Dear Editor,
In 2009, an outbreak of an unknown infectious disease occurred in rural areas of Hubei Province, China, affecting 17 persons, five of whom died. It was initially suspected to be human anaplasmosis (Zhang et al.2008). However, the laboratory evidence was insufficient to verify the diagnosis in most cases. In March 2011, a new virus was isolated from a patient’s blood and named severe fever with thrombocytopenia syndrome (SFTS) virus (SFTSV) in the Bunyaviridae (Yu et al.2011). The main signs and symptoms of SFTS are fever, thrombocytopenia, leukopenia, and a decreased platelet count (Yu et al.2011; McMullan et al.2012; Zhang et al.2017; Hu et al.2018). Similar viruses have recently been identified in the United States, South Korea, and Japan (McMullan et al.2012; Kim et al.2013; Takahashi et al.2014; Zhan et al.2017a, b). The disease has a high mortality rate (12%–30%) (Yu et al.2011; Liu et al.2013). Based on the detection of SFTSV in Haemaphysalis longicornis ticks (Yu et al.2011; Park et al.2014), it was believed to be transmitted by ticks. Transmission may also occur from person to person through exposure to infected blood (Wang et al.2014). As humans are often in close contact with domestic animals and may encounter rodents when they work outdoors, transmission between animals and humans is another possible main transmission route. Studies have examined SFTSV in 15 animal species, including cattle, sheep, chickens, pigs, elk, deer, geese, and rodents (Chen et al.2019). Sheep and cattle had the highest seroprevalence at 75%–95% and 57%–80%, respectively, and SFTSV RNA was detected in 11 animal species, with carriage rates varying from 0.23% to 26.31% (Chen et al.2019). However, the role of domesticated animals and rodents in the circulation and transmission of SFTSV remains unclear, and the natural reservoir hosts of SFTSV have not been determined. This study explored whether animal hosts play an essential role in the circulation and transmission of SFTSV by collecting samples from domesticated animals and rodents.
Samples were collected from several domestic animals and rodents from SFTSV epidemic areas (Suizhou, Macheng) and non-epidemic areas (Yicheng, Qianjiang, Xiantao) in Hubei Province, China, from July 2010 to November 2016, and were submitted mainly for routine surveillance purposes (Supplementary Figure S1). A total of 544 serum samples were collected, including 98 sheep, 58 cattle, 19 dogs, and 369 rodents. In addition, the heart, liver, spleen, lungs, and kidneys of each rodent were collected.
Antibodies against SFTSV in sheep, cattle, dogs, and rodents were detected using a double-antigen sandwich enzyme-linked immunosorbent assay (ELISA; Niu et al.2013; Jiao et al.2012; Zhan et al.2017a, b). A His-tagged affinity-chromatography-purified recombinant nucleocapsid protein (NP) of SFTSV (strain HB29) expressed in Escherichia coli was used for serosurveillance. Micro-titer plates were coated with 0.2 μg/well SFTSV NP, and incubated with serum samples at a dilution of 1:10, followed by detection with horseradish peroxidase (HRP)-conjugated N protein. 3,3′,5,5′-Tetramethylbenzidine dihydrochloride (TMB) peroxidase substrate was used for color development. The optical density (OD) was detected with an incidence wave length of 450 nm and reference wave length of 620 nm using an enzymatic marker (PHOMO, Autobio Company). Cut-off values for the assay were determined as the mean of the negative control serum samples plus 0.1. Samples with an optical density (OD) ≥ cut-off value were considered positive. Antibodies were detected in serum samples from 80 (81.63%) of 98 sheep, 39 (67.24%) of 58 cattle, 12 (63.16%) of 19 dogs, and 4 (1.08%) of 369 rodents in Suizhou, Macheng, Yicheng, Xiantao, and Qianjiang Counties (Table 1).
Table 1.
