Abstract
The ability to determine full-length nucleotide composition of individual RNA molecules is essential for understanding the architecture and function of a transcriptome. However, experimental approaches capable of capturing the sequences of both 5’ and 3’ termini of the same transcript remain scarce. Here we present SEnd-seq—a high-throughput and unbiased method that simultaneously maps transcription start and termination sites with single-nucleotide resolution. Using this method, we obtain a comprehensive view of the Escherichia coli transcriptome, which displays an unexpected level of complexity. SEnd-seq significantly expands the catalog of transcription start sites and termination sites, defines unique transcription units, and detects prevalent antisense RNA. Strikingly, our results unveil widespread overlapping bidirectional terminators located between opposing gene pairs. We further show that convergent transcription is a major contributor to highly efficient bidirectional termination both in vitro and in vivo. This finding highlights an underappreciated role of RNA polymerase conflicts in shaping transcript boundaries and suggests an evolutionary strategy for modulating transcriptional output by arranging gene orientation.
It has become widely appreciated that RNA is not merely the messenger that relays genetic information from DNA to protein, but also itself carries out diverse regulatory roles in cell physiology1. The function of an RNA transcript is fundamentally determined by its constituent sequence elements, including those residing at the 5’ and 3’ ends. Next-generation RNA sequencing (RNA-seq) is a revolutionary tool for profiling a transcriptome—the set of all RNA molecules in a cell2. However, Illumina-based short-read RNA-seq—the most commonly used platform for transcriptomic analysis—requires strand fragmentation, which decouples the 5’-end sequence of an RNA molecule from its 3’-end sequence. As such, the resultant transcriptome map reports ensemble RNA levels, but information on the end-to-end nucleotide composition of individual transcripts is inevitably lost. Although various methods have been developed to delineate the 5’ or 3’ extremities of transcripts3–9, they cannot concomitantly sequence both ends of RNA. On the other hand, single-molecule long-read sequencing platforms possess the ability to read an RNA molecule from one end to the other. Nonetheless, their read depth, error rate, and cost still compare unfavorably with the Illumina platform10.
Prokaryotic transcriptomes were once considered simple due to their small size and limited splicing. This view is rapidly changing due to the growing list of RNA-based gene regulatory mechanisms found in prokaryotes11,12. However, counterintuitively, prokaryotic transcriptomic analyses have lagged behind eukaryotic counterparts. In particular, transcription termination sites, which mark the 3’ ends of primary transcripts, have remained incompletely annotated even in model organisms such as Escherichia coli13. Intramolecular ligation of 5’ and 3’ RNA termini allows for simultaneous capture of the sequences of both ends, thereby representing a promising strategy for inferring full-length sequences of prokaryotic RNA. However, existing methods that employ this strategy suffer from strong length bias14,15. In addition, they are not readily applicable to prokaryotic transcripts because of their reliance on 3’-end polyadenylate tails for the generation of complementary DNA (cDNA). Thus, a method capable of comprehensively profiling full-length transcripts in prokaryotes is still urgently needed.
In this work, we developed a method—termed simultaneous 5’ and 3’ end sequencing (SEnd-seq)—to concurrently read both ends of cellular transcripts. This method enabled us to determine the correlated occurrence of transcription start sites (TSS) and termination sites (TTS) with single-nucleotide resolution across the whole transcriptome. Using SEnd-seq, we identified a large number of previously unannotated TSS and TTS in E. coli. Strikingly, SEnd-seq unveiled prevalent occurrence of overlapping bidirectional TTS between head-to-head gene pairs or between a gene and an opposing non-coding RNA (ncRNA). We further conducted in vitro and in vivo experiments to support the model in which convergent transcription is an important contributor to highly efficient bidirectional termination.
Results
Simultaneous 5’- and 3’-end capture by SEnd-seq.
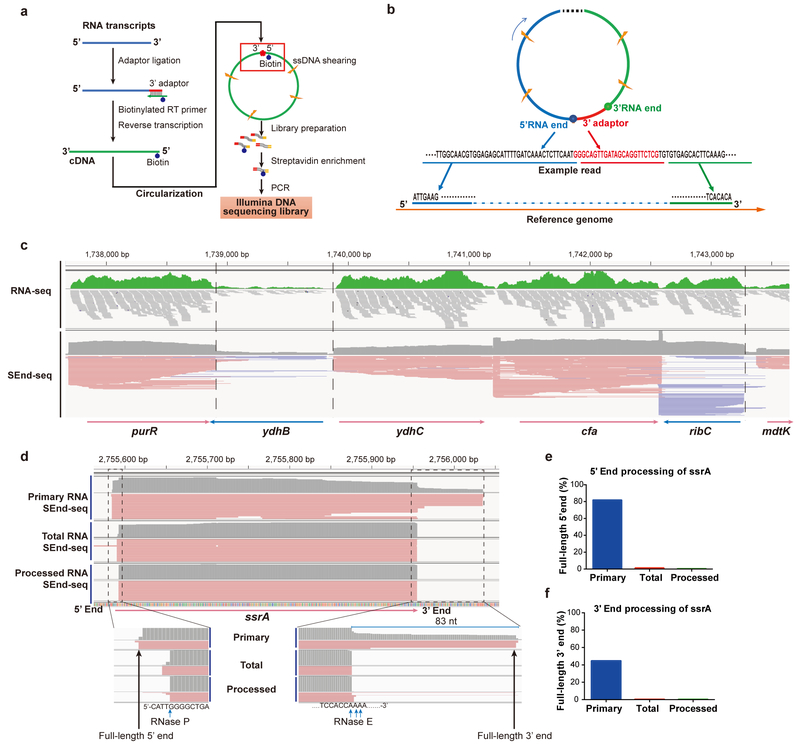
The general workflow of SEnd-seq is depicted in Fig. 1a. The key step is the circularization of cDNA by a single-stranded ligase that strongly favors intramolecular ligation16 (Supplementary Fig. 1a). Importantly, this step circularizes DNA of varying lengths with uniformly high efficiencies (Supplementary Fig. 1b). After fragmentation, the biotin-labeled pieces containing the 5’−3’ junction are isolated and prepared for paired-end sequencing. The 5’- and 3’-end sequences are extracted and mapped to the reference genome (Fig. 1b). The full-length composition of individual transcripts is then inferred by connecting the two termini (Supplementary Fig. 1c). Besides total RNA SEnd-seq, we also developed workflows to selectively enrich primary (5’ triphosphorylated) or processed (5’ monophosphorylated) transcripts (Supplementary Fig. 2).
Fig. 1 |. Simultaneous capture of 5’- and 3’-end sequences of bacterial transcripts by SEnd-seq.
a, Workflow of SEnd-seq. See Methods for details. b, An example read illustrating how to infer the full-length sequence of individual transcripts by extracting correlated 5’- and 3’-end sequences and mapping them to the reference genome. c, A sample data track of the log-phase E. coli transcriptome showing the comparison between standard RNA-seq and SEnd-seq. Dashed lines highlight the sharp boundaries of transcripts delineated by SEnd-seq, which are obscured in standard RNA-seq. d, SEnd-seq reads mapped to the ssrA gene in primary, total and processed RNA datasets. e, Ratio of ssrA transcripts with an intact, unprocessed 5’ end in different datasets. f, Ratio of ssrA transcripts with an intact 3’ end in different datasets.
Evaluation of the performance of SEnd-seq.
We applied SEnd-seq to E. coli cells collected under different growth conditions (Supplementary Fig. 3a,b). The read coverage on each gene is highly correlated between SEnd-seq replicates (Supplementary Fig. 3c), and between SEnd-seq and standard RNA-seq (Supplementary Fig. 3d). The transcriptome dataset yielded by SEnd-seq exhibits no severe nucleotide bias at either the 5’ or 3’ end of RNA (Supplementary Fig. 3e,f).
The advantage of SEnd-seq over standard RNA-seq in mapping the boundaries of individual RNA molecules is apparent in a direct comparison of their respective data tracks (Fig. 1c). To assess the ability of SEnd-seq to reproduce the precise ends of input transcripts, we added a mixture of in vitro synthesized RNA to the cellular RNA and subjected them together to SEnd-seq analysis. Correct lengths were recovered for all tested spike-in RNA species (Supplementary Fig. 4a–c), indicating minimal sample deterioration during the procedure. The read coverage on each spike-in RNA species matches the ratio at which it was added to the mixture (Supplementary Fig. 4d), arguing against any significant length bias of SEnd-seq.
