Abstract
Optogenetics is a technology that allows targeted, fast control of precisely defined events in biological systems as complex as freely moving mammals. By delivering optical control at the speed (millisecond-scale) and with the precision (cell type–specific) required for biological processing, optogenetic approaches have opened new landscapes for the study of biology, both in health and disease.
Optogenetics1 is the combination of genetic and optical methods to achieve gain or loss of function of well-defined events in specific cells of living tissue. In the broadest sense2, optogenetics encompasses a core technology—targetable control tools that respond to light and deliver effector function—and enabling technologies for (i) delivering light into tissues under investigation, (ii) targeting the control tools to cells of interest and (iii) obtaining compatible readouts and performing analysis, such as targeted imaging or electrical recording of evoked activity.
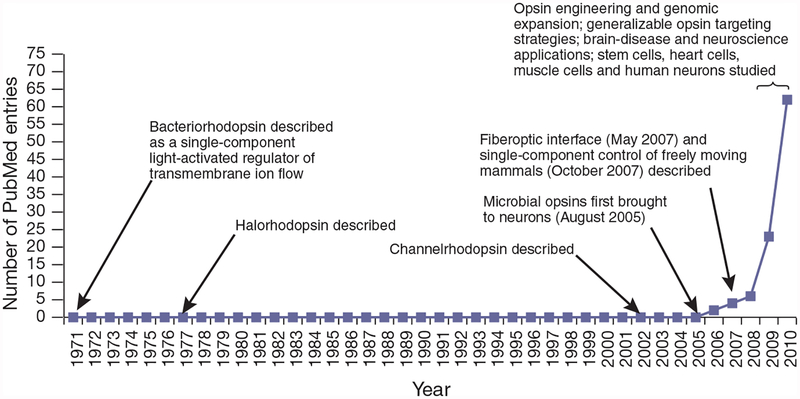
Certain elements have been known to exist in earlier forms and in other contexts, though not conceptualized or developed as a control technology, as far back3 as 1971, with their fundamental transition to the emergence of optogenetics beginning in 2005 (Fig. 1) triggered by the demonstration of single-component control tools in neuroscience: microbial opsin genes that could safely confer to neurons both light-detection capability and defined high-speed effector function in a single readily targetable module4.
Figure 1 |.
Graphical illustration of ‘optogenetics’ emerging in the scientific literature. Demonstration of single-component optogenetic control of neurons with microbial opsins4 was followed by corresponding optogenetic terminology2 in October 2006, and corresponding optogenetic control of freely moving mammals using microbial opsins and the fiberoptic neural interface9,10. Also marked are identifications of bacteriorhodopsin3, halorhodopsin5 and channelrhodopsin6, all of which were much later (2005–2010) shown to function as fast, single-component optogenetic tools in neurons. Numbers indicate only publications searchable by ‘optogenetics’ or derivatives thereof on 1 December 2010.
Although it arose from neuroscience, optogenetics addresses a much broader unmet need in the study of biological systems: the need to control defined events in defined cell types at defined times in intact systems. Such analyses are important because cellular events are typically meaningful only in the context of other events occurring in the rest of the tissue, the organism and the environment as a whole.
Single-component optogenetics
In 1979 Francis Crick suggested that the major challenge facing neuroscience was the need to control one type of cell in the brain while leaving others unaltered. As electrodes cannot be used to precisely target defined cells and drugs act much too slowly, Crick later speculated that light might have the properties to serve as a control tool, but at the time neuroscientists knew of no clear strategy to make specific cells responsive to light.
Yet 40 years ago microbial biologists knew that some microorganisms produce visible light–gated proteins that directly regulate the flow of ions across the plasma membrane. In 1971, Stoeckenius and Oesterhelt discovered that bacteriorhodopsin acts as an ion pump that can be rapidly activated by visible-light photons3. And this original theme from 1971 of single-gene, single-component control continued with the later identification of other members of this family: halorhodopsin in 1977 by Matsuno-Yagi and Mukohata5, and channelrhodopsin in 2002 by Hegemann, Nagel and their colleagues6.