Prevalence rates of SFTSV viral RNA and specific antibodies in domesticated animals and rodents
| Species | Macheng | Suizhou | Qianjiang | Xiantao | Yicheng | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Antibody | RNA | No. | Antibody | RNA | No. | Antibody | RNA | No. | Antibody | RNA | No | Antibody | RNA | No. | Antibody | RNA | |
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | |||||||
| Positive | Positive | Positive | Positive | Positive | Positive | Positive | Positive | Positive | Positive | Positive | Positive | |||||||
| Sheep | 80 | 66 (82.5) | 3 (3.75) | 18 | 14 (77.78) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 98 | 80 (81.63) | 3 (3.06) |
| Cattle | 53 | 35 (66.04) | 3 (5.66) | 5 | 4 (80) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 58 | 39 (67.24) | 3 (5.17) |
| Dogs | 8 | 6 (75) | 0 | 11 | 6 (54.55) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 12 (63.16) | 0 |
| Rodents | 5 | 0 | 0 | 31 | 1 (3.23) | 0 | 146 | 2 (1.37) | 0 | 87 | 0 | 0 | 100 | 1 (1) | 0 | 369 | 4 (1.08) | 0 |
| Total | 146 | 107 (73.29) | 6 (4.11) | 65 | 25 (38.46) | 0 | 146 | 2 (1.37) | 0 | 87 | 0 | 0 | 100 | 1 (1) | 0 | 544 | 135 (24.82) | 6 (1.1) |
Viral RNA was extracted from serum using a QIAamp Viral RNA Mini Kit (52904, QIAGEN). The tissues from the rodents were ground and viral RNA was extracted using RNA Mini Kits (74106, QIAGEN). TaqMan quantitative real-time reverse transcription PCR (qRT-PCR) was performed on all animal serum and tissue samples using a certified qRT-PCR kit (4387391, ABI). The extracted RNA was used as a template for qRT-PCR to amplify SFTSV RNA using primers derived from the small RNA (sRNA) segment of the virus: F-5′-GGGTCCCTGAAGGAGTTGTAAA-3′ and R-5′-TGCCTTCACCAAGACTATCAATGT-3′. The probe was FAM-5′-TTCTGTCTT GCTGGCTCCGCGC-3′-TAMRA. qRT-PCR was performed for an initial 30 min at 50 °C for reverse transcription, followed by 2 min at 95 °C for denaturation. Next, 45 cycles of 15 s at 95 °C and 40 s at 60 °C were performed with the fluorescent signals measured at 60 °C. The cut-off cycle threshold for a positive sample was 35 cycles. Viral RNA was detected in serum samples from 3 (3.75%) of 80 sheep and 3 (5.66%) of 53 cattle in Suizhou and Macheng Counties (Table 1). The overall positive rate of viral RNA in the five counties was 3.06% and 5.17% for sheep and cattle, respectively. No viral RNA was detected in rodent tissue samples from the five counties.
All viral RNA-positive serum samples were used for virus isolation. Vero cells were maintained in complete Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mmol/L l-glutamine, 1.5 g/L sodium bicarbonate, 1.0 mmol/L sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin. The cells were cultured at 37 °C in 5% CO2. A 100 μL volume of each SFTSV RNA-positive serum was inoculated into a six-well cell culture plate with 2% FBS for 2 h. The supernatants were replaced with fresh DMEM with FBS and incubated at 37 °C in 5% CO2 for 8 days. The culture supernatants were harvested when obvious cytopathogenic effects occurred. After three cell passages, the virus was harvested, and all cell debris was removed from the centrifuged supernatant through three freeze-thaw cycles, and was identified using qRT-PCR. Positive strains were stored at − 80 °C for further analyses. We successfully isolated virus from sheep with RNA-positive serum samples from Macheng County and named it HB3-sheep03 (Supplementary Table S1). We failed to isolate virus from cattle with RNA-positive serum samples. During the sampling period, serum samples from SFTS patients in Hubei Province were also collected for virus isolation and genetic analysis. Among those samples, three SFTSV strains (HB154/155/156) were isolated from Macheng County in 2011. Furthermore, twenty-one strains of SFTSV were successfully isolated from other areas of Hubei Province. Their complete genomes were sequenced and submitted to GenBank (Supplementary Table S1).