We also demonstrated that SEnd-seq is able to recover the boundaries of endogenous transcripts with single-nucleotide resolution. For example, intact 5’ and 3’ ends of the 452-nt ssrA RNA precursor were enriched in the primary RNA sample, whereas the processed and total RNA datasets predominantly yielded the mature 365-nt ssrA species with its termini exactly corresponding to the known RNase cleavage sites17 (Fig. 1d–f). As another example, the 1,861-nt 16S rRNA precursor and the major intermediates in its maturation pathway were successfully detected by SEnd-seq (Supplementary Fig. 5).
Identification of transcription start sites.
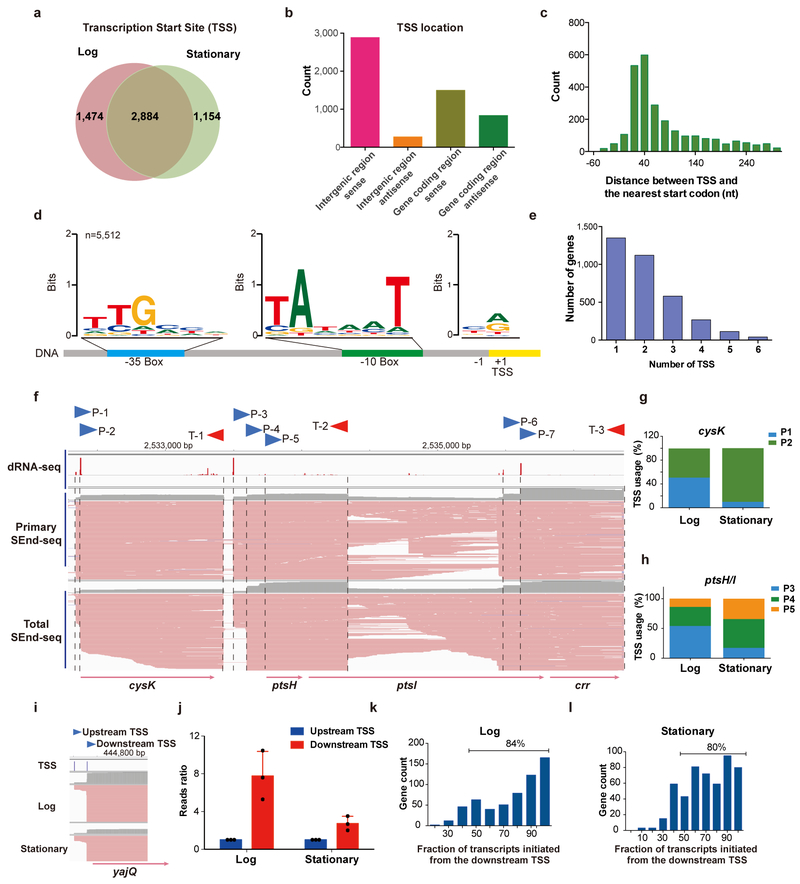
The single-nucleotide resolution afforded by SEnd-seq allows us to precisely annotate transcription start sites (TSS) and termination sites (TTS) in the same assay. Using primary RNA datasets, we identified 4,358 and 4,038 TSS for log-phase and stationary-phase E. coli cells respectively, among which 2,884 are common sites (Fig. 2a, Supplementary Table 1). These sites are located both within intergenic regions and inside gene bodies (Fig. 2b,c). Most of them display a characteristic bacterial promoter sequence in the 5’ flank18 (Fig. 2d).
Fig. 2 |. Identification of transcription start sites (TSS).
a, Venn diagram showing the number of TSS identified by SEnd-seq for E. coli cells growing in log phase versus stationary phase. b, Number of TSS located within intergenic regions or inside annotated genes (either in the sense orientation or in the antisense orientation). c, Distribution of the distance between an identified TSS and the start codon of its nearest annotated coding region (cutoff is 300 nt). d, Motif analysis of the +1 site, −10 element and −35 element from all TSS detected by SEnd-seq in log phase E. coli cells. e, Distribution of the number of alternative TSS for a given annotated gene. f, Log-phase SEnd-seq data track for the cysK-ptsH-ptsI-crr operon that shows multiple TSS (P-1 to P-7) and TTS (T-1 to T-3). TSS identified by dRNA-seq is shown on the top for comparison51. g,h, Bar graphs displaying the differential usage of alternative TSS for the cysK (g) and ptsH/I (h) genes during different growth stages. i, SEnd-seq data track showing two TSS controlling the expression of the yajQ gene. j, Bar graphs displaying the amount of yajQ transcripts initiated from the upstream versus downstream TSS. Values are normalized to the upstream TSS transcript level for each experimental replicate. Data are mean ± s.d. from three independent replicates. k,l, Histogram of the percentage of detected transcripts initiated from the most downstream TSS for any gene that employ multiple TSS using cells harvested from the log phase (k) or stationary phase (l) of growth.
SEnd-seq not only reproduced the vast majority of TSS previously annotated by other 5’-end mapping methods19,20, but also identified thousands of TSS unknown until now (Supplementary Fig. 6). A subset of these start sites was selected and validated by primer-extension assays (Supplementary Fig. 7). We found 2,133 genes that feature alternative TSS upstream of their coding regions (Fig. 2e,f), indicating that those genes are each controlled by multiple promoters. In many cases, the usage of alternative TSS is dependent on the growth condition (Fig. 2g,h and Supplementary Fig. 8). For the genes that employ multiple TSS, we analyzed the fraction of transcripts initiated from the upstream TSS versus the downstream TSS [e.g., yajQ (Fig. 2i,j)]. We found that the most downstream TSS (i.e., the one closest to the start codon) tends to make the largest contribution to the overall RNA expression level (Fig. 2k,l). The upstream and downstream TSS regions share a similar bacterial promoter −10 element, while exhibiting minor differences in the −35 element (Supplementary Fig. 9).
Identification of transcription termination sites.
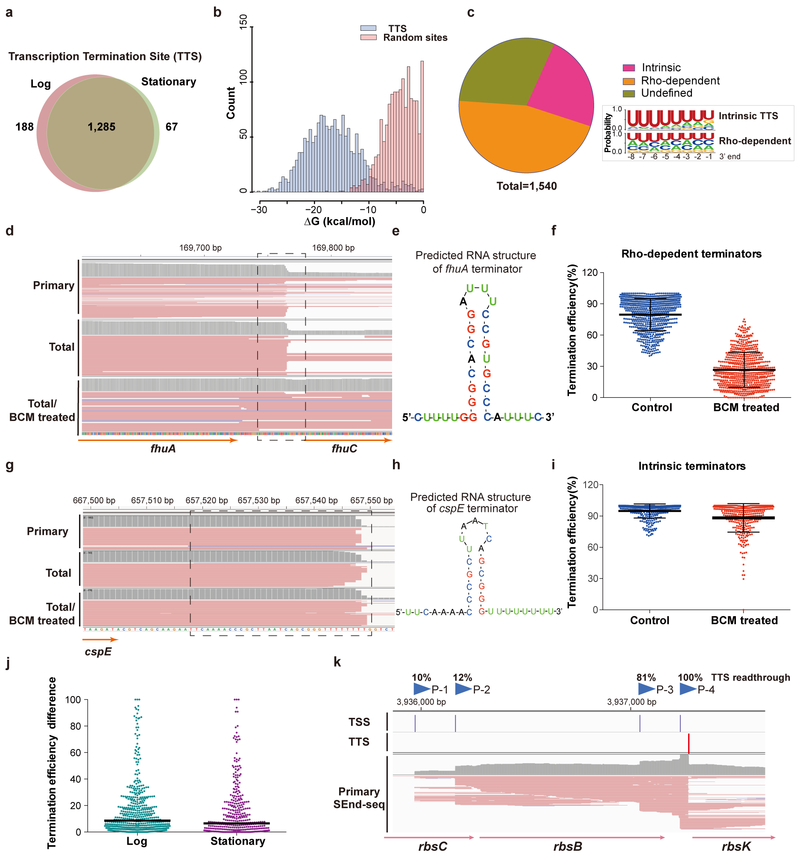
Two major transcription termination mechanisms have been well documented in bacteria: intrinsic termination that is mediated by a hairpin structure formed in the nascent RNA followed by a U-rich tract, and factor-dependent termination that relies on the Rho ATPase21. The identification of TTS is more challenging than TSS because of the lack of chemical distinction between bona fide termination sites and processed 3’ ends, resulting in much fewer annotated TTS in the existing database. To exclude post-processing cleavage sites, we created single-deletion E. coli strains in which each of the three genes (pnp, rnb, rnr) that encodes a major 3’−5’ exoribonuclease is knocked out22. Only those RNA 3’ ends that were not affected by any of these knockouts were annotated as TTS, notwithstanding the caveat that these RNases likely play redundant roles. We identified 1,285 TTS that are common between log-phase and stationary-phase E. coli cells, as well as 255 growth-stage-specific ones (Fig. 3a, Supplementary Table 2). SEnd-seq recaptures most of the TTS annotated by other 3’-end mapping methods20,23, but also finds a large number of previously unknown sites (Supplementary Fig. 10). We found that TTS predominantly reside within intergenic regions (89%), although there are cases where termination occurs prematurely within the 5’ untranslated region (UTR) of a gene (Supplementary Fig. 11).
