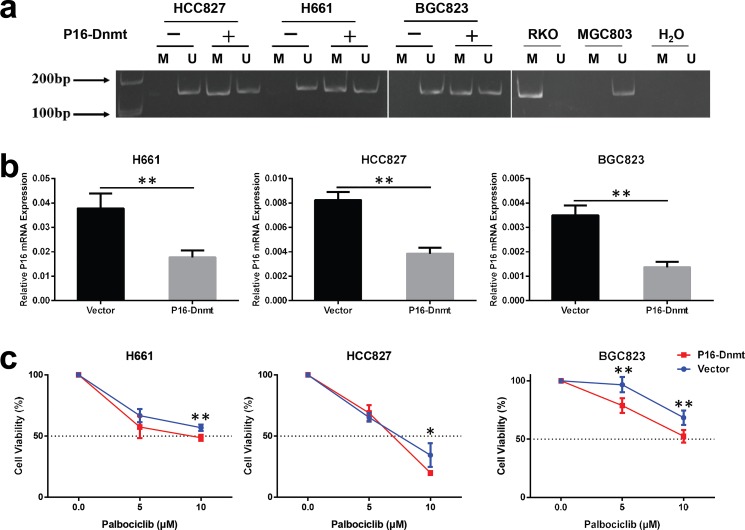
To determine whether P16 methylation directly increases the sensitivity of cancer cells to palbociclib, we stably transfected three P16-U cell lines (including two lung cancer cell lines, H661 and HCC827, and one gastric cancer cell line, BGC823) with the P16-Dnmt vector and induced P16-Dnmt expression in these cells by doxycycline treatment for 2 weeks. P16 methylation was successfully induced in these P16-Dnmt cells, as evidenced by MSP (Fig 2A). qRT-PCR analysis revealed that the level of P16 expression was markedly decreased in these P16-Dnmt cells (Fig 2B). Then, these cells were treated with palbociclib at different concentrations (0, 5, 10 μM), and cell viability (survival rate) was assessed. The results showed that epigenetic inactivation of the P16 gene by P16-Dnmt significantly decreased the viability of palbociclib-treated cells (Fig 2C). Forty-eight hrs post treatment with palbociclib (final concentration, 10 μM), the survival rate of P16-Dnmt cells was significantly decreased compared to that of vector control cells: 48.6% vs. 56.9% for H661 cells, 19.9% vs. 34.5% for HCC827 cells, and 52.5% vs. 68.3% for BGC823 cells. These results indicate that P16-specific methylation can directly increase the sensitivity of cancer cells to palbociclib.
Fig 2. Effect of P16-specific methylation on the sensitivity of cancer cells to palbociclib.
(a) Methylation of the CpG P16 island in three cancer cell lines stably transfected with an engineered P16-specific DNA methyltransferase (P16-Dnmt) by MSP. Genomic DNA samples from RKO and MGC803 cells were used as the positive and negative controls for P16 methylation, respectively. (b) Effect of P16-specific methylation on the levels of P16 expression in cell lines stably transfected with P16-Dnmt, as assessed by qRT-PCR. (c) Effect of P16-specific methylation on the viability (survival rate) of cells treated with palbociclib for 48 hrs, as assessed with the IncuCyte ZOOM system. Each point represents the mean ± SD of 4 wells. Student’s t-test, *p<0.05, **p<0.01.