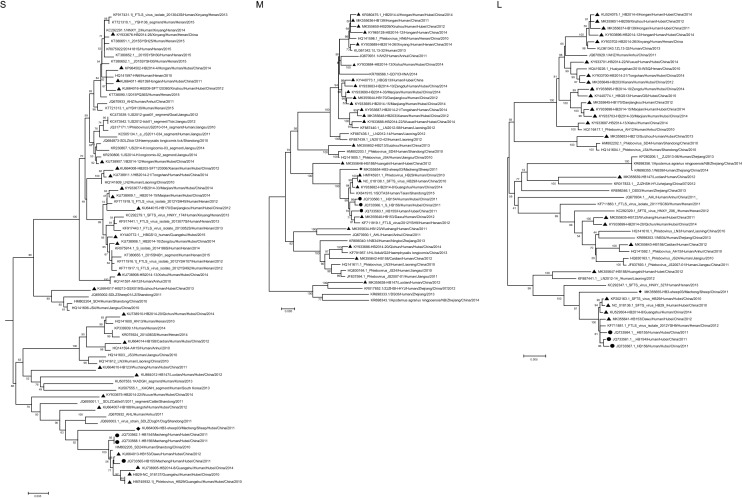
To perform a phylogenetic analysis of SFTSV strains, the available sequences were edited using the EditSeq program in DNAStar to obtain formatted sequences. Phylogenetic analysis was conducted based on the S, L, and M segments of SFTSV. The complete sequences were aligned using DNAStar software with the ClustalW method. Multiple sequence files were converted into a merged file in Clustal format by the program SeqVerter. Phylogenetic trees were constructed using the maximum likelihood method with MEGA ver. 6.06 and tested with 2000 bootstrap replicates. We compared the homology of HB3-sheep03 with the viruses isolated from animals and patients from different places (Fig. 1, Supplementary Fig. S1). The virus isolated from sheep (HB3-sheep03) was genetically close to the viruses isolated from three SFTS patients (HB154/HB155/HB156) from the same county in 2011, and was in the same subfamily. The nucleotide and amino acid identities of the S fragment were 97.6%–97.7% and 94.2%–94.4%, those of the M fragment were 98.6%–98.7% and 98.9%–99%, and those of the L fragment were 98.1% and 96.8%–97%, respectively. Compared with viruses from Shandong, Henan, and Anhui Provinces, the nucleotide and amino acid identities of the S fragment were 94%–100% and 86%–100%, 92.8%–100% and 96%–100% for the M fragment, and 95.0%–100% and 95.2%–100% for the L fragment, respectively. The nucleotide identity of the S fragment was 94.4%–96.3% and the amino acid identity was 85.8%–93.3% with viruses isolated from sheep, cattle, dogs, and ticks from Shandong and Jiangsu Provinces. We found that SFTSV strains isolated from Hubei Province were on different branches, suggesting multiple co-prevalent genotypes. We concluded that SFTSV strains from Hubei Province exhibit most of the genetic diversity found in China, and genetic evolutionary distance analysis showed that all sequences of the isolates from domesticated animals and human patients shared high homology, which indicated that there was a close evolutionary relationship among those viruses isolated from domesticated animals, rodents, ticks, and SFTS patients. These findings suggest that livestock are potential natural hosts and probably play an important role in the transmission of SFTSV.
Fig. 1.
Phylogenetic analysis of severe fever with thrombocytopenia syndrome virus (SFTSV) isolates from sheep (S, L, M segment). The evolutionary relationship of S, M, L segments of SFTSV isolated from domesticated animals, SFTS patients and ticks was calculated by using the ML method with MEGA 6.06, and tested by the bootstrap method with 2000 replicates. Rhombuses indicate the original sequences of SFTSV strains obtained from sheep; black circles indicate the original sequences of SFTSV strains obtained from SFTS patients around sheep in same disease-endemic county; black triangles indicate the sequences of SFTSV strains obtained from SFTS patients in Hubei province, China. Scale bar indicates nucleotide substitutions per site.