Fig. 3 |. Identification of transcription termination sites (TTS).
a, Venn diagram showing the number of identified TTS for log versus stationary phase E. coli cells. b, Distribution of the RNA folding energy for identified TTS sequences (blue bars) compared with that for sequences of identical length randomly selected from the E. coli genome (red bars). c, (left) Pie chart showing the fraction of intrinsic and Rho-dependent terminators identified by SEnd-seq. (right) Nucleotide profiles for the 3’-end sequences of intrinsic and Rho-dependent TTS. Data are representative of two independent experiments. d, SEnd-seq data track for an example Rho-dependent terminator located downstream of the fhuA gene. When treated with the Rho inhibitor bicyclomycin (BCM), the fraction of readthrough transcripts significantly increased. e, Predicted secondary structure of the fhuA terminator. f, Average termination efficiency of all identified Rho-dependent terminators without or with BCM treatment. n = 709 (number of terminators analyzed). Error bars denote s.d. Data are representative of two independent experiments. g, SEnd-seq data track for an example intrinsic terminator located downstream of the cspE gene. h, Predicted secondary structure of the cspE terminator. i, Average termination efficiency of all identified intrinsic terminators without or with BCM treatment. n = 357. Error bars denote s.d. Data are representative of two independent experiments. j, Scatter plot showing the span of termination efficiency for each TTS that is linked to multiple TSS. For example, a data point at 50% means that, for this TTS, the maximal termination efficiency and the minimal efficiency—depending on the choice of TSS—differ by 50%. n = 520 for the log-phase dataset and 395 for the stationary-phase dataset. The black bars indicate median values. k, An example SEnd-seq data track illustrating that the alternative usage of TSS can induce differential termination efficiencies at the same TTS. The fractions of readthrough transcripts initiated from any given TSS (P-1 to P-4) are indicated.
TTS sites identified here tend to form stable secondary structures (Fig. 3b). The termination efficiency, derived from the level of readthrough transcripts across the termination site, varies widely (Supplementary Fig. 12a). We assigned 709 TTS as Rho-dependent terminators based on their sensitivity to the Rho-specific inhibitor bicyclomycin (BCM)24 (Fig. 3c–f). Among the other TTS, which are less sensitive to BCM treatment, many display sequence characteristics of an intrinsic terminator (a GC-rich hairpin followed by a 7–8 nt U-rich tract)21 (Fig. 3g–i). As the number of uridines decreases, the termination efficiency drops—consistent with previous results25—and can be further reduced by Rho inhibition (Supplementary Fig. 12b). This result suggests that the intrinsic and Rho-dependent termination mechanisms are not mutually exclusive and can act on the same site. Alternatively, such apparent overlap could result from RNase trimming following Rho action downstream of the hairpin23,26, despite that the aforementioned exonuclease knockout did not substantially change the 3’-end pattern of these sites.
Taking advantage of the ability of SEnd-seq to simultaneously determine the 5’ and 3’ ends of the same transcript, we asked whether the TSS selection—especially for those genes that employ multiple start sites—influences the termination efficiency at the corresponding TTS. We found 71 TTS whose termination efficiency alters by at least 40% depending on the choice of TSS (Fig. 3j,k), implying crosstalk between the two termini as previously proposed27,28.
Annotation of transcription units and antisense transcripts.
The concomitant mapping of TSS and TTS enabled us to define 3,578 unique transcription units (TU) in the E. coli transcriptome (Supplementary Fig. 13a,b; Supplementary Table 3). Most TU have their boundaries located within intergenic regions. We did detect 323 TU with TSS in a gene-coding region, yielding a shorter RNA product (Supplementary Fig. 13c). We also found 452 TU with an intragenic TSS that drives transcription of a downstream gene (Supplementary Fig. 13d).
The ability of SEnd-seq to comprehensively profile full-length RNA of different sizes also allowed us to analyze the genome-wide distribution of antisense transcripts, whose prevalence and importance in bacteria are increasingly being appreciated29,30. We found that a substantial fraction of transcripts (~15%) are derived from the complementary strand of protein-coding genes. These antisense transcripts are mostly located toward the 3’ end of a coding region or within a 3’ UTR, and have a wide range of lengths (Supplementary Fig. 14).
Prevalent overlapping bidirectional TTS revealed by SEnd-seq.
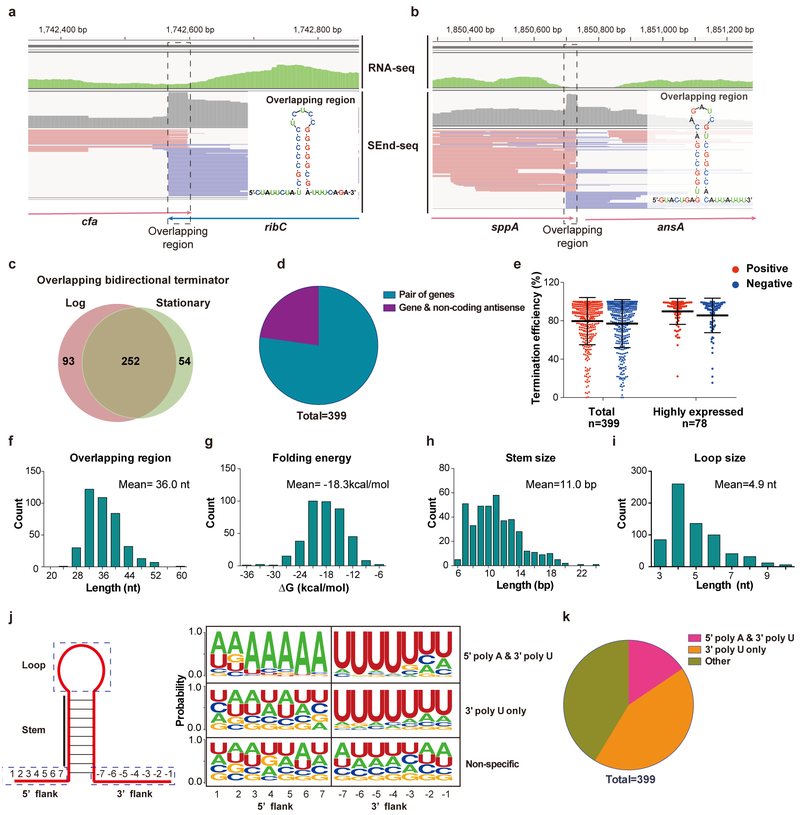
As demonstrated above, SEnd-seq provides an unprecedented inventory of the E. coli transcriptome. In the following we focus on one of the most striking findings that emerged from the SEnd-seq dataset. There are 658 pairs of neighboring genes in E. coli that are orientated in a head-to-head manner (Supplementary Fig. 15). Unexpectedly, we discovered that two opposing TTS frequently overlap with each other between a pair of convergent genes (284 out of 658 pairs) (Fig. 4a–d; Supplementary Table 4). In addition, we found 115 cases in which TTS of an unopposed gene overlaps with that of an antisense RNA (Fig. 4b,d). These overlapping regions are largely hidden from the standard RNA-seq dataset due to its lack of coverage around RNA ends (Fig. 4a,b).
Fig. 4 |. Pervasive bidirectional overlapping TTS revealed by SEnd-seq.
a, SEnd-seq data track for an example convergent gene pair (cfa-ribC) exhibiting overlapping TTS. Standard RNA-seq data track is shown in green for comparison. (inset) Predicted secondary structure for the overlapping region. Data are representative of three independent experiments. b, SEnd-seq data track and predicted secondary structure of an example overlapping TTS between a coding gene (sppA; red reads) and a non-coding antisense RNA (blue reads). Data are representative of three independent experiments. c, Venn diagram showing the number of overlapping bidirectional terminators identified for log versus stationary phase E. coli cells. d, Pie chart showing the fraction of overlapping TTS located between a gene pair or between a gene and an antisense ncRNA. e, (left) Average termination efficiency for all identified overlapping bidirectional terminators in either orientation (positive direction in red; negative direction in blue). n = 399. (right) Average termination efficiency for those bidirectional TTS that are located between a pair of highly expressed genes. n = 78. Error bars denote s.d. Data are representative of two independent experiments. f-i, Distributions of the length (f), folding energy (g), predicted stem size (h) and loop size (i) for the overlapping TTS. j, (left) Schematic of the stem-loop structure formed in the overlapping region. (right) Nucleotide profiles for the 5’ and 3’ flanking sequences of the stem-loop within an overlapping region. Such profiling allows for classification of the overlapping TTS into three categories. k, Pie chart showing the fraction of each category described in (j).