But it took more than 30 years for neuroscientists to bring the two fields together because such an approach was thought to be very unlikely to work. Instead, scientists considered multicomponent strategies7,8 that involved no microbial opsin genes at all but rather cascades of different genes or combinations of custom-synthesized chemicals and genes. It was also likely that these foreign membrane proteins would be toxic to fragile mammalian neurons, with photo-currents that would also likely be too slow and weak to be useful. Moreover, scientists did not believe that this approach would lead to the long-sought single-component strategy (a perception that had also slowed the development and application of green fluorescent protein), as microbial opsins require a chemical cofactor, all-trans retinal, to absorb photons.
An August 2005 report, however, described that upon introduction of a microbial opsin gene without any other parts, chemicals or components, neurons became precisely responsive to light4. Several additional reports followed over the next year, and by 2010 channelrhodopsin, bacteriorhodopsin and halorhodopsin all had proved capable of turning neurons on or off, rapidly and safely, in response to diverse colors of light (Fig. 2). Vertebrate tissues contain natural all-trans retinal—the cofactor essential for photonic control of microbial opsins—and as a result researchers showed that optogenetic control was feasible even in intact mammalian brain tissue2 and freely moving mammals9,10. In a fundamental shift from earlier approaches, microbial opsin genes therefore provided a single-component strategy.
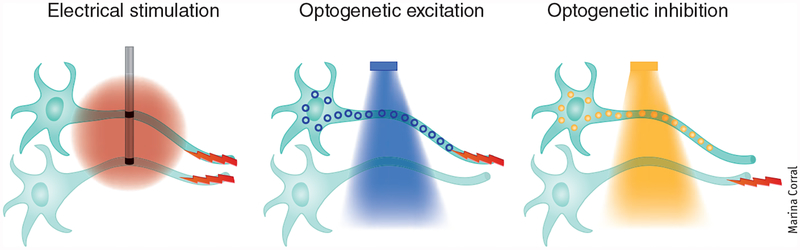
Figure 2 |.
Principle of optogenetics in neuroscience. Targeted excitation (as with a blue light–activated channelrhodopsin) or inhibition (as with a yellow light–activated halorhodopsin), conferring cellular specificity and even projection specificity not feasible with electrodes while maintaining high temporal (action-potential scale) precision.
Optogenetic tools have now changed the way neuroscience is conducted owing to a convergence over the past two years of the intrinsic tractability of the single-component tools with rapid advances in the associated enabling technologies. Obtaining precise causal control in intact systems as complex as behaving mammals is certainly important in neuroscience, just as in other fields of biology, but historically, it has not been possible to deliver causal, temporally precise gain or loss of function in one type of brain cell or in a defined projection from one brain region to another.
Now, in addition to conferring temporal precision and applicability to freely moving mammals, the single-component character of the microbial opsin system has enabled generalizable targeting. Latest-generation Cre recombinase–dependent opsin-expressing viruses have dovetailed with the extensive and growing resource of mouse lines selectively expressing Cre recombinase in defined cell types; optogenetic control can now be delivered to defined cells in freely moving mice with substantial versatility11. Likewise, a strategy of illuminating axons far from the opsin-transduced cell body (capitalizing on trafficking of light-sensitive molecules down the axon itself) enables projection-based control of cells12,13 with-out requiring any genetic knowledge at all, which is important for versatile optogenetic control in less genetically tractable species such as rats and primates. These and other general-purpose targeting strategies heavily depend on the single-component property.