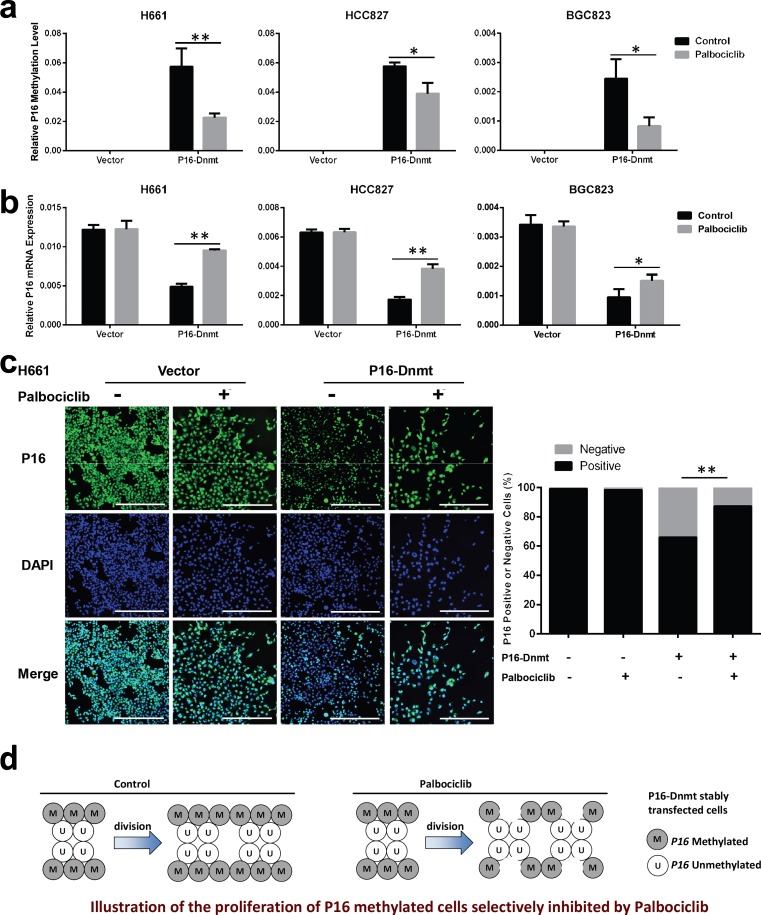
Selective inhibition of the proliferation of P16-methylated cancer cells by palbociclib in vitro
As described above, P16 inactivation by methylation was not detected in approximately one/third of P16-Dnmt cells (Fig 2B). Therefore, we further investigated whether palbociclib treatment could selectively inhibit the proliferation of the P16-methylated subpopulation of P16-Dnmt cells. As expected, the population of P16-methylated cells was significantly decreased by palbociclib treatment in the H661, HCC827, and BGC823 cell lines stably transfected with P16-Dnmt (Fig 3A). In contrast, the P16 mRNA level was significantly increased (Fig 3B) by palbociclib treatment, while the P16 mRNA level in the vector control cells was not affected. Furthermore, immunofluorescence and confocal microscopy analyses confirmed the qRT-PCR results. An increase in the number of P16 protein-positive cells was observed in these P16-Dnmt cell lines treated with palbociclib (Fig 3C, S1 and S2 Figs). Such a change was not observed in vector control cells. These results show that the proliferation of P16-methylated (or P16-negative) cells is selectively inhibited by palbociclib treatment (Fig 3D).
Fig 3. Selective inhibition of the proliferation of P16-methylated cancer cells by palbociclib in vitro.
After palbociclib treatment (H661 and BGC823 cells: 10 μM; HCC827 cells: 7 μM) for 48 hrs, cells were harvested at 70–80% confluence. (a and b) Effect of palbociclib treatment on the levels of P16-methylated alleles and P16 mRNA in cell lines stably transfected with P16-Dnmt, as assessed by MSP and qRT-PCR assays, respectively. The data are presented as the means ± SDs. (c) The results of immunofluorescence and confocal microscopy analyses to directly detect changes in the subpopulation of cells staining positive for P16 protein within the H661 cell line stably transfected with P16-Dnmt. (d) Illustration of the proliferation of P16 methylated cells selectively inhibited by palbociclib treatment. Scale bar, 400 μm. Student’s t-test, *p<0.05, **p<0.01.