Overlapping bidirectional TTS are on average ~80% efficient in both directions. The termination efficiency tends to be even higher for the sites that are sandwiched between two highly expressed genes (Fig. 4e). The length of the overlapping region ranges from 18 to 60 nt (Fig. 4f). The vast majority of these overlapping sequences are predicted to form RNA stem-loop structures (Fig. 4g–i). However, only a minor fraction (~16%) exhibit features of a canonical bidirectional intrinsic terminator25,31, i.e., a short GC-rich hairpin flanked by an A-tract and a U-tract on either side (Fig. 4j,k). Most overlapping regions feature a nonspecific flanking sequence on at least one side of the hairpin. Moreover, the stems tend to be longer than those of typical intrinsic terminators and often contain mismatches and bulges (Supplementary Fig. 16). These bidirectional terminators do not appear to be primarily Rho-dependent either, as the BCM inhibitor only confers a minor effect on their termination efficiency (Supplementary Fig. 17).
We found that the patterns of these overlapping regions in the RNase-knockout strains (Δpnp, Δrnb, Δrnr) are largely similar to those in the wildtype strain (Supplementary Fig. 18), suggesting that the boundaries of these regions are genuine termination sites rather than products of RNase trimming. In further support of this notion, the overlapping sequences identified here almost always contain single-stranded regions flanking the stem loop, unlike decay products that are usually processed until the edge of the protective hairpin stem32.
Convergent transcription drives bidirectional termination in vitro.
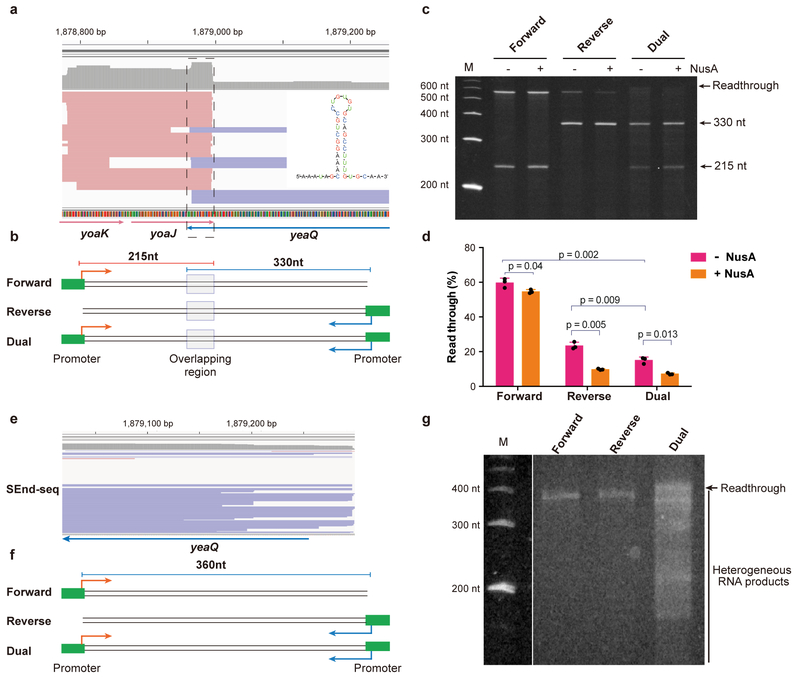
Since neither intrinsic termination nor Rho-mediated termination can fully explain the widespread occurrence of overlapping TTS between convergent TU pairs, we postulated that head-on collisions between opposing transcription machineries may cause termination in both directions. To test this hypothesis, we performed in vitro transcription assays with E. coli RNA polymerase (RNAP) on synthetic DNA templates harboring a convergent gene pair. We copied the genomic sequence around the yoaJ-yeaQ locus into the template (Fig. 5a,b). This region contains a 34-nt overlapping TTS sequence and displays strong bidirectional termination in vivo. When a T7A2 promoter that controls transcription initiation by E. coli RNAP was placed at one end of the template, unidirectional transcription was permitted, which resulted in significant readthrough (Fig. 5c,d). This result confirms the notion that the overlapping TTS sequence alone cannot cause efficient termination. In comparison, in vitro transcription using a strong intrinsic terminator yielded much lower readthrough (Supplementary Fig. 19).
Fig. 5 |. Convergent transcription is required for bidirectional termination in vitro.
a, SEnd-seq data track for the yoaJ-yeaQ gene pair showing an overlapping TTS. Data are representative of three independent experiments. b, Schematic of DNA templates harboring the yoaJ-yeaQ overlapping TTS region that are used for the in vitro transcription assay. c, Gel showing the RNA products transcribed from the different templates shown in (b) in the absence or presence of NusA. Data are representative of three independent experiments. d, Quantification of the fraction of readthrough transcripts for the different templates. Data are mean ± s.d. from three independent experiments. P values were determined by two-sided unpaired Student’s t-tests. e,f, SEnd-seq data track for part of the yeaQ gene (e) and DNA templates derived from this region that lacks a terminator sequence (f). The templates contain either one or two promoters to allow unidirectional or convergent transcription, respectively. g, Gel showing predominant readthrough for unidirectional transcription (Forward and Reverse templates) and heterogeneous RNA products for convergent transcription (Dual template). Data are representative of three independent experiments.
Importantly, when a promoter was incorporated in both ends of the template in order to support convergent transcription, the readthrough level was significantly reduced (Fig. 5c,d). The sizes of the RNA products are consistent with termination occurring at positions demarcating the overlapping region. Similar results were obtained with sequences taken from other convergent gene pairs (Supplementary Fig. 20). These in vitro results strongly suggest that RNAP conflicts alone—without other cellular factors—can induce bidirectional termination.
NusA is known to stimulate bacterial transcription termination33. We examined the influence of NusA on convergent transcription and found that NusA further enhanced the bidirectional termination efficiency (Fig. 5c,d and Supplementary Fig. 20). Therefore, the effect of NusA and the effect of RNAP conflicts on the termination efficiency can be additive.
How do transcription complexes originating from stochastic initiation events always meet at the overlapping region? Given that the formation of RNA hairpins often contributes to RNAP pausing34, we posited that the stem-loop structures formed in the overlapping regions—although they do not lead to termination per se—cause RNAP to pause for an extended period of time such that another polymerase traveling from the opposite direction causes interference at the pausing site. To test this idea, we conducted in vitro transcription assays with DNA templates that lack an overlapping TTS sequence (Fig. 5e,f). As expected, unidirectional transcription yielded predominantly readthrough transcripts (Fig. 5g). Interestingly, when convergent transcription was allowed, readthrough decreased but the RNA products were heterogeneous in length (Fig. 5g), indicating promiscuous collision sites. This is in contrast to the uniform RNA products released from templates harboring an overlapping TTS sequence (Fig. 5c). Therefore, the overlapping TTS sequence—and hence the pausing signal—is required for synchronizing the converging transcription complexes, causing them to interfere with each other at well-defined positions.
Convergent transcription contributes to bidirectional termination in vivo.
