Abstract
Purpose:
Pitt Hopkins Syndrome (PTHS) is a rare genetic disorder caused by mutations of a specific gene, transcription factor 4 (TCF4), located on chromosome 18. PTHS results in individuals that have moderate to severe intellectual disability, with most exhibiting psychomotor delay. PTHS also exhibits features of autistic spectrum disorders, which are characterized by the impaired ability to communicate and socialize. PTHS is comorbid with a higher prevalence of epileptic seizures which can be present from birth or which commonly develop in childhood. Attenuated or absent TCF4 expression results in increased translation of peripheral ion channels Kv7.1 and Nav1.8 which triggers an increase in after-hyperpolarization and altered firing properties.
Methods:
We now describe a high throughput screen (HTS) of 1280 approved drugs and machine learning models developed from this data. The ion channels were expressed in either CHO (KV7.1) or HEK293 (Nav1.8) cells and the HTS used either 86Rb+ efflux (KV7.1) or a FLIPR assay (Nav1.8).
Results:
The HTS delivered 55 inhibitors of Kv7.1 (4.2% hit rate) and 93 inhibitors of Nav1.8 (7.2% hit rate) at a screening concentration of 10 μM. These datasets also enabled us to generate and validate Bayesian machine learning models for these ion channels. We also describe a structure activity relationship for several dihydropyridine compounds as inhibitors of Nav1.8.
Conclusions:
This work could lead to the potential repurposing of nicardipine or other dihydropyridine calcium channel antagonists as potential treatments for PTHS acting via Nav1.8, as there are currently no approved treatments for this rare disorder.
Keywords: high throughput screen, Kv7.1, Nav1.8, Pitt Hopkins Syndrome, repurposing
Introduction
Pitt Hopkins Syndrome (PTHS) is a rare genetic disorder affecting a specific gene in chromosome 18, called transcription factor 4 (TCF4). The connection to TCF4 was not established until 2007 when it was found to play an essential role in the development of the nervous system and the brain (1–3). PTHS is characterized by developmental delay, breathing problems of episodic hyperventilation and/or breath-holding while awake (55%−60%), recurrent seizures/epilepsy (40%−50%), gastrointestinal issues, lack of speech, and distinctive facial features. As more is learned about PTHS the developmental spectrum of the disorder is widening to include anxiety, attention deficit hyperactivity disorder (ADHD), and sensory disorders. PTHS was first described by D. Pitt and I. Hopkins in the Australian Pediatric Journal in 1978 (4). Any functional deficiency of TCF4 greatly affects how a child develops over time. More than 500 PTHS patients have been identified worldwide, but there are likely many who remain undiagnosed. PTHS is considered an autism spectrum disorder, and some patients have also been diagnosed with autism, with atypical autistic characteristics, and/or sensory integration dysfunction. There are currently no FDA approved treatments available for PTHS, so a treatment for this disease would completely alter clinical practice and have a transformational effect on these patients and their families.
In a recent 2016 article using cortical neurons from a Pitt Hopkins mouse (5), Maher and colleagues have shown: 1. TCF4 loss of function alters the intrinsic excitability of prefrontal neurons; 2. TCF4-dependent excitability deficits are rescued by SCN10a and KCNQ1 antagonists; and 3. that intact Tcf4 represses expression of SCN10a and KCNQ1 genes, in central neurons. Therefore, the pathological expression of these ion channels in the central nervous system (CNS) creates a unique opportunity to target these channels with therapeutic small molecule agents, and it is speculated that targeting these ions channels may ameliorate cognitive deficits observed in PTHS. Over the past two decades various voltage-gated sodium and potassium channels have been confirmed as therapeutically desirable targets (6) and recent work has focused on delivering subtype-selective modulators.
A variety of sodium channel modulating drugs have been applied to the treatment of clinical conditions caused by abnormal cell excitability (7). In particular, they have been applied to CNS conditions acting as anticonvulsants (carbamazepine) and epilepsy therapy (phenytoin) via modulation of sodium channels expressed in the brain. Antiarrhythmics such as (mexiletine) and flecainide rectify cardiac arrhythmia by acting on sodium channels in the heart. Finally, local anesthetics (lidocaine and bupivacaine) have been established as injectable or topical agents for the treatment of pain via the blockade of sodium channels in peripheral nerves. These compounds are largely subtype unselective within the sodium channel family leading to the potential for undesirable side effects which severely limit their application for certain chronic indications.
Nav1.8 is a sodium ion channel subunit that in humans is encoded by the SCN10A gene (8–11). Nav1.8-containing channels are a voltage-gated channel subtype that are tetrodotoxin (TTX)-resistant. Nav1.8 is expressed in peripheral sensory neurons. In the dorsal root ganglion(DRG), the channel is expressed in unmyelinated, small-diameter sensory neurons called C-fibers, and is involved in nociception (12, 13). C-fibers can be activated by noxious thermal or mechanical stimuli and thus can carry pain messages. The specific location of Nav1.8 in sensory neurons of the DRG have made it a key therapeutic target for the development of new analgesics (14) as well as for the treatment of chronic pain (15).
It was previously thought that the voltage gated potassium channel KV7.1 encoded by the KCNQ1 gene was expressed only in peripheral epithelial cells and in the cell membranes of cardiac tissue (16–18). Curiously however, it has since been observed that mutation of KV7.1 is linked to sudden unexplained death in epilepsy (SUDEP) which is a catastrophic complication of human idiopathic epilepsy with an estimated prevalence of greater than 18%.(19) Finally, in 2011 Roepke, et al. demonstrated that KV7.1 is expressed in forebrain neuronal networks and brainstem nuclei, regions in which a defect in the ability of neurons to repolarize after an action potential can produce seizures and dysregulate autonomic control of the heart (20). Targeted CNS control of the KV7.1 ion channel has potential to be advantageous in the treatment of PTHS as well as other seizure related idiopathic illness. Although there is a potential cardiac liability due to the role of KV7.1 in repolarization of cardiomyocytes, an acceptable safety window could possibly be achieved if only a small fraction of Kv7.1 block is required to restore normal neuronal excitability.
