Abstract
Background
Berberine (BBR) from the widely used Chinese herbal medicine Huanglian has an array of pharmacological and biochemical properties, including anti-neoplastic activity. However, the specific mechanisms underlying these properties are unknown. The aim of this study was to explore the anti-tumor mechanisms of BBR in non-small cell lung cancer (NSCLC).
Methods
The effects of BBR on NSCLC tumor development and programmed cell death were investigated both in vivo and in vitro. Luciferase reporter assays were used to determine whether tissue factor (TF) was a target of miR-19a.
Results
BBR suppressed NSCLC growth and promoted apoptosis in NSCLC cells by modulating miR-19a and TF expression. Luciferase assays showed that TF was a direct inhibitory target of miR-19a in NSCLC cells. BBR induced apoptosis through the miR-19a/TF/MAPK axis.
Conclusion
The results suggest that BBR induces apoptosis of NSCLC cells via the miR-19a/TF/MAPK signaling pathway.
Keywords: non-small-cell lung carcinoma, NSCLC, berberine, miR-19a, tissue factor, TF, MAPK
Introduction
Despite remarkable clinical advances in the treatment of lung cancer, its global morbidity and mortality remain high. Approximately 30 of every 100 cancer-related deaths are lung cancer associated.1 Histologically, 4/5 lung cancer cases are non-small-cell lung cancer (NSCLC), including squamous cell carcinoma (SCC), adenocarcinoma and large-cell carcinoma. Up to 70% of NSCLC cases are not confirmed until terminal stages, and NSCLC patients have relatively low five-year survival rates (about 15%).2 Chemotherapy is the mainstay lung cancer treatment, but its efficacy is limited.The median overall survival (OS) of cancer patients receiving chemotherapy is about 8–11 months.3 Although the median OS ranged from 27.7 to 46.9 months for EGFR mutation-positive advanced NSCLC patients, the 5-year survival for these patients treated with erlotinib or gefitinib remains 14.6%.4 Effective natural anticancer drugs with low toxicity are urgently required. Recent therapeutic studies have shifted their attention from conventional chemotherapy and immunotherapy drugs to traditional Chinese medicine.
Berberine (BBR), or Huanglian is an important Chinese herbal medicine which is often used for intestinal infections, particularly bacterial diarrhea.5 BBR has been shown to possess antitumor activity against a range of human cancers. Liu et al6 showed that BBR inhibits senescent human glioblastoma cells by down regulating EGFR-MEK-ERK signaling. Wang et al7 showed that BBR inhibits liver cancer cells by inducing apoptosis and autophagic cell death. No specific mechanisms underlying these effects were reported.
MicroRNAs (miRNAs) are small noncoding RNAs of 18–25 nucleotides in length that are expressed in all cells. miRNAs negatively regulate target gene expression via direct binding to 3ʹ untranslated regions (3ʹUTRs)and inhibiting protein translation.8 Protein dysfunction due to abberant miRNA expression contributes to tumorigenesis in NSCLC.9,10 miR-19a correlates with NSLC prognosis11,12 and the aberrant expression of tissue factor (TF) in solid tumors.13 Volm and Koomagi14 were the first to study the expression of TF using immunohistochemical methods. TF was synthesized in 32% of the identified tumors. TF contributes to tumor metastasis in some NSCLC cases.15 TF has also been reported to activate multiple signaling cascades including PI3K/AKT and MAPK.16
In this study, we aimed to clarify the mechanism of action of BBR on miR-19a and evaluated its biological functions in lung cancer. We show that BBR inhibits NSCLC growth by up-regulating the miR-19a induced down-regulation of TF expression, leading to apoptotic induction and decreased NSCLC cell proliferation. These findings suggest that BBR is a cancer suppressor that induces apoptosis through miR-19a/TF/MAPK signaling.
Materials and methods
Reagents
BBR and dimethyl sulfoxide (DMSO) were obtained from Sigma Chemicals. Anti-β-actin, anti-Bcl-2, anti-Bax, anti-TF, anti-Ki67, anti -p38, anti-p-p38, anti -JNK, anti-p-JNK and anti-β-actin were purchased from Abcam. Other chemicals and experimental materials were provided by BioRad.
Cell culture
The human lung cancer cell lines A549, PC9, H460, H1299, Beas-2b and 293T cells (Cell Bank of Shanghai Institute of Cell Biology, Shanghai, China) were cultured in RPMI 1640 (Gibco) containing 1% penicillin/streptomycin (Hyclone) and 10% fetal bovine serum (FBS, TBD, Tianjin) at a constant temperature of 37 °C and 5 % CO2.
Cell transfection
miR-19a mimics, miR-19a inhibitor, miRNA negative controls (miR-NC), and TF overexpression plasmids were purchased from Genepharm (Shanghai, China). PC9 and A549 cells were transfected with Lipofectamine3000 (Invitrogen) according to the manufacturer’s protocols.