Future directions: beyond neuroscience
Molecular engineering has also enabled optogenetic control of well-defined biochemical events. Early in 2009, capitalizing on the retinoid content of mammalian brain tissue and the low activity of retinal-based signaling modules in the dark, researchers described optical control of distinct G protein–coupled receptor (GPCR) biochemical pathways in freely moving mammals using vertebrate rhodopsin-GPCR chimeras (optoXRs), which recruit pathways that are governed by distinct heterotrimeric G-protein pathways (Gs and Gq)14. Subsequently optical control over small GTPases (with resulting changes in cellular shape and motility) was achieved in cultured cells using novel strategies from several different laboratories15,16. The GTPases were activated by recruitment to the plasma membrane via optically modulated protein-protein interactions, a method that may ultimately become generally suitable for controlling additional classes of biochemical signal transduction (particularly if the method can be shown to operate in single-component fashion in intact mammalian tissues, as seems likely in cases in which relevant flavin or biliverdin chromophores are present). In this issue, Lim and colleagues17 discuss strategies for development of biochemical optogenetic control. Finally, microbial adenylyl cyclases with low activity in the dark have been described that represent a marked advance over earlier microbial cyclases from the optogenetic perspective, use a flavin chromophore and, as optoXRs, are suitable for single-component optical control in neurons18. Together, these experiments opened the door to optogenetics in essentially every cell and tissue whether electrically excitable or not.
Going forward, applications of these and other tools to tissues and systems throughout biology will become a major theme. Such tools have already been applied to nonneuronal systems, including glial, muscle, cardiac and embryonic stem cells, and the temporal precision provided by light may continue to be crucial in this regard by providing a defined event in a defined cell population at specific times relative to environmental events.
Different speeds of operation are required for different cells and tissues; for example, cardiac optogenetics may not always require the millisecond-scale precision of tools used in fast-spiking central nervous system neurons because the heart in some ways operates on slower timescales, though faster experimental methods may unlock preciously unanticipated regimes of fast tissue computation and adaptation. And even the microbial opsins can act by biochemical rules as well; the trace Ca2+ flux of channelrhodopsin has been used to drive activation of nonexcitable glial cells19 in the central nervous system and could be used to drive this ubiquitous second messenger in a rich array of Ca2+-modulated cells ranging from insulin-secreting pancreatic beta cells to T lymphocytes, in intact endocrine-exocrine and immunologic tissues. In stem cell biology and engineering, biochemical or electrical drive can now be delivered independently and selectively to niche cells, stem cells or stem cell progeny, even with intact tissue and animals.
In general, optogenetic tools can now be selected that are appropriate for the target tissue of choice, with regard to electrical or biochemical effector function, speed and other properties.
Future directions: expanding the toolkit
Initially researchers noted that the fidelity of optogenetic control via opsin gene expression was not optimal, with noise in the system characterized by occasional extra action potentials or missing action potentials4. Subsequent optimization of microbial opsins addressed this problem for optogenetics, returning conceptually to the 1971 discovery of bacteriorhodopsin and building upon mutagenesis of the bacteriorhodopsin gene in many laboratories over decades. As a result, modified opsins for optogenetics now include fast and slow mutants that variously enable high-fidelity control over high-frequency action potential trains20, bistable changes in excitability21 and orders-of-magnitude increased light sensitivity21. Additionally, stronger opsin expression and other improved properties have resulted from creating chimeras between different opsins in combination with mutagenesis22. In this issue, Hegemann and Möglich23 discuss the interface between bacteriorhodopsin mutagenesis and the optimization of optogenetic tools.
Going forward, molecular tools will rapidly prove important for optogenetics in several other ways as well, beyond targeted mutagenesis of known genes. Certainly subcellular targeting of optogenetic tools is of great interest, and a new frontier consists of delivering optical control (whether biochemical or electrical) to well-defined subcellular domains or intracellular (for example, membranous) compartments. Screens for optical tools that modulate protein-protein interactions may open the door to optogenetic control of kinases and transcription factors. And molecular engineering will deliver optogenetic tools with altered chromophore dependence (for example, enabling new uses of endogenous chromophores such as biliverdin or flavin) as well as altered effector function.