We now describe high throughput screens of a drug library to identify potential molecules for repurposing for PHTS. We have additionally used the data from the Nav1.8 and Kv7.1 HTS to generate and validate machine learning models. These models could help to understand the fundamental structure activity relationship for these ion channels and the identification and development of additional novel inhibitors that are selective for each channel. Such compounds may also be more broadly applicable for other neurological disorders leading to further research on therapeutics targeting these proteins.
Materials and Methods
Compounds
The Prestwick chemical library was purchased from Prestwick (Illkirch, France). Individual dihydropyridine calcium channel antagonists were purchased from Sigma.
Ion Channel Screening
The Prestwick chemical library was prepared as 10 mM DMSO stocks and screened at 10 μM test concentration (n=2 each sample) on either 86Rb+ efflux (Kv7.1) or FLIPR (Nav1.8) platforms. Ion channels were recombinantly expressed as clonal cell lines in either CHO (KV7.1) or HEK293 (Nav1.8) cells.
KV7.1 86Rb+ HTS
All experimental measurements were performed on the Wallac Microbeta (Perkins Elmer, Shelton, CT). Counts per minute (CPM) values were collected over a 1 minute read time per well. The day before the assay was to be run, human KV7.1 containing CHO cells were plated according to tissue culture protocols (40,000 cells per well in 96-well tissue culture treated plates).16–24 hours prior to running, 50 μl of media containing 1 μCi/ml 86Rb+ was added to all wells and incubated at 37°C, 5% CO2. Prior to the start of the assay, the 86Rb spiked media was removed from the plate and cells were washed 3X with 150 μl of Earle’s balanced salt solution (EBSS). After the last wash, the buffer was removed, replaced with 100 μl of EBSS with 1X compound or controls, and incubated for 5 minutes. After this initial incubation, the buffer was removed and replaced with 100 μl of 70mM KCl EBSS with 100 μM Niflumic Acid buffer containing test compound or controls and incubated for an additional 3 minute time period. After the stimulation incubation, the assay buffer was removed from the plates and transferred to a counting plate. The cells were then lysed with 100 μl of 0.1% SDS and the lysate transferred to a second counting plate. Both counting plates were then read on the Wallac Microbeta to 86Rb+ content. From the rubidium CPM data, % 86Rb+ efflux values were calculated for all wells in Excel. Percent efflux values were calculated as % Efflux = ((CPM efflux buffer) / (CPM efflux buffer + CPM cell lysate))*100. Percent efflux values were then normalized to baseline and max stim controls using the following equation: % inhibition = 100-(((% efflux test well – 10μM chromanol 293B control) / (70mM KCl+ niflumic Acid control response – 10μM chromanol 293B control response))*100)
Nav1.8 FLIPR HTS
All studies were performed using the FLIPR TETRA (Molecular Devices, San Jose, CA) 96 well fluorescence imaging platform. Test agent effects on Nav1.8 activity were measured using the following protocols. ANG-2 max excitation was 517nm with max emission at 540nm. For these experiments, the TETRA excitation filter was 470–495nm paired with the 565–625nm emission filter. The day prior to testing, cells were harvested in growth media and plated on 96 well, PDL-coated black walled with clear bottom microplates. Prior to testing, growth media was removed from the plate and 50μl of 5μM Asante Natrium Green (Teflabs) was added (mixed with equal volume of 20% Pluronic F127 first, then added to EBSS). Cells were then incubated for 60–90 minutes at room temperature, protected from light. After incubation, the dye was removed from the plates and replaced with 50μl of 2mM KCl EBSS (145mM NaCl, 0.8mM MgCl, 1.8mM CaCl2, 2mM KCl, 10mM HEPES, 5mM Glucose, pH = 7.4, osmolarity = 300mOsm). Cell and assay plates were loaded onto the FLIPR and the run using 5 min/15 min protocol. Test compound was applied for 5 min and recorded at which point 10 μM deltamethrin was added to activate Nav1.8 channels for an additional 15 min. Max control wells contained 10 μM deltamethrin. Min control wells contained 10μM deltamethrin + 30μM tetracaine. Data were exported as area under curve over the 15 minute addition and fluorescence units (RFU) are normalized to negative and positive controls using the following equation: % inhibition = (((RFU test well – RFU media control) / (10μM deltamethrin control – media control))*100). Because the data are normalized to positive and negative control wells, test compounds that block more than 30 μM tetracaine readout as >100% inhibition.
Nav1.8 Automated Patch Clamp
Selected hits were confirmed on the PatchXpress automated patch clamp platform (Molecular Devices, San Jose, CA) using 5 point concentration-responses to determine IC50. The test protocol was as follows: custom scripts were employed to determine the Vhalf of inactivation for each cell from the fit of a steady-state inactivation curve prior to compound testing. The voltage protocol was then run from a holding potential of −100 mV at a frequency of 0.1Hz as depicted in Figure 1. Automated online statistical analysis was used for steady-state determination prior to compound addition and wash. Peak steady state current was determined from the 0 mV activating step for control, compound and wash conditions. Currents in response to test compound were normalized to the average of the control + wash currents to determine percent inhibition. The mean ±SEM inhibition values were fit to a 4 parameter logistic equation in Prism 7.0 (GraphPad)
Figure 1.

Automated patch clamp voltage protocol.
Machine learning
The Assay Central project uses the source code management system Git to gather and store molecular datasets from diverse sources, in addition to scripts for curating well-defined structure-activity datasets. This machine learning approach has been recently described in detail and compared with other approaches (21–28). These scripts employ a series of rules for the detection of problem data that is corrected by a combination of automated structure standardization (including removing salts, neutralizing unbalanced charges, and merging duplicate structures) with finite activities and human re-curation. The output is a high-quality dataset and a Bayesian model which can be conveniently used to predict activities for proposed compounds. Each model in Assay Central includes the following metrics for evaluative predictive performance: Recall, Precision, Specificity, F1-Score, Receiver Operating Characteristic (ROC) curve, Cohen’s Kappa, and the Matthews Correlation Coefficient (MCC). We utilized Assay Central to prepare and merge datasets collated in Molecular Notebook (29), as well as generate Bayesian models using extended-connectivity fingerprints of maximum diameter 6 (ECFP6) descriptors (30, 31).