Selective inhibition of the growth of P16-methylated cancer cells by palbociclib in nude mice
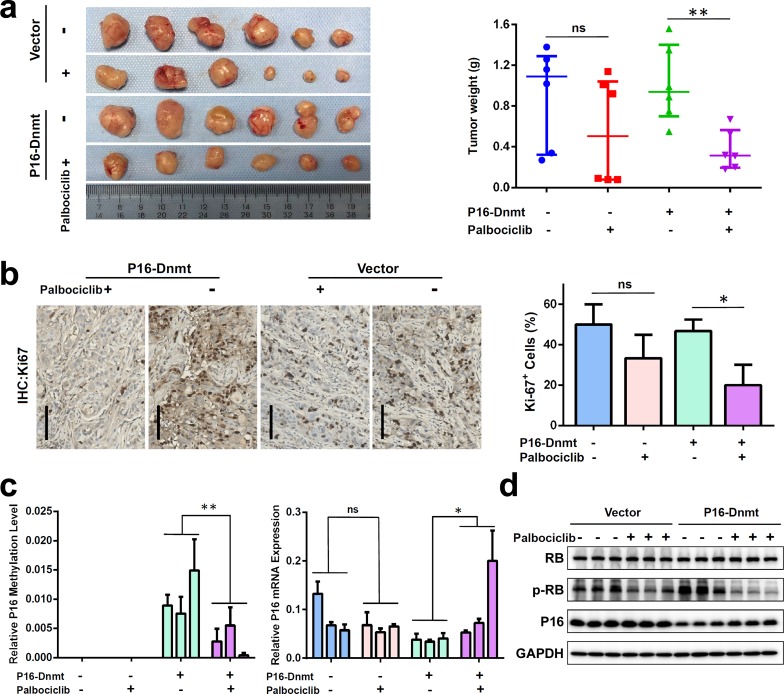
To verify the selective inhibition of P16-methylated cancer cells by palbociclib described above, HCC827 cells stably transfected with P16-Dnmt were subcutaneously transplanted into nude mice. When the xenograft tumor size reached approximately 200 mm3, mice were randomized and treated with palbociclib (100 mg/kg b.w., i.g.) or control buffer for three weeks. No adverse event was observed in these mice. Compared with control buffer, palbociclib significantly inhibited the growth of P16-Dnmt tumors (p<0.01; Fig 4A). Consistent with this result, the proportion of Ki67-positive cells was significantly decreased in P16-Dnmt tumors treated with palbociclib but not in control vector tumors (Fig 4B).
Fig 4. Effect of palbociclib treatment on the growth of tumors derived from HCC827 cancer cells with and without stable transfection of P16-Dnmt in nude mice.
(a) Images of HCC827 xenograft tumors in different groups of mice (6 mice/group). These mice were treated with palbociclib (100 mg/kg b.w., i.g.) or control buffer for 3 weeks. A tumor weight chart is inserted on the right side. The data are expressed as the medians ± interquartile ranges; Mann–Whitney test, **p<0.01. (b) Immunohistochemical staining for Ki-67 in HCC827 xenograft tumors in the different groups (scale bar, 100 μm). The percentage of Ki-67+ cells is presented as the mean ± SD of three sections. Ki-67+ cells were counted in 5 random fields per section. (c) The levels of methylated P16 alleles (left chart) and P16 expression (middle chart) in three representative tumors from each group, as assessed by MethyLight and qRT-PCR assays. (d) The levels of RB, phosphorylated RB (p-RB), and P16 proteins in three representative tumors from each group, as assessed by Western blot. Student’s t-test, *p<0.05, **p<0.01.
Moreover, the level of methylated P16 alleles was significantly decreased in three representative P16-Dnmt tumors treated with palbociclib, while the level of P16 mRNA was significantly increased in these tumors (Fig 4C). Changes in the P16 mRNA level in control vector tumors could not be induced by palbociclib treatment. In addition, more P16 protein was detected in P16-Dnmt tumors treated with palbociclib than in those treated with control buffer, as evidenced by Western blot analysis (Fig 4D). Such a difference was not observed between control vector tumors with and without palbociclib treatment. In contrast, the p-RB levels in P16-Dnmt tumors treated with palbociclib were lower than those in the control vector tumors treated with palbociclib. Together, these results indicate that palbociclib can selectively inhibit the growth of P16-methylated cells in vivo.