Moroever, rapidly accelerating molecular genomics efforts will continue to expand the optogenetics toolkit—which now ranges across and beyond the visible spectrum—a process that began with the discovery of a red-shifted channelrhodopsin24 for combinatorial control in 2008. Although most microbial opsin genes do not express well in mammalian neurons25,26, it has been found that the major underlying problem is one of membrane trafficking25. This cell-biological concept led to the identification of membrane trafficking motifs that, when added to specific locations on microbial opsin genes, confer robust expression and optogenetic control to opsins that are otherwise problematic to express or express poorly, including the original microbial opsin gene, bacteriorhodopsin25. These molecular principles will provide a wealth of diverse light sensitivity and effector function properties, unlocking the potential of thousands of microbial opsin genes that occur throughout the major kingdoms of life.
Future directions: reverse engineering
As another distinct advantage, fast and specific optical control opens a new landscape for systems physiology by permitting simultaneous input-output interrogation of excitable tissue. Electrical recording was never fully possible with simultaneous electrical stimulation at the same site owing to electrical artifacts that can now be avoided with optical stimulation. As simultaneous readout measures for optogenetically controlled systems become more rich and complex, the concept of ‘reverse engineering’ of biology will be taken further. This will allow us to infer computational roles of biological tissues, based on how they transform the information we provide and how these transformations are altered in complex disease states (in much the same way that reverse engineering is carried out on computer chips to determine the underlying processing).
Novel devices and systems are required to advance this vision. Beginning in 2007, soon after fiberoptic and laser-diode tools9 enabled optogenetic control even deep in the brains of freely moving mammals10, closely related hybrids of fiberoptics and electrodes13 (‘optrodes’) allowed high-speed simultaneous readouts that kept pace with the high-speed inputs of optogenetics19.
A major area of future work will be expansion of the capabilities of these mixed-modality devices with regard to (i) output channel number and type, (ii) smooth temporal and spatial integration with increasingly complex optical input channel number and type, and (iii) closed-loop control. (Whereas many studies have been published describing light-triggered behavior or light-triggered physiology, only a few have emerged on behavior-triggered light or physiology-triggered light, and systematically closing the control loop will enable real-time bidirectional communication between input and output streams.)
Cell-type specificity is still not readily enabled for electrical recording, with the exception of targeted microelectrode- or patch clamp–accessible preparations such as brain slices or surface brain structures2,12. Although in some ways the orthogonality of electrical and optical methods suggests a special value for the mixed-modality, optrode-style approach, readouts may also be achieved with genetically encoded optical measures of activity such as genetically encoded Ca2+ indicators and voltage sensors. Certainly all-optical interrogation of neural circuits has already been carried out using Ca2+ dyes or voltage-sensitive dyes for output that are spectrally compatible with microbial opsins for input26. In recent years, researchers have made great strides in achieving specific genetically encoded readouts, which Peron and Svoboda27 disscuss in this issue, along with complementary light-input strategies for superficial neural structures.
Conclusion
As discussed here, efforts to expand the capabilities of microbial-opsin optogenetics since 2005 have spanned genomic tool discovery, molecular engineering, opsin targeting and optical-device development. The importance of optogenetics as a research tool continues to grow rapidly, and it is now used in more than 800 laboratories around the world. In this context, it is intriguing to note that a membrane trafficking–enhanced microbial opsin (in this case, a halorhodopsin25,26, eNpHR2.0) has recently been delivered to living human neural tissue (the ex vivo retina) with potent optogenetic functionality28.
Yet the most fundamental impact of optogenetics, even on human health, does not arise from direct introduction of opsins into human tissue but rather from use as a research tool to obtain insights into complex tissue function, as has already been the case for Parkinson’s disease19. Owing to technological limitations in probing intact neural circuits with cellular precision, our current understanding of brain disorders does not do full justice to the brain as a high-speed cellular circuit. Rather than conceptualizing the brain as a mix of neurotransmitters, ideally we would be able to move toward a circuit-engineering approach, in which devastating symptoms of disease are understood to causally result from specific spatiotemporal patterns of aberrant circuit activity relating to specific neuronal populations. But technology has been lacking for the requisite high-speed, targeted, causal control of intact neural circuit function, and this challenge extends to basic neuroscience and other biological systems as well. Although optogenetics meets this challenge, much work remains, including advancing the genomic expansion of optogenetic tools, refining the molecular engineering for optimized functionality and developing light and genetic targeting strategies for various biological systems and animal models.