SFTSV is thought to circulate in an enzootic tick–vertebrate–tick cycle. Although there is no high-quality evidence that SFTSV causes disease in animals, we found that the seroprevalence of SFTSV in domestic animals was higher than in rodents. However, viral RNA, typically at low levels, was detected only in a small proportion of the domesticated animals studied (3.06%–5.17%). We conducted a retrospective investigation for nucleic-acid-positive animals. The results showed that there were no obvious symptoms in nucleic-acid-positive sheep or in cattle and sheep with a high serum positive rate. These findings suggest that domestic animals act as reservoir hosts of SFTSV, which plays an important role in the spread of SFTSV by feeding the ticks.
Previous studies demonstrated that rodents in China were seronegative for SFTSV (Lu et al.2011; Cui et al.2013). But our serology results also detected SFTSV infection in rodents with a seropositive rate of 1.08%, which was lower than a previous study reporting 7% in Apodemus agrarius and 8% in Mus musculus and Rattus norvegicus (Wang et al.2013). No SFTSV-specific viral RNA was detected in the dissected tissues of rodents, in part because either too few rodents were examined or the rodents were collected from sites where SFTSV was not endemic. It is also possible that the primer for the S segment of SFTSV used in our study for qRT-PCR was not as sensitive as that for the M segment. Liu et al. (2013) reported that 0.7% (3/440) of rodents were viral-RNA positive using primers for reverse transcription PCR designed from the M segment of the SFTSV genome.
According to our survey of serum antibodies and nucleic acids, SFTSV is widely found in domestic cattle and sheep. In the epidemic areas and during the SFTSV season, these domestic animals may play a key role in the circulation of SFTSV. Further studies need to examine the carriage of virus nucleic acids in serum from infected animals, the change in serum antibody titers, whether the virus proliferates in the animals, and its association with the people around animals and ticks. Our findings suggest that sheep and cattle act as natural hosts and might play an important role in the transmission of SFTSV. Given the high serum antibody and nucleic acid positive rates in sheep, sheep may be the key to preventing and controlling SFTSV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the China Mega-Project for Infectious Diseases of the Ministry of Science and Technology and Ministry of Health of the People’s Republic of China (2018ZX10201002009007/2017ZX10103005003), a grant from Science and Technology Department of Hubei Province (2018CFB630), and a grant from the Opening Research Fund Program of the State Key Laboratory of Virology of China, Wuhan University (2017KF003), and a grant from Open Fund for Occupational Hazard and Identification in key Laboratory of Hubei Province (OHIC2017G02). We thank Prof. Mifang Liang and Prof. Quanfu Zhang (National Institute for Viral Disease Control and Prevention, Chinese Centers for Disease Control and Prevention) for guidance and technical support. We also thank all workers involved in clinical sample collection.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This research was approved by the Ethics Committee of the Centers for Disease Control and Prevention of Hubei province, which uses international guidelines for the care and use of animals have been followed. Informed consent was obtained from all study participants.
Contributor Information
Shuang Rong, Phone: +86-18571727264, Email: rongshuang@wust.edu.cn.
Jianbo Zhan, Phone: +86-027-87652009, Email: jbzhan8866@163.com.