Discussion
On the basis of its good performance in clinical trials, the specific CDK4/6 inhibitor palbociclib has been approved by the FDA and EMA for HR-positive and HER2-negative advanced breast cancer. The P16 protein encoded by the CDKN2A gene is an endogenous CDK4/6 inhibitor. This gene is most frequently inactivated in human cancer genomes by copy number deletion, with a frequency of about 10%, and DNA methylation, with a frequency of approximately 30%. It has been reported that CDK4/6 inhibitors can effectively inhibit processes in tumor cells that lose endogenous inhibition of CDK4/6 because of P16 gene deletion [14, 24]. However, it is not known whether P16 inactivation by DNA methylation may similarly increase the sensitivity of cancer cells to these CDK4/6 inhibitors. In the present study, we found, for the first time, that the proliferation and growth of P16-methylated cancer cells can be selectively inhibited by palbociclib in vitro and in vivo.
It has been reported recently that high CDK4 target engagement by palbociclib in cells with knockout of CDKN2A gene and attenuated target engagement when CDKN2A/P16 is abundant [37]. Our bioinformatics analysis via mining public pharmacogenomic datasets indicates that the IC50 value of palbociclib is positively correlated with the transcription level of the CDKN2A gene in 522 cancer cell lines. As P16 methylation can directly inactivate gene transcription [29], we found that similar to cell lines with P16 deletion, P16-methylated cancer cell lines were more sensitive to palbociclib than P16-unmethylated cell lines. Notably, P16-specific methylation by P16-Dnmt directly increased the sensitivity of lung cancer cells to palbociclib in vitro and in vivo. Furthermore, we observed that palbociclib selectively inhibited the proliferation and growth of P16-methylated cells in P16-Dnmt tumors. Lower protein levels of p-RB were detected in P16-Dnmt tumors than in control vector tumors in nude mice treated with palbociclib. These phenomena strongly suggest that P16-methylated cells have increased sensitivity to CDK4/6 inhibitor drugs.
In the phase II clinical study PALOMA-1, it was found that patient selection based on CCDN1 amplification and/or P16 loss (CDKN2A deletion) did not improve outcomes of the administration of palbociclib plus letrozole in HR-positive breast cancer patients [38]. In the phase III PALOMA-2 trial, although the expression levels of genes in the Cyclin D-CDK4/6-RB pathway did not correlate with a benefit from palbociclib plus letrozole, 59 breast cancer patients without P16 expression exhibited a benefit [39, 40]. In another phase I study of the CDK4/6 inhibitor ribociclib (LEE011), CCND1 amplification vs. CDKN2A and CDKN2B codeletion trended toward a longer vs. shorter treatment duration [12]. We recently reported that P16 methylation could increase the resistance of lung (and stomach) cancers to paclitaxel treatment in a clinical trial [41]. In contrast, here, we reveal that P16 methylation can increase the sensitivity of lung and stomach cancers to palbociclib. It is worth performing clinical trials to assess whether P16 methylation could be used to predict the therapeutic efficacy of CDK4/6 inhibitors such as palbociclib in patients with lung and stomach cancers.
Conclusion
This study demonstrates that P16 methylation can increase the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib. P16 methylation may be used as a predictive marker for sensitivity to CDK4/6 inhibitors.
Supporting information
The results of immunofluorescence and confocal microscopy analyses to directly detect alterations in the subpopulation of cells staining positive for P16 protein within HCC827 lung cancer cells stably transfected with P16-Dnmt. Scale bar, 400 μm. Student’s t-test, **p<0.01.
(TIF)
The results of immunofluorescence and confocal microscopy analysis to directly detect alterations of the subpopulation of cells staining positive for P16 protein within the BGC823 gastric cancer cells stably transfected with P16-Dnmt. Scale bar, 400 μm. Student’s t-test, **p<0.01.
(TIF)
(PDF)
(PDF)
Acknowledgments
This work was financially supported by a grant from the Beijing Municipal Commission of Health and Family Planning (PXM2018_026279_00005) to DD.