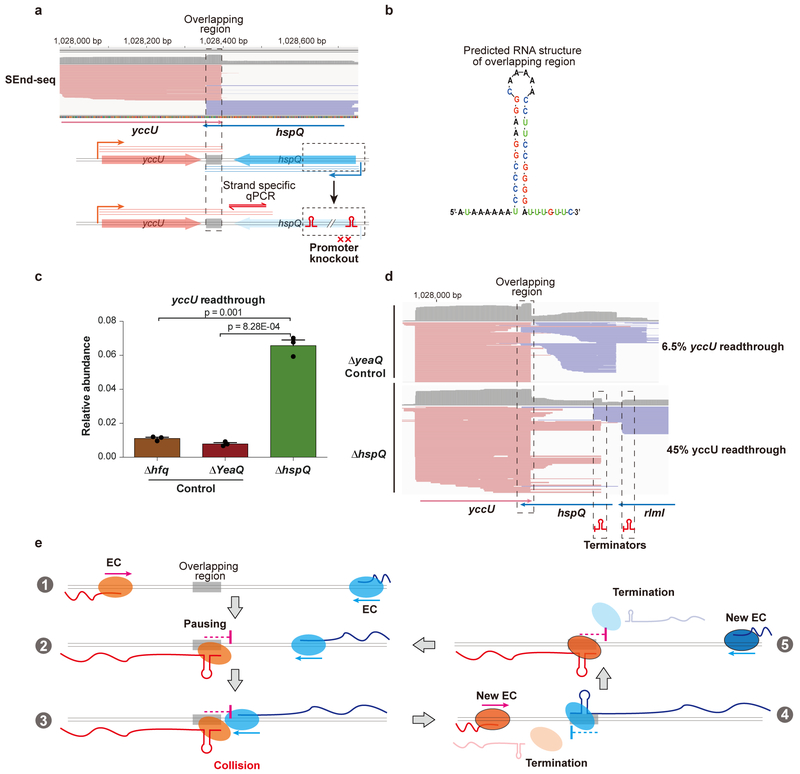
To seek further evidence that converging transcription elongation complexes contribute to their own termination inside the cell, we performed in vivo genome editing to disrupt transcription from one direction in an opposing gene pair. We targeted the yccU-hspQ convergent pair, which displays a 40-nt overlapping TTS (Fig. 6a,b). To disrupt hspQ transcription, we created the ΔhspQ strain by deleting the promoter sequence for hspQ and inserting two strong intrinsic terminators around the original TSS of hspQ. We then assessed the extent of yccU readthrough across the overlapping region with strand-specific qPCR. As predicted, the ΔhspQ strain showed a significant increase in the abundance of yccU readthrough transcripts (Fig. 6c). Disrupting the transcription of other genes at distal genomic locations did not confer the same effect on yccU readthrough (Δhfq and ΔyeaQ in Fig. 6c). Furthermore, we performed SEnd-seq with the ΔhspQ strain and examined the transcript profile around the yccU-hspQ region (Fig. 6d). First of all, hspQ transcription was indeed abolished. Secondly, the yccU readthrough level markedly increased in the ΔhspQ dataset compared to the control dataset (45% vs. 6.5%). Similar results were obtained from genome editing experiments on other convergent gene pairs (Supplementary Fig. 21).
Fig. 6 |. Convergent transcription contributes to bidirectional termination in vivo.
a, SEnd-seq data track (top) and schematic of in vivo genomic modification (bottom) for the yccU-hspQ convergent gene pair. To disrupt hspQ transcription, we replaced the promoter and part of the gene body of hspQ with two strong intrinsic terminators. Data are representative of three independent experiments. b, Predicted secondary structure for the overlapping TTS between yccU and hspQ. c, qPCR results showing the relative abundance of yccU readthrough transcripts across the overlapping region when hspQ transcription is abolished (ΔhspQ). We also edited genes outside the convergent pair with the same procedure (Δhfq and ΔyeaQ) as controls. Data are mean ± s.d. from three independent experiments. P values were determined by two-sided unpaired Student’s t-tests. d, SEnd-seq data track around the yccU-hspQ region for the ΔyeaQ (top) or ΔhspQ strain (bottom). The fraction of yccU readthrough transcripts for each strain is indicated. Data are representative of two independent experiments. e, Model illustrating that head-on collisions between converging RNA polymerases drive bidirectional termination. The overlapping region produces an RNA hairpin that traps the transcription machinery, which is dislodged by another elongation complex traveling from the opposite direction—either through direct physical interaction or via torsional stress accumulated in the DNA. This process occurs repeatedly, resulting in highly efficient termination in both directions.
Together, these in vitro and in vivo results support a model in which the stem-loop structure formed near the 3’ ends of two converging transcription units causes pausing of the elongation complex and, subsequently, transcription termination when an opposite elongation complex collides into it (Fig. 6e). This model predicts that RNAP occupancy is enriched at the overlapping bidirectional TTS due to pausing. We thus performed RNAP ChIP-seq experiments using antibodies against the β or β’ subunit. Indeed, stronger ChIP signals were observed around the overlapping TTS sites compared to nearby regions (Supplementary Fig. 22).
Discussion
Despite the reinvigorated interest of the scientific community in RNA biology and the myriad RNA-seq technologies, methods capable of defining the boundaries of all transcripts in a transcriptome still remain scarce. TIF-seq, which was developed to analyze eukaryotic transcript isoforms15, ligates the termini of dsDNA—as opposed to ssDNA in SEnd-seq—and displays a strong bias toward short transcripts. Recently, a method based on PacBio long-read sequencing was reported20. But this method involves size-selection steps that remove any RNA shorter than 1,000 nt, and therefore is blind to all small RNA and a significant fraction of mRNA. In contrast, SEnd-seq comprehensively profiles RNA of different sizes in a single assay with reduced length bias. It is worth noting that the conversion from RNA to full-length cDNA in SEnd-seq is critically dependent on the performance of reverse transcription. A highly processive reverse transcriptase was used in this study35. Continued enzyme engineering could further enhance the transcriptome coverage of SEnd-seq.
SEnd-seq enabled us to determine the correlated occurrence of TSS and TTS and to discern the crosstalk between promoters and terminators that control the same transcript. Future experiments are needed to elucidate the origin of such crosstalk. Our method uses the sequences of 5’ and 3’ termini to infer the full-length composition of each distinct transcript. Thus it is most ideally suited for studying organisms with limited splicing. SEnd-seq could also be employed for meta-transcriptomics analysis with RNA pooled from multi-species communities.
The sharp transcript boundaries defined by SEnd-seq led us to identify a widespread but previously underappreciated mechanism of transcription termination driven by head-on interference between transcription complexes. The unique ability of SEnd-seq to determine the 5’-end origin of terminated RNA and the full sequence of the overlapping region helped to uncover this mechanism. Transcriptional interference resulting from convergent promoters has been well documented in bacteria36–38. However, studies of transcriptional interference have thus far mainly focused on its negative impact on gene activity due to promoter occlusion or random RNAP collisions during elongation39. The present work shows that such interference can be exploited to precisely terminate transcription, thereby limiting undesired readthrough and fine-tuning the transcriptional output. Moreover, although overlapping bidirectional terminators have been reported for a few individual genes40,41, the extent to which they occur genome-wide was unexplored. Here we show that this phenomenon is pervasive, which raises the intriguing scenario that head-to-head gene pairs are functionally related, akin to co-directional genes within the same polycistronic operon. In the cases where an opposing gene is absent, antisense transcription can also suppress the readthrough of sense transcription, which adds to the functional repertoire of non-coding RNA.
In this work we used the strong T7A2 promoter for the in vitro transcription experiments, where we observed efficient bidirectional termination. In vivo, the likelihood of RNAP head-on encounter is influenced by additional factors, notably the promoter strength42. For highly expressed convergent gene pairs, the frequent physical interference between RNAP is likely a major contributor to the bidirectional termination, although we do not exclude alternative, but not mutually exclusive, mechanisms that may play a role in shaping the transcript 3’ boundaries, such as antisense-RNA-mediated attenuation43. Moreover, given the known effect of ribosome movement on RNAP pause release44,45, the uncoupling between transcription and translation downstream of the stop codon may enhance RNAP pausing and termination at intergenic bidirectional TTS. With regard to RNAP collisions, further studies are required to elucidate whether termination is induced by direct contacts between the converging motors or by the accumulation of torsional stress in DNA when they approach46,47. Finally, considering that convergent genes and polymerase conflicts are also found in eukaryotes48–50, it will be interesting to investigate whether the transcription termination mechanism documented here is conserved across kingdoms of life.
Methods
Bacterial strains and growth conditions.
E. coli K-12 MG1655 and K-12 SIJ_488 (Addgene #68246; a gift from Alex Nielsen) were cultured in LB media (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl, pH 7.4) under aerobic conditions at 37 °C. To inhibit Rho activity, cells were cultured in LB media with 50 μg/ml bicyclomycin (Santa Cruz, sc-391755) at 37 °C for 15 min at indicated growth condition. Δpnp, Δrnb and Δrnr strains were generated using a previously reported protocol based on the arabinose inducible lambda Red recombineering system and the rhamnose inducible flippase recombinase52. PCR primers listed in Supplementary Table 5 were used to amplify the kanamycin‐resistant gene in pKD13 and the DNA product was transformed into the K-12 SIJ_488 strain. After selection for positive colonies, the inserted kanamycin‐resistant gene was excised by culturing with L-rhamnose. To knockout a gene in a convergent gene pair, two strong intrinsic terminators were put into the insert DNA to replace the promoter region of the target gene.