References
- Chen C, Li P, Li KF, Wang HL, Dai YX, Cheng X, Yan JB. Animals as amplificationhosts in the spread of severe fever with thrombocytopenia syndrome virus: a systematic review and meta-analysis. Int J Infect Dis. 2019;79:77–84. doi: 10.1016/j.ijid.2018.11.017. [DOI] [PubMed] [Google Scholar]
- Cui F, Cao HX, Wang L, Zhang SF, Ding SJ, Yu XJ, Yu H. Clinical and epidemiological study on severe fever with thrombocytopeni syndrome in Yiyuan County, Shandong Province, China. Am J Trop Med Hyg. 2013;88:510–512. doi: 10.4269/ajtmh.11-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Cai K, Liu M, Li WJ, Xu JQ, Qiu F, Zhan JB. Laboratory detection and molecular phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in Hubei Province, central China. Arch Virol. 2018;163:3243–3254. doi: 10.1007/s00705-018-3993-5. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zeng X, Guo X, Qi X, Zhang X, Shi Z, Zhou M, Bao C, Zhang W, Xu Y, Wang H. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus nucleocapsid protein for detection of total antibodies in human and animal sera by double-antigen sandwich enzyme-linked immunosorbent assay. J Clin Microbiol. 2012;50:372–377. doi: 10.1128/JCM.01319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, Choe PG, Kim NJ, Lee JK, Oh MD. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis. 2013;219:1892–1894. doi: 10.3201/eid1911.130792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JW, Wen HL, Fang LZ, Zhang ZT, He ST, Xue ZF, Ma DQ, Zhang XS, Wang T, Yu H, Zhang Y, Zhao L, Yu XJ. Prevalence of SFTSV among Asian House Shrews and Rodents, China, January–August 2013. Emerg Infect Dis. 2013;12:2126–2128. doi: 10.3201/eid2012.141013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YN, Dou XF, Wang XM, Tian LL, Yang YS, Wang HY, Li LQ, Zhang XC, Sun YL, Guan ZZ, Li XY, Huang H, Wang QY. Preliminary investigation on the carriage status of the novel bunyavirus among animals and ticks in Beijing area. Int J Virol. 2011;18:33–36. [Google Scholar]
- McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- Niu GY, Li JD, Liang MF, Jiang XL, Jiang M, Yin HY, Wang ZD, Li C, Zhang QF, Jin C, Wang XJ, Ding SJ, Zheng Xing, Wang S, Bi ZQ, Li DX. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg Infect Dis. 2013;19:756–763. doi: 10.3201/eid1905.120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Song BG, Shin EH, Yun SM, Han MG, Park MY, Park C, Ryou J. Prevalence of severe fever with thrombocytopenia syndrome virus in Haemaphysalis longicornis ticks in South Korea. Ticks Tick Borne Dis. 2014;5:975–977. doi: 10.1016/j.ttbdis.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M. The first identification and retrospective study of severe Fever with thrombocytopenia syndrome in Japan. J Infect Dis. 2014;209:816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QK, Ge HM, Hu JL, Zhang ZY, Wang YP, Jiao YJ, Li ZF, Hu SM, Lu DJ, Wang XH, Liu HZ. Surveillance of vectors and host animals of severe fever with thrombocytopenia syndrome virus in Donghai, China in 2010–2011. Chin J Vector Biol Control. 2013;4:313–316. [Google Scholar]
- Wang YL, Deng BC, Zhang J, Cui W, Yao WQ, Liu P. Person-to-person asymptomatic infection of severe fever with thrombocytopenia syndrome virus through blood contact. Intern Med. 2014;53:903–906. doi: 10.2169/internalmedicine.53.1164. [DOI] [PubMed] [Google Scholar]
- Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan JB, Cheng J, Hu B, Li J, Pan RG, Yang ZH, Zou WJ, Zhan FX, Guo DY. Pathogens and epidemiologic feature of severe fever with thrombocytopenia syndrome in Hubei Province, China. Virus Res. 2017;232:63–68. doi: 10.1016/j.virusres.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan JB, Wang Q, Cheng J, Hu B, Li J, Zhan FX, Song Y, Guo DY. Current status of severe fever with thrombocytopenia syndrome in China. Virol Sin. 2017;32:51–62. doi: 10.1007/s12250-016-3931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, Wan K, Li D, Liang G, Jiang X, Jing H, Run J, Luan M, Fu X, Zhang J, Yang W, Wang Y, Dumler JS, Feng Z, Ren J, Xu J. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. 2008;300:2263–2270. doi: 10.1001/jama.2008.626. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Shen S, Shi JM, Su ZY, Li MY, Zhang WJ, Li MG, Hu ZH, Peng C, Zheng X, Deng F. Isolation, characterization, and phylogenic analysis of three new severe fever with thrombocytopenia syndrome bunyavirus strains derived from Hubei Province, China. Virol Sin. 2017;32:89–96. doi: 10.1007/s12250-017-3953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.