The single point HTS binary data described previously was utilized for modelling of Nav1.8 and KV7.1 activity. The 1280 compound library was reduced to 1276 total, after removal of mixtures (Prestw-160 and Prestw-265) and metal-coordinating compounds (Prestw-433) as well as merging duplicate compounds after salt removal (Prestw-820 and Prestw-873). All removed and merged compounds were inactive, maintaining the number of active compounds from the HTS results.
Results
KV7.1 Results
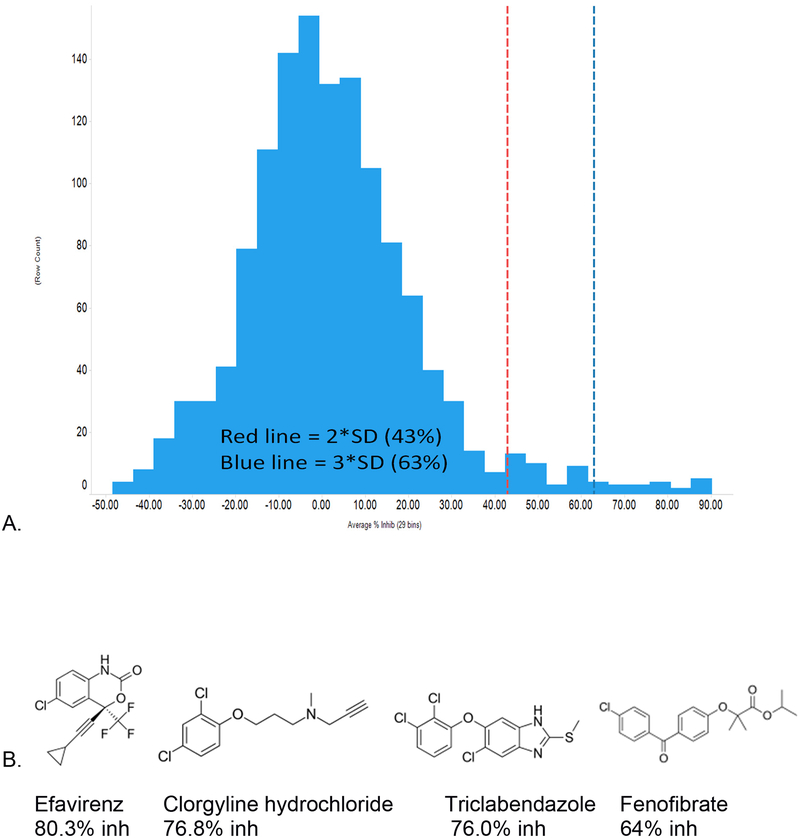
The KV7.1 screen identified 55 hits (4.2% hit rate, Table 1) from the 1280 compound Prestwick chemical library at >2SD from the mean response. >2SD and >3SD cut offs were 43% and 63% inhibition, respectively. The Z prime for the assay was 0.6 and the data appear normally distributed. Screen statistics and selected hits are shown in Figure 2. Several of the FDA approved drugs appear promising and were unexpected such as efavirenz (HIV antiviral) and clorgyline (MAO inhibitor). Fenofibrate (PPARalpha activator) has previously been shown to inhibit KV7.1 (IC50 of 1μM) and thus represents validation that our screen is able to retrieve this compound (32).
Table 1.
KCNQ1 percent inhibition data.
| Drug Name | Average percent inhibition at 10 μM |
|---|---|
| Diethylstilbestrol | 90.2 |
| Ethynodiol diacetate | 88.0 |
| Tibolone | 87.6 |
| Ethaverine hydrochloride | 86.9 |
| Econazole nitrate | 85.6 |
| Hexestrol | 82.0 |
| Lithocholic acid | 80.8 |
| Efavirenz | 80.3 |
| Miconazole | 79.8 |
| Clorgyline hydrochloride | 76.8 |
| Triclabendazole | 76.0 |
| Ethinylestradiol | 75.8 |
| Triclosan | 74.0 |
| Troglitazone | 73.0 |
| Tolnaftate | 69.4 |
| Cyclopenthiazide | 68.3 |
| Butoconazole nitrate | 67.0 |
| Equilin | 64.6 |
| Fenofibrate | 64.0 |
| Latanoprost | 63.7 |
| Liranaftate | 62.8 |
| Dienestrol | 61.3 |
| Isoconazole | 61.2 |
| Paroxetine Hydrochloride | 59.9 |
| Phenothiazine | 59.8 |
| Idebenone | 59.0 |
| Sertaconazole nitrate | 58.6 |
| Benzbromarone | 58.1 |
| Oxybenzone | 57.4 |
| Thiethylperazine dimalate | 56.9 |
| Estradiol-17 beta | 56.3 |
| Tioconazole | 53.5 |
| Dehydroisoandosterone 3-acetate | 52.9 |
| Etofenamate | 51.8 |
| Benidipine hydrochloride | 51.1 |
| Nicardipine hydrochloride | 50.7 |
| Famprofazone | 50.7 |
| Mitotane | 49.5 |
| Tribenoside | 48.8 |
| Cyclosporin A | 48.7 |
| Nisoldipine | 48.0 |
| Nimesulide | 47.9 |
| Dicyclomine hydrochloride | 47.5 |
| Racecadotril | 46.4 |
| Clotrimazole | 46.3 |
| Nitrendipine | 46.1 |
| Cyclizine hydrochloride | 45.9 |
| Papaverine hydrochloride | 45.6 |
| Benazepril hydrochloride | 44.8 |
| Topiramate | 44.3 |
| Stanozolol | 43.6 |
| Homochlorcyclizine dihydrochloride | 43.5 |
| Piperacillin sodium salt | 43.5 |
| Raloxifene hydrochloride | 43.3 |
| Cinnarizine | 43.3 |
Figure 2.