Cell proliferation and viability assays
Cell proliferation was using the Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan). Approximately of 2 × 103 cells were seeded into 96-well plates and treated with various concentrations of BBR (0, 20, 40, 80, 120, and 160 μM). CCK-8 reagent was added to the wells 24, 48 and 72 h, followed by 2 h incubation at 37 °C. Optical densities (OD) were measured at 450 nm using a spectrophotometer (BioTek, USA).
Colony formation assays
Single cell proliferation was assessed through colony formation assays. Cells plated in 6-well plates were treated with BBR (0, 40, 80, and 120 μM), After 48h, cells were washed with PBS and trypsinized. Then cells (2×103/mL) were plated in 6-well plates. The cultures were maintained in a 37 °C, 5% CO2 incubator for 10 days. The medium was discarded and the cells washed with pre-cooled PBS, fixed in methanol for 20 min, and stained with crystal violet for 15 min. Cells were washed with ddH2O and cell colonies were counted using Image J software.
Wound healing and invasion assays
Wound healing assays were performed to assess cell migration. Transfected cells were seeded into 6-well plates and cultured until 95% confluent. Scratches were generated using sterile 200 μL pipette tips. Cells were cultured in FBS-free RPMI-1640 following the addition of different concentrations of BBR (0, 40, 80 and 120 μM) for 24 and 48 h. Cells were imaged on a phase-contrast microscope (× 100) (DMIRB, Leica, Germany). For invasion assays, 5×104 cells were added to the upper wells of matrigel-coated invasion chambers in serum free media, and 600 μL RPMI 1640 containing 10% FBS was added to the lower chamber as a chemoattractant. After 36 h incubation at 37 °C, non-invading cells were removed with a cotton swab. Migrating cells were fixed in 95% ethanol for 20 min, stained with 0.1% crystal violet for 10 min, washed with ddH2O three times, and air dried. Five spots in each well were randomly selected, imaged, and the number of colonies calculated using a light microscope (DMIRB).
Flow cytometry analysis
For apoptosis assays, A549 and PC9 cells were pre-treated with various concentrations (0, 40, 80 and 120 μmol/L) of BBR for 48 h. Cells were washed with pre-cooled PBS, and stained with Annexin V-FITC/PI (BD, San Jose, CA) in the dark for 20 min. Cells were analyzed by flow cytometry (BD Biosciences, USA).
RNA extraction and quantitative real-time PCR
RNA was extracted from cultured cells and frozen tissues using TRIzol reagent (Invitrogen Ltd). RNA purity and concentrations were determined on an ND-2000 spectrophotometer (Thermo Fisher Scientific, USA). Using 1 μg of RNA, cDNA was synthesized to detect miR-19a using One Step Prime script miRNA cDNA Synthesis Kits (Qiangen, Valencia, USA). Quantitative real-time PCR (qRT-PCR) was performed using KAPA SYBR FAST qPCR Kits (Kapa Biosystems, USA). MiR-19a expression was normalized to U6 expression levels. PrimeScript RT Reagent Kits (TaKaRa, Japan) were used to synthesize cDNA for the detection of TF mRNA. qRT-PCR was performed using KAPA SYBR FAST qPCR Kits (Kapa Biosystems, USA). mRNA levels were normalized to GAPDH mRNA. The primers used for qRT-PCR analysis were as follows:
MiR-19a forward primer: 5ʹ-GCGTGTGCAAATCTATGCAA-3ʹ;
MiR-19a reverse primer: 5ʹ-AGTGCAGGGTCCGAGGTATT-3ʹ;
U6 forward primer: 5ʹ-TGCGGGTGCTCGCTTCGCAGC-3ʹ;
U6 reverse primer: 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ;
TF forward primer: 5ʹ-CCGAACAGTTAACCGGAAGA-3ʹ;
TF reverse primer: 5ʹ-TCAGTGGGGAGTTCTCCTTC-3ʹ;
GAPDH forward primer: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ;
GAPDH reverse primer: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ;
For cDNA amplification, thermal cycling was performed for 10 min at 95 °C, 10-sec 45-cycle duration at 95 °C and 30-sec extension at 60 °C. The number of threshold cycles were carefully recorded. The 2-ΔΔCt method was applied for data analysis. Results are shown as the mean ± standard deviation (SD).
Luciferase reporter assays
TF and mutant 3ʹ-UTR segments were PCR amplified and ligated into psiCHECK-2 vectors (Promega, USA). For luciferase assays, HEK293T cells were seeded into 24 well plates and transfected with 100 nM miR-NC or miR-19a with 100 ng luciferase reporter vector. After 48 h incubation, luciferase activity was measured using the luciferase reporter assay system (Promega, USA) according to the manufacturers instructions.