Abbreviations
- 7ZFP
seven zinc finger protein
- BSA
bovine serum albumin
- CCLE
Cancer Cell Line Encyclopedia
- CDK
cyclin-dependent kinase
- GDSC
Genomics of Drug Sensitivity in Cancer
- HER2
human epidermal growth factor receptor 2
- HR
hormone receptor
- IC50
half-maximal inhibitory concentration
- MSP
methylation-specific PCR
- P16-Dnmt
P16-specific methyltransferase
- PFS
progression-free survival
- RB
retinoblastoma protein
- RCN
relative copy number
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by a grant from the Beijing Municipal Commission of Health and Family Planning (PXM2018_026279_00005) to DD. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144(5):646–74. 10.1016/j.cell.2011.02.013 PubMed PMID: WOS:000288007100007. [DOI] [PubMed] [Google Scholar]
- 2.Nurse P. Cyclin dependent kinases and cell cycle control (Nobel lecture). Chembiochem. 2002;3(7):596–+. PubMed PMID: WOS:000176625200001. [DOI] [PubMed] [Google Scholar]
- 3.Lim SH, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–93. 10.1242/dev.091744 PubMed PMID: WOS:000321864900002. [DOI] [PubMed] [Google Scholar]
- 4.Malumbres M, Barbacid M. To cycle or not to cycle: A critical decision in cancer. Nature Reviews Cancer. 2001;1(3):222–31. 10.1038/35106065 PubMed PMID: WOS:000180397200014. [DOI] [PubMed] [Google Scholar]
- 5.Macaluso M, Montanari M, Giordano A. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 2006;25(38):5263–7. 10.1038/sj.onc.1209680 PubMed PMID: WOS:000240064100011. [DOI] [PubMed] [Google Scholar]
- 6.Harbour JW, Luo RX, Santi AD, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–69. 10.1016/s0092-8674(00)81519-6 PubMed PMID: WOS:000082679200012. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24(11):1770–83. 10.1200/JCO.2005.03.7689 PubMed PMID: WOS:000236783900017. [DOI] [PubMed] [Google Scholar]
- 8.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nature Reviews Drug Discovery. 2015;14(2):130–46. 10.1038/nrd4504 PubMed PMID: WOS:000348968100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade MA, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Molecular Cancer Therapeutics. 2004;3(11):1427–37. PubMed PMID: WOS:000225070800010. [PubMed] [Google Scholar]
- 10.DeMichele A, Clark AS, Tan KS, Heitjan DF, Gramlich K, Gallagher M, et al. CDK 4/6 Inhibitor Palbociclib (PD0332991) in Rb+ Advanced Breast Cancer: Phase II Activity, Safety, and Predictive Biomarker Assessment. Clinical Cancer Research. 2015;21(5):995–1001. 10.1158/1078-0432.CCR-14-2258 PubMed PMID: WOS:000351982800011. [DOI] [PubMed] [Google Scholar]
- 11.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. New Engl J Med. 2016;375(18):1738–48. 10.1056/NEJMoa1609709 PubMed PMID: WOS:000387007300007. [DOI] [PubMed] [Google Scholar]
- 12.Infante JR, Cassier PA, Gerecitano JF, Witteveen PO, Chugh R, Ribrag V, et al. A Phase I Study of the Cyclin-Dependent Kinase 4/6 Inhibitor Ribociclib (LEE011) in Patients with Advanced Solid Tumors and Lymphomas. Clinical Cancer Research. 2016;22(23):5696–705. 10.1158/1078-0432.CCR-16-1248 PubMed PMID: WOS:000389438100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patnaik A, Rosen LS, Tolaney SM, Tolcher AW, Goldman JW, Gandhi L, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer discovery. 2016;6(7):740–53. 10.1158/2159-8290.CD-16-0095 PubMed PMID: WOS:000383354500025. [DOI] [PubMed] [Google Scholar]
- 14.Ingham M, Schwartz GK. Cell-Cycle Therapeutics Come of Age. J Clin Oncol. 2017;35(25):2949–+. 10.1200/JCO.2016.69.0032 PubMed PMID: WOS:000408568300015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nature reviews Clinical oncology. 2016;13(7):417–30. Epub 2016/04/01. 10.1038/nrclinonc.2016.26 . [DOI] [PubMed] [Google Scholar]