A. KV7.1 percent inhibition data distribution and B. select KV7.1 hits. The 2SD and 3SD cutoffs were 43% and 63% inhibition, respectively. 55 compounds produced >2SD inhibition and 20 compounds were >3SD.
Nav1.8 Results
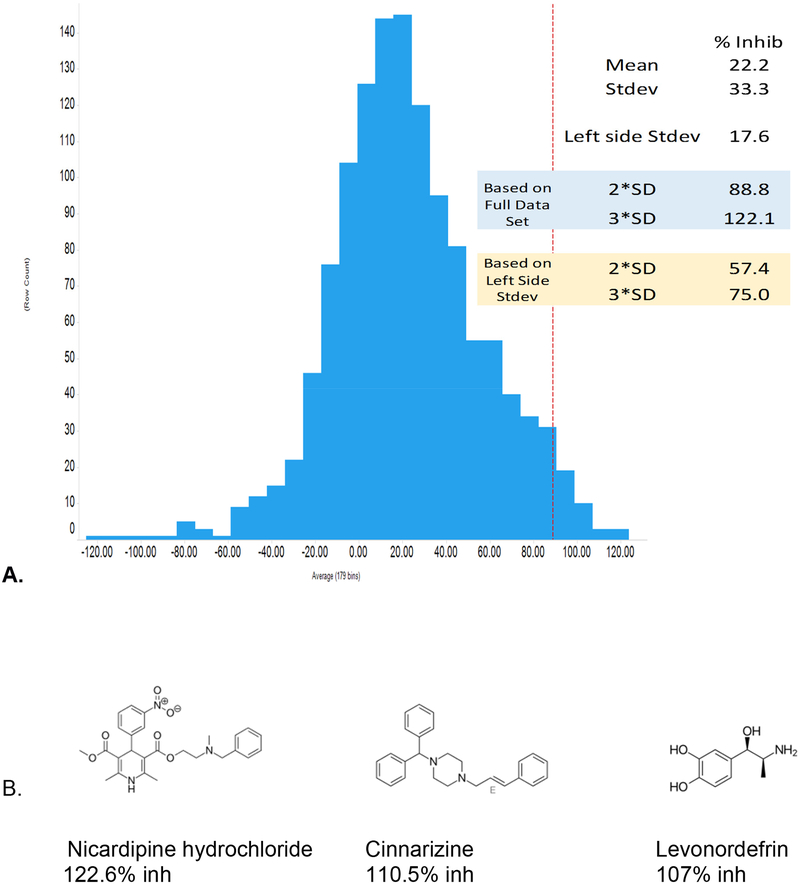
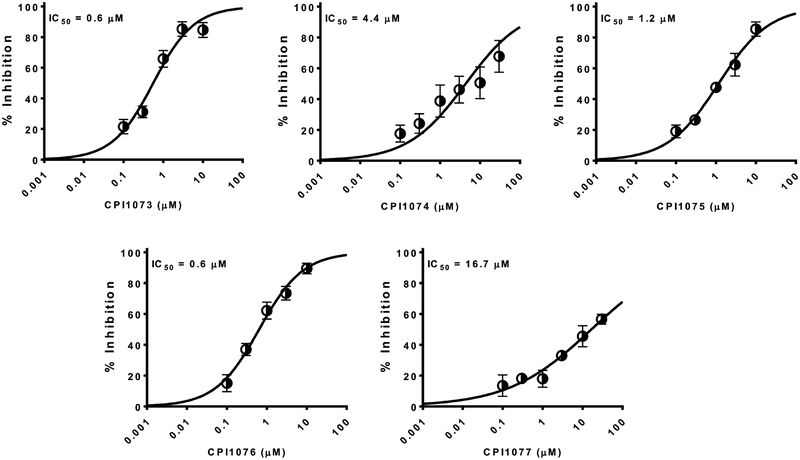
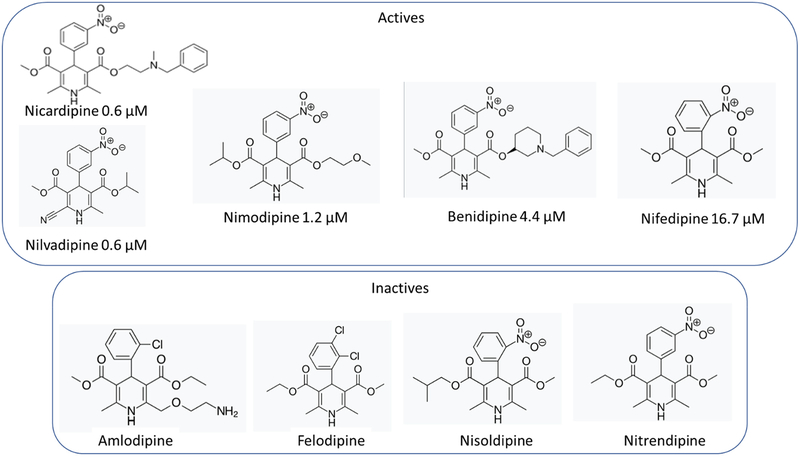
The Nav1.8 screen identified 93 hits (7.2% hit rate, Table 2) from the 1280 compound Prestwick chemical library at >2SD from the mean response. >2SD and >3SD cut offs were 88 % and 122.1 % inhibition, respectively. Because percent inhibition are calculated relative to both negative (DMSO) and positive control wells (30 μM tetracaine), >100% inhibition represents compounds that blocked to a great extent than tetracaine. The Z prime for the assay was 0.6. Screen statistics and selected hits are shown in the Figure 3. This screen identified many known dihydropyridine calcium channel inhibitors which represent a class of compounds that are used frequently and relatively safe. These represented a potential interesting class of compounds for which there was also plentiful preclinical and clinical data. We have also selected several interesting inhibitors that appear to have not been identified previously as Nav1.8 inhibitors. These should all be CNS active based on their known uses. For example, cinnarizine, is a known antihistamine. The five dihydropyridine calcium channel antagonists with activity against Nav1.8 were then followed up with electrophysiology dose response analysis to generate a definitive potency using whole cell voltage-clamp on the PatchXpress automated HTEP platform with 5-point concentration response curves. Fresh compound was purchased for this analysis. Nicardipine and nilvadipine were found to be the most potent with IC50 = 0.6 μM. followed by nimodipine (1.2 μM), benidipine (4.4 μM) and nifedipine (16.7 μM) (Figure 4).