SEnd-seq pipeline.
Cellular RNA isolation.
The overnight culture medium was diluted 1:50 into fresh media and grown to an OD600 of 0.4 to 0.6 for the log phase sample or an OD600 over 2.0 for the stationary phase sample. E. coli cells were quenched by adding 0.5× vol of cold Stop Buffer (5% phenol in ethanol) to the culture medium immediately before harvest and placed on ice for 15 min. Cell pellets were collected by centrifugation (6,000 rpm for 5 min at 4 °C), thoroughly resuspended in 100 μl of lysozyme solution [2 mg/ml in TE buffer (10 mM Tris-HCl and 1 mM EDTA)], and incubated for 2 min. The cells were then immediately lysed by adding 1 ml of TRIzol Reagent (Invitrogen, 15596) and subsequently pipetted vigorously until the solution was clear. After incubation for 5 min at room temperature, 200 μl of chloroform was added and the sample was gently inverted several times until reaching homogeneity. The sample was then incubated for 15 min at room temperature before centrifugation at 12,000 g for 10 min. The upper phase (~600 μl) was gently collected and mixed at a 1:1 ratio with 100% isopropanol. The sample was incubated for 1 hr at −20 °C and then centrifuged at 14,000 rpm for 10 min at 4 °C. The pellet was washed twice with 1 ml of 75% ethanol, air dried for 5 min, and dissolved in nuclease-free water. RNA integrity was assessed with 1% agarose gel and Agilent 2100 Bioanalyzer System.
3’ adaptor ligation.
RNA with or without 5’ adaptor ligation (see below) was subjected to 3’ adaptor ligation by mixing 12 μl of RNA (<5 μg) with 1 μl of 100 μM 3’ adaptor (Supplementary Table 5), 0.5 μl of 50 mM ATP, 2 μl of dimethyl sulfoxide, 5 μl of 50% PEG8000, 1 μl of RNase Inhibitor (New England BioLabs, M0314), and 1 μl of High Concentration T4 RNA Ligase 1 (New England BioLabs, M0437). After incubation at 23 °C for 5 hr, the reaction was diluted to 40 μl with water and purified twice with 1.5× vol of Agencourt RNAClean XP beads (Beckman Coulter, A63987) to remove excess RNA adaptors. The sample was subsequently eluted in 12 μl of water.
rRNA removal and reverse transcription.
The eluted RNA was subjected to an optional step of rRNA removal with Ribo-Zero rRNA Removal Kit (Illumina, MRZB12424). The RNA was then recovered by ethanol precipitation. 11.5 μl of eluted RNA was incubated with 0.5 μl of 100 μM biotinylated reverse transcription primer (Supplementary Table 5) and 1 μl of 10 mM Deoxynucleotide Solution Mix (dNTPs) (New England BioLabs, N0447) at 65 °C for 5 min, and then placed on ice for 2 min. 1 μl of the maturase reverse transcriptase from Eubacterium rectale (recombinantly purified from E. coli, a gift from Anna Marie Pyle, Yale University)35, 4 μl of 5× maturase buffer, 2 μl of 100 mM DTT and 0.5 μl of RNase Inhibitor were added to the reaction and incubated at 42 °C for 90 min. The reaction was then terminated by incubation at 85 °C for 10 min. Following reverse transcription, 10 μl of 1 N NaOH solution was added and incubated at 70 °C for 15 min to remove the RNA templates. After neutralization by adding 10 μl of 1 N HCl solution, the reaction was diluted to 100 μl with TE buffer and cleaned twice with 100 μl of TE-saturated phenol:chloroform:isoamyl alcohol (25:24:1, vol/vol) (Thermo Fisher Scientific, 15593031). The cDNA was purified by ethanol precipitation, dissolved in TE buffer and cleaned once with 1.5× vol of Agencourt AMPure XP beads (Beckman Coulter, A63881). The cDNA was then eluted with 30 μl of water and subjected to 5’ phosphorylation by adding 2 μl of T4 Polynucleotide Kinase (New England BioLabs, M0201), 4 μl of PNK Reaction Buffer and 4 μl of 10 mM ATP. After incubation at 37 °C for 60 min and 65 °C for 20 min, the cDNA was cleaned with 1.5× vol of AMPure beads again and eluted with 20 μl of 0.1× TE buffer. The cDNA concentration was determined by the Qubit ssDNA Assay Kit (Invitrogen, Q10212).
Enrichment of primary transcripts.
Primary transcripts were enriched following a protocol adapted from a previously published method8. 5 μg of total RNA was mixed with 5 μl of 10× VCE Buffer (New England BioLabs, M2080) in a total volume of 50 μl, incubated for 2 min at 70 °C, and then placed on ice. 5 μl of 3’-Desthiobiotin-GTP (New England BioLabs, N0761) and 5 μl of Vaccinia virus Capping Enzyme (New England BioLabs, M2080) were added to the reaction and incubated at 37 °C for 30 min. After purification with 1.5× RNAClean beads, the capped RNA was eluted and subjected to 3’ adaptor ligation as described above. The RNA was cleaned twice with 1.5× RNAClean beads and then enriched with Hydrophilic Streptavidin Magnetic Beads (New England BioLabs, S1421). After washing thoroughly four times with Binding Buffer (10 mM Tris‐HCl pH 7.5, 2 M NaCl, 1 mM EDTA) and three times with Washing Buffer (10 mM Tris‐HCl pH 7.5, 0.25 M NaCl, 1 mM EDTA), the RNA was eluted with 26 μl of Biotin Buffer (10 mM Tris‐HCl pH 7.5, 0.5 M NaCl, 1 mM EDTA, 1 M biotin) and incubated at 37 °C for 25 min on a rotator. Then 14 μl of Binding Buffer was added and incubated for another 4 min. The RNA was cleaned with 1.5× RNAClean beads and eluted in 12 μl of H2O. The 5’ capped and 3’ ligated RNA was reverse transcribed by the maturase as described above.
Enrichment of processed transcripts.
Processed RNA in a total RNA sample was selectively ligated to a 5’ adaptor. Briefly, 5 μg of total RNA was incubated for 2 min at 70 °C and then placed on ice. 1 μl of 100 μM 5’ adaptor (Supplementary Table 5), 0.5 μl of 50 mM ATP, 2 μl of dimethyl sulfoxide, 5 μl of 50% PEG8000, 1 μl of RNase Inhibitor and 1 μl of High Concentration T4 RNA Ligase 1 were added to the sample. After incubation at 23 °C for 5 hr, the sample was diluted with water and cleaned twice with 1.5× vol of Agencourt RNAClean XP beads. After the SEnd-seq pipeline, we used a custom shell script to search for the adaptor-labeled reads, thereby specifically extracting processed RNA ends.
Circularization.
50 ng of cDNA was mixed with 2 μl of CutSmart Buffer (New England BioLabs, B7204), 2 μl of 50 mM MnCl2, 2 μl of 0.1 M DTT, 2 μl of 5 M betaine (Affymetrix, 77507) and 2 μl of TS2126 RNA Ligase I (a gift from Kevin Ryan, City College of New York)16. The reaction was incubated at 37 °C for 5–16 hr. Subsequently, the reaction was supplemented with 1 μl of 10 mM dNTPs and diluted to 100 μl with TE buffer and 0.1% SDS. Then 100 μl of TE-saturated phenol:chloroform:isoamyl alcohol (25:24:1, vol/vol) was added and incubated for 1 hr with occasional vortexing. After centrifugation, the water phase was cleaned again with phenol:chloroform:isoamyl alcohol. Finally, the circularized cDNA was ethanol precipitated and dissolved in 130 μl of TE buffer.
Library preparation.
Circularized cDNA was fragmented by acoustic shearing in microTUBE (Covaris, 520045) with Covaris S220 Focused-ultrasonicator under the condition of Peak145 for 90 sec. After ethanol precipitation, the ssDNA was converted to dsDNA by the Second Strand cDNA Synthesis Kit (New England BioLabs, E6114) at 16 °C for 2 hr. The product was cleaned with 1.8× vol of AMPure beads and eluted in 50 μl of 0.1× TE buffer. The DNA ends were prepared and ligated to the Illumina sequencing adaptor with the NEBNext Ultra II DNA Library Prep Kit (New England BioLabs, E7645). The ligated product was cleaned twice with 1× vol of AMPure beads and eluted in 50 μl of 0.1× TE buffer. Biotin-labeled DNA strands were bound to the Dynabeads M-280 Streptavidin (Invitrogen, 11205D) and cleaned four times with Washing Buffer (5 mM Tris-HCl pH 7.5, 1 M NaCl, 0.5 mM EDTA) and twice with TE buffer. The beads were re-suspended thoroughly with the Q5 High-Fidelity 2× Master Mix (New England BioLabs, M0492). The DNA library was then amplified for 13 (total RNA SEnd-seq) to 17 cycles (primary RNA SEnd-seq) following the manufacturer’s protocol. The final library was cleaned twice with 1× vol (50 μl) of AMPure beads, and its concentration and size distribution were determined with Agilent 2200 TapeStation (Agilent, 5067–5576).