Western blot analysis
Cells were lysed in RIPA buffer supplemented with protease inhibitors (NCM Biotech). Protein concentrations were determined via BCA assays (Google Bio). Equal amounts of protein were resolved on 10% SDS-PAGE gels (Haochen Biotechnology Co., Ltd., Shanghai, China), and transferred to nitrocellulose membranes. Membranes were blocked in 3% BSA (Google Bio) for 1 h and probed with primary antibodies targeting anti-β-actin (1:2000), anti-p-JNK, anti –JNK, anti-p-p38, anti -p38, anti-TF, anti-Bax, anti-Bcl-2, anti-β-actin and anti-TF (1:1000) antibodies overnight at 4 °C. Membranes were collected, rinsed three times with PBST and labeled with secondary antibodies at room temperature for 1 h. Proteins were visualized on an Odyssey scanner (LI-COR Biosciences, USA).
Immunohistochemical (IHC) analysis
For IHC, samples were paraffin-embedded and incubated in 3% hydrogen peroxide for 30 min to suppress endogenous peroxidase activity. Citrate buffer was used to infiltrate the sections, followed by 10-min heating in a microwave for antigen retrieval. Sections were labeled overnight with primary antibodies (anti-ki67, 1:2000) and treated with peroxidase-labeled goat anti-mouse secondary antibodies at room temperature for 1 h. Sections were stained with hematoxylin and 3ʹ-diaminobenzidine tetrahydrochloride (DAB) and visualized.
Nude mice xenograft tumor assays
In nude mice xenograft tumor assays, a total of 1×107 A549 cells in 100 μL of serum-free media were implanted into female BALB/c nude mice aged 4 weeks via subcutaneous injection through the right flank. One day post-tumor cell inoculation, 16 experimental mice were equally randomized into two groups: the saline vehicle group and a BBR (500 mg/kg) group. Treatments were employed on alternative days for a month. Changes in body weight, and vertical and horizontal tumor diameters (W and L) were measured and recorded. Tumor sizes were determined using the equation: V (volume) = (LW2)/2, where W stands for the smallest horizontal diameter and L stands for the largest vertical diameter. All studies were approved by the Animal Care Committee of Tongji University in compliance with the Institutional Animal Care and Use Committee guidelines.
Biostatistical analysis
Variance analysis was performed to examine deviations in miRBase in one of the 24 features. P-values ≤0.05 were considered statistically significant. Each feature was considered on a distinctive scale, and all features centered on zero with the application to unit variance, corresponding to z-scores. After standardization, data were applied using Principal Component Analysis (PCA) and clustering for multivariate analysis. Complete linkage hierarchical clustering was applied to the 24 features to cluster the miRBase versions. Z-scores were cut at an absolute threshold of 3 to reduce the impact of single features. Low dimensional representation of miRBase versions was produced via PCA. For the calculation of miRNA precursors, the standard deviation and mean for each feature were calculated, and z-scores were assessed by the number of standard deviations below or above the precursor set mean. Absolute z-scores were cut at 3 to reduce the impact of single features. Z-score’s and average absolute values were also calculated. Statistical programming environment R (version 3.0.2) was used to perform all calculations.
Statistical analysis
Statistical analysis was performed using GraphPad software. Data were expressed as the mean ± standard deviation (SD). Student’s t-test and one-way ANOVAs were used to calculate the experimental data. *P<0.05, **P<0.001 indicated significant differences from vehicle controls (0.006% DMSO).
Results
BBR suppresses NSCLC cell proliferation, migration and invasion and promotes apoptosis in vitro
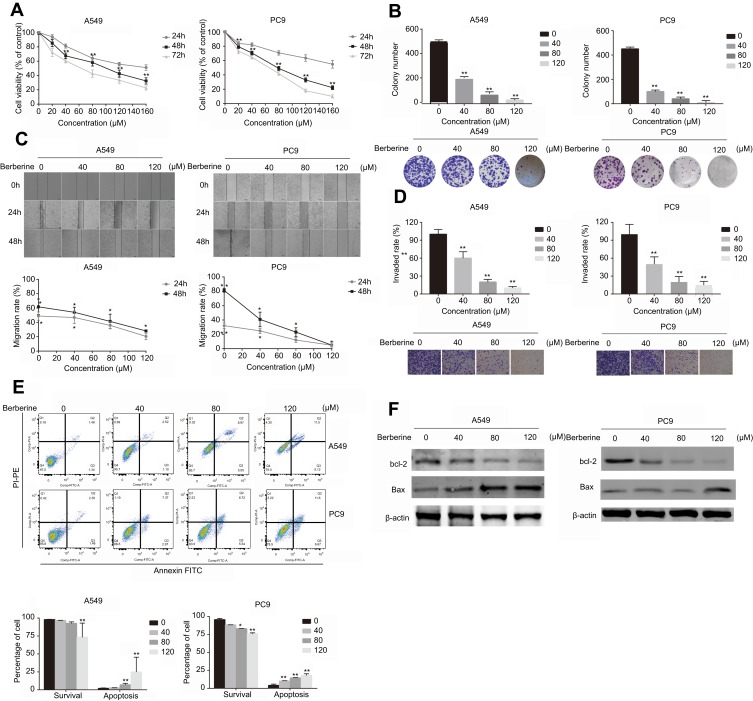
To determine the antitumor effects of BBR, cell viability was assessed by CCK-8 assays. The percentage of viable cells decreased after BBR treatment in a time- and dose-dependent manner particularly at 80 μM for 48 h (A549=58.33±4.96, PC9=49.00±2.87) (Figure 1A). To further define how BBR influences cell proliferation in lung cancer, colony formation assays were performed. After exposure to different concentrations of BBR for 48 h, colony formation was inhibited in both A549 and PC9 cells (Figure 1B). BBR also inhibited NSCLC cell proliferation. Wound healing assays showed that BBR inhibited NSCLC cell migration in a time-dependent manner (Figure 1C). Invasion assays showed that BBR significantly suppressed NSCLC cell invasion in a dose-dependent manner (Figure 1D). Flow cytometry assays on A549 and PC9 cells showed that the proportion of both early and late apoptotic cells in the BBR group were significantly higher than the control group, indicating that BBR increases the number of apoptotic cells in a dose-dependent manner (Figure 1E). Western blot analysis showed that the levels of Bax increased whilst the levels of Bcl-2 decreased after BBR treatment (Figure 1F). These results confirmed that BBR treatment induces A549 and PC9 cell apoptosis and inhibits NSCLC cell proliferation.