Table 2.
Nav1.8 percent inhibition data.
| Drug Name | Average percent inhibition at 10 μM |
|---|---|
| Carmofur | 123.6 |
| Nicardipine hydrochloride | 122.6 |
| Oxiconazole Nitrate | 117.1 |
| Benidipine hydrochloride | 113.2 |
| Cinnarizine | 110.5 |
| Levonordefrin | 107.0 |
| Carvedilol | 103.7 |
| Proparacaine hydrochloride | 103.3 |
| Dehydroisoandosterone 3-acetate | 100.7 |
| Pimozide | 100.6 |
| Fluspirilen | 100.1 |
| Dioxybenzone | 100.0 |
| Salmeterol | 99.6 |
| Diethylstilbestrol | 99.5 |
| (−)-Isoproterenol hydrochloride | 99.2 |
| Dyclonine hydrochloride | 99.0 |
| Isoetharine mesylate salt | 98.5 |
| Tegaserod maleate | 98.3 |
| Flunarizine dihydrochloride | 96.6 |
| Nimodipine | 96.1 |
| Naftifine hydrochloride | 96.1 |
| Levopropoxyphene napsylate | 95.1 |
| Racecadotril | 94.8 |
| Suloctidil | 93.8 |
| Vinpocetine | 93.4 |
| Oxethazaine | 92.9 |
| Saquinavir mesylate | 92.8 |
| Lidoflazine | 92.7 |
| Diperodon hydrochloride | 92.2 |
| Ketanserin tartrate hydrate | 91.9 |
| Hexestrol | 91.6 |
| Naftopidil dihydrochloride | 91.4 |
| Fendiline hydrochloride | 91.3 |
| Lomerizine hydrochloride | 91.0 |
| Terbutaline hemisulfate | 90.6 |
| Adiphenine hydrochloride | 90.4 |
| Perospirone | 89.7 |
| Amodiaquin dihydrochloride dihydrate | 89.6 |
| Bepridil hydrochloride | 89.1 |
| (+)-Isoproterenol (+)-bitartrate salt | 89.0 |
| Doxepin hydrochloride | 88.7 |
| Trimebutine | 88.7 |
| Nefazodone hydrochloride | 88.5 |
| Pimethixene maleate | 88.5 |
| Raloxifene hydrochloride | 88.4 |
| Promazine hydrochloride | 88.1 |
| Nilvadipine | 87.9 |
| Triclosan | 87.8 |
| (R)-Duloxetine hydrochloride | 87.7 |
| Droperidol | 87.6 |
| GBR 12909 dihydrochloride | 87.0 |
| Thiethylperazine dimalate | 86.6 |
| Tribenoside | 86.2 |
| Luteolin | 86.2 |
| Phenoxybenzamine hydrochloride | 86.1 |
| Nortriptyline hydrochloride | 85.9 |
| Methiothepin maleate | 85.6 |
| Pentolinium bitartrate | 85.6 |
| Ethopropazine hydrochloride | 85.5 |
| Levalbuterol hydrochloride | 85.3 |
| Benperidol | 84.2 |
| Benoxinate hydrochloride | 83.7 |
| Benzydamine hydrochloride | 83.6 |
| Nebivolol hydrochloride | 82.7 |
| Nicergoline | 82.3 |
| Clemastine fumarate | 82.2 |
| Methyl benzethonium chloride | 82.0 |
| Spiperone | 82.0 |
| Benzethonium chloride | 81.4 |
| Protriptyline hydrochloride | 80.0 |
| Benzonatate | 79.9 |
| (+,−)-Synephrine | 79.6 |
| Dibucaine | 79.4 |
| Perhexiline maleate | 79.2 |
| Trifluoperazine dihydrochloride | 79.1 |
| Trimeprazine tartrate | 79.0 |
| Felbamate | 78.6 |
| Promethazine hydrochloride | 78.5 |
| Anethole-trithione | 78.5 |
| Metaproterenol sulfate, orciprenaline sulfate | 78.5 |
| Fenoterol hydrobromide | 78.3 |
| Estrone | 78.2 |
| Salbutamol | 78.0 |
| Florfenicol | 77.5 |
| Chlorprothixene hydrochloride | 77.4 |
| Oxybutynin chloride | 77.3 |
| Dilazep dihydrochloride | 76.9 |
| Drofenine hydrochloride | 76.8 |
| Lopinavir | 76.8 |
| Doxazosin mesylate | 76.1 |
| Moricizine hydrochloride | 75.8 |
| Dimethisoquin hydrochloride | 75.5 |
Figure 3.
A. Nav1.8 percent inhibition data distribution and B. select hits. The mean of the Nav1.8 activity distribution was 22.2% inhibition with a right side SD of 33.3. This skewed distribution was from the larger number of hits relative to library size. For this reason we used the left side SD of 17.6 for statistical analysis. This resulted in 2SD and 3SD cutoffs of 57.4% and 75.0% inhibition, respectively. 92 compounds produced >3SD inhibition.
Figure 4.
Concentration-response relationships for dihydropyridine compounds with Nav1.8 activity determined using automated patch clamp. Nicardipine (CPI1073) and nilvadipine (CPI1076) were found to be the most potent with IC50 = 0.6 μM, followed by nimodipine (CPI1075, 1.2 μM), benidipine (CPI1074, 4.4 μM) and nifedipine (CPI1077, 16.7 μM).