Spike-in RNA preparation.
A T7 promoter sequence was incorporated upstream of four DNA sequences with different lengths taken from the bacteriophage λ genome. After PCR amplification and gel excision/cleanup, the DNA templates were subjected to in vitro transcription by T7 RNA Polymerase (New England BioLabs, M0251). DNA was removed by adding 1 μl of TURBO DNase (Life Technologies, AM2238) and incubated at 37 °C for 15 min. Full-length RNA products were purified by polyacrylamide gel electrophoresis. After cleanup and concentration measurement, all spike-in RNA species were pooled together. Typically the spike-in RNA mix was added to the total bacterial RNA at a mass ratio of 1:1000.
RNA-seq.
For standard RNA-seq, ~5 μg of RNA was treated with TURBO DNase and recovered by ethanol precipitation. Ribosomal RNA was depleted with the Ribo-Zero rRNA Removal Kit. The sequencing library was prepared with the TruSeq Stranded mRNA Library Prep Kit (Illumina, RS-122–2101) following the manufacturer’s instructions.
RNAP ChIP-seq.
The ChIP-seq workflow is adapted from a previously published ChIP-microarray study53. Briefly, cells were grown to the stationary stage and crosslinked by the addition of formaldehyde (1% final concentration) with continued shaking at 37 °C for 10 min before quenching with glycine (100 mM final concentration). Cells were then lysed and DNA was sheared by sonication followed by treatment with micrococcal nuclease (New England BioLabs, M0247S) and RNase A (Thermo Fisher Scientific, EN0531). Antibodies against the RNAP β or β’ subunit (BioLegend 663903 or 662904) were used for immunoprecipitation. RNAP-DNA crosslinks were enriched by protein A/G beads (Thermo Fisher Scientific, 26159). Enriched immunoprecipitated DNA and input DNA sequencing libraries were prepared with NEBNext Ultra II DNA Library Prep Kit.
Primer extension assay.
~5 μg of RNA was treated with TURBO DNase, cleaned three times with phenol:chloroform:isoamyl alcohol (25:24:1, vol/vol), and recovered by ethanol precipitation. Subsequently the RNA was denatured at 70 °C for 2 min and then treated with Terminator 5’-Phosphate-Dependent Exonuclease (Illumina, TER51020) at 30 °C for 1 hr. After ethanol precipitation, the recovered RNA was treated with RppH (New England BioLabs, M0356S) at 37 °C for 1 hr. The RNA was cleaned by 1.5× vol of Agencourt RNAClean XP beads (Beckman Coulter, A63987) and ligated to a 5’ adaptor as described above. After reaction, the RNA was cleaned with 1.5× vol of Agencourt RNAClean XP beads. The eluted RNA was then reverse transcribed to cDNA with pooled RT primers by the maturase. Subsequently, 10 μl of 1 N NaOH solution was added and incubated at 70 °C for 15 min to remove the RNA templates. The second strand DNA was synthesized with an oligo complementary to the 5’ adaptor. The resultant dsDNA was used for sequencing library preparation and sequencing was performed on MiSeq.
Data analysis.
Sequencing data collection and processing.
SEnd-seq data were collected by the Illumina MiSeq or NextSeq 500 platform in a paired-end mode (150 nt ×2). After quality filter and adaptor trimming, the paired-end reads were merged to single-end reads by using the FLASh software. The correlated 5’-end and 3’-end sequences were extracted by the custom script fasta_to_paired.sh. The full-length sequences were inferred by mapping to the reference E. coli genome NC_000913.3 by using Bowtie 2. Reads with an insert length greater than 10,000 nt were discarded. For each sample we obtained over 2 million usable reads (i.e., those harboring at least 15 nucleotides on each end of the same transcript). RNA-seq and ChIP-seq data were collected by the Illumina MiSeq or NextSeq 500 platform in a paired-end mode (75 nt ×2). After quality filter, the sequencing data were analyzed by the Rockhopper software54. The wig files and SAM files were further analyzed by custom Perl scripts. The results were visualized with the Integrative Genome Viewer (IGV).
Gene coverage quantification.
For SEnd-seq data, each read was first mapped to the genome. Each position within the intervening region of the read (from the start site to the end site) was considered as effective coverage. For RNA-seq data, the coverage of each nucleotide position was directly extracted from the wig files generated by the Rockhopper software54. Gene coverage was quantified by summing the coverage of all nucleotide positions spanned by each gene. Only genes longer than 200 nt were used for the correlation analysis between SEnd-seq and RNA-seq.
TSS identification.
Transcription start sites (TSS) were identified from the primary transcript SEnd-seq data with a custom Perl script. Only positions with more than 10 reads starting at that position and with an increase of at least 50% in read coverage from its upstream to its downstream were retained. Candidate TSS positions within 5 bases in the same orientation were clustered together, and the position with the largest amount of read increase was used as the representative TSS position. Motif analysis around the TSS regions (−40 nt to +1 nt) was performed by MEME55.
TTS identification.
Based on previous work56 and our observation that transcripts with intact, unprocessed 3’ termini are enriched in the primary RNA SEnd-seq dataset, we reasoned that transcription termination sites (TTS) should be reproducible between the total RNA and primary RNA datasets. In practice, we first identified from the total RNA SEnd-seq data positions with more than 10 reads ending at that position (outside of rRNA genes) and with a reduction of more than 40% in read coverage from its upstream to its downstream. We then cross-checked the site in the primary SEnd-seq dataset and with RNase-knockout strains (2206pnp, Δrnb, Δrnr). Candidate TTS positions within 5 bases in the same orientation were clustered together, and the position with the largest amount of read reduction was used as the representative TTS position. Only the TTS sites identified from at least two samples were used for further analysis. The terminators are classified into Rho-dependent terminators (those showing a readthrough percentage increase of > 30% upon BCM treatment), intrinsic terminators (those showing a readthrough percentage of < 30% in the control sample, a readthrough percentage increase of < 15% upon BCM treatment, and harboring at least five uracils out of the eight bases in the 3’ flank region of the terminator hairpin), or undefined.
Overlapping bidirectional TTS identification.
Overlapping bidirectional termination sites were identified by screening for two opposing TTS with a custom Perl script. Only those with an overlapping region shorter than 60 nt and yielding a stem-loop structure were retained for further analysis. Highly expressed convergent gene pairs were defined as those with > 20 read counts for each gene in the pair.
RNA secondary structure analysis.
The sequence from 45-nt upstream to 9-nt downstream of an identified TTS was used for RNA secondary structure prediction with RNAfold57 combined with custom Perl scripts.
Motif analysis.
The −45 nt to +9 nt TTS regions and overlapping bidirectional TTS regions were used for motif analysis. Nucleotide logos around TTS were generated by WebLogo58.
Transcription unit annotation.
Transcription units were identified by a custom Perl script based on the defined TSS, TTS and read coverage. Only those with a continuous coverage of more than 5 reads were retained for further analysis. We also excluded units with a length shorter than 80 nt.
ChIP-seq data analysis.
The RNAP ChIP-seq signal at each nucleotide position was calculated and normalized to the input sample data using a custom script. The normalized ChIP/input ratio was used for downstream analysis.
Previously deposited datasets.
dRNA-seq datasets (SRR1411276 and SRR1411277 for log and stationary phase E. coli RNA, respectively)19 and SMRT-Cappable-seq dataset (GSE117273)20 were used for comparison with the SEnd-seq results from this study.
In vitro transcription.