Figure 1.
Berberine suppresses the invasion, migration and proliferation of NSCLC cells and induces apoptosis in vitro. (A) Berberine inhibits A549 and PC9 cell proliferation. (B) Berberine inhibits colony formation of A549 and PC9 cells. (C) Berberine inhibits A549 and PC9 cell migration. (D) Berberine inhibits A549 and PC9 cell invasion. (E) Flow cytometry was used to quantify early and late apoptosis in A549 and PC9 cells in various concentrations of berberine for 48 h. (F) Western blot analysis was performed to examine the effects of berberine on the expression of apoptosis-related proteins. Mean values of three independent experiments are shown. Values are the mean ± SD, *P<0.01; **P<0.001, compared to vehicle controls (0.008% DMSO).
BBR inhibits the growth of NSCLC tumors and promotes the apoptosis of NSCLC cells
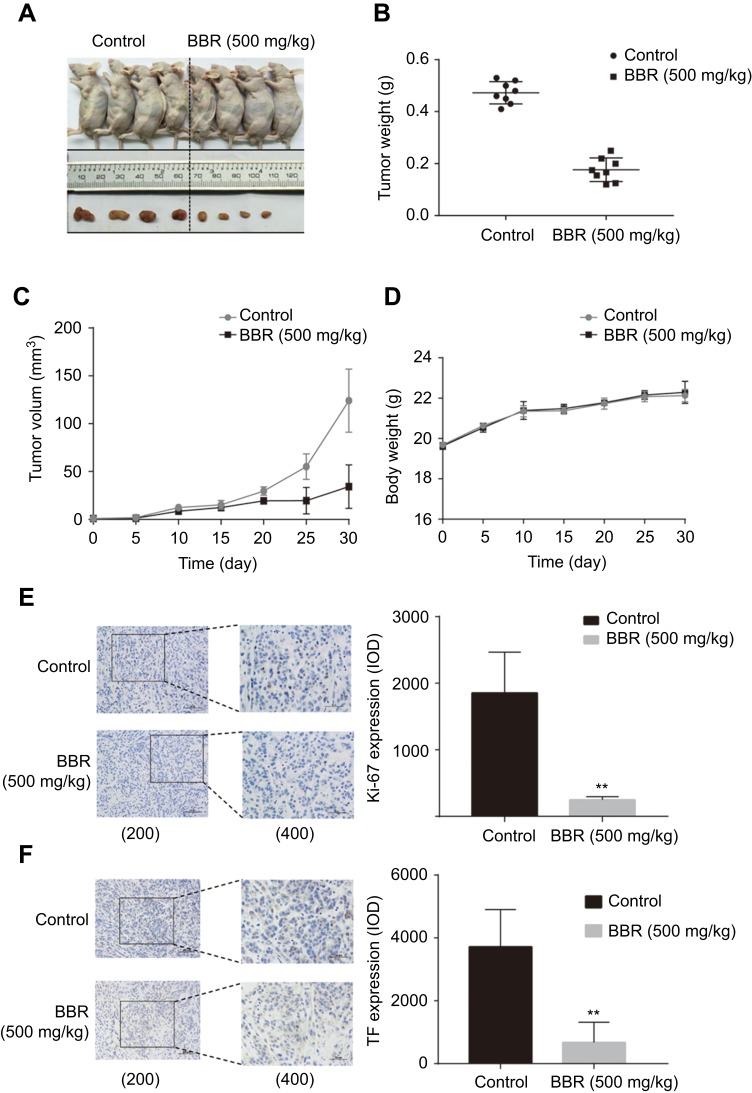
To confirm the potential of BBR as an effective treatment for NSCLC, we established a heterologous A549 cell tumor model. The growth rates of A549 cells treated with BBR were significantly lower than the saline group. After 30-day incubation, the mean tumor size of the BBR group was 34.30±22.68 mm3 vs 124.01±33.03 mm3 in the saline group (Figure 2A–C). The weight of mice treated with BBR changed insignificantly compared to mice in the saline group, indicating that BBR had mild adverse effects in vivo (Figure 2D). In addition, Ki67 levels were used to assess the proliferation of solid tumor cells and BBR group suppressed its levels in comparison to the saline group (Figure 2E). The expression of TF in the cancer tissues of the BBR group were more prominent than the saline group (Figure 2F). These findings suggest that BBR suppresses A549 cell proliferation and induces cell apoptosis.