The challenges faced today in the study of diverse intact biological systems conceptually parallel the challenge faced by neuroanatomy more than a hundred years ago, with the common theme being the need to link information across spatial scales. At that time in history, microscopy had defined small cellular elements of the brain and large architectonic divisions, but the broad mesoscale of cellular connections in intact neuronal circuitry was largely inaccessible until Ramón y Cajal and his students and colleagues used the Golgi technology to systematically map local circuit relationships with cellular precision yet still within the intact system. Inferences from even these descriptive anatomical methods still reverberate through neuroscience. Optogenetics has targeted the analogous need for causal control of defined small-scale events occurring in specified cellular populations while these populations still remain embedded and functioning within larger intact-tissue systems, at appropriate spatial and temporal resolution and under normal or pathological conditions.
Footnotes
COMPETING FINANCIAL INTERESTS
The author declares no competing financial interests.
References
- 1.Deisseroth K Sci. Am 303, 48–55 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Deisseroth K et al. J. Neurosci 26, 10380–10386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oesterhelt D & Stoeckenius W Nat. New Biol 233, 149–152 (1971). [DOI] [PubMed] [Google Scholar]
- 4.Boyden ES, Zhang F, Bamberg E, Nagel G & Deisseroth K Nat. Neurosci 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Matsuno-Yagi A & Mukohata Y Biochem. Biophys. Res. Commun 78, 237–243 (1977). [DOI] [PubMed] [Google Scholar]
- 6.Nagel G et al. Science 296, 2395–2398 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Zemelman BV, Lee GA, Ng M & Miesenbock G Neuron 33, 15–22 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Banghart M, Borges K, Isacoff E, Trauner D & Kramer RH Nat. Neurosci 7, 1381–1386 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravanis AM et al. J. Neural Eng 4, S143–S156 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K & de Lecea L Nature 450, 420–424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai HC et al. Science 324, 1080–1084 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petreanu L, Huber D, Sobczyk A & Svoboda K Nat. Neurosci 10, 663–668 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Gradinaru V et al. J. Neurosci 27, 14231–14238 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Airan RD, Thompson KR, Fenno LE, Bernstein H & Deisseroth K Nature 458, 1025–1029 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Levskaya A, Weiner OD, Lim WA & Voigt CA Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YI et al. Nature 461, 104–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toettcher JE, Voigt CA, Weiner OD & Lim WA Nat. Methods advance online publication, doi: 10.1038/nmeth.f.326 (20 December 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stierl M et al. J. Biol. Chem published online 28 October 2010. (doi: 10.1074/jbc.M110.185496). [DOI] [Google Scholar]
- 19.Gradinaru V, Mogri M, Thompson KR, Henderson JM & Deisseroth K Science 324, 354–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunaydin LA et al. Nat. Neurosci 13, 387–392 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Berndt A, Yizhar O, Gunaydin LA, Hegemann P & Deisseroth K Nat. Neurosci 12, 229–234 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Lin JY, Lin MZ, Steinbach P & Tsien RY Biophys. J 96, 1803–1814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegemann P & Möglich A Nat. Methods advance online publication, doi: 10.1038/nmeth.f.327 (20 December 2010). [DOI] [PubMed] [Google Scholar]
- 24.Zhang F et al. Nat. Neurosci 11, 631–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradinaru V et al. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F et al. Nature 446, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Peron S & Svoboda K Nat. Methods advance online publication, doi: 10.1038/nmeth.f.325 (20 December 2010). [DOI] [PubMed] [Google Scholar]
- 28.Busskamp V et al. Science 329, 413–417 (2010). [DOI] [PubMed] [Google Scholar]