- 16.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer discovery. 2016;6(4):353–67. 10.1158/2159-8290.CD-15-0894 PubMed PMID: WOS:000375843800020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roskoski R. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacological Research. 2019;139:471–88. 10.1016/j.phrs.2018.11.035 PubMed PMID: WOS:000458709000047. [DOI] [PubMed] [Google Scholar]
- 18.Beaver JA, Amiri-Kordestani L, Charlab R, Chen W, Palmby T, Tilley A, et al. FDA Approval: Palbociclib for the Treatment of Postmenopausal Patients with Estrogen Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Clinical Cancer Research. 2015;21(21):4760–6. 10.1158/1078-0432.CCR-15-1185 PubMed PMID: WOS:000364488100005. [DOI] [PubMed] [Google Scholar]
- 19.Shah A, Bloomquist E, Tang SH, Fu WT, Bi YW, Liu Q, et al. FDA Approval: Ribociclib for the Treatment of Postmenopausal Women with Hormone Receptor-Positive, HER2-Negative Advanced or Metastatic Breast Cancer. Clinical Cancer Research. 2018;24(13):2999–3004. 10.1158/1078-0432.CCR-17-2369 PubMed PMID: WOS:000437270800005. [DOI] [PubMed] [Google Scholar]
- 20.Corona SP, Generali D. Abemaciclib: a CDK4/6 inhibitor for the treatment of HR+/HER2-advanced breast cancer. Drug Des Dev Ther. 2018;12:321–30. 10.2147/Dddt.S137783 PubMed PMID: WOS:000425328800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schettini F, De Santo I, Rea CG, De Placido P, Formisano L, Giuliano M, et al. CDK 4/6 Inhibitors as Single Agent in Advanced Solid Tumors. Front Oncol. 2018;8 10.3389/fonc.2018.00608 PubMed PMID: WOS:000453070700002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teh JLF, Aplin AE. Arrested Developments: CDK4/6 Inhibitor Resistance and Alterations in the Tumor Immune Microenvironment. Clinical Cancer Research. 2019;25(3):921–7. 10.1158/1078-0432.CCR-18-1967 PubMed PMID: WOS:000457395900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao R, Choi BY, Lee MH, Bode AM, Dong ZG. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine. 2016;8:30–9. 10.1016/j.ebiom.2016.04.017 PubMed PMID: WOS:000378622200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong Y, Sheng X, Wu X, Yan J, Ma M, Yu J, et al. Frequent Genetic Aberrations in the CDK4 Pathway in Acral Melanoma Indicate the Potential for CDK4/6 Inhibitors in Targeted Therapy. Clin Cancer Res. 2017. Epub 2017/08/24. 10.1158/1078-0432.CCR-17-0070 . [DOI] [PubMed] [Google Scholar]
- 25.Konecny GE, Winterhoff B, Kolarova T, Qi JW, Manivong K, Dering J, et al. Expression of p16 and Retinoblastoma Determines Response to CDK4/6 Inhibition in Ovarian Cancer. Clinical Cancer Research. 2011;17(6):1591–602. 10.1158/1078-0432.CCR-10-2307 PubMed PMID: WOS:000288435300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cen L, Carlson BL, Schroeder MA, Ostrem JL, Kitange GJ, Mladek AC, et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 2012;14(7):870–81. Epub 2012/06/20. 10.1093/neuonc/nos114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young RJ, Waldeck K, Martin C, Foo JH, Cameron DP, Kirby L, et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res. 2014;27(4):590–600. Epub 2014/02/06. 10.1111/pcmr.12228 . [DOI] [PubMed] [Google Scholar]