Figure 2.
Berberine inhibits the growth of NSCLC and promotes apoptosis in vivo. (A) Berberine inhibits NSCLC growth in vivo. (B) Tumor weights in berberine and control groups. (D) Weights of mice in berberine and control groups. (E-F) Berberine had inhibitory effects on solid tumor cell proliferation shown by immunohistochemistry of Ki67, bcl-2. Values in each group are expressed as mean ± SD. **P<0.001.
BBR induces NSCLC apoptosis by modulating the expression of miR-19a
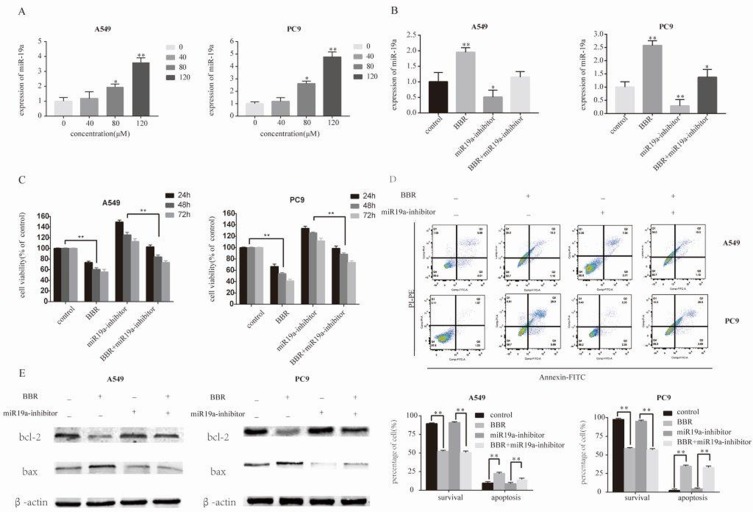
We observed a dose-dependent decrease in the expression of miR-19a following BBR treatment, which was most significant in 80 μM and 120 μM BBR groups (Figure 3A). To investigate the regulatory effects of miR-19a on the tumorigenicity of NSCLC, we silenced miR-19a in A549 and PC9 cells through transfection of the miR-19a inhibitor. qRT-PCR analysis showed that the miR-19a levels dramatically decreased in miR-19a inhibitor transfected groups compared to controls, and were further reduced in the presence of 80 μM BBR (Figure 3B). We sought to determine whether miR-19a down-expression mediated the repressive effects of BBR on the tumorigenicity of NSCLC. BBR treatment of A549 and PC9 cells was performed in the presence and absence of miR-19a inhibitors to examine whether the restoration of miR-19a impaired the effects of BBR on tumorigenicity. We found BBR significantly decreased cell viability in A549 and PC9 cells in a time-dependent manner, compared with the control group. However, the loss in cell viability was less significant in BBR-treated A549 and PC9 cells following the down regulation of miR-19a (Figure 3C). Flow cytometry showed a larger number of apoptotic cells in BBR-treated groups compared to controls. Transfection with the miR-19a inhibitor blunted the effects of BBR on the apoptosis of A549 and PC9 cells (Figure 3D). The results were consistent with those observed in WB analysis (Figure 3E).
Figure 3.
Berberine induces apoptosis by regulating miR-19a in NSCLC. (A) Berberine up-regulates miR19a expression in A549 and Pc9 cells in a dose-dependent manner. (B) qRT-PCR analysis of the miR-19a expression levels in miR-NC and miR-19b inhibitor transfection groups treated with berberine (80μM) for 48 h. (C) Percentage of relative cell viability after berberine treatment following transfection of the miR-19a inhibitor for 48 h. (D) Suppression of miR-19a by berberine treatment results in a significant increase in apoptotic cells. (E) Western blot analysis was performed to demonstrates that berberine induces apoptosis through miR-19a. Mean values of at least three independent experiments are shown. Values are expressed as mean ± SD, *P<0.05; **P<0.001, compared to vehicle controls (0.008% DMSO).