Machine learning
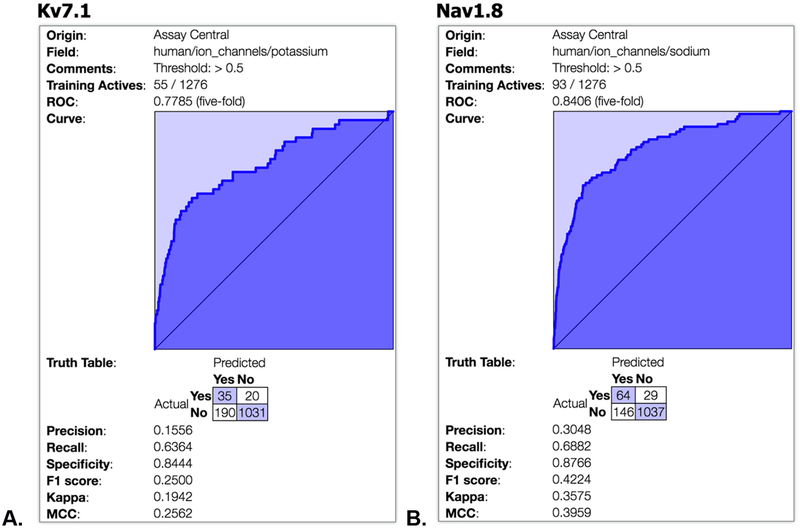
The results of our Bayesian machine learning 5-fold cross validation for KV7.1 (ROC of 0.77, Figure 5A) and Nav1.8 (ROC of 0.84, Figure 5B) suggest reasonable preliminary models considering they are highly imbalanced in favor of inactives. These models can be used in future to score additional libraries of compounds and suggest further molecules for testing.
Figure 5.
Bayesian machine learning results for A. KV7.1 and B. Nav1.8 models in Assay Central.
Discussion
Over the past two decades various sodium and potassium channels have been confirmed as therapeutically desirable targets and recent work has focused on delivering subtype selective modulators. In the current study, the 1280 compound Prestwick library was successfully screened (10 μM, n=2) against human Nav1.8 and KV7.1 channels. The Prestwick chemical library is a well-regarded small molecule library of approved drugs which has been widely used in many other studies ranging from discovering treatments for cancers, cardiovascular disease, antivirals, antibacterials etc. (33). The screens in our study were robust with Z’ of 0.62 for NaV1.8 and 0.58 for KV7.1. For NaV1.8, 93 compounds produced >3 SD inhibition (75% inhibition) when adjusted for high hit rate. The KV7.1 screen produced 55 compounds with >2 SD inhibition (43%) and 20 that were >3 SD inhibition (63%). These represent good hit rates on a par with other published HTS.
The NaV1.8 screen produced several known dihydropyridine calcium channel antagonists as potent inhibitors, a very interesting discovery which has not previously been described by others. In addition, there were several compounds in this class that also had less or no effect (Figure 6) and this may point to important structural characteristics for binding to this channel. One of the most active compounds was nicardipine which is widely used to treat angina and hypertension. It has also been used for neuroprotection of microglia (34) and increasing clearance of beta amyloid (35). This compound may represent a very promising lead for further evaluation, but there are still several questions that need to be addressed. For example: from in vitro blood/brain barrier predictions, what dose is required to engage Nav1.8 in the brain? What are the known adverse events at that dose? Nicardipine is prescribed as an anti-hypertensive but does it lower blood pressure in normo-tensive subjects such as PTHS patients? What is the behavioral effect of nicardipine in a Pitt-Hopkins vs normal mouse? If there is an effect, does it differentiate from other pan-Nav channel blockers such as carbamazepine, mexiletine and/or lamotrigine? Addressing these questions is outside the scope of the present study.
Figure 6.
Structure activity relationship for dihydropyridine compounds against NaV1.8.
The Nav1.8 channel, which unlike Kv7.1 (20) is normally not expressed in the brain, are upregulated in the Pitt Hopkins mouse. In future studies we will need to test the efficacy of the Nav1.8 inhibitors we have identified here such as nicardipine in this model to assess their ability to reverse the Pitt Hopkins phenotype. A battery of neurobehavioral tests could then be used to validate the phenotype of B6;129-Tcf4tm1Zhu/J mice (8–12 weeks old) after i.p. dosing. The neurobehavioral tests could include the Open Field Test, the Novel Spatial Recognition Task (30 min delay memory task), and the Holeboard Learning and Memory Task.
We have previously performed drug repurposing (36) using machine learning methods to identify FDA and EMA approved drugs for Ebola (37) and Chagas disease (38). Most recently we have been actively constructing Bayesian models for absorption, distribution, metabolism and excretion (ADME) properties such as aqueous solubility, mouse liver microsomal stability, and Caco-2 cell permeability (30), complementing earlier ADME machine learning work (39–47). This has led to models with acceptable ROC scores > 0.7 (30). We have recently developed a Bayesian machine learning software called Assay Central (31, 48–50) and applied it in several studies (21–28) and in the current study with ECFP6 descriptors. These studies suggest we can build very promising models that have good statistics and ROC values and can identify chemical features that contribute to activity or lack thereof. To create novel intellectual property around KV7.1 and Nav1.8 inhibitors we can learn from the complete HTS datasets already generated using these machine learning models to virtually screen large libraries of to prioritize compounds for testing in vitro. The models developed in Assay Central can also be shared with others via a customized executable (21–28).
An alternative approach to that described in this study would be to take a known molecule as a starting point and try to modify it. For example, Pfizer and other companies have published on Nav1.8 compounds such as PF-06305591 which was run in safety and proof of concept trials at the same time. The pharmacokinetics was reported to be acceptable and no obvious safety issues were seen. It was however terminated because the pain efficacy was underwhelming (51). This compound is available from Sigma and chemistry is tractable so one could propose developing analogs of this or another related compound (52). The machine learning models described herein could be used to score these potential analogs prior to synthesis.
Over the past two decades various sodium and potassium channels have been confirmed as therapeutically desirable targets and recent work has focused on delivering subtype selective modulators. In summary, we have now used a repurposing screening approach to identify potential compounds for testing as treatments for PTHS that act at these ion channels. We have also used this new data to generate Bayesian machine learning models which could further increase the efficiency of identifying additional ion channel inhibitors. A great strength of the combined HTS and machine learning strategy is its potential wide applicability for other rare diseases which need treatments.