- 28.Wiedemeyer WR, Dunn IF, Quayle SN, Zhang JH, Chheda MG, Dunn GP, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. P Natl Acad Sci USA. 2010;107(25):11501–6. 10.1073/pnas.1001613107 PubMed PMID: WOS:000279058000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui C, Gan Y, Gu L, Wilson J, Liu Z, Zhang B, et al. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome biology. 2015;16:252 10.1186/s13059-015-0819-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. 10.1038/nature11003 PubMed PMID: WOS:000302006100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–U87. 10.1038/nature11005 PubMed PMID: WOS:000302006100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. P Natl Acad Sci USA. 1996;93(18):9821–6. 10.1073/pnas.93.18.9821 PubMed PMID: WOS:A1996VF61400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Cao J, Lu ZM, Liu HW, Deng DJ. A 115-bp MethyLight assay for detection of p16 (CDKN2A) methylation as a diagnostic biomarker in human tissues. Bmc Med Genet. 2011;12 10.1186/1471-2350-12-67 PubMed PMID: WOS:000291947000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang BZ, Xiang SY, Zhong QM, Yin YR, Gu LK, Deng DJ. The p16-Specific Reactivation and Inhibition of Cell Migration Through Demethylation of CpG Islands by Engineered Transcription Factors. Hum Gene Ther. 2012;23(10):1071–81. 10.1089/hum.2012.070 PubMed PMID: WOS:000310364400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gan Y, Ma WR, Wang XH, Qiao JL, Zhang BZ, Cui CH, et al. Coordinated transcription of ANRIL and P16 genes is silenced by P16 DNA methylation. Chinese J Cancer Res. 2018;30(1):93–+. 10.21147/j.issn.1000-9604.2018.01.10 PubMed PMID: WOS:000426439800010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian W, Du YT, Ma YW, Gu LK, Zhou J, Deng DJ. MALAT1-miR663a negative feedback loop in colon cancer cell functions through direct miRNA-lncRNA binding. Cell Death & Disease. 2018;9 10.1038/s41419-018-0925-y PubMed PMID: WOS:000443087500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green JL, Okerberg ES, Sejd J, Palafox M, Monserrat L, Alemayehu S, et al. Direct CDKN2 Modulation of CDK4 Alters Target Engagement of CDK4 Inhibitor Drugs. Mol Cancer Ther. 2019;18(4):771–9. Epub 2019/03/07. 10.1158/1535-7163.MCT-18-0755 . [DOI] [PubMed] [Google Scholar]
- 38.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. 10.1016/S1470-2045(14)71159-3 PubMed PMID: WOS:000346912800042. [DOI] [PubMed] [Google Scholar]
- 39.Finn R, Jiang Y, Rugo H, Moulder SL, Im SA, Gelmon KA, et al. Biomarker analyses from the phase 3 PALOMA-2 trial of palbociclib (P) with letrozole (L) compared with placebo (PLB) plus L in postmenopausal women with ER +/HER2-advanced breast cancer (ABC). Annals of Oncology. 2016;27 10.1093/annonc/mdw435.5 PubMed PMID: WOS:000393913000694. [DOI] [Google Scholar]
- 40.Finn RS, Liu Y, Martin M, Rugo HS, Dieras V, Im SA, et al. Comprehensive gene expression biomarker analysis of CDK 4/6 and endocrine pathways from the PALOMA-2 study. Cancer Research. 2018;78(4). PubMed PMID: WOS:000425489400257. [Google Scholar]
- 41.Liu Z, Lin H, Gan Y, Cui C, Zhang B, Gu L, et al. P16 Methylation Leads to Paclitaxel Resistance of Advanced Non-Small Cell Lung Cancer. J Cancer. 2019;10(7):1726–33. Epub 2019/06/18. 10.7150/jca.26482 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The results of immunofluorescence and confocal microscopy analyses to directly detect alterations in the subpopulation of cells staining positive for P16 protein within HCC827 lung cancer cells stably transfected with P16-Dnmt. Scale bar, 400 μm. Student’s t-test, **p<0.01.
(TIF)
The results of immunofluorescence and confocal microscopy analysis to directly detect alterations of the subpopulation of cells staining positive for P16 protein within the BGC823 gastric cancer cells stably transfected with P16-Dnmt. Scale bar, 400 μm. Student’s t-test, **p<0.01.
(TIF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.