The miR-19a/TF axis regulates BBR inhibition on NSCLC
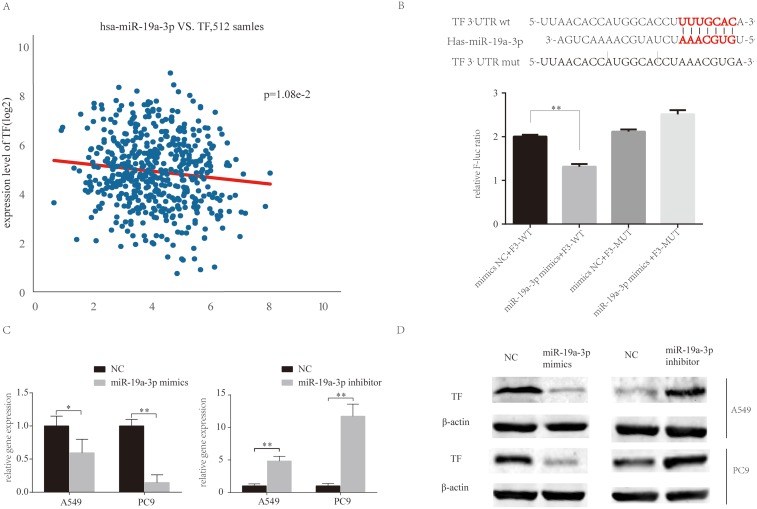
The miRBase predicted that miR-19a directly targets TF (Figure 4A). HEK293T cells were transfected with TF luciferase reporters with miR-19a or agomiR-NC in 293T cells and luciferase activity was asseessed. The results showed that miR-19a overexpression reduced luciferase activity of the wild-type TF reporter, but did not infleucne the activity of mutant-type TF reporters (Figure 4B). Additionally, the over-expression or inhibition of miR-19a influenced TF expression at the protein (Figure 4D) and mRNA level (Figure 4C) in both PC9 and A549 cells. These findings suggest that miR-19a directly targets TF to post-transcriptionally suppress gene expression.
Figure 4.
MiR-19a directly targets TF in NSCLC. (A) MiR-19a directly targets TF in the miRBase. (B) Luciferase activity after transfection of F3-WT or F3-MUT with miR-19a-3p mimics or mimics-NC. (C) qRT-PCR analysis of the relative expression of TF in A549 and PC9 cells transfected with miR-19a mimics or miR-19a inhibitors, with or without berberine treatment. (D) Western blot analysis of the relative TF expression in A549 and PC9 cells transfecred with miR-19a mimics or miR-19a inhibitors, with or without berberine treatment. β-Actin was used as a loading control. Values are expressed as mean ± SD, *P<0.05; **P<0.001, compared to vehicle controls (0.008% DMSO).
BBR induces apoptosis by regulating the miR-19a/TF/MAPK axis in NSCLC
Reverse transcription-PCR assays showed that TF mRNA could be detected within all cell lines, with particularly high expression levels in PC9cells (a highly invasive cell line) compared to Beas-2b cells (Figure 5A). We observed a dose-dependent decrease in the expression of TF following BBR treatment, with the most significant increase in cells treated with 80 μM BBR (Figure 5B). We next examined the function of TF in A549 and PC9 cells. There was a dose-dependent decrease in the expression of miR-19a following BBR treatment (Figure 5B). To examine the role of TF in regulating the tumorigenicity of NSCLC, we over-expressed TF in A549 and PC9 cells and which was confirmed by qRT-PCR. TF expression was inhibited in the presence of 80 μM BBR (Figure 5C). We next determined whether TF overexpression mediates the repressive effects of BBR on the tumorigenicity of NSCLC. BBR significantly decreased cell viability in A549 and PC9 cells in a time-dependent manner. However, the inhibiton of cell viability was less significant in BBR -treated A549 and PC9 cells overexpressing TF (Figure 5D). Flow cytometry analysis showed a higher number of apoptotic cells in the BBR treatment group compared to the control group, whilst transfection of TF blunted the effects of BBR on the apoptosis of A549 and PC9 cells (Figure 5E).The result were comparable by WB analysis (Figure 5F). Furthermore, we found that BBR increased the phosphorylation of JNK and p38 MAPK by regulating the expression of TF (Figure 5F).
Figure 5.
Berberine induces apoptosis by regulating the miR-19a/TF/MAPK axis in NSCLC. (A) qRT-PCR analysis of TF expression in BEAS-2B, A549, Pc9, H1299 and H460 cell lines. (B) Berberine up-regulates TF expression in A549 and Pc9 cells in a dose-dependent manner. (C) qRT-PCR analysis of TF expression in control and TF overexpression groups ± berberine (80 μM) for 48 h. (D) Percentage of relative cell viability after berberine treatment and TF overexpression for 48 h. (E) Suppression of TF overexpression by berberine significantly increases apoptosis. (F) Western blot analysis was performed to assess the effects of miR-19a on the expression of apoptosis-related proteins in response to berberine. Mean values of at least three independent experiments are shown. Values are expressed as mean ± SD, *P<0.05; **P<0.001, compared with the vehicle control (0.008% DMSO).
Discussion
In this study, we found that BBR exerts anti-tumor effects in NSCLC by up-regulating miR-19a and down-regulating TF. We identified TF as a target gene of miR-19a that activates MAPK signaling and promotes tumor cell apoptosis. In addition, NSCLC apoptosis was induced by BBR through upregualitng the expression of miR-19a, and downregulating TF, activating MAPK signaling.
As a major pharmacological component of Huanglian, BBR has cholesterol-lowering, anti-angiogenesic, anti-diabetic and anti-inflammatory activities.5,17 Additionally, recent studies suggest that BBR has antitumor activity against a range of tumor cells. Wang et al14 reported that the expression of miR-23a induced by BBR led to anti-HCC effects that were mediated through p53. You et al15 found that BBR reduced the sensitivity of cisplatin in human gastric cancer cells by upregulating miR-203. In this study, we found that BBR could significantly inhibited the viability of A549 and PC9 cells (Figure 1) and promoted apoptosis in NSCLC cells by upregulating the expression of miR-19a (Figure 3).
Current studies18,19 report that the majority of NSCLC cases are in advanced stages with distant metastasis or invasion at the time of diagnosis. TF was used as a prognosis marker of lung cancer.20 miRs control TF expression at the post-transcriptional level. Chuang et al21 showed that miR-106b and miR-93 effectively regulated the expression of total TF in leiomyosarcoma cells. In other studies, Zhang et al22 demonstrated that miR-19 reduced total TF expression by directly binding to TF mRNA in breast cancer cells. Although miR-19a plays an oncomiR role in NSCLC and the overexpression of serum miR-19a are considered as independent prognostic factors for NSCLC patients.12 Li et al found that miR-19a reduced proliferation of A549 and HCC827 in vitro and in vivo.23 Cao et al exploded that the overexpression of miR-19a predicted a slightly better prognosis by analyzing NSCLC patients from the TCGA database, Meanwhile, they found that miR-19a overexpression inhibited the ability of NSCLC cells, such as A549, PC9, to migrate and promoted apoptosis by decreasing c-Met expression.24 This is consistent with our findings that BBR promote miR-19a expression by downregulating the expression of TF.
The interaction between FVIIa and TF has been reported to activate multiple signaling pathways including p44/42 MAPK,25 p38 MAPK, and JNK.26 Extracellular signal-regulated kinase (ERK) is involved in growth factor-induced cell proliferation, whilst SAPK/JNK or p38 MAPK contribute to stress-induced apoptosis.27,28 Our study discovered that BBR induced apoptosis by regulating the TF/MAPK axis in NSCLC.
Conclusion
This study demonstrated that BBR enhances the expression of miR-19a and decreases TF expression in NSCLC cells. MiR-19a played a key role in the ability of BBR to inhibit cell apoptosis. The TF/MAPK pathway played a mediatory role in the apoptotic induction induced by miR-19a in PC9 and A549 cells. Taken together, the miR-19a/TF/MAPK axis may improve our understanding of the anticancer effects of BBR, though multi-center clinical trials are required to confirm the clinical value of BBR for the treatment of NSCLC.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (81473469, 31770131, 81472579, 81472180); the Scientific Research Projects of Shanghai Science and Technology Commission (14411971200); and the Scientific Research Projects of Shanghai Municipal Commission of Health and Family Planning (201840056). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
This research program was approved by the Ethics Committee of Shanghai 10th People’s Hospital of Tongji University. All participants signed the informed consent forms.
Abbreviations
NSCLC, non-small cell lung cancer; miRNAs, microRNAs; TF, tissue factor; MAPK, mitogen-activated protein kinase; SCC, squamous cell carcinoma; CT, chemotherapy; OS, overall survival; BBR, berberine; EGFR, epidermal growth factor receptor; MEK, extracellular regulated protein kinases; ERK, extracellular regulated protein kinases; PI3, phosphatidylinositol 3-kinase; PKB, protein kinases B; DMSO, dimethyl sulfoxide; miR-NC, miRNA negative control; CCK-8, cell counting kit-8; OD, optical density; PBS, phosphate buffer saline; SD, standard deviation; DAB, diaminobenzidine tetrahydrochloride; V, volume; PCA, principal component analysis; qRT-PCR, Real-time Polymerase Chain Reaction.
Availability of data and materials
Datasets are available from the corresponding authors on reasonable request.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5(4):263–274. doi: 10.1038/nrc1586 [DOI] [PubMed] [Google Scholar]
- 4.Inoue A, Yoshida K, Morita S, et al. Characteristics and overall survival of EGFR mutation-positive non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors: a retrospective analysis for 1660 Japanese patients. Jpn J Clin Oncol. 2016;46(5):462–467. doi: 10.1093/jjco/hyw014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, Feng Y, Tsao S, Wang N, Curtain R, Wang Y. Berberine and Coptidis rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. J Ethnopharmacol. 2009;126(1):5–17. doi: 10.1016/j.jep.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Xu X, Zhao M, et al. Berberine induces senescence of human glioblastoma cells by downregulating the EGFR-MEK-ERK signaling pathway. Mol Cancer Ther. 2015;14(2):355–363. doi: 10.1158/1535-7163.MCT-14-0634 [DOI] [PubMed] [Google Scholar]
- 7.Wang N, Feng Y, Zhu M, et al. Berberine induces autophagic cell death and mitochondrial apoptosis in liver cancer cells: the cellular mechanism. J Cell Biochem. 2010;111(6):1426–1436. doi: 10.1002/jcb.22869 [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300(1):10–19. doi: 10.1016/j.canlet.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Cao Y, He Z, et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232(2):85–95. doi: 10.1620/tjem.232.85 [DOI] [PubMed] [Google Scholar]
- 12.Lin Q, Chen T, Lin Q, et al. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2013;107(7):767–771. doi: 10.1002/jso.23312 [DOI] [PubMed] [Google Scholar]
- 13.Callander NS, Varki N, Rao LV. Immunohistochemical identification of tissue factor in solid tumors. Cancer. 1992;70(5):1194–1201. doi: [DOI] [PubMed] [Google Scholar]
- 14.Wang N, Zhu M, Wang X, Tan HY, Tsao SW, Feng Y. Berberine-induced tumor suppressor p53 up-regulation gets involved in the regulatory network of MIR-23a in hepatocellular carcinoma. Biochim Biophys Acta 2014;1839(9):849–57. doi: 10.1016/j.bbagrm.2014.05.027 [DOI] [PubMed] [Google Scholar]
- 15.You HY, Xie XM, Zhang WJ, Zhu HL, Jiang FZ. Berberine modulates cisplatin sensitivity of human gastric cancer cells by upregulation of miR-203. In Vitro Cell Dev Biol Anim 2016;52(8):857–63. doi: 10.1007/s11626-016-0044-y [DOI] [PubMed] [Google Scholar]
- 16.Han X, Guo B, Li Y, Zhu B. Tissue factor in tumor microenvironment: a systematic review. J Hematol Oncol. 2014;7:54. doi: 10.1186/s13045-014-0054-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong W, Wei J, Abidi P, et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10(12):1344–1351. doi: 10.1038/nm1135 [DOI] [PubMed] [Google Scholar]
- 18.Regina S, Valentin JB, Lachot S, Lemarie E, Rollin J, Gruel Y. Increased tissue factor expression is associated with reduced survival in non-small cell lung cancer and with mutations of TP53 and PTEN. Clin Chem. 2009;55(10):1834–1842. doi: 10.1373/clinchem.2009.123695 [DOI] [PubMed] [Google Scholar]
- 19.Regina S, Rollin J, Blechet C, Iochmann S, Reverdiau P, Gruel Y. Tissue factor expression in non-small cell lung cancer: relationship with vascular endothelial growth factor expression, microvascular density, and K-ras mutation. J Thorac Oncol. 2008;3(7):689–697. doi: 10.1097/JTO.0b013e31817c1b21 [DOI] [PubMed] [Google Scholar]
- 20.Goldin-Lang P, Tran QV, Fichtner I, et al. Tissue factor expression pattern in human non-small cell lung cancer tissues indicate increased blood thrombogenicity and tumor metastasis. Oncol Rep. 2008;20(1):123–128. [PubMed] [Google Scholar]
- 21.Chuang TD, Luo X, Panda H, Chegini N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol Endocrinol. 2012;26(6):1028–1042. doi: 10.1210/me.2012-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Yu H, Lou JR, et al. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J Biol Chem. 2011;286(2):1429–1435. doi: 10.1074/jbc.M110.146530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Yang S, Yan W, et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95(9):1056–1070. doi: 10.1038/labinvest.2015.76 [DOI] [PubMed] [Google Scholar]
- 24.Cao X, Lai S, Hu F, et al. miR-19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Sci Rep. 2017;7(1):2939. doi: 10.1038/s41598-017-01153-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camerer E, Gjernes E, Wiiger M, Pringle S, Prydz H. Binding of factor VIIa to tissue factor on keratinocytes induces gene expression. J Biol Chem. 2000;275(9):6580–6585. doi: 10.1074/jbc.275.9.6580 [DOI] [PubMed] [Google Scholar]
- 26.Camerer E, Rottingen JA, Gjernes E, et al. Coagulation factors VIIa and Xa induce cell signaling leading to up-regulation of the egr-1 gene. J Biol Chem. 1999;274(45):32225–32233. doi: 10.1074/jbc.274.45.32225 [DOI] [PubMed] [Google Scholar]
- 27.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23(16):2838–2849. doi: 10.1038/sj.onc.1207556 [DOI] [PubMed] [Google Scholar]
- 28.Seger R, Krebs EG. The MAPK signaling cascade. Faseb J. 1995;9(9):726–735. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets are available from the corresponding authors on reasonable request.