Acknowledgments
The work at Icagen was funded by the Pitt Hopkins Research Foundation. SE kindly acknowledges NIH funding R43GM122196 and R44GM122196–02A1 “Centralized assay datasets for modelling support of small drug discovery organizations” from NIH/NIGMS which supported development of Assay Central. We also kindly thank Dr. Alex Clark for assistance with Assay Central and Dr. Mary Lingerfelt, Dr. Aaron McMurtray and Dr. Kimberly Goodspeed for discussions.
Footnotes
Competing interests:
S.E. is owner, and K.M.Z., are employees of Collaborations Pharmaceuticals Inc. J.G. was an intern at Collaborations Pharmaceuticals Inc. A.G., B.A. and Z.L. are employees of Icagen.
References
- 1.Brockschmidt A, Todt U, Ryu S, Hoischen A, Landwehr C, Birnbaum S, Frenck W, Radlwimmer B, Lichter P, Engels H, Driever W, Kubisch C, Weber RG. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Human molecular genetics. 2007;16(12):1488–1494. [DOI] [PubMed] [Google Scholar]
- 2.Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, Reardon W, Saraiva J, Cabral A, Gohring I, Devriendt K, de Ravel T, Bijlsma EK, Hennekam RC, Orrico A, Cohen M, Dreweke A, Reis A, Nurnberg P, Rauch A. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). American journal of human genetics. 2007;80(5):994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, Colleaux L. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. American journal of human genetics. 2007;80(5):988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitt D, Hopkins I. A syndrome of mental retardation, wide mouth and intermittent overbreathing. Australian paediatric journal. 1978;14(3):182–184. [DOI] [PubMed] [Google Scholar]
- 5.Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, Phan BN, Hyde TM, Kleinman JE, Shin JH, Jaffe AE, Weinberger DR, Maher BJ. Psychiatric Risk Gene Transcription Factor 4 Regulates Intrinsic Excitability of Prefrontal Neurons via Repression of SCN10a and KCNQ1. Neuron. 2016;90(1):43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagal SK, Marron BE, Owen RM, Storer RI, Swain NA. Voltage gated sodium channels as drug discovery targets. Channels (Austin). 2015;9(6):360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanwick RS, Pristera A, Okuse K. The trafficking of Na(V)1.8. Neurosci Lett. 2010;486(2):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Entrez Gene: sodium channel. Available from: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=6336.
- 9.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57(4):411–425. [DOI] [PubMed] [Google Scholar]
- 10.Plummer NW, Meisler MH. Evolution and diversity of mammalian sodium channel genes. Genomics. 1999;57(2):323–331. [DOI] [PubMed] [Google Scholar]
- 11.Rabert DK, Koch BD, Ilnicka M, Obernolte RA, Naylor SL, Herman RC, Eglen RM, Hunter JC, Sangameswaran L. A tetrodotoxin-resistant voltage-gated sodium channel from human dorsal root ganglia, hPN3/SCN10A. Pain. 1998;78(2):107–114. [DOI] [PubMed] [Google Scholar]
- 12.Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379(6562):257–262. [DOI] [PubMed] [Google Scholar]
- 13.Akopian AN SV, England S, Okuse K, Ogata N, Ure J, Smith A, Kerr BJ, McMahon SB, Boyce S, Hill R, Stanfa LC, Dickenson AH, Wood JN The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nature Neuroscience. 1999;2:541–548. [DOI] [PubMed] [Google Scholar]
- 14.Cummins TR, Sheets PL, Waxman SG. The roles of sodium channels in nociception: Implications for mechanisms of pain. Pain. 2007;131(3):243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nardi A, Damann N, Hertrampf T, Kless A. Advances in targeting voltage-gated sodium channels with small molecules. ChemMedChem. 2012;7(10):1712–1740. [DOI] [PubMed] [Google Scholar]
- 16.Georgijevic Milic L. [Molecular genetics in the hereditary form of long QT syndrome]. Med Pregl. 2000;53(1–2):51–54. [PubMed] [Google Scholar]
- 17.Harmer SC, Tinker A. The role of abnormal trafficking of KCNE1 in long QT syndrome 5. Biochem Soc Trans. 2007;35(Pt 5):1074–1076. [DOI] [PubMed] [Google Scholar]
- 18.Peroz D, Rodriguez N, Choveau F, Baro I, Merot J, Loussouarn G. Kv7.1 (KCNQ1) properties and channelopathies. J Physiol. 2008;586(7):1785–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman AM, Glasscock E, Yoo J, Chen TT, Klassen TL, Noebels JL. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Sci Transl Med. 2009;1(2):2ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roepke TK, Kanda VA, Purtell K, King EC, Lerner DJ, Abbott GW. KCNE2 forms potassium channels with KCNA3 and KCNQ1 in the choroid plexus epithelium. FASEB J. 2011;25(12):4264–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anantpadma M, Lane T, Zorn KM, Lingerfelt MA, Clark AM, Freundlich JS, Davey RA, Madrid PB, Ekins S. Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads. ACS Omega. 2019;4(1):2353–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalecki AG, Zorn KM, Clark AM, Ekins S, Narmore WT, Tower N, Rasmussen L, Bostwick R, Kutsch O, Wolschendorf F. High-throughput screening and Bayesian machine learning for copper-dependent inhibitors of Staphylococcus aureus. Metallomics. 2019;11(3):696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez HW, Soeung M, Zorn KM, Ashoura N, Mottin M, Andrade CH, Caffrey CR, de Siqueira-Neto JL, Ekins S. High Throughput and Computational Repurposing for Neglected Diseases. Pharmaceutical research. 2018;36(2):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane T, Russo DP, Zorn KM, Clark AM, Korotcov A, Tkachenko V, Reynolds RC, Perryman AL, Freundlich JS, Ekins S. Comparing and Validating Machine Learning Models for Mycobacterium tuberculosis Drug Discovery. Molecular pharmaceutics. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo DP, Zorn KM, Clark AM, Zhu H, Ekins S. Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Molecular pharmaceutics. 2018;15(10):4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval PJ, Zorn KM, Clark AM, Ekins S, Wright SH. Assessment of Substrate-Dependent Ligand Interactions at the Organic Cation Transporter OCT2 Using Six Model Substrates. Molecular pharmacology. 2018;94(3):1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang PF, Neiner A, Lane TR, Zorn KM, Ekins S, Kharasch ED. Halogen Substitution Influences Ketamine Metabolism by Cytochrome P450 2B6: In Vitro and Computational Approaches. Molecular pharmaceutics. 2019;16(2):898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zorn KM, Lane TR, Russo DP, Clark AM, Makarov V, Ekins S. Multiple Machine Learning Comparisons of HIV Cell-based and Reverse Transcriptase Data Sets. Molecular pharmaceutics. 2019;16(4):1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark AM. Molecular Notebook. Available from: http://molmatinf.com/MolNote/.