DNA templates for T7 RNAP were amplified from the FLuc Control Template (New England BioLabs, E2040S). DNA templates for E. coli RNAP were prepared by PCR from the E. coli genomic DNA with indicated primer sets (Supplementary Table 5). The T7A2 promoter sequence was incorporated at one or both ends of the template. Purified E. coli RNAP and sigma factor σ70 holoenzyme (a gift from the Darst Lab at The Rockefeller University) was used for in vitro transcription reactions. The reaction mixture included 4 μl of 5× Reaction Buffer (200 mM Tris-HCl, 600 mM KCl, 40 mM MgCl2, 4 mM DTT, 0.04% Triton X-100, pH 7.5 at 25 °C), 0.5 μl of RNase Inhibitor, 0.5 pmol of DNA template and 2 pmol of E. coli RNAP holoenzyme. When applicable, 20 pmol of NusA (a gift from the Landick Lab at University of Wisconsin-Madison) was added to the reaction mixture. The mixture was incubated at 37 °C for 30 min before rNTPs (50 μM each) were added to initiate transcription. After 5 min of reaction (unless noted otherwise), reinitiation of transcription was prevented by adding heparin (Sigma-Aldrich, H4784) to a final concentration of 100 μg/ml. After incubation with 0.3 μl of TURBO DNase for 10 min, the RNA was separated by 5% urea polyacrylamide gel electrophoresis, stained by SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific, S11494), scanned by Axygen Gel Documentation System (Corning, GD1000), and quantified by ImageJ (National Institutes of Health).
Quantitative PCR.
First-strand cDNA was reverse transcribed from the total RNA of indicated samples with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813) and strand-specific RT primers (Supplementary Table 5). Control cDNA was reverse transcribed with random primers and the same amount of input RNA. Quantitative RT-PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems, 4309155) and QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The relative abundance of RNA is represented as the signal ratio between the target transcript and the reference rnpB gene from the same sample using the formula: 2−(ΔCT) (ΔCT = CTtarget - CTrnpB; CT stands for cycle threshold).
Statistics.
Data are shown as mean ± s.d. unless noted otherwise. P values were determined by two-sided unpaired Student’s t-tests using GraphPad Prism 6. The difference between two groups was considered statistically significant when the P value is less than 0.05 (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
Data availability.
SEnd-seq and standard RNA-seq datasets from this study have been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE117737. The custom scripts used in this study are available on Github (https://github.com/LiuLab-codes/SEnd_seq_analysis). Other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank S. Darst and E. Campbell for help with the in vitro transcription experiments and critical reading of the manuscript; K. Ryan, A. Pyle and R. Landick for sharing reagents; and E. Cheng for help with data analysis. This work was supported by a C. H. Li Memorial Scholar Fund Award (X.J.), the Robertson Foundation, the Quadrivium Foundation, a Monique Weill-Caulier Career Scientist Award, a March of Dimes Basil O’Connor Starter Scholar Award, a Kimmel Scholar Award, and NIH grants R00GM107365 and DP2HG010510 (S.L.).
Footnotes
Competing interests
The Rockefeller University has filed a provisional patent application encompassing aspects of the SEnd-seq technology on which S.L. and X.J. are listed as inventors.
References
- 1.Morris KV & Mattick JS The rise of regulatory RNA. Nat Rev Genet 15, 423–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M & Snyder M RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma CM et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–5 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Wurtzel O et al. A single-base resolution map of an archaeal transcriptome. Genome Res 20, 133–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dar D et al. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352, aad9822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babski J et al. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics 17, 629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalanne JB et al. Evolutionary Convergence of Pathway-Specific Enzyme Expression Stoichiometry. Cell 173, 749–761 e38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ettwiller L, Buswell J, Yigit E & Schildkraut I A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics 17, 199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matteau D & Rodrigue S Precise Identification of Genome-Wide Transcription Start Sites in Bacteria by 5’-Rapid Amplification of cDNA Ends (5’-RACE). Methods Mol Biol 1334, 143–59 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Goodwin S, McPherson JD & McCombie WR Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17, 333–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hor J, Gorski SA & Vogel J Bacterial RNA Biology on a Genome Scale. Mol Cell 70, 785–799 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Guell M, Yus E, Lluch-Senar M & Serrano L Bacterial transcriptomics: what is beyond the RNA horiz-ome? Nat Rev Microbiol 9, 658–69 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Gama-Castro S et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44, D133–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan X & Ruan Y Genome wide full-length transcript analysis using 5’ and 3’ paired-end-tag next generation sequencing (RNA-PET). Methods Mol Biol 809, 535–62 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Pelechano V, Wei W & Steinmetz LM Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497, 127–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lama L & Ryan K Adenylylation of small RNA sequencing adapters using the TS2126 RNA ligase I. RNA 22, 155–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin-Chao S, Wei CL & Lin YT RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc Natl Acad Sci U S A 96, 12406–11 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruff EF, Record MT Jr. & Artsimovitch I Initial events in bacterial transcription initiation. Biomolecules 5, 1035–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conway T et al. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio 5(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan B, Boitano M, Clark TA & Ettwiller L SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat Commun 9, 3676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray-Soni A, Bellecourt MJ & Landick R Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu Rev Biochem 85, 319–47 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Hui MP, Foley PL & Belasco JG Messenger RNA degradation in bacterial cells. Annu Rev Genet 48, 537–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dar D & Sorek R High-resolution RNA 3’-ends mapping of bacterial Rho-dependent transcripts. Nucleic Acids Res 46, 6797–6805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwiefka A, Kohn H & Widger WR Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry 32, 3564–70 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Chen YJ et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 10, 659–64 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Wang X et al. Processing generates 3’ ends of RNA masking transcription termination events in prokaryotes. Proc Natl Acad Sci U S A (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goliger JA, Yang XJ, Guo HC & Roberts JW Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J Mol Biol 205, 331–41 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Telesnitsky AP & Chamberlin MJ Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol 205, 315–30 (1989). [DOI] [PubMed] [Google Scholar]
- 29.Thomason MK et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197, 18–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dornenburg JE, Devita AM, Palumbo MJ & Wade JT Widespread antisense transcription in Escherichia coli. MBio 1(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters JM, Vangeloff AD & Landick R Bacterial transcription terminators: the RNA 3’-end chronicles. J Mol Biol 412, 793–813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dar D & Sorek R Extensive reshaping of bacterial operons by programmed mRNA decay. PLoS Genet 14, e1007354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondal S, Yakhnin AV, Sebastian A, Albert I & Babitzke P NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol 1, 15007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J & Landick R A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem Sci 41, 293–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao C, Liu F & Pyle AM An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24, 183–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callen BP, Shearwin KE & Egan JB Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 14, 647–56 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Horowitz H & Platt T Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res 10, 5447–65 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elledge SJ & Davis RW Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev 3, 185–97 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Shearwin KE, Callen BP & Egan JB Transcriptional interference--a crash course. Trends Genet 21, 339–45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sameshima JH, Wek RC & Hatfield GW Overlapping transcription and termination of the convergent ilvA and ilvY genes of Escherichia coli. J Biol Chem 264, 1224–31 (1989). [PubMed] [Google Scholar]
- 41.Postle K & Good RF A bidirectional rho-independent transcription terminator between the E. coli tonB gene and an opposing gene. Cell 41, 577–85 (1985). [DOI] [PubMed] [Google Scholar]
- 42.Sneppen K et al. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J Mol Biol 346, 399–409 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Brantl S & Wagner EG An antisense RNA-mediated transcriptional attenuation mechanism functions in Escherichia coli. J Bacteriol 184, 2740–7 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landick R, Carey J & Yanofsky C Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci U S A 82, 4663–7 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proshkin S, Rahmouni AR, Mironov A & Nudler E Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma J, Bai L & Wang MD Transcription under torsion. Science 340, 1580–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crampton N, Bonass WA, Kirkham J, Rivetti C & Thomson NH Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res 34, 5416–25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hobson DJ, Wei W, Steinmetz LM & Svejstrup JQ RNA polymerase II collision interrupts convergent transcription. Mol Cell 48, 365–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prescott EM & Proudfoot NJ Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A 99, 8796–801 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eszterhas SK, Bouhassira EE, Martin DI & Fiering S Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol Cell Biol 22, 469–79 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Creecy JP & Conway T Quantitative bacterial transcriptomics with RNA-seq. Curr Opin Microbiol 23, 133–40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen SI, Lennen RM, Herrgard MJ & Nielsen AT Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress. Sci Rep 5, 17874 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters JM et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A 106, 15406–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClure R et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41, e140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Celesnik H, Deana A & Belasco JG Initiation of RNA decay in Escherichia coli by 5’ pyrophosphate removal. Mol Cell 27, 79–90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lorenz R et al. ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SEnd-seq and standard RNA-seq datasets from this study have been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE117737. The custom scripts used in this study are available on Github (https://github.com/LiuLab-codes/SEnd_seq_analysis). Other data that support the findings of this study are available from the corresponding author upon request.