- 30.Clark AM, Dole K, Coulon-Spector A, McNutt A, Grass G, Freundlich JS, Reynolds RC, Ekins S. Open source bayesian models: 1. Application to ADME/Tox and drug discovery datasets. Journal of chemical information and modeling. 2015;55:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark AM, Ekins S. Open Source Bayesian Models: 2. Mining A “big dataset” to create and validate models with ChEMBL. Journal of chemical information and modeling. 2015;55:1246–1260. [DOI] [PubMed] [Google Scholar]
- 32.Bajwa PJ, Alioua A, Lee JW, Straus DS, Toro L, Lytle C. Fenofibrate inhibits intestinal Cl- secretion by blocking basolateral KCNQ1 K+ channels. American journal of physiology Gastrointestinal and liver physiology. 2007;293(6):G1288–1299. [DOI] [PubMed] [Google Scholar]
- 33.Publications mentioning the Prestwick Chemical library. Available from: http://www.prestwickchemical.com/libraries-publications.html
- 34.Huang BR, Chang PC, Yeh WL, Lee CH, Tsai CF, Lin C, Lin HY, Liu YS, Wu CY, Ko PY, Huang SS, Hsu HC, Lu DY. Anti-neuroinflammatory effects of the calcium channel blocker nicardipine on microglial cells: implications for neuroprotection. PloS one. 2014;9(3):e91167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachmeier C, Beaulieu-Abdelahad D, Mullan M, Paris D. Selective dihydropyiridine compounds facilitate the clearance of beta-amyloid across the blood-brain barrier. Eur J Pharmacol. 2011;659(2–3):124–129. [DOI] [PubMed] [Google Scholar]
- 36.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug Disc Today. 2011;16:298–310. [DOI] [PubMed] [Google Scholar]
- 37.Ekins S, Freundlich J, Clark A, Anantpadma M, Davey R, Madrid P. Machine learning models identify molecules active against Ebola virus in vitro. F1000Research. 2015;4:1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekins S, Lage de Siqueira-Neto J, McCall L-I, Sarker M, Yadav M, Ponder EL, Kallel EA, Kellar D, Chen S, Arkin M, Bunin BA, McKerrow JH, Talcott C. Machine Learning Models and Pathway Genome Data Base for Trypanosoma cruzi Drug Discovery PLoS neglected tropical diseases. 2015;9(6):e0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekins S. Progress in computational toxicology. Journal of pharmacological and toxicological methods. 2014;69(2):115–140. [DOI] [PubMed] [Google Scholar]
- 40.Dong Z, Ekins S, Polli JE. Structure-activity relationship for FDA approved drugs as inhibitors of the human sodium taurocholate cotransporting polypeptide (NTCP). Molecular pharmaceutics. 2013;10(3):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astorga B, Ekins S, Morales M, Wright SH. Molecular Determinants of Ligand Selectivity for the Human Multidrug And Toxin Extrusion Proteins, MATE1 and MATE-2K. The Journal of pharmacology and experimental therapeutics. 2012;341(3):743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y, Li L, Kim G, Ekins S, Wang H, Swaan PW. Identification and Validation of Novel hPXR Activators Amongst Prescribed Drugs via Ligand-Based Virtual Screening. Drug metabolism and disposition: the biological fate of chemicals. 2011;39:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zientek M, Stoner C, Ayscue R, Klug-McLeod J, Jiang Y, West M, Collins C, Ekins S. Integrated in silico-in vitro strategy for addressing cytochrome P450 3A4 time-dependent inhibition. Chemical research in toxicology. 2010;23(3):664–676. [DOI] [PubMed] [Google Scholar]
- 44.Ekins S, Williams AJ, Xu JJ. A Predictive Ligand-Based Bayesian Model for Human Drug Induced Liver Injury. Drug metabolism and disposition: the biological fate of chemicals. 2010;38:2302–2308. [DOI] [PubMed] [Google Scholar]
- 45.Diao L, Ekins S, Polli JE. Quantitative Structure Activity Relationship for Inhibition of Human Organic Cation/Carnitine Transporter. Molecular pharmaceutics. 2010;7:2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X, Ekins S, Raufman JP, Polli JE. Computational models for drug inhibition of the human apical sodium-dependent bile acid transporter. Molecular pharmaceutics. 2009;6(5):1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekins S, Kortagere S, Iyer M, Reschly EJ, Lill MA, Redinbo MR, Krasowski MD. Challenges predicting ligand-receptor interactions of promiscuous proteins: the nuclear receptor PXR. PLoS computational biology. 2009;5(12):e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anon. Assay Central video. Available from: https://www.youtube.com/watch?v=aTJJ6Tyu4bY&feature=youtu.be.
- 49.Anon. Assay Central Website. Available from: www.assaycentral.org.
- 50.Clark AM, Sarker M, Ekins S. New target predictions and visualization tools incorporating open source molecular fingerprints for TB Mobile 2.0. Journal of cheminformatics. 2014;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anon. PF-06305591 Available from: https://clinicaltrials.gov/ct2/results?cond=&term=PF06305591&cntry1=&state1=&SearchAll=Search+all+studies&recrs=.
- 52.Bagal SK, Brown AD, Kemp MI, Klute W, Sanz LM, Marron BE, Miller DC, Skerrat E, Suto MJ, West CW. Chemical Compounds. In: US, editor.: Pfizer Limited; 2013. [Google